Abstract
Immune cell activation occurs concurrently with metabolic reprogramming. As important components of the tumor microenvironment, monocytic myeloid-derived suppressor cells (M-MDSCs) are featured by their potent immunosuppressive abilities on anti-tumor effector cells. However, little is known about the contribution of metabolic adaptations to their suppressive roles. In this study, we found that tumor-infiltrating M-MDSCs had the same phenotype with splenic M-MDSCs. Compared with splenic M-MDSCs, tumor-infiltrating M-MDSCs exhibited stronger suppressive activities which was accompanied by higher glycolysis. Inhibition of glycolysis impaired the suppressive function of tumor M-MDSCs. Meanwhile, the results demonstrated that mTOR was responsible for this function regulation. mTOR inhibition by rapamycin decreased the glycolysis and reduced the suppressive activities of these cells. Furthermore, rapamycin treatment inhibited the tumor growth and reduced the percentage of M-MDSCs in 3LL tumor bearing mice. These results demonstrated that modulation of metabolism in immune cells can be an effective way to enhance anti-tumor effects.
Electronic supplementary material
The online version of this article (10.1007/s00262-018-2177-1) contains supplementary material, which is available to authorized users.
Keywords: MDSC, Glycolysis, mTOR, Immunotherapy
Introduction
Myeloid-derived suppressor cells (MDSCs) are a heterogeneous regulatory cell population featured by their myeloid origin, immature state and remarkable immunosuppressive functions. MDSCs are comprised of monocytic (M-MDSC, with a phenotype CD11b+Ly6ChighLy6G−) and polymorphonuclear (PMN-MDSC, with a phenotype CD11b+Ly6CintLy6G+) subpopulations which are distinguished phenotypically and morphologically [1, 2]. On a per cell basis, M-MDSC is more suppressive compared with PMN-MDSC and is the dominant immunosuppressive subset [3]. As one of the major suppressive stromal cell populations in the tumor microenvironment, MDSCs not only suppress antitumor immune response and protect tumor cells from chemotherapy and radiotherapy [4], but also induce the production of regulatory T cells, and secrete immunosuppressive molecules such as IL-10 and TGF-β, further strengthening the suppressive microenvironment [5–8].Thus, blocking the immunosuppressive activity of M-MDSCs would be an effective way to greatly improve the efficacy of tumor immunotherapy [9–13].
Available data showed that MDSCs’ functions in the peripheral lymphoid tissue were different from that in tumors [14]. Gabrilovich et al. demonstrated that tumor-infiltrating MDSCs expressed higher HIF-1α and were more suppressive than splenic MDSCs via upregulation of Arginase and NOS2 [15]. Rodriguez et al. found that tumor-induced stress reinforced the suppressive activity of MDSCs in tumor [16]. These results suggested that tumor M-MDSCs and splenic M-MDSCs may use different mechanisms to regulate their suppressive activities. It is well known that MDSCs can produce many enzymes such as Arg1 (arginase I) and IDO (indoleamine 2,3-dioxygenase) to modulate amino acid metabolism to regulate the functions of other cells [17–19]. But little is known about how metabolic activities in MDSCs influence the functions of MDSCs themselves.
As one of the most important energy sources in organisms from bacteria to humans, glucose plays an indispensable role in physiological as well as pathological processes. Glucose metabolism regulation may influence the function of cancer cells as well as immune cells. Sukumar et al. found that inhibiting glycolytic metabolism enhanced CD8 + T cell memory and antitumor function [20–22]. However, little is known about the glucose metabolism in MDSCs.
In this study, we aim to elucidate the differences of glucose metabolism pathways used by the M-MDSCs in different tissues but with the same phenotype, and identify new therapeutic targets specific to tumor M-MDSCs to enhance antitumor immune response.
Materials and methods
Tumor model
1 × 106 3LL tumor cells were inoculated subcutaneously into the C57BL/6 mice at 8–10 weeks of age. Mice were killed 15 days after inoculation. Spleens and tumors were excised and made into single cell suspension. Cells were labeled with cell surface markers for flow cytometry detection.
Cell culture
T cells were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum (Gibco), 20 mM L-glutamine (Gibco), 100units/mL penicillin–streptomycin (Gibco), 100 mM HEPES (Gibco) and 10 mM sodium pyruvate (Gibco). For suppression assays, T cells were activated by plate-bound anti-CD3 antibody (5 µg/mL) and soluble anti-CD28 antibody (2 µg/mL) in flat-bottom 48-well plates. For the glucose uptake assays, MDSC were cultured in RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum. For coculture assays of MDSC and T cells, T cell culture media were used. Both T cells and MDSC were incubated at 37 °C in 5% CO2.
Flow cytometry
Antibodies anti-CD45 (48–0451,25–0451, ebioscience), anti-CD11b (11–0112, ebioscience), anti-Ly6C (25-5932,12-5932, ebioscience), anti-IL-6 (48-7061, ebioscience), anti-IL-10 (12-7101, ebioscience), anti-TGF-β (565638, BD Pharmingen™), anti-CD69 (11–0691, ebioscience), anti-Glut1 (ab115730, abcam), anti-Ly6G (560599, BD Pharmingen™), and anti-IFN-γ (554413, BD Pharmingen™) were used for flow cytometry according to the manufacturers’ instructions.
Quantitative RT-PCR
Total RNA was prepared using RNAiso reagent (9109, Takara) and first-strand cDNA was synthesized using PrimeScript ™ RT reagent kit with gDNA Eraser (RR047, Takara) following the instructions of the manufacturer. Real-time PCR was conducted on Applied Biosystems® 7500(ThermoFisher, America) using SYBR Green reagent (RR820A, Takara). Gene expression was normalized to actin using 2-∆∆CT method. All primer sequences are listed in supplementary table 1.
T cell and MDSC isolation
T cells were isolated from spleen of WT C57/B6 mice by MACS using EasySep™ Mouse T cell isolation kit (19851, Stem Cell). To isolate splenic M-MDSCs and tumor M-MDSCs, single cells from spleen and tumor of 3LL tumor-bearing mice were labeled with the cell surface markers for M-MDSCs, defined as CD45+CD11b+Ly6Chigh and Ly6G−. Splenic M-MDSCs and tumor M-MDSCs were sorted by FACS (Moflo, Beckman Coulter). The purity of T cells was greater than 90% while the purity of M-MDSC was greater than 95%.
Western blot
MDSCs were sorted and lysed in RIPA buffer (9806, CST), supplemented with protease inhibitors and phosphatase inhibitors (04906845001, Roche) and then centrifuged in a microcentrifuge at 12,000g for 15 min at 4 °C to collect the supernatant. The concentration of cleared lysate was determined by the Bradford protein assay reagent (Bio-Rad, USA) and 30 µg of protein was separated by SDS-polyacrylamide gel electrophoresis (PAGE) after boiling for 15 min in 1 × SDS loading buffer (P0015L, Beyotime Biotechnology) at 80 V for 30 min and followed by 120V for 1 h. The proteins were then transferred to a polyvinylidene difluoride membrane (0.2um, Millipore, USA) by a transfer apparatus at 300 mA for 2 h. The membrane was blocked with 5% non-fat dry milk for 1 h before incubation with primary antibodies. After incubation of primary antibodies overnight at 4 °C, membranes were incubated with horseradish peroxidase-conjugated secondary antibody (CST 7074 anti-rabbit IgG, HRP-linked antibody) and were visualized using an enhanced chemiluminescence reagent (34095, Thermo Pierce). The primary antibodies were β-actin (M1210-5, Huaan), Akt antibody (9272, CST), phospho-Akt (Ser473) antibody (4058, CST), phospho-NF-κB p65 (Ser536) antibody (3033, CST), NF-κB p65 antibody (8242, CST), phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) antibody (4370, CST), p44/42 MAPK (Erk1/2) antibody (4695, CST), phospho-p38 MAPK (Thr180/Tyr182) antibody (4511, CST), p38 MAPK antibody (9212, CST), phospho-SAPK/JNK (Thr183/Tyr185) antibody (9251, CST), SAPK/JNK antibody (9252, CST) and mTOR (2972, CST).
CFSE assay
Flat-bottom plates were coated with 100 µL per well (96-well plate) of CD3 (e06299-1631, ebioscience) at 5 µg/mL concentration overnight at 4 °C. T cells were labeled with 5 µM CFSE (ThermoFisher) and cocultured with the M-MDSC at the indicated ratio for 3 days in 37 °C. T cell culture was supplemented with CD28 (e06392-1633, ebioscience) at 2 µg/mL concentration.
Glucose uptake assay
2NBDG (N13195, ThermoFisher) was used as a fluorescent indicator to directly measure glucose uptake. For in vitro assay, M-MDSCs were sorted and incubated with 100 µg/mL 2NBDG in 37 °C for 2 h and detected by FACS. For in vivo assay, mice were killed after injection of 200 µg 2NBDG for 4 h. Spleens and tumors were excised and made into single-cell suspension. Spleen M-MDSCs and tumor M-MDSCs were detected after staining with certain cell surface markers.
Metabolomic analysis by liquid chromatography–tandem mass spectrometry (LC/MS)
Chromatographic separation of targeted glycolytic and TCA intermediates was performed based on the methods of Hinder et al. (2012) [23]. Briefly, hydrophilic interaction liquid chromatography was performed using a Phenomenex Luna NH2 column (5 µm, 50 × 2.0 mm) on a SHIMADAZU LC20AB coupled to an ABSCIEX 4000QTRAP mass spectrometer. Mobile phase A was 5 mM ammonium acetate in water. Mobile phase B was acetonitrile. The instrument was operated in multiple-reaction monitoring (MRM) mode using MS/MS transitions previously optimized by analysis of authentic standards. The following parameters were implemented: flow rate was 0.3 mL/min, column temperature 25 °C and injection volume 10 µL. The ratio of each metabolite peak area to the closest matching standard was calculated. Relative quantification of metabolites was compared by peak intensity using software HemI 1.0 according to the instructions.
Statistical analysis
GraphPad Prism 6 software was used for all statistical analyses. The means of two groups were compared using the Student’s unpaired t test or unpaired t test with Welch’s correction. Differences were considered statistically significant at p < 0.05.
Results
Tumor M-MDSC was more suppressive than splenic M-MDSC
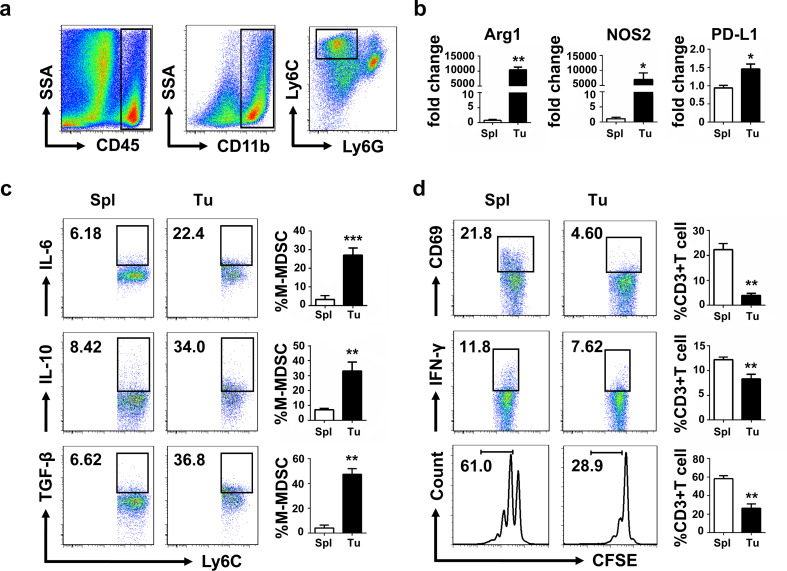
Tumor-derived factors promoted the expansion and accumulation of MDSCs [24]. To explore the functions of splenic M-MDSCs and tumor M-MDSCs, these cells from 3LL tumor-bearing mice were analyzed and sorted according to the cell surface marker CD45+CD11b+Ly6ChighLy6G− (Fig. 1a). Results of qPCR showed that tumor M-MDSCs displayed significantly increased expression of suppression-associated genes such as Arg1, NOS2 and PD-L1 than splenic M-MDSCs (Fig. 1b). Intracellular flow cytometry demonstrated that tumor M-MDSCs secreted more suppressive cytokines including IL-6, IL-10 and TGF-β than splenic M-MDSCs (Fig. 1c). Activated T cells were cocultured with splenic M-MDSCs or tumor M-MDSCs to analyze suppressive activity of M-MDSCs, by detecting the expression of activation markers CD69 and IFN-γ of T cells, and also their proliferation (Fig. 1d). Results showed that tumor M-MDSCs expressed more suppressive factors than splenic M-MDSCs, and T cells cocultured with tumor M-MDSCs expressed lower activation markers and proliferated slower than those cocultured with splenic M-MDSCs. These results demonstrated that tumor M-MDSCs had stronger suppressive activities as shown by the upregulation of Arg1, NOS2 and PD-L1, as well as increased suppression on activated T cells.
Fig. 1.
Tumor M-MDSCs were more suppressive than splenic M-MDSCs. a Gating strategy to identify tumor M-MDSC. It is the same with splenic M-MDSC. b Quantitative RT-PCR analysis of suppression-associated genes Arg1, NOS2 and PD-L1. c IL-6, IL-10 and TGF-β expression of tumor M-MDSCs and splenic M-MDSCs. d CD3+ T cells were isolated by MACS and cocultured with tumor M-MDSCs and splenic M-MDSCs, respectively, in a 48-well plate precoated with anti-CD3 (5 µg/mL) and anti-CD28 (2 µg/mL). CD69 and IFN-γ expression of CD3+T cells were measured 24 h later and proliferation of T cells labeled with CFSE was measured 72 h later (M-MDSC: T ratio = 1:4). Data are pooled from three independent experiments each with 3–5 mice and expressed as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001
Increased glycolysis was observed in tumor M-MDSC
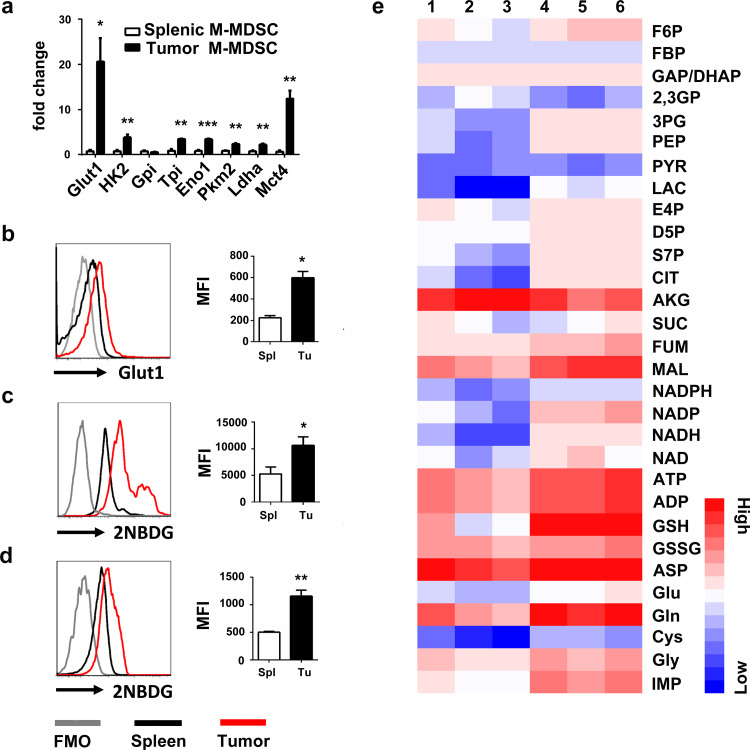
We aimed to find out the distinguishing characteristics of glucose metabolism between splenic M-MDSC and tumor M-MDSC and determine whether they could regulate the suppressive effects of these cells. Glycolysis is the first step of the glucose catabolism and is almost the universal pathway that converts glucose into pyruvate. Results showed that tumor M-MDSCs displayed a significantly increased expression of genes associated with glycolysis, including Glut1, Hk2, Gpi, Tpi, Eno1, Pkm2, Ldha and MCT4 (Fig. 2a, b). Furthermore, both in vitro (Fig. 2c) and in vivo (Fig. 2d) studies showed that, compared with splenic M-MDSCs, tumor M-MDSCs had a remarkably increased absorption of 2NBDG, a fluorescent indicator to measure glucose uptake. LC/MS (liquid chromatography–mass spectrometry) was used to analyze glucose metabolites and the results showed that tumor M-MDSC had more glucose metabolites than splenic M-MDSC. (Fig. 2e).
Fig. 2.
Tumor M-MDSCs have higher glycolysis than splenic M-MDSCs. a Quantitative RT-PCR analysis of glycolysis-associated genes including Glut1, Hk2, Gpi, Tpi, Eno1, Pkm2, Lhda and Mct4. b Cell surface Glut1 levels of tumor M-MDSCs and splenic M-MDSCs were detected by flow cytometry. 2NBDG incorporations of tumor M-MDSC and splenic M-MDSC were tested c in vitro and d in vivo. e LC/MS was used to detect the glycolytic metabolites and relative expressions were shown with heat map analyzed by hemI 1.0. Lines 1, 2 and 3 were metabolites of splenic M-MDSCs while lines 4, 5 and 6 were metabolites of tumor M-MDSCs. Full name of these metabolites are listed in supplementary table 2. FMO, Fluorescence Minus One. Data are from one experiment each with 3–10 mice and expressed as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001
mTOR phosphorylation level was high in tumor M-MDSC
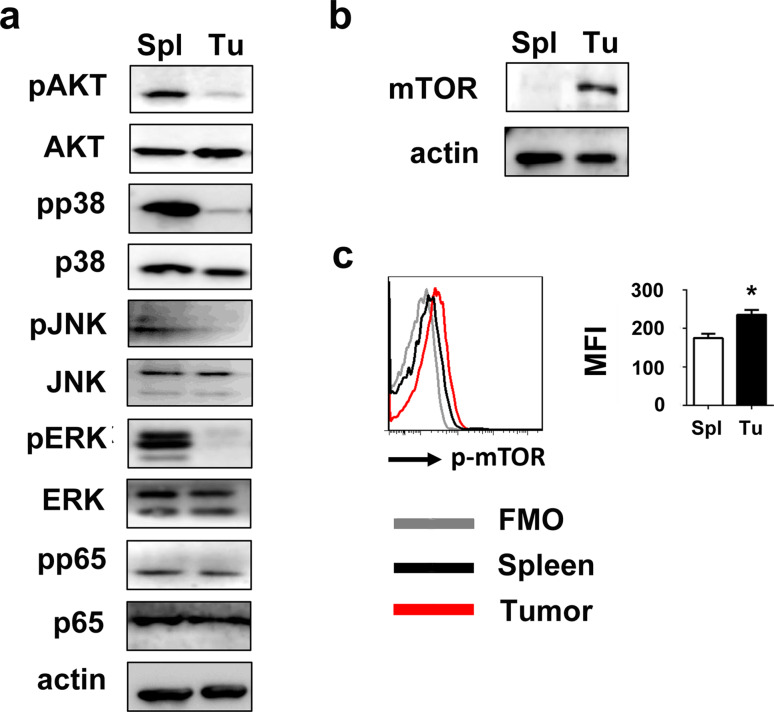
As a basic activity of cells, glycolysis is regulated by many pivotal signaling pathways, which helps to control the metabolic processes by molecular modifications step by step. We hypothesized a causality between upregulated glycolysis of tumor M-MDSCs and increased activation of signaling pathways relating to their suppressive activities. To our surprise, western blot showed that tumor M-MDSC had dramatically lower phosphorylation of Akt and MAPK pathway including ERK1/2, JNK and p38 than splenic M-MDSCs (Fig. 3a). However, we found that although many crucial signaling pathways were downregulated in tumor M-MDSCs, while mTOR, which was the target of rapamycin and one of the key nutritional sensors at the cellular and organismal levels, was upregulated (Fig. 3b). mTOR phosphorylation was also upregulated (Fig. 3c).
Fig. 3.
Tumor M-MDSCs downregulated Akt, NF-κB and MAPK but upregulated mTOR a Activation of Akt, NF-κB and MAPK pathways and b mTOR expression of tumor M-MDSCs and splenic M-MDSCs were detected by western blot. c mTOR phosphorylation was detected by flow cytometry. Data are pooled from three independent experiments each with three mice and expressed as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001
Suppressive activity of tumor M-MDSC was dependent on glycolysis
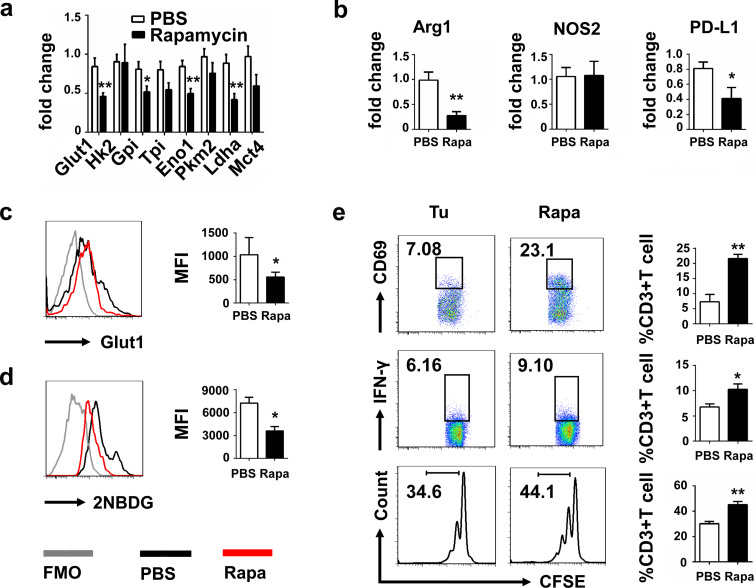
There are increasing evidences suggesting that mTOR is crucial for many major cellular behaviors such as survival, growth and aging, but how mTOR inhibition influences the physiology and suppressive function of MDSCs, especially tumor M-MDSCs, has not been extensively investigated. So, we explored the effects of mTOR inhibition on the functions of tumor M-MDSCs sorted from tumor tissue. Results showed that genes associated with glycolysis (Fig. 4a) and inhibitory molecules including Arg1 and PD-L1 (Fig. 4b) of tumor M-MDSCs were significantly downregulated after rapamycin treatment. Rapamycin pretreatment reduced the expression of Glut1 (Fig. 4c) and the absorption of 2NBDG (Fig. 4d). Rapamycin pretreatment also reduced their suppressive effects on T cells, resulting in enhanced T cell proliferation and upregulation of CD69 and IFN-γ expression (Fig. 4e). Because of the extensive role of mTOR in regulating cellular basic activities [25], we used 2-DG, which was the well-characterized glycolytic inhibitor via targeting the hexokinase, to test the effects of glycolysis inhibition on the suppressive functions of tumor M-MDSCs. After treatment of 2-DG for 24 h, tumor M-MDSCs were washed and cocultured with activated CFSE-labeled T cells. Similar results that suppressive effects of tumor M-MDSCs treated with 2-DG were also decreased, manifesting by decreased suppressive molecules and enhanced activation of cocultured T cells were obtained (supplementary Fig. 1). However, observed cardiac side effects (prolongation of the Q–T interval in the heart’s electrical cycle) in a clinical trial of this reagent limit its further clinical use [26]. On the contrary, rapamycin was approved by the US Food and Drug Administration to prevent organ transplant rejection. Its safety has been demonstrated in clinical use [27–29].
Fig. 4.
Rapamycin inhibited the glycolysis and reduced the suppressive ability of tumor M-MDSCs. a Quantitative RT-PCR analysis of glycolysis-associated genes including Glut1, Hk2, Gpi, Tpi1, Eno1, Pkm2, Lhda and Mct4 and suppression-associated genes b Arg1, NOS2 and PD-L1 of tumor M-MDSCs after treatment of rapamycin for 24 h. c Tumor M-MDSCs were isolated by FACS and treated with 100 µg/mL rapamycin for 24 h. Glut1 expressions were detected by flow cytometry. d 2NBDG incorporations of tumor M-MDSC and tumor M-MDSC treated with rapamycin were tested in vitro. e CD3+ T cells were isolated by MACS and cocultured with rapamycin pretreated or PBS pretreated tumor M-MDSCs, respectively, in a 48-well plate precoated with anti-CD3 (5 µg/mL) and anti-CD28 (2 µg/mL). CD69 and IFN-γ expressions of CD3+T cells were measured 24 h later and proliferation of T cells labeled with CFSE was measured 72 h later (M-MDSC: T ratio = 1:4). Data are pooled from three independent experiments and expressed as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001
Glycolysis inhibition in vivo reduced the tumor growth and the suppressive activity of tumor M-MDSC
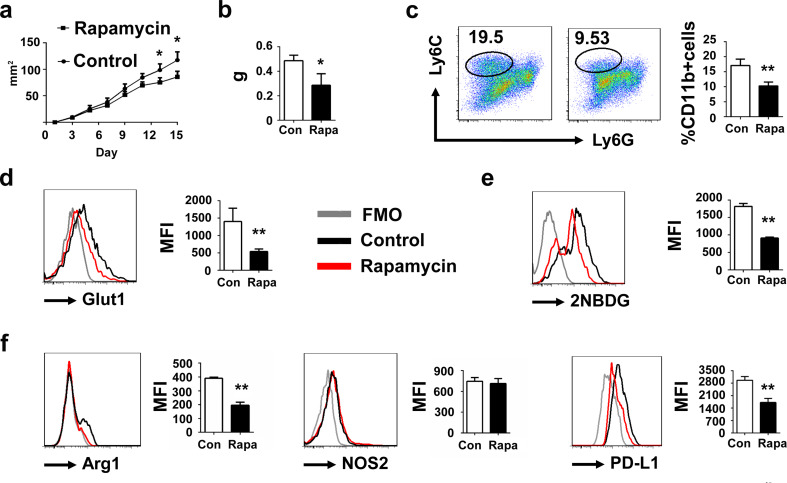
We next investigated whether rapamycin treatment reduced the tumor growth. Rapamycin were injected in situ with indicated time (Fig. 5a). 16 days later, mice were killed and tumors were harvested. We observed that rapamycin significantly reduced the tumor weight (Fig. 5b). There are also significant differences in size and tumor M-MDSC population when tumors were larger. (Supplementary data 2). To elucidate the effects of rapamycin on tumor M-MDSC, M-MDSCs in tumor were analyzed. We found that percentage of M-MDSCs in tumor was declined (Fig. 5c). Glut1 expression (Fig. 5d) and the absorption of 2NBDG (Fig. 5e) of tumor M-MDSC were also downregulated in rapamycin-treated group, indicating that rapamycin reduced the glycolysis of tumor M-MDSC. Suppression-associated genes such as Arg1, NOS2 and PD-L1 were also detected by flow cytometry. Results showed that rapamycin significantly reduced the expression of Arg1, PD-L1 but not NOS2 (Fig. 5f). We also detected the expression of cytokines including IL6, IL-10 and TGF-β after rapamycin treatment. We observed that rapamycin decreased the expression of these cytokines but there is no statistically significant difference in our experiments (supplementary Fig. 3). We thought this phenomenon may be due to the diverse sensitivities to rapamycin between different suppressive factors of tumor M-MDSC.
Fig. 5.
Glycolysis inhibition reduced the tumor growth. a C57BL/6 mice were implanted with 1 × 106 3LL tumor cells and treated with 10 µg rapamycin (solved in 2% DMSO + 30% PEG 300 + 5% Tween 80 + ddH2O) in situ every 2 days for five 5 times from day 3 when the tumors were palpable. Tumor areas were monitored every 2 days. b Tumor weight was compared between the control group (treated with 2% DMSO + 30% PEG 300 + 5% Tween 80 + ddH2O) and rapamycin-treated group. c Percentage of tumor M-MDSC was detected. d Glut1 expression and e 2NBDG incorporation of tumor M-MDSC from control group and rapamycin-treated group were detected by flow cytometry. f Arg1, NOS2 and PD-L1 expressions of tumor M-MDSC from control group and rapamycin-treated group were detected by flow cytometry. Data are pooled from three independent experiments and expressed as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001
Discussion
Here, we report that tumor M-MDSCs exhibit higher glycolytic rate and higher suppressive activities but dramatically lower phosphorylation of crucial signaling pathways including Akt and MAPK than splenic M-MDSCs. Instead, mTOR phosphorylation of tumor M-MDSC was upregulated. Inhibition of mTOR phosphorylation reduced the suppressive activity of tumor M-MDSCs and limited tumor growth. These observations are rather surprising because Akt and MAPK pathways are often aberrantly activated in cancer cells, which can lead to higher metabolic activities and contribute to tumor cell survival, tumorigenesis as well as metastasis [30]. Protein kinases play an important role in the activation of key components in signal transduction pathway, which makes their inhibitors promising for cancer therapy. However, there are still lots of patients failing to respond to treatment of kinase inhibitors [31]. As important suppressive cells in the tumor microenvironment, tumor M-MDSCs often contribute to chemotherapy or radiotherapy resistance. Our results showed that tumor M-MDSC exhibited stronger suppressive activities and increased glucose metabolism than splenic M-MDSC. But some of the crucial signal pathways were actually less activated in tumor M-MDSC. These findings indicate that certain pathways may not play important roles in the activities of tumor M-MDSC and inhibitors targeting these pathways may be unable to inhibit functions of these cells, which may result in resistance of cancer therapy.
Hossain et al. reported that tumor MDSCs increased fatty acid uptake and FAO (fatty acid β-oxidation) accompanied by an increase in both OCR (oxygen consumption rate) and ECAR (extracellular acidification rate) [32]. But the molecular pathways to regulate glucose metabolism, as well as the potential implication for the development of targeted therapeutics, remain unclear. Wu et al. showed that rapamycin significantly inhibited M-MDSC immunosuppressive function via iNOS pathway. Most of their results were focused on the peripheral MDSCs or cytokine-induced MDSCs [33]. Therefore, whether glucose metabolism is involved in tumor M-MDSCs’ function and the specific mechanisms of its regulation still needs more investigation. In this study, mTOR expression was demonstrated as a distinguishing characteristic between tumor M-MDSC and splenic M-MDSC, and may at least partially contribute to the enhanced function of tumor M-MDSC. However, factors upregulating the mTOR activity of tumor MDSCs remain unknown.
Rapamycin is considered an inducer of autophagy, and it is possible that by blocking glycolysis the cells would seek to preserve themselves via another means. Lin et al. found that autophagy decreased the level of glycolysis through ubiquitin-mediated selective degradation of HK2, a key enzyme in glycolysis, which demonstrated that glycolytic activity was negatively correlated with autophagy level in liver cancer. Their investigation also indicated that impaired autophagy contributed to substantial concomitant enhancement of glycolysis [34]. According to the results of Lin et al., elevated autophagy level may decrease the glycolysis. On the contrary, high level of glycolysis may be accompanied by low level of autophagy. Tumor M-MDSC had higher glycolysis level compared with splenic M-MDSC, which indicated that tumor M-MDSC may have the lower level of autophagy than splenic M-MDSC. Autophagy has closely relationship with glycolysis. In this study, we cannot exclude the possibility that rapamycin promoted the autophagy of MDSC. Furthermore, we speculated that enhancement of autophagy induced by rapamycin may be responsible for the glycolysis decrease of M-MDSC, which was in accordance with our results that rapamycin inhibited the glycolysis and reduced the suppressive activities of M-MDSC.
In this study, rapamycin reduced Arg1 and PD-L1 but not NOS2. The expression of inducible nitric oxide synthase (NOS2) is complex and is regulated by many different compounds including Toll-like receptors such as bacterial lipopolysaccharide (LPS) and stimulatory cytokines such as interferon-g (IFN-g), interleukin-1β(IL-1β), interleukin-6 (IL-6), tumor necrosis factor-a(TNF-a). These molecules induced NOS2 expression through different signaling pathways. According to the previous reports, there are many transcription factors involved in NOS2 expression including NF-κB, IRF-1, AP-1, C/EBP. Transcription factors bind to the promoter region to regulate the mRNA expression. Despite of the promoter activity, NOS2 mRNA and protein stability are also regulated by many pathways. Due to the complexity of NOS2 induction, different activators/inhibitors for one specific signal pathway often resulted in controversial data because of the cell type and species specificity. One of the possible reasons is that mTOR may not be the most important key regulator responsible for NOS2 expression of M-MDSC in this model. NOS2 expression and degradation may be regulated by other pathways. Another possibility is that rapamycin influenced the expression of NOS2 but there are compensatory signal transduction pathways neutralizing the effects of rapamycin. However, we still need more investigation to illuminate the exact reasons why rapamycin had no effects on NOS2 expression in M-MDSC. We also found that metabolites produced by both glycolysis and TCA cycle (tricarboxylic acid cycle) were increased in tumor M-MDSCs, which suggests a possibility that tumor M-MDSCs systemically upregulate their nutrition metabolism to support the suppressive activities of these cells. Because TCA cycle is a series reactions used by cells to release energy through the oxidation of acetyl-CoA derived from carbohydrates, fats and proteins into carbon dioxide [35]. Thus, more studies are needed to determine the mechanisms behind the phenomenon observed in this research. However, this study suggests a promising tactic to target both the cancer cells and the metabolism of tumor M-MDSCs to enhance antitumor effects. Furthermore, this study emphasizes the significance of studying immune cell metabolism because cells with the same phenotype may have diverse functions resulted from different metabolic patterns in specific microenvironment. A recent study found that inhibition of STAT3 resulted in depletion of MDSC in spleen but not in tumor, which also demonstrates the difference between tumor MDSC and splenic MDSC [36]. Most studies of MDSCs were focused on the peripheral MDSCs or MDSCs induced by cytokines because of the technical difficulties to isolate MDSC from tumor tissue. This study also implies that we should be cautious when trying to interpret the functions of MDSCs by in vitro experiments. Because MDSCs induced in vitro by different methods may not resemble closely enough the cells isolated from tumor-bearing hosts.
In summary, we find the suppressive activities of tumor M-MDSCs are associated with their glycolysis, and mTOR pathway plays a crucial role in this process, which further illuminates the molecular and cellular mechanisms to use rapamycin for cancer therapy and support the possibility to combine rapamycin with conventional therapies to enhance anti-tumor effects. These results may shed new light on development of new therapeutics against cancer by modulating metabolism of immune cells.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Abbreviations
- 2-DG
2-Deoxy-d-glucose
- 2NBDG
2-(N-(7-Nitrobenz-2-oxa-1,3-diazol-4-yl) amino)-2-deoxyglucose
- Arg1
Arginase I
- ECAR
Extracellular acidification rate
- Eno1
Enolase 1
- FMO
Fluorescence minus one
- HK2
Hexokinase 2
- Glut1
Glucose transporter 1
- Gpi
Glucose-6-phosphate isomerase
- LC/MS
Liquid chromatography–mass spectrometry
- Ldha
Lactate dehydrogenase A
- Mct4
Monocarboxylate transporter 4
- mTOR
Mammalian target of rapamycin
- NOS2
Nitric oxide synthase 2
- OCR
Oxygen consumption rate
- PD-L1
Programmed death ligand 1
- Pkm2
Pyruvate kinase muscle isozyme
- TCA cycle
Tricarboxylic acid cycle
- Tpi
Triosephosphate isomerase
Author contributions
Conception and design: YC, YD and JY; development of methodology: YD, JY and FL; acquisition of data: YC, YD, JY, FL, JQ, HY, DZ and RL; analysis and interpretation of data: YC, YD, JY and FL; writing, review, and/or revision of the manuscript: YC, YD, JY, FL and RL. All authors have read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China under Grants 31570892, 81730045 and 91527305.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest. 3LL Lewis Lung Carcinoma cells were purchased from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences. C57BL/6 mice were purchased from the Shanghai SLAC Laboratory Animal Co., Ltd. (Shanghai, China).
Animal rights
All experiments were approved by the Institutional Animal Care and Use Committee of Fudan University (20140226067) following the Guidelines for the Care and Use of Laboratory Animals (No. 55 issued by Ministry of Health, China on January 25th, 1998).
Footnotes
Yuting Deng and Jiao Yang contributed equally to this manuscript and should be considered as co-first authors.
References
- 1.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9(3):162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Filipazzi P, Huber V, Rivoltini L. Phenotype, function and clinical implications of myeloid-derived suppressor cells in cancer patients. Cancer Immunol Immunother. 2012;61(2):255–263. doi: 10.1007/s00262-011-1161-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haverkamp JM, Smith AM, Weinlich R, Dillon CP, Qualls JE, Neale G, Koss B, Kim Y, Bronte V, Herold MJ, Green DR, Opferman JT, Murray PJ. Myeloid-derived suppressor activity is mediated by monocytic lineages maintained by continuous inhibition of extrinsic and intrinsic death pathways. Immunity. 2014;41(6):947–959. doi: 10.1016/j.immuni.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Zhao Y, Wu T, Shao S, Shi B, Zhao Y. Phenotype, development, and biological function of myeloid-derived suppressor cells. Oncoimmunology. 2015;5(2):e1004983. doi: 10.1080/2162402X.2015.1004983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui TX, Kryczek I, Zhao L, Zhao E, Kuick R, Roh MH, Vatan L, Szeliga W, Mao Y, Thomas DG, Kotarski J, Tarkowski R, Wicha M, Cho K, Giordano T, Liu R, Zou W. Myeloid-derived suppressor cells enhance stemness of cancer cells by inducing microRNA101 and suppressing the corepressor CtBP2. Immunity. 2013;39(3):611–621. doi: 10.1016/j.immuni.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu X, Meng Q, Erben U, Wang P, Glauben R, Kuhl AA, Wu H, Ma CW, Hu M, Wang Y, Sun W, Jia J, Wu X, Chen W, Siegmund B, Qin Z. Myeloid-derived suppressor cells promote B-cell production of IgA in a TNFR2-dependent manner. Cell Mol Immunol. 2017;14(7):597–606. doi: 10.1038/cmi.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ostrand-Rosenberg S, Sinha P. Myeloid-derived suppressor cells: linking inflammation and cancer. J Immunol. 2009;182(8):4499–4506. doi: 10.4049/jimmunol.0802740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang HF, Ning F, Liu ZC, Wu L, Li ZQ, Qi YF, Zhang G, Wang HS, Cai SH, Du J. Histone deacetylase inhibitors deplete myeloid-derived suppressor cells induced by 4T1 mammary tumors in vivo and in vitro. Cancer Immunol Immunother. 2016 doi: 10.1007/s00262-016-1935-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sawant A, Schafer CC, Jin TH, Zmijewski J, Tse HM, Roth J, Sun Z, Siegal GP, Thannickal VJ, Grant SC, Ponnazhagan S, Deshane JS. Enhancement of antitumor immunity in lung cancer by targeting myeloid-derived suppressor cell pathways. Cancer Res. 2013;73(22):6609–6620. doi: 10.1158/0008-5472.CAN-13-0987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Mitri D, Toso A, Alimonti A. Molecular pathways: targeting tumor-infiltrating myeloid-derived suppressor cells for cancer therapy. Clin Cancer Res. 2015 doi: 10.1158/1078-0432.CCR-14-2261. [DOI] [PubMed] [Google Scholar]
- 12.Spinetti T, Spagnuolo L, Mottas I, Secondini C, Treinies M, Ruegg C, Hotz C, Bourquin C. TLR7-based cancer immunotherapy decreases intratumoral myeloid-derived suppressor cells and blocks their immunosuppressive function. Oncoimmunology. 2016;5(11):e1230578. doi: 10.1080/2162402X.2016.1230578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trikha P, Plews RL, Stiff A, Gautam S, Hsu V, Abood D, Wesolowski R, Landi I, Mo X, Phay J, Chen CS, Byrd J, Caligiuri M, Tridandapani S, Carson W. Targeting myeloid-derived suppressor cells using a novel adenosine monophosphate-activated protein kinase (AMPK) activator. Oncoimmunology. 2016;5(9):e1214787. doi: 10.1080/2162402X.2016.1214787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maenhout SK, Thielemans K, Aerts JL. Location, location, location: functional and phenotypic heterogeneity between tumor-infiltrating and non-infiltrating myeloid-derived suppressor cells. Oncoimmunology. 2014;3(10):e956579. doi: 10.4161/21624011.2014.956579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corzo CA, Condamine T, Lu L, Cotter MJ, Youn JI, Cheng P, Cho HI, Celis E, Quiceno DG, Padhya T, McCaffrey TV, McCaffrey JC, Gabrilovich DI. HIF-1alpha regulates function and differentiation of myeloid-derived suppressor cells in the tumor microenvironment. J Exp Med. 2010;207(11):2439–2453. doi: 10.1084/jem.20100587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thevenot PT, Sierra RA, Raber PL, Al-Khami AA, Trillo-Tinoco J, Zarreii P, Ochoa AC, Cui Y, Del Valle L, Rodriguez PC. The stress-response sensor chop regulates the function and accumulation of myeloid-derived suppressor cells in tumors. Immunity. 2014;41(3):389–401. doi: 10.1016/j.immuni.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gantt S, Gervassi A, Jaspan H, Horton H. The role of myeloid-derived suppressor cells in immune ontogeny. Front Immunol. 2014;5:387. doi: 10.3389/fimmu.2014.00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chandra D, Gravekamp C. Myeloid-derived suppressor cells: Cellular missiles to target tumors. Oncoimmunology. 2013;2(11):e26967. doi: 10.4161/onci.26967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Condamine T, Ramachandran I, Youn JI, Gabrilovich DI. Regulation of tumor metastasis by myeloid-derived suppressor cells. Annu Rev Med. 2014 doi: 10.1146/annurev-med-051013-052304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sukumar M, Liu J, Ji Y, Subramanian M, Crompton JG, Yu Z, Roychoudhuri R, Palmer DC, Muranski P, Karoly ED, Mohney RP, Klebanoff CA, Lal A, Finkel T, Restifo NP, Gattinoni L. Inhibiting glycolytic metabolism enhances CD8 + T cell memory and antitumor function. J Clin Invest. 2013;123(10):4479–4488. doi: 10.1172/JCI69589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang C-H, Qiu J, O’Sullivan D, Buck Michael D, Noguchi T, Curtis Jonathan D, Chen Q, Gindin M, Gubin Matthew M, van der Windt Gerritje JW, Tonc E, Schreiber Robert D, Pearce Edward J, Pearce Erika L. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell. 2015;162(6):1229–1241. doi: 10.1016/j.cell.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinder LM, Vivekanandan-Giri A, McLean LL, Pennathur S, Feldman EL. Decreased glycolytic and tricarboxylic acid cycle intermediates coincide with peripheral nervous system oxidative stress in a murine model of type 2 diabetes. J Endocrinol. 2012;216(1):1–11. doi: 10.1530/JOE-12-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar V, Patel S, Tcyganov E, Gabrilovich DI. The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends Immunol. 2016 doi: 10.1016/j.it.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raez LE, Papadopoulos K, Ricart AD, Chiorean EG, Dipaola RS, Stein MN, Rocha Lima CM, Schlesselman JJ, Tolba K, Langmuir VK, Kroll S, Jung DT, Kurtoglu M, Rosenblatt J, Lampidis TJ. A phase dose-escalation trial of 2-deoxy-d-glucose alone or combined with docetaxel in patients with advanced solid tumors. Cancer Chemother Pharmacol. 2013;71(2):523–530. doi: 10.1007/s00280-012-2045-1. [DOI] [PubMed] [Google Scholar]
- 27.Waldner M, Fantus D, Solari M, Thomson AW. New perspectives on mTOR inhibitors (rapamycin, rapalogs and TORKinibs) in transplantation. Br J Clin Pharmacol. 2016;82(5):1158–1170. doi: 10.1111/bcp.12893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baroa-Mazo A, Revilla-Nuin B, Ramirez P, Pons JA. Immunosuppressive potency of mechanistic target of rapamycin inhibitors in solid-organ transplantation. World J Transplant. 2016;6(1):183–192. doi: 10.5500/wjt.v6.i1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saunders RN, Metcalfe MS, Nicholson ML. Rapamycin in transplantation: a review of the evidence. Kidney Int. 2001;59(1):3–16. doi: 10.1046/j.1523-1755.2001.00460.x. [DOI] [PubMed] [Google Scholar]
- 30.Populo H, Lopes JM, Soares P. The mTOR signalling pathway in human cancer. Int J Mol Sci. 2012;13(2):1886–1918. doi: 10.3390/ijms13021886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu P, Nielsen TE, Clausen MH. FDA-approved small-molecule kinase inhibitors. Trends Pharmacol Sci. 2015;36(7):422–439. doi: 10.1016/j.tips.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 32.Hossain F, Al-Khami AA, Wyczechowska D, Hernandez C, Zheng L, Reiss K, Valle LD, Trillo-Tinoco J, Maj T, Zou W, Rodriguez PC, Ochoa AC. Inhibition of fatty acid oxidation modulates immunosuppressive functions of myeloid-derived suppressor cells and enhances cancer therapies. Cancer Immunol Res. 2015;3(11):1236–1247. doi: 10.1158/2326-6066.CIR-15-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu T, Zhao Y, Wang H, Li Y, Shao L, Wang R, Lu J, Yang Z, Wang J, Zhao Y. mTOR masters monocytic myeloid-derived suppressor cells in mice with allografts or tumors. Sci Rep. 2016;6:20250. doi: 10.1038/srep20250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiao L, Zhang HL, Li DD, Yang KL, Tang J, Li X, Ji J, Yu Y, Wu RY, Ravichandran S, Liu JJ, Feng GK, Chen MS, Zeng YX, Deng R, Zhu XF. Regulation of glycolytic metabolism by autophagy in liver cancer involves selective autophagic degradation of HK2 (hexokinase 2) Autophagy. 2017 doi: 10.1080/15548627.2017.1381804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fernie AR, Carrari F, Sweetlove LJ. Respiratory metabolism: glycolysis, the TCA cycle and mitochondrial electron transport. Curr Opin Plant Biol. 2004;7(3):254–261. doi: 10.1016/j.pbi.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 36.Kumar V, Cheng P, Condamine T, Mony S, Languino LR, McCaffrey JC, Hockstein N, Guarino M, Masters G, Penman E, Denstman F, Xu X, Altieri DC, Du H, Yan C, Gabrilovich DI. CD45 phosphatase inhibits STAT3 transcription factor activity in myeloid cells and promotes tumor-associated macrophage differentiation. Immunity. 2016;44(2):303–315. doi: 10.1016/j.immuni.2016.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.