Abstract
Class I and class II HLA proteins, respectively, have been associated with subsets of V(D)J usage resulting from recombination of the T-cell receptor (TCR) genes. Additionally, particular HLA alleles, in combination with dominant TCR V(D)J recombinations, have been associated with several autoimmune diseases. The recovery of TCR recombination reads from tumor specimen exome files has allowed rapid and extensive assessments of V(D)J usage, likely for cancer resident T-cells, across relatively large cancer datasets. The results from this approach, in this report, have permitted an extensive alignment of TCR-β VDJ usage and HLA class I and II alleles. Results indicate the correlation of both better and worse cancer survival rates with particular TCR-β, V and J usage-HLA allele combinations, with differences in median survival times ranging from 7 to 130 months, depending on the cancer and the specific TCR-β V and J usage/HLA class allele combination.
Keywords: T-cell receptor, V and J segment usage, HLA class I and class II, Cancer survival.
Introduction
Cancer development has long been connected with a failure of the T-cell arm of the adaptive immune system, with the observation that immunosuppressive drugs known to impact T-cell replication lead to adventitious tumor development [1, 2]. And, CD4 T-cell depletion in AIDS is associated with cancer development [3]. Numerous experimental, and even therapeutic approaches to cancer have substantiated those initial observations [4]. However, the role of the interaction of specific antigen presenting molecules, i.e., HLA allelic variants, with the protein coding results of particular TCR V(D)J recombinations, in cancer development, has not yet been established or well-researched. There have been several approaches describing the TCR-HLA binding process, particularly with experimental manipulation of small numbers of TCR-HLA class I binding partners [5, 6]. And recently, there have been big-data indications that particular TCR V(D)J recombinations are associated with particular HLA class I or class II molecules [7, 8]; and that particular TCR V(D)J recombinations and HLA class I alleles are associated with certain autoimmune diseases [9] or viral infections [10]. In the cancer setting, there have been TCR V(D)J usage, HLA connections in melanoma, but without significant survival distinctions [11]. Here we report that, for a wide variety of cancer types, the combination of particular HLA class I or II alleles, and TCR-β VDJ recombinations, are associated with either a better or worse survival rate, depending on the cancer and the TCR/HLA binding partners involved. The analyses and conclusions reported here have been facilitated by the efficiency of recovering TCR-β VDJ recombination reads from tumor specimen exome files (WXS) [12–15], and the simultaneous use of those files for determining HLA types.
Methods
Recovery of TCR-β recombination reads from tumor specimen whole exome files (WXS)
TCR-β recombination reads were recovered from WXS files using minor modifications of the algorithm described previously [12]. All candidate, TCR recombination reads were processed by the IMGT web tool for identification of productive TRB (TCR-β) recombinations. Only reads indicating a productive TRB coding region, based on 20 V-nucleotide and 20 J-nucleotide matches, were set aside for further study.
Determination of HLA class I and class II alleles for the cancer Genome Atlas (TCGA) SKCM (melanoma), LUAD (lung adenocarcinoma), STAD (stomach adenocarcinoma), HNSC (head and neck cancer), and OV (ovarian cancer) datasets
WXS files downloaded via TCGA approved project #6300 were used to determine the HLA-A, HLA-B, and HLA-C alleles for barcodes (samples) in the TCGA SKCM (469 samples), LUAD (573 samples), STAD (416 samples), HNSC (490), and OV (386) datasets. These WXS sequence files were downloaded as .BAM files from http://gdc-portal.nci.nih.gov. HLA-A, HLA-B, and HLA-C alleles were determined using Optitype [16] installed at USF research computing from a source at github.com. To check the accuracy of the program as well as for errors in post processing, RNASeq files were selected randomly to match corresponding WXS samples and processed via the Optitype protocol. Results for TCGA barcodes using these independent TCGA files types (WXS and RNASeq) were compared and were identical. The HLA class II alleles were obtained via the software described by ref [17].
Matching patient HLA class I alleles with T-cell receptor (TCR) beta recombinations
HLA data were matched with corresponding productive VDJ recombinations. This was accomplished using an original script termed, VDJ-AlleleMatcher.pl. Only the J segment categories, J1 and J2, were considered in this analysis. The sub-segments, e.g., J2-1, were stripped off by the script, i.e., TRBJ2-1 was considered as “TRBJ2”. All TRBV segments were included in the analyses; however, TRBV alleles were not differentiated. After completion of this process, specific TCR-β VDJ-HLA class I allele combinations were counted and recorded for subsequent analysis.
Overall survival and disease-free survival assessments for most frequently observed TCR-β VDJ segments and HLA allele combinations
Barcodes for patients representing various TCR-β VDJ-HLA class I allele combinations were assessed for survival patterns using Kaplan–Meier (KM) curves, using the cBioPortal.org web tool, as well as via independent analyses from downloaded, raw clinical data followed by use of the IBM SPSS software.
Raw data and computer code
A very large amount of raw data, including large numbers of TCR-β recombination reads, HLA class I allele verification data, and KM curve p value calculations were generated in the course of this project. These data are available upon request to corresponding author. In particular, the results below describe combinations of TCR-β V or J usage and HLA alleles representing distinct survival rates. In most cases, these survival rates represent cases where neither the TCR-β gene segment usage, nor the HLA allele, was independently associated with a significant survival distinction. The data that illustrate lack of survival distinctions, where noted below, are included in the supplemental data available from the corresponding author. Additionally, the computer scripts used for WXS file mining, and for HLA class I allele typing, are available upon request. In addition, all of the above indicated raw data and computer scripts can be accessed at http://www.universityseminarassociates.com/Supporting_online_material_for_scholarly_pubs.php.
Results
We evaluated the disease-free and overall survival rates of the TCGA melanoma dataset, SKCM, with respect to TCR-β VDJ usage. The barcode group (patients) representing usage of any one of the TRBJ2 gene segments, as detected by recovery of TCR-β VDJ recombination reads from the SKCM WXS files [12, 14], represented better overall survival (p < 0.032; Table 1). The better survival rate for the barcode group representing recovery of TRBJ2 gene segments, in the recombinations reads, was in comparison to a barcode group representing all remaining samples in the SKCM dataset, i.e., a combination of: (1) samples where only TRBJ1 recombination reads were recovered; and (2) samples with no recovery of TCR-β recombination reads.
Table 1.
Survival patterns of TCGA-SKCM barcode groups representing TCR-β VDJ recombinations paired with specific HLA class I alleles
| Number of barcodes | Overall survival in comparison to all other barcodes, p value | Disease-free survival in comparison to all remaining barcodes, p value | Overall survival outcome | Disease-free survival outcome | |
|---|---|---|---|---|---|
| TCR-β J2 segment paired with HLA class I allele | |||||
| TRBJ2 HLA-A*01:01 | 46 | 0.0061 | 0.18 | Significantly better | Trends better |
| TRBJ2 HLA-A*03:01 | 29 | 0.13 | 0.043 | Trends better | Significantly better |
| TRBJ2 HLA-A*24:02 | 28 | 0.26 | 0.032a | Trends worse | Significantly worsea |
| TRBJ2 HLA-B*08:01 | 33 | 0.0086 | 0.091 | Significantly better | Trends better |
| TRBJ2 HLA-C*07:01 | 44 | 0.034 | 0.48 | Significantly better | No difference |
| TCR-β VDJ2-HLA class I allele combinations not associated with a survival outcome | |||||
| TRBJ2 HLA-A*02:01 | 65 | ||||
| TRBJ2 HLA-A*11:01 | 20 | ||||
| TRBJ2 HLA-B*07:02 | 35 | ||||
| TRBJ2 HLA-C*07:02 | 38 | ||||
| TRBJ2 HLA-C*04:01 | 28 | ||||
aBoth the TCR-β gene segment and the HLA class I allele are required for a statistically significant association with the indicated outcome. Lack of “a”, with a significant p value, indicates that it is not possible to rule out a significant association via either the TCR-β gene segment or HLA class I allele alone
Within the TRBJ2 barcode group, several HLA class I allele combinations maintained the increased survival rates, in comparison to all remaining SKCM barcodes, and several did not (Table 1). However, the sample sizes were insufficient to establish a clear association of a single TRBJ2-HLA class I allele combination with increased survival rates, in comparison with a distinct TRBJ2 barcode group. For example, the TRBJ2-HLA-A*01:01 combination maintains the better overall survival rate, in comparison to the barcode group representing all remaining SKCM samples (Table 1). However, there are too few HLA-A*01:01 samples lacking TRBJ2 to determine whether the HLA-A*01:01 allele, in the absence of TRBJ2, is sufficient for a better survival rate. While a comparison of p values for different combinations of TRBJ2 and HLA class I allele provides some hint that a single, specific TRBJ2-HLA class I allele combination represents better survival rates than TRBJ2 alone, overall, data available here do not firmly establish any such results.
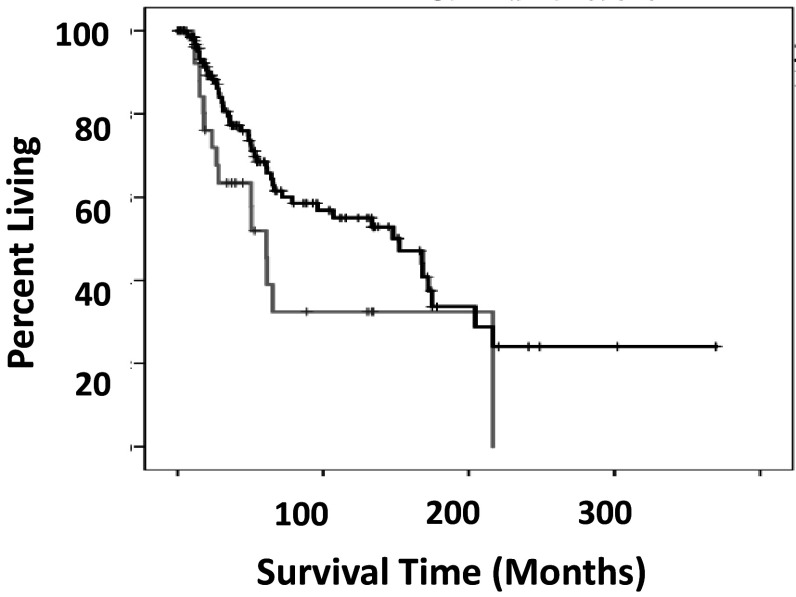
One of the identified SKCM TRBJ2-HLA class I combinations, TRBJ2-HLA-A*24:02, indicated a reduced disease-free survival rate in comparison to all remaining samples (Table 1). As noted above, recovery of any one of the TRBJ2 gene segments alone represents increased disease-free survival rates; and the HLA-A*24:02 allele in the absence of a TRBJ2 gene segment, in comparison to all remaining samples, does not indicate any survival rate distinction. Finally, TRBJ2-HLA-A*24:02 barcode group has worse overall survival rates in comparison to a barcode group representing all barcodes where there was recovery of a TRBJ2 gene segment in the recombination reads (Fig. 1).
Fig. 1.
Overall survival comparison of SKCM TRBJ2-HLA-A*24:02 and all remaining barcodes with TRBJ2. Grey line indicates barcodes with TRBJ2-HLA-A*24:02. Black line indicates barcodes that contain TRBJ2 with barcodes of TRBJ2-HLA-A*24:02 removed. The difference in the overall survival medians was 141.9 months, where TRBJ2-HLA-A*24:02 had a reduced survival rate compared to all remaining barcodes with TRBJ2. (Log rank p < 0.039)
The LUAD dataset yielded four significant TCR-β VDJ-HLA-class I allele combinations (Table 2). Three of these combinations had statistically significant, decreased disease-free survival rates in comparison to all remaining barcodes in the LUAD dataset. One identified combination, TRBJ2-HLA-C*07:02, represented increased overall survival in comparison to all remaining barcodes. For the TCR-β VDJ2-HLA class I allele combinations in this LUAD dataset, indicated above, neither component (the TCR-β VDJ2 segment or the HLA class I allele) was found to have a statistically significant association with either disease-free survival or overall survival, i.e., statistically significant survival associations were only found when assessing the TCR-β VDJ-HLA class I alleles as combinations noted above (Table 2).
Table 2.
Survival patterns of LUAD, STAD, HNSC, OV barcode groups representing TCR-β VDJ recombinations paired with specific HLA class I alleles
| Cancer | TRBV or TRBJ usage paired with HLA allele | Number of bar-codes | Overall survival in comparison to all other barcodes, p value | Disease-free survival in comparison to all other barcodes, p value | Overall survival outcome | Disease-free survival outcome |
|---|---|---|---|---|---|---|
| LUAD | ||||||
| TRBV5 HLA-A*02:01 | 37 | 0.14 | 0.023a | Trends worse | Significantly worsea | |
| TRBV6 HLA-A*03:01 | 34 | 0.34 | 0.044a | Trends worse | Significantly worsea | |
| 1 | TRBV7 HLA-A*03:0 | 24 | 0.88 | 0.036a | Trends worse | Significantly worsea |
| TRBJ2 HLA-C*07:02 | 63 | 0.03a | 0.14 | Significantly bettera | Trends better | |
| STAD | ||||||
| TRBJ1 HLA-A*24:02 | 36 | 0.0068 | 0.017 | Significantly better | Significantly better | |
| TRBJ1 HLA-C*07:01 | 31 | 0.04a | 0.084 | Significantly bettera | Trends better | |
| TRBJ1 HLA-C*06:02 | 26 | 0.41 | 0.035a | No difference | Significantly bettera | |
| HNSC | ||||||
| TRBJ1 HLA-A*02:01 | 85 | 0.014 | 0.59 | Significantly better | No difference | |
| TRBJ2 HLA-B*07:02 | 29 | 0.01a | 0.11 | Significantly bettera | Trends better | |
| TRBJ2 HLA-C*07:02 | 34 | 0.01a | 0.31 | Significantly bettera | No difference | |
| TRBV12 HLA-A*02:01 | 27 | 0.021 | 0.092 | Significantly better | No difference | |
| OV | ||||||
| TRBJ1 HLA-A*01:01 | 21 | 0.35 | 0.034a | No difference | Significantly bettera | |
| TRBJ1 HLA-A*02:01 | 47 | 0.37 | 0.017a | Trends better | Significantly bettera | |
| TRBJ2 HLA-A*02:01 | 38 | 0.33 | 0.012 | Trends better | Significantly better | |
| TRBJ2 HLA-A*03:01 | 16 | 0.05a | 0.079 | Significantly bettera | Trends better | |
| TRBV6 HLA-A*02:01 | 21 | 0.69 | 0.0063 | Trends better | Significantly better | |
aBoth the TCR-β gene segment and the HLA class I allele are required for a statistically significant association with the indicated outcome. Lack of “a”, with a significant p value, indicates that it is not possible to rule out a significant association via either the TCR-β gene segment or HLA class I allele alone
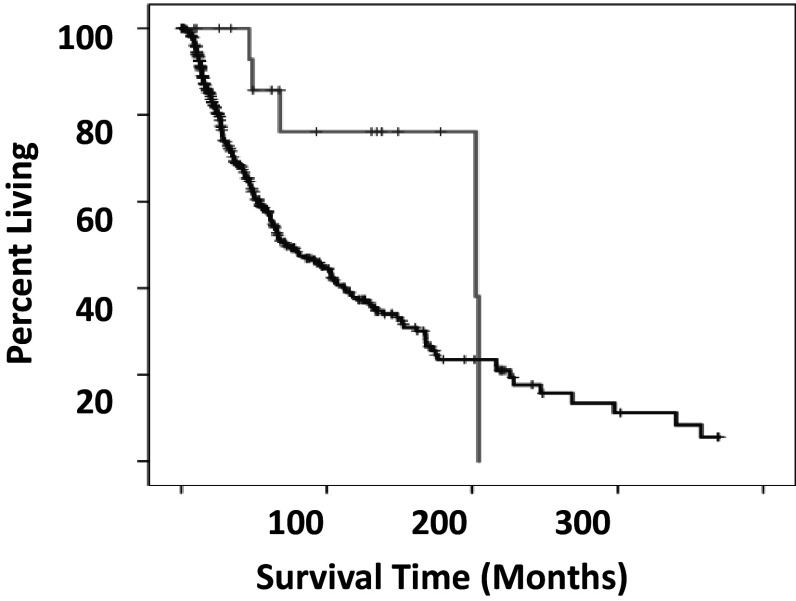
The STAD dataset revealed two significant TCR-β VDJ1-HLA class I allele combinations (Table 2), where a statistically significant association with a survival outcome required both components of the TCR-β VDJ-HLA class I allele combination (Fig. 2). These two combinations represented statistically significant associations with either disease-free survival or overall survival, respectively, in comparison to all remaining barcodes. One additional TCR-β VDJ1-HLA class I allele combination, TRBJ1-HLA-A*24:02, represented a statistically significant association for both disease-free and overall survival. However, there is a significant association with an increase in both disease-free and overall survival associated for all STAD barcodes with the HLA-A*24:02 allele in comparison with all remaining barcodes. This trend may be explained by the fact that a large number of the HLA-A*24:02 alleles are present in combination with TCR-β VDJ segment J1, however, that possibility cannot be verified with the current, STAD dataset.
Fig. 2.
Overall survival of STAD, TRBJ1-HLA-C*7:01 barcodes versus all remaining barcodes that lack a statistically significant, VDJ-HLA class I combination associating with outcome. Grey line indicates barcodes with TRBJ1-HLA-C*7:01. Black line indicates all remaining barcodes, except with TRBJ1-HLA-C*7:01, TRBJ1 HLA-A*24:02, and TRBJ1 HLA-C*06:02 barcodes removed. The difference in the mean overall survival times was 18.37 months. (Log rank p < 0.028)
The initial analyses of TCGA-HNSC revealed four TCR-β VDJ-HLA class I allele combinations where there was an association with either disease-free or overall survival, when comparing the TCR-β-HLA class I allele combination barcodes to all remaining barcodes in the HNSC dataset (Table 2). In the case of the TRBJ1-HLA-A*02:01 combination, the TRBJ1 segments were found to have an association with overall survival when compared with barcodes that do not have J1 segments, i.e., those containing a TRBJ2 segment. In the case of the TRBV12-HLA-A*02:01 combination, the TRBV12 segment was found to have a significant overall survival result when compared against barcodes that do not have a V12 segment. (A comparison of TRBV12 HLA-A*02:01 barcodes with a TRBV12 segment matched with other HLA class I alleles was not feasible due to an insufficiency of remaining barcodes containing a V12 segment.)
When examining the TCGA OV dataset one TCR-β VDJ-HLA class I allele combination, TRBJ2-HLA-A*03:01, was significantly associated with overall survival, and in this case, neither the TRBJ2 nor the HLA-A*03:01 component of the combination was significantly, independently associated with overall survival. However, barcode groups representing the TCR-β VDJ2 and V6 gene segments were statistically, significantly associated with disease-free survival, i.e., this dataset did not permit an opportunity to associate any TCR-β J2 or V6 HLA class I allele combinations with disease-free survival. Two TCR-β VDJ1-HLA class I allele combinations were found to be statistically, significantly associated with disease-free survival when compared against all other barcodes (Table 2). In these latter two cases, neither the TCR-β nor the HLA class I allele component had a statistically significant, independent association with disease-free survival.
To further illustrate the survival patterns and the possible importance of TCR-β VDJ-HLA class I allele combinations, we examined the median and mean survival time differences for barcodes representing above TCR-β-HLA combinations, versus all remaining barcodes (Table 3). For example, LUAD patients with the TRBV5-HLA-A*02:01 combination (Tables 2, 3) had a median disease-free survival time that was 19 months shorter than patients without that combination. As another example, HNSC showed that patients with the TRBJ2 HLA-C*07:02 combination lived 32.4 months longer than patients with other TCR-β VDJ-HLA class I combinations (Table 3).
Table 3.
Estimated survival time differences for TCR-β VDJ-HLA class I allele combinations, versus remaining barcodes, where V and J usage or HLA class I alleles do not independently demonstrate any survival rate associations. This list of survival associations matches the “&”-list of Table 2
| TCR-β VDJ-HLA class I allele combination | Difference between indicated TCR-HLA combination and remaining barcodes for overall survival, median time (months), if significant | Difference between indicated TCR-HLA combination and remaining barcodes for disease-free survival, median time (months), if significant | |
|---|---|---|---|
| (A) Dataset | |||
| SKCM | TRBJ2 HLA-A*24:02 | − 7.1 | |
| LUAD | TRBV5 HLA-A*02:01 | − 19.06 | |
| LUAD | TRBV6 HLA-A*03:01 | − 4.92 | |
| LUAD | TRBV7 HLA-A*03:01 | − 16.76 | |
| LUAD | TRBJ2 HLA-C*07:02 | 11.3 | |
| HNSC | TRBJ2 HLA-B*07:02 | 32.69 | |
| HNSC | TRBJ2 HLA-C*07:02 | 34.9 | |
| (B) Dataset | |||
| STAD | TRBJ1 HLA-C*07:01 | 17.77 | |
| STAD | TRBJ1 HLA-C*06:02 | 8.943 | |
| OV | TRBJ1 HLA-A*01:01 | 35.45 | |
| OV | TRBJ1 HLA-A*02:01 | 17 | |
(A) Selected barcodes represent TCR-β VDJ-HLA combinations where neither the TCR nor HLA arm represented barcodes that were independently significant against remaining barcodes. For these selections, data are considered normal where barcode survival data are available across a sufficient amount of time. For these combinations median values are considered
(B) Selected barcodes represent TCR-β VDJ-HLA combinations where neither the TCR nor the HLA arm represented barcodes that were independently significant against remaining barcodes. For these selections data are not available across a sufficient amount of time for reporting of median values. For these combinations, mean values are reported
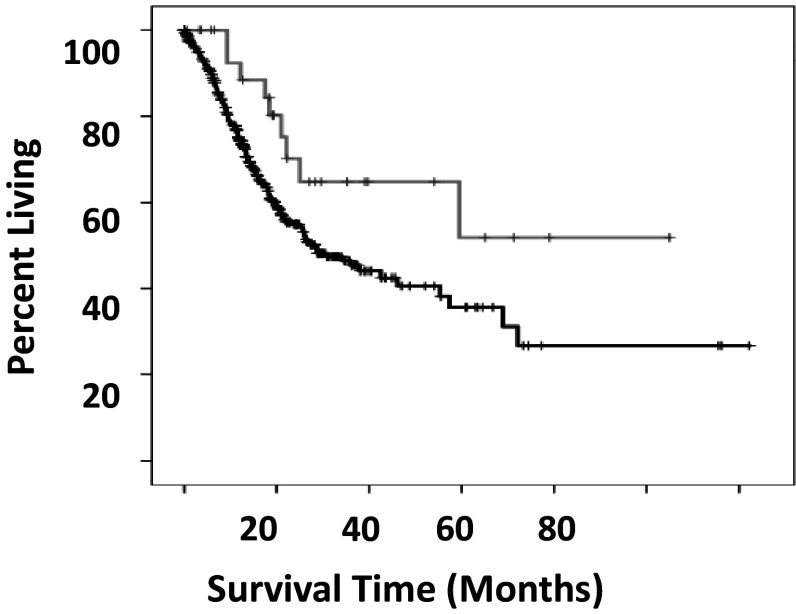
With regard to TCR-β V or J usage and HLA class II alleles, barcodes containing the TRBV6-HLA-DQB1*03:01 combination had significantly better overall survival times when compared to barcodes that do not contain this combination (Fig. 3). Strikingly, patients with this combination survived with median survival times 130 months longer than patients without this combination. The TRBV6 component of this combination did not have a significant overall survival pattern on its own. The DQB1*03:01 component of this combination also did not have a significant survival rate difference in comparison with all remaining barcodes, after removing the barcodes that had the TRBV6-HLA-DQB1*03:01 combination.
Fig. 3.
Overall survival of SKCM TRBV6-HLA-DQB1*3:01 versus all other barcodes without a significant TCR-β VDJ-HLA class II survival association. Grey line indicates barcodes with TRBV6-HLA-DQB1*03:01. Black line indicates all remaining barcodes. The difference in the median overall survival times was 130.5 months. (Log rank p < 0.025)
Discussion
The data provided here, linking HLA alleles and TCR-β V and J usage to distinct cancer outcomes, as well as closely related data in the cases of infection and autoimmunity, reported by other groups [9, 10, 13], indicate one reason why there is an upper limit on the use of HLA alleles in assessing disease prospects: without knowledge of TCR-β V or J usage dominance in the disease setting, the full impact of a particular HLA allele may not be apparent. This is likely to be an even stronger conclusion with future big-data correlations with other, more detailed aspects of the TCR-HLA interactions, such as usage of particular J sub-segments and D-regions; and generation of CDR3 (complementarity determining-3) sequences. Currently, a limitation of the above approach is the lack of enough large datasets to replicate particular HLA, V or J usage combinations within a given cancer. Presumably the above results will facilitate database expansions to add more rigor to statistically challenging questions and settings, or the above results will facilitate a related goal, to focus on particular, apparent dominant TCR gene segment usage, HLA allele combinations as test combinations in comparison to control combinations.
In many of the cases reported here, the binning of the J1 and J2, TCR-β segment sets, was done to establish statistically significant J usage and HLA allele combinations, due to the lack of sufficient numbers of cases to work with individual J1 and J2 sub-segments. The data reported above strongly indicate that the J1 and J2 groups, respectively, have sub-segment features in common that affect their binding capacities for allelic variant, HLA polypeptides. In particular, there is an LTVV amino acid motif at the 3′ end of three of the six J1 group segments that does not appear in any J2 group segment. And, there is an LTVL amino acid motif at the 3′ end of four of the J2 segments that does not appear in any J1 segment.
However, it is important to point out that HLA-VDJ association features may not be limited to the molecular features of the binding of the J1 or J2 segment polypeptides and particular HLA allelic, variant polypeptides. Other possibilities could include indirect manifestations of molecular processes, such as a variable cellular stability, or lifetime maintenance of certain of TCR-β VDJ recombinant polypeptides when complexed with particular HLA allelic variant polypeptides, especially keeping in mind the variable cell types represented by the different cancer types and variable infection or immunization histories of the cancer patients. Nevertheless, at least from the standpoint of a correlation, it is possible to identify the indicated, “J-grouped” associations, and the most likely possibility is that the associations do reflect a specific feature of the binding of J region polypeptides with particular HLA antigen presenting molecules.
Cancer development is associated with failures of the immune system to recognize and destroy cells that have become malignant [18]. Over the last several decades, it has become clear that the adaptive arms of the immune system, namely T cells and B cells, are important components of the immune activity that prevent cancer metastasis and death due to cancer. However, there are contradictory roles for the immune system, including the adaptive immune system, in cancer development. For example, acquired immunodeficiency disease, which negatively impacts T-cells, is associated with the development of Kaposi’s sarcoma. However, Crohn’s disease, associated with particular alleles of the HLA genes [19, 20], is a risk factor for colon cancer [21]. Considering autoimmunity and TCR/HLA combinations, it is likely that a self-antigen bound to an HLA antigen presenting molecule increases the binding affinity of the TCR and the HLA molecule-antigen complex, leading to T-cell activation and normal tissue destruction. The presumption has been that certain HLA allelic variants bind a particular self-antigen well, and after some initiating event, such as a viral induced amplification of a T cell with a particular V(D)J recombination, and a particular CDR3, a TCR-HLA interaction that was previously weak becomes sufficiently strong to lead to autoimmunity. But as noted above, the cancer setting is more complicated. Thus, the above data indicating that certain TCR-β V or J usage is associated with better survival does not automatically indicate that a specific V or J usage leads to a stronger TCR-β-HLA binding. Given the correlations between autoimmune inflammation and cancer development, it is possible that the V or J usage associations reported here represent reduced levels of inflammation and thereby slower, terminal cancer development. Alternatively, the V or J usage, HLA allele combinations may reflect HLA allelic variants that bind tumor antigens or neoantigens in such a way as to facilitate a stronger T-cell activation and more efficient tumor cell killing. Future work, either via in vitro binding studies or with molecular modeling approaches, with the HLA allele, TCR-β V and J combinations indicated here, and many other such combinations soon to be revealed, will likely provide a resolution to this question.
Furthermore, there are other antigen presentation factors that could be at play, independently of the TCR/antigen/HLA binding efficiency characteristics, such as more efficient interactions of certain HLA polypeptides with components of the antigen presentation proteins. In other words, just as an example, the effectiveness of the HLA allele and particular VDJ usage could be dependent on high-level HLA protein binding to components of the antigen processing machinery for the best possible, combined HLA-VDJ impact. In addition, a combination of available HLA-VDJ combinations may be associated with best possible survival possibilities, such as one HLA class II-VDJ combination for antigen presentation and one HLA class I-VDJ combination for tumor cell killing. Regardless, the TCR-β V and J usage, HLA allele combinations reported here provide a large selection of immune-biomarkers over multiple cancers with the very likely opportunity to provide information regarding dramatic differences in survival times (Table 3; Fig. 3).
Additional avenues for future work include a more detailed assessment of the TCR receptors involved in associations with survival rate distinctions, also a goal that would likely require much larger datasets. For example, the current datasets did not reveal any skewed usage of the TRAV and TRAJ segments, either due to the very large numbers of these gene segments preventing such detection, or due to the fact that there is no such association, given that the majority of antigen-specificity is thought to reside with TRBV and J segments, and the TRB CDR3 region [22, 23]. However, it should be noted that studies have indicated the potential for an overwhelming TCR clonotype heterogeneity [24, 25], and the above study, despite extensive considerations, did not reveal any CDR3/clonotype associations with survival outcomes. Additionally, the lack of TRAV and TRAJ results, which could have revealed invariant NKT, TRAV24 and TRAJ18 usage segments, as well as the lack of any invariant NKT-specific, TRBV11 associations with outcome, in the above study, raise questions about the absence of invariant NKT cells in the microenvironment [26–28].
Acknowledgements
Authors would like to acknowledge the support of University of South Florida research computing. The whole exome files for this project were made available via database of Genotypes and Phenotypes approved project #6300, to George Blanck.
Abbreviations
- CDR3
Complementarity determining region-3
- dbGaP
Database of genotypes and phenotypes
- HLA
Human leukocyte antigen
- HNSC
Head and neck cancer
- LUAD
Lung adenocarcinoma
- KM
Kaplan–Meier
- NKT
Natural killer T-cell
- OV
Ovarian cancer
- SKCM
Skin, cutaneous melanoma
- SPSS
Statistical package for the social sciences
- STAD
Stomach adenocarcinoma
- TCGA
The cancer genome atlas
- TCR
T-cell receptor
- WXS
Whole exome sequence
Author contributions
Blake M. Callahan contributed to the design of the project and conducted most of the original, basic assessment of TCR and HLA combinations in the whole exome files; and contributed to manuscript preparation. Blake M. Callahan, John M. Yavorski, and Jacob C. Kinskey contributed extensively to the KM analyses. Yaping N. Tu, Wei Lue Tong and Timothy J. Fawcett contributed extensively to the code-writing and use of USF research computing. Kendall R. Clark contributed TCR data for a subset of cancer types. George Blanck contributed to the design of the project, supervised the project, and participated extensively in manuscript preparation.
Funding
This work was supported by the taxpayers of the State of Florida. Blake M. Callahan was a recipient of a Bonati research stipend.
Compliance with ethical standards
Conflict of interest
Authors have no conflicts of interests to declare.
References
- 1.Green CJ. Immunosuppression with cyclosporin A: a review. Diagn Histopathol. 1981;4:157–174. [PubMed] [Google Scholar]
- 2.Rao A. NFATp, a cyclosporin-sensitive transcription factor implicated in cytokine gene induction. J Leukoc Biol. 1995;57:536–542. doi: 10.1002/jlb.57.4.536. [DOI] [PubMed] [Google Scholar]
- 3.Taff ML, Siegal FP, Geller SA. Outbreak of an acquired immunodeficiency syndrome associated with opportunistic infections and Kaposi’s sarcoma in male homosexuals: an epidemic with forensic implications. Am J Forensic Med Pathol. 1982;3:259–264. doi: 10.1097/00000433-198209000-00013. [DOI] [PubMed] [Google Scholar]
- 4.Skornick Y, Topalian S, Rosenberg SA. Comparative studies of the long-term growth of lymphocytes from tumor infiltrates, tumor-draining lymph nodes, and peripheral blood by repeated in vitro stimulation with autologous tumor. J Biol Response Mod. 1990;9:431–438. [PubMed] [Google Scholar]
- 5.Rudolph MG, Wilson IA. The specificity of TCR/pMHC interaction. Curr Opin Immunol. 2002;14:52–65. doi: 10.1016/S0952-7915(01)00298-9. [DOI] [PubMed] [Google Scholar]
- 6.Miles JJ, Bulek AM, Cole DK, Gostick E, Schauenburg AJ, Dolton G, Venturi V, Davenport MP, Tan MP, Burrows SR, Wooldridge L, Price DA, Rizkallah PJ, Sewell AK. Genetic and structural basis for selection of a ubiquitous T cell receptor deployed in Epstein-Barr virus infection. PLoS Pathog. 2010;6:e1001198. doi: 10.1371/journal.ppat.1001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klarenbeek PL, Doorenspleet ME, Esveldt RE, van Schaik BD, Lardy N, van Kampen AH, Tak PP, Plenge RM, Baas F, de Bakker PI, de Vries N. Somatic variation of T-cell receptor genes strongly associate with HLA class restriction. PLoS One. 2015;10:e0140815. doi: 10.1371/journal.pone.0140815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glanville J, Huang H, Nau A, Hatton O, Wagar LE, Rubelt F, Ji X, Han A, Krams SM, Pettus C, Haas N, Arlehamn CSL, Sette A, Boyd SD, Scriba TJ, Martinez OM, Davis MM. Identifying specificity groups in the T cell receptor repertoire. Nature. 2017;547:94–98. doi: 10.1038/nature22976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Sante G, Tolusso B, Fedele AL, Gremese E, Alivernini S, Nicolo C, Ria F, Ferraccioli G. Collagen specific T-cell repertoire and HLA-DR alleles: biomarkers of active refractory rheumatoid arthritis. EBioMedicine. 2015;2:2037–2045. doi: 10.1016/j.ebiom.2015.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng G, Huang Y, Huang Y, Lyu Z, Lesniak D, Randhawa P. Antigen-specificity of T cell infiltrates in biopsies with T cell-mediated rejection and BK polyomavirus viremia: analysis by next generation sequencing. Am J Transplant. 2016;16:3131–3138. doi: 10.1111/ajt.13911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miles JJ, Douek DC, Price DA. Bias in the alphabeta T-cell repertoire: implications for disease pathogenesis and vaccination. Immunol Cell Biol. 2011;89:375–387. doi: 10.1038/icb.2010.139. [DOI] [PubMed] [Google Scholar]
- 12.Gill TR, Samy MD, Butler SN, Mauro JA, Sexton WJ, Blanck G. Detection of productively rearranged TcR-alpha V-J sequences in TCGA exome files: implications for tumor immunoscoring and recovery of antitumor T-cells. Cancer Inform. 2016;15:23–28. doi: 10.4137/CIN.S35784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tong WL, Tu YN, Samy MD, Sexton WJ, Blanck G. Identification of immunoglobulin V(D)J recombinations in solid tumor specimen exome files: Evidence for high level B-cell infiltrates in breast cancer. Hum Vaccin Immunother. 2017;13:501–506. doi: 10.1080/21645515.2016.1246095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Samy MD, Tong WL, Yavorski JM, Sexton WJ, Blanck G. T cell receptor gene recombinations in human tumor specimen exome files: detection of T cell receptor-beta VDJ recombinations associates with a favorable oncologic outcome for bladder cancer. Cancer Immunol Immunother. 2017;66:403–410. doi: 10.1007/s00262-016-1943-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tu YN, Tong WL, Fawcett TJ, Blanck G. Lung tumor exome files with T-cell receptor recombinations: a mouse model of T-cell infiltrates reflecting mutation burdens. Lab Invest. 2017;97:1516–1520. doi: 10.1038/labinvest.2017.80. [DOI] [PubMed] [Google Scholar]
- 16.Szolek A, Schubert B, Mohr C, Sturm M, Feldhahn M, Kohlbacher O. OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics. 2014;30:3310–3316. doi: 10.1093/bioinformatics/btu548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie C, Yeo ZX, Wong M, Piper J, Long T, Kirkness EF, Biggs WH, Bloom K, Spellman S, Vierra-Green C, Brady C, Scheuermann RH, Telenti A, Howard S, Brewerton S, Turpaz Y, Venter JC. Fast and accurate HLA typing from short-read next-generation sequence data with xHLA. Proc Natl Acad Sci USA. 2017;114:8059–8064. doi: 10.1073/pnas.1707945114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Disis ML. Immune regulation of cancer. J Clin Oncol. 2010;28:4531–4538. doi: 10.1200/JCO.2009.27.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahdi BM. Role of HLA typing on Crohn’s disease pathogenesis. Ann Med Surg (Lond) 2015;4:248–253. doi: 10.1016/j.amsu.2015.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slade JD, Luskin AT, Gewurz H, Kraft SC, Kirsner JB, Zeitz HJ. Inherited deficiency of second component of complement and HLA haplotype A10,B18 associated with inflammatory bowel disease. Ann Intern Med. 1978;88:796–798. doi: 10.7326/0003-4819-88-6-796. [DOI] [PubMed] [Google Scholar]
- 21.Parrish RA, Karsten MB, McRae AT, Moretz WH. Segmental Crohn’s colitis associated with adenocarcinoma. Am J Surg. 1968;115:371–375. doi: 10.1016/0002-9610(68)90163-3. [DOI] [PubMed] [Google Scholar]
- 22.Valkenburg SA, Day EB, Swan NG, Croom HA, Carbone FR, Doherty PC, Turner SJ, Kedzierska K. Fixing an irrelevant TCR alpha chain reveals the importance of TCR beta diversity for optimal TCR alpha beta pairing and function of virus-specific CD8 + T cells. Eur J Immunol. 2010;40:2470–2481. doi: 10.1002/eji.201040473. [DOI] [PubMed] [Google Scholar]
- 23.Yoshikai Y, Kimura N, Toyonaga B, Mak TW. Sequences and repertoire of human T cell receptor alpha chain variable region genes in mature T lymphocytes. J Exp Med. 1986;164:90–103. doi: 10.1084/jem.164.1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gong Q, Wang C, Zhang W, Iqbal J, Hu Y, Greiner TC, Cornish A, Kim JH, Rabadan R, Abate F, Wang X, Inghirami GG, McKeithan TW, Chan WC. Assessment of T-cell receptor repertoire and clonal expansion in peripheral T-cell lymphoma using RNA-seq data. Sci Rep. 2017;7:11301. doi: 10.1038/s41598-017-11310-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shugay M, Bagaev DV, Zvyagin IV, Vroomans RM, Crawford JC, Dolton G, Komech EA, Sycheva AL, Koneva AE, Egorov ES, Eliseev AV, Van Dyk E, Dash P, Attaf M, Rius C, Ladell K, McLaren JE, Matthews KK, Clemens EB, Douek DC, Luciani F, van Baarle D, Kedzierska K, Kesmir C, Thomas PG, Price DA, Sewell AK, Chudakov DM. VDJdb: a curated database of T-cell receptor sequences with known antigen specificity. Nucleic Acids Res. 2018;46:D419-D427. doi: 10.1093/nar/gkx760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cameron G, Pellicci DG, Uldrich AP, Besra GS, Illarionov P, Williams SJ, La Gruta NL, Rossjohn J, Godfrey DI. Antigen specificity of type I NKT cells is governed by tcr beta-chain diversity. J Immunol. 2015;195:4604–4614. doi: 10.4049/jimmunol.1501222. [DOI] [PubMed] [Google Scholar]
- 27.Arase H, Arase N, Ogasawara K, Good RA, Onoe K. An NK1.1 + CD4 + 8- single-positive thymocyte subpopulation that expresses a highly skewed T-cell antigen receptor V beta family. Proc Natl Acad Sci U S A. 1992;89:6506–6510. doi: 10.1073/pnas.89.14.6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lantz O, Bendelac A. An invariant T cell receptor alpha chain is used by a unique subset of major histocompatibility complex class I-specific CD4 + and CD4-8- T cells in mice and humans. J Exp Med. 1994;180:1097–1106. doi: 10.1084/jem.180.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]