Abstract
As a major component of the microenvironment of solid tumors, tumor-associated macrophages (TAMs) facilitate tumor progression. Intermediate-sized hyaluronan (INT-HA) fragments have an immunological function in cell differentiation; however, their role in promoting the polarization of non-activated macrophages to an M2-like TAM phenotype has not been characterized, and the underlying mechanisms remain unclear. Here, we used a miRNA microarray to find that some miRNAs (especially miR-935) were differentially regulated in INT-HA-induced M2-like macrophages. According to RT-qPCR and Western blot, there was an association between miR-935 and C/EBPβ, that control the polarization of macrophages. Moreover, we found that INT-HA induced an M2-like phenotype via the TLR4 receptor. In our study, there was a negative correlation between plasma HA and miR-935 in monocytes from the peripheral blood of patients with solid tumors. There was also a negative correlation between miR-935 and M2-like macrophage markers in monocytes. These findings suggest that HA fragments interact with TLR4 and educate macrophage polarization to an M2-like phenotype via miR-935. Therefore, this study provides new insight into the role of miR-935 in INT-HA-induced M2-like polarization, and suggests a potential therapeutic target for antitumor treatment.
Electronic supplementary material
The online version of this article (10.1007/s00262-018-2261-6) contains supplementary material, which is available to authorized users.
Keywords: Hyaluronan, MiR-935, TLR4, CD44, Tumor-associated macrophage
Introduction
Macrophages, an important immunologic cell type, play a vital role in host defense against viral, bacterial, and parasitic infections. Macrophages are a heterogeneous cell population under different conditions and possess diverse functional programs [1]. As an important cellular component of the tumor microenvironment, macrophages can differentiate into M1-like and M2-like tumor-associated macrophages (TAMs) [2]. M1-like macrophages produce pro-inflammatory cytokines and can kill microorganisms and tumor cells [1]. In contrast, M2-like macrophages suppress inflammatory signaling and play an important role in tumor progression [3].
The biosynthesis and degradation of hyaluronan (HA), a major constituent in the extracellular matrix (ECM), have been proposed to play an important role in angiogenesis, cell proliferation and tumor progression [4]. In most tissues, native HA (high molecular weight hyaluronan, HMW-HA, >106Da) is composed of repeating disaccharides of (β, 1–4)-glucuronic acid and (β, 1–3)-N-acetylglucosamine. Increasing the breakdown of HMW-HA by HYALs induces the accumulation of low molecular weight hyaluronan (LMW-HA), which has different functions than its precursors [5, 6]. HMW-HA is known to maintain water homeostasis and matrix structure and inhibit tumor growth [7, 8]. However, small- (6–20 kDa) or intermediate-sized HA (INT-HA) (50–1000 kDa) fragments activate dendritic cells and macrophages via CD44 or TLR, whereas HMW-HA do not [9–11]. As a HA receptor, CD44 has been shown to play an important role in signal transduction to control cell growth, differentiation, and survival [12, 13]. Previous studies have suggested that INT-HA fragments play an important role in the polarization of macrophages via CD44 [14–16]. However, toll-like receptor 4 (TLR4) also has a signal transduction effect because it promotes macrophage differentiation toward a distinct activation pattern [13, 17]. Furthermore, there are many hyaluronan receptors, but only TLR4 and CD44 are distinctly expressed in monocytes/macrophages [18]. Various transcriptional factors, such as STAT1, STAT6, PPARγ and C/EBPβ, control the polarization of macrophages [13]. For example, activating the PPARγ/STAT6 or C/EBPβ pathway could promote the expression of M2-associated genes, such as mannose receptor 1 (Mrc1), arginase 1 (Arg1), IL10, CCL17, and CCL18 [19, 20]. More importantly, there is a close relationship between TLR4 and C/EBPβ in macrophage polarization [13, 21]. Therefore, these findings suggest that whether hyaluronan induce macrophage polarization via TLR4 or not needs to be investigated.
MicroRNAs (miRNAs) are small, non-coding RNAs that suppress gene expression at the post-transcriptional level. It is increasingly clear that miRNAs participate in the regulation of most biological and physiological processes, including differentiation, cell proliferation, apoptosis and metabolism. Furthermore, recent studies have revealed that microRNAs are important mediators of the macrophage activation process [22–24]. For example, miR-155 has been reported to promote classical macrophage activation (M1-like polarization) by down-regulating the inhibitor SOCS1 by binding to its 3′-UTR [25]. Recent data also indicated that several distinct miRNAs, such as miR-142-5p, miR-let-7c, and miR-511-3p, are induced upon alternative macrophage activation (M2-like polarization) [26–28]. However, whether miRNAs contribute to the M2-like polarization of hyaluronan-activated macrophages has not been explored.
In the present study, we first explored which hyaluronan molecular weight could induce macrophage polarization. Then, we screened the miRNAs expression levels in macrophages using a microRNA microarray, and found that miR-935 expression was decreased through TLR4 in hyaluronan-treated macrophages. Moreover, we also investigated the relationships among the monocyte miR-935 levels, M2-like macrophages, and plasma HA concentrations in patients with solid tumors.
Method
Acquisition of INT-HA
HMW-HA was purchased from Sigma-Aldrich (USA). LMW-HA (10 kDa) was purchased from Lifecore (USA). INT-HA fragments were prepared by digesting HMW-HA as previously described [17, 29]. In our study, HMW-HA was digested with 30U of testicular hyaluronidase for 40 min at 37 °C. Thereafter, the digested samples were adjusted to a neutral pH and filtered through Detoxi-Gel Endotoxin Removing Columns (Thermo Scientific, USA) to remove endotoxin; then, the samples were used for cell stimulation. The endotoxin levels in all samples were measured, and the endotoxin concentration in the working fluid was less than 0.0005 EU/ml. The sizes of the INT-HA fragments were determined by 0.5% agarose gel electrophoresis and images were collected (Supplementary Figure 1).
Monocyte isolation and culture
Peripheral blood monocytes from healthy donors and patients were isolated from frozen PBMCs by a CD14 microbead mediated sorting system (Miltenyi Biotec, USA). Then, the purified monocytes were cultured in T-25 plates (Corning, USA) at 5 × 105 cells/ml in RPMI 1640 medium (Gibco, China) with 10% FBS (Gibco, BRL) at 37 °С and 5% CO2; the cells were then allowed to adhere overnight. Over 90% of the adherent cells were monocytes as determined by flow cytometric analyses. In some experiments, the monocytes were incubated with different concentrations and molecular weights of hyaluronan for 72 h.
Cell line
The human monocyte THP-1 cell line was maintained in RPMI 1640 medium at 37 °C and 5% CO2; the cells were allowed to differentiate during an overnight incubation with 320 nM PMA (Merck Millipore, Germany). The PMA-containing medium was removed after incubation and the adherent cells were treated with hyaluronan in fresh culture medium for 72 h to generate M2-like polarized macrophages.
RT-qPCR
RNA was isolated from monocytes and THP-1-derived macrophages using TRIzol (Takara, Japan). RNA quantity was measured using a NanoDrop 2000 spectrophotometer (Thermo Scientific, USA). Quantitative real-time PCR (RT-qPCR) was conducted with an ABI 7500 instrument (ABI, USA) and SYBR Premix Ex Taq™ (Takara, Japan) according to the manufacturer’s instructions. The primers sequences for the selected genes are listed in Supplementary Table 1.
ELISA
The concentrations of M2-related cytokines (Human CCL18, CCL22, TGF-β and IL10) in the culture supernatants were measured by ELISA kits (R&D, USA) according to the manufacturer’s instructions.
Plasma concentrations of HA
The concentrations of plasma total hyaluronan (Maglumi 2000, China) were measured by the chemiluminescence method according to the manufacturer’s instructions.
MicroRNA expression
RNA from human primary macrophages treated with or without 50 µg/ml INT-HA for 72 h was isolated using TRIzol (Takara, Japan) and purified with an RNeasy mini kit (QIAGEN, Germany) according to the manufacturer’s instructions. RNA quality and quantity were determined using a NanoDrop 2000 spectrophotometer and RNA integrity was measured by gel electrophoresis. MiRNA microarray analysis was performed by the KangChen Corporation (Shanghai, China) and used to compare the miRNA expression profiles. Briefly, the samples were labeled using the miRCURY™ Hy3TM/Hy5™ Power labelling kit and hybridized on a miRCURY LNA Array (v.19.0; Exiqon, Vedbaek, Denmark) according to the array manual. The slides were washed and scanned using an Axon GenePix 4000B microarray scanner (Axon Instruments, Foster City, CA). The scanned images were then imported into GenePix Pro 6.0 software (Axon) for grid alignment and data extraction. miRNA replicates were averaged, and miRNA with intensities ≥ 30 for all samples were chosen to calculate the normalization factor. The expressed data were normalized using the median normalization. After normalization, significant differentially expressed miRNAs between the two groups were identified by fold change calculations. RT-qPCR was performed using the miRcute Plus miRNA First-Strand cDNA Synthesis Kit and miRcute Plus miRNA qPCR Detection Kit (Tiangen, China) using an ABI 7500. U6 was used as an internal control and the expression levels of mature miRNAs were calculated. Each sample was measured in triplicate. The Gene Expression Omnibus database accession number is GSE110339 for the miRNA microarray.
Bioinformatics methods
Potential miRNA targets were identified using online software. TargetScan (http://www.targetscan.org/index.html), miRDB (http://www.mirdb.org/), and miRWalk (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk/index.html) were used to analyze the potential targets.
Transfection of miRNAs
miRNA mimics, inhibitors and their negative controls were purchased from Ribobio (China). THP-1 derived macrophages were transfected with 20 nM miRNA mimics (miR10004978, sense: 5′-CCAGUUACCGCUUCCGCUACCGC-3′, antisense: 5′-GCGGUAGCGGAAGCGGUAACUGG-3′) and negative control (miR01101, sense: 5′-UUUGUACUACACAAAAGUACUG-3′, antisense: 5′-CAGUACUUUUGUGUAGUACAAA-3′), 40 nM miRNA inhibitors (miR20004978, sense: 5′-GCGGUAGCGGAAGCGGUAACUGG-3′) and negative control (miR02101, sense:5′-CAGUACUUUUGUGUAGUACAAA-3′). Transient transfections were performed using riboFECT™ CP reagent (Ribobio, China) according to the manufacturer’s instructions.
Western blotting
Cells were lysed in cell lysate buffer (RIPA), and the cell lysates were incubated on ice for 45 min and centrifuged at 12,000×g at 4 °C for 10 min, then the supernatant was collected. Protein levels were measured using a bicinchoninic acid (BCA) protein quantitation kit (Thermo Scientific, USA). Equal amounts of proteins from each cell lysate were subjected to SDS-PAGE electrophoresis before transferred to PVDF membranes. After blocking with 5% skimmed milk powder, membranes were incubated with primary antibody against CEBPβ (1:1000 dilution, CST, USA) overnight at 4 °C, followed by incubation with horseradish peroxidase-linked goat anti-rabbit secondary antibody (1:5000 dilution, liankebio, China) at room temperature for 1 h. Subsequently, protein on the membranes was visualized by enhanced chemiluminescence assay (ECL, Thermo Scientific, USA).
Statistical analysis
Normally distributed data are expressed as the mean ± SD, which was determined by two-tailed unpaired Student’s t test between two groups, and multiple comparisons were conducted using one-way ANOVA and Tukey tests. Non-normally distributed data are expressed as a median (P25-P75), which was determined by Mann–Whitney U test. All statistical analyses were carried out using GraphPad Prism Software Version 7.0 (GraphPad Software Inc., La Jolla, CA, USA). A p value of < 0.05 was considered statistically significant.
Results
INT-HA induces human macrophage polarization toward the M2-like phenotype
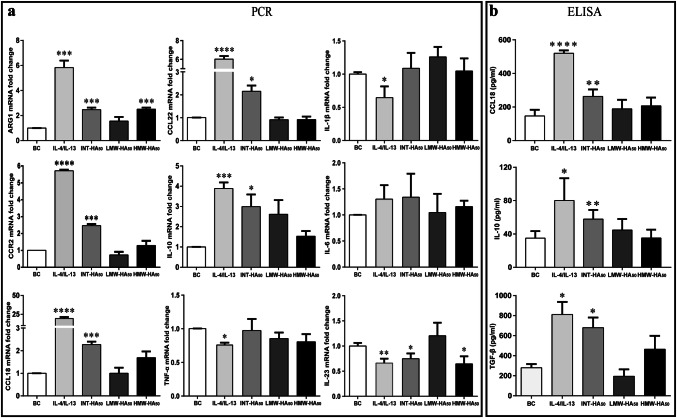
To investigate the effects of hyaluronan on the macrophage polarization, we used hyaluronan with different molecular weights and different concentrations to stimulate primary human monocytes. As shown in Fig. 1 and Supplementary Figure 2, compared with positive control by IL-4/IL-13 treatment, we found that the M2-like polarization of macrophages induced by 50 µg/ml INT-HA fragments was also particularly evident, whereas the effects of HMW-HA and LMW-HA were not obvious. In contrast to untreated monocytes (M0), INT-HA-treated monocytes exhibited higher mRNA levels of M2 phenotype markers such as arginase 1 (ARG1), IL10, CCL18, CCL22, and CCR2, and produced higher levels of TGF-β in culture supernatant. However, the M1 phenotype markers, such as TNF-α, IL-1β, and IL-6 remain unchanged (Fig. 1 and Supplementary Figure 2).
Fig. 1.
Gene expression and cytokine secretion in human monocytes in response to hyaluronan of different molecular weights. Primary human monocytes (1 × 106) were treated with 50 µg/ml low molecular weight HA (LWM-HA50), intermediate-sized HA (INT-HA50) or high molecular weight HA (HMW-HA50) for 72 h. Moreover, the treatment by IL-4/IL-13 was used as a positive control. The results are expressed as the means ± SD (n = 3). mRNA expression levels of M2 or M1 macrophage markers were assessed by RT-qPCR and normalized to GAPDH expression. Gene expression levels of Arg1, CCL18, CCL22, CCR2, and IL10 steadily increased with INT-HA50 treatment (a). Furthermore, gene expression levels of TNFα, IL-1β, and IL-6 were not changed with INT-HA50 treatment, but IL-23 level was decreased (a). The secretion of CCL18, IL10, and TGF-β by macrophages into the culture medium was measured by ELISA; adding INT-HA50 increased the secretion levels (b). *, **, ***, and ****: significantly different from the blank control (BC) with p < 0.05, 0.01, 0.001, and 0.0001, respectively
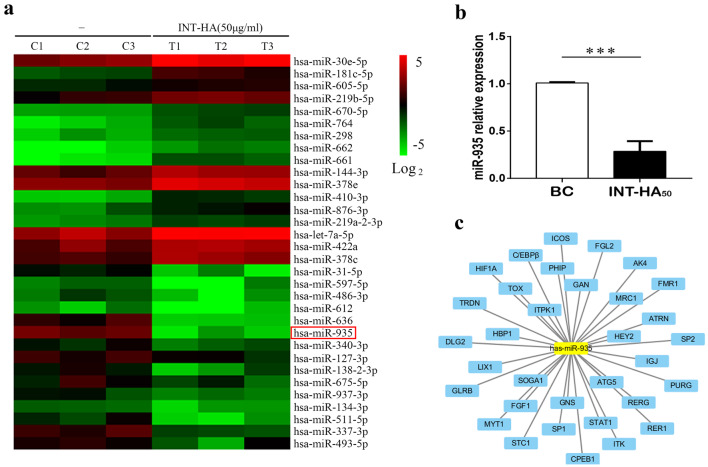
MiR-935 is down-regulated in INT-HA-induced M2-like macrophages
To screen for the miRNAs whose abundance was changed significantly following the incubation of monocytes under INT-HA conditions, we compared miRNA expression levels in the untreated cells versus the INT-HA treated monocytes by miRNA microarray analysis. Clustering analysis revealed significant alterations in the expression of 52 miRNAs (32 increased and 20 decreased, respectively, Fig. 2a). Next, we searched for and identified miRNAs that might be involved in the hyaluronan-driven polarization of human monocytes to M2-like macrophages. We focused our investigation on miR-935, which was a markedly down-regulated miRNA in this analysis. RT-qPCR validated the dramatic decrease in miR-935 expression upon the INT-HA-induced differentiation of monocytes into macrophages (Fig. 2b). Furthermore, according to online analysis software, we found that the potential target genes of miR-935 in macrophage polarization were STAT1/C/EBPβ (Fig. 2c). The transcriptional factor STAT1 controls macrophage polarization from M0 to M1, but C/EBPβ drives macrophages toward M2 polarization. Therefore, there are intricate mechanisms involved in the regulation of macrophage polarization via miR-935.
Fig. 2.
INT-HA50 alters the miRNA expression profiles in monocyte-derived macrophages. a Primary human macrophages derived from peripheral blood monocytes (n = 3) were treated with 50 µg/ml INT-HA for 72 h, and the miRNA expression profiles were analyzed by microarray. b Relative quantification of miR-935 expression at 72 h following INT-HA50 treatment as detected by RT-qPCR (mean ± SD, ***p < 0.001, compared with untreated macrophages). c Potential targets of miR-935 according to the following online software: TargetScan, miRDB, and miRWalk
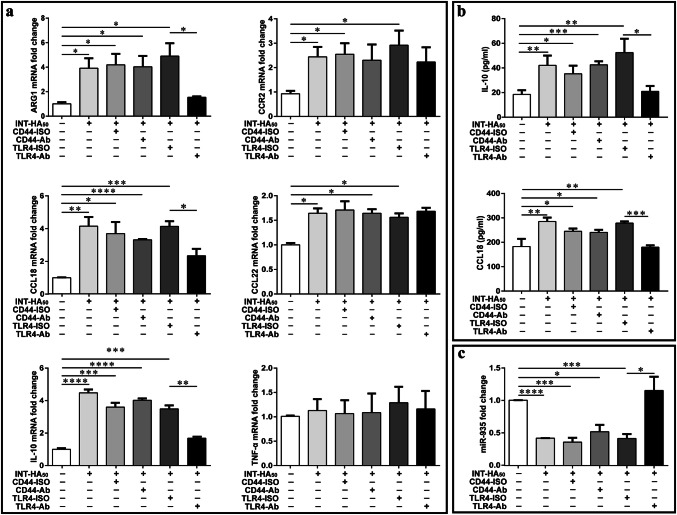
Down-regulation of miR-935 in M2 macrophages is dependent on HA–TLR4 interactions
To elucidate the mechanism through which INT-HA induces TAM, we investigated the interactions between INT-HA and CD44 and TLR4 in THP-1-derived macrophages; CD44 and TLR4 were chosen because they are the main hyaluronan receptors on the surface of monocytes or macrophages. THP-1-derived macrophages were pretreated overnight with CD44 and TLR4 blocking antibodies (20 µg/ml), followed by INT-HA for 3 days. In the presence of the TLR4 blocking antibody, INT-HA induced an approximately threefold decrease in Arg1 expression, a twofold decrease in IL10 expression, and up to a twofold decrease in CCL18 expression (Fig. 3a, b). This decrease was statistically significant (p < 0.001). Moreover, there were no obvious changes in CCR2 and CCL22 after using the TLR4 blocking antibody. The isotype IgG2 control had no effect on Arg1/IL10/CCL18 levels. However, following treatment with the CD44 blocking antibody, we did not find any obvious changes in Arg1, IL10, CCL18, CCL22, or CCR2 expression levels with INT-HA treatment. According to the previous studies, LPS induces the differentiation of macrophages to M1 phenotype via the TLR4-NF-κB/AP1/IRF3 pathway [13]. In contrast to INT-HA, LPS did not drive macrophage differentiate into M2-like phenotype (Supplementary Figure 3). Therefore, INT-HA induced M2-like macrophage differentiation via TLR4, which might be different from the signal pathway activated by LPS.
Fig. 3.
Effects of CD44 and TLR4 blocking antibodies on INT-HA50-stimulated THP-1-derived macrophage. a Gene expression levels of Arg1, CCL18, CCL22, CCR2 and IL10 were measured by RT-qPCR following the exposure of THP-1-derived macrophage to INT-HA50 for 72 h after pretreatment with CD44 (10 µg/ml, clone IM7, eBioscience) and TLR4 (20 µg/ml, clone HTA125, Biolegend) blocking antibodies. Reduced transcription levels of Arg1, CCL18, and IL10 were found after TLR4 blocking antibody treatment, while CCL22 and CCR2 were hardly affected by anti-TLR4 treatment. b CCL18 and IL10 levels in the corresponding culture media were also measured by ELISA; these levels were down-regulated after TLR4 blocking antibody treatment. c Increased expression levels of miR-935 were seen in the presence of the TLR4 blocking antibody but were not affected by anti-CD44 treatment. (*p < 0.05, **p < 0.01, ***p < 0.001)
We next examined whether miR-935 down-regulation in THP-1-derived macrophages was due to the TLR4-mediated regulation induced by INT-HA treatment. Similar to monocytes, we also observed that INT-HA could decrease the expression of miR-935 in THP-1-derived macrophages (Fig. 3c), which further indicated that TLR4 blocking antibody treatment effectively inhibited the HA-mediated decrease in miR-935 expression. In contrast, the CD44-blocking antibody and non-immune IgG control did not block the HA-mediated down-regulation of miR-935. These results indicate that miR-935 down-regulation in macrophages is HA-dependent and TLR4-specific.
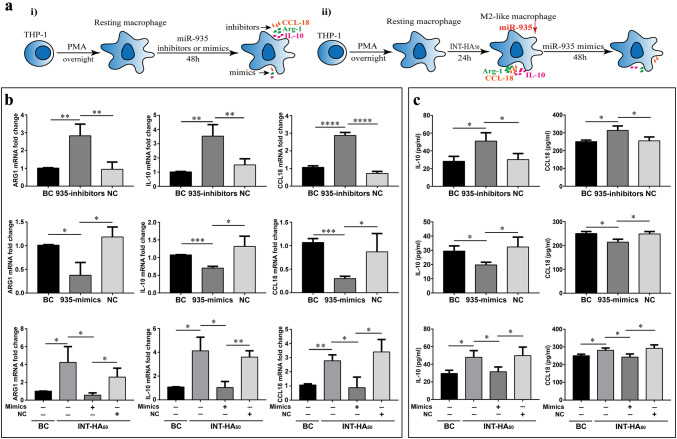
MiR-935 is a negative regulator in hyaluronan-induced M2 polarized macrophages
To confirm the roles of miR-935 in macrophage polarization, we transfected THP-1-derived macrophages with miR-935 mimics to overexpress of miR-935 and inhibitors to suppress miR-935 expression. The expression of miR-935 in M0 macrophages and INT-HA-induced M2-like macrophages after transfection of miRNA mimics and inhibitors were shown in Supplementary Figure 4. We measured the expression and secretion of ARG1, IL10 and CCL18 in macrophages transfected with the miR-935 mimics or inhibitors. We found that transduction of the macrophages with the miR-935 mimics reduced CCL18 and IL10 secretion and expression (Fig. 4b-c). Moreover, we also demonstrated an opposite effect in M2 macrophages transfected with miR-935 inhibitors. To further investigate the roles of miR-935 in INT-HA-induced M2 polarized macrophages, we transduced M2 macrophages (INT-HA treated for 72 h) with miR-935 mimics. Our results showed that the expression and secretion of Arg1, CCL18 and IL10 were decreased in INT-HA-induced M2-like macrophages. Therefore, transfecting THP-1-derived macrophages with miR-935 mimics significantly reduced the expression of M2 markers induced by INT-HA. Overall, these results indicate that miR-935 may play an important role in M2 macrophage polarization which was activated by INT-HA.
Fig. 4.
MiR-935 regulates INT-HA50-induced M2-polarized THP-1 macrophages. a Schematics of the approach. i). THP-1 derived macrophages were transfected with a negative control (NC), miR-935 inhibitors (40 nM) or mimics (20 nM) for 48 h. ii). THP-1-derived macrophages were treated with INT-HA50 for 24 h; then, the cells were transduced with NC and mimics for 48 h. b Gene expression levels of Arg1, CCL18 and IL10 in macrophages as determined by RT-qPCR. The representative histograms and quantification of the fold changes are shown (mean ± SD, n = 3 independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001). c Cytokine levels of CCL18 and IL10 in the media from macrophages (mean ± SD, n = 3 independent experiments. *p < 0.05)
MiR-935 modulates macrophage polarization by inhibiting C/EBPβ expression
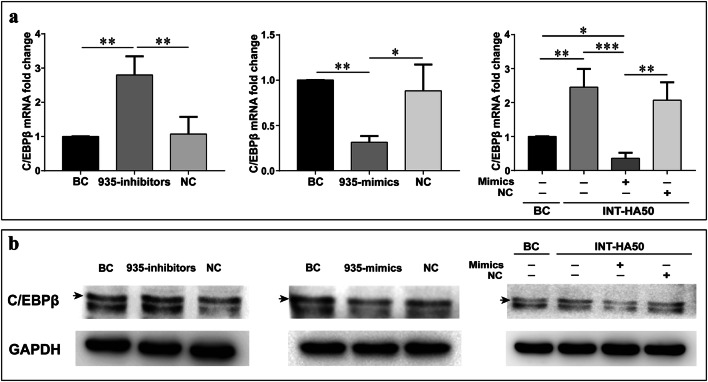
In our study, the potential target genes of miR-935 in macrophage polarization were STAT1 and C/EBPβ. According to RT-qPCR, we did not find that INT-HA treatment down-regulated the gene expression of STAT1 (Supplementary Figure 5). Moreover, transfection of the miR-935 mimics or miR-935 inhibitors did not regulate the expression of STAT1. To further demonstrate whether miR-935 represses C/EBPβ expression in INT-HA-induced M2-like macrophages, we performed RT-qPCR analysis and observed that the C/EBPβ mRNA level was decreased significantly through the transfection of miR-935 in macrophages after INT-HA treatment, while the transfection of miR-935 inhibitor resulted in a significant increase in the C/EBPβ mRNA level (Fig. 5a). Additionally, Western blot analysis revealed that transfection of the miR-935 mimics brought on a down-regulated C/EBPβ protein level in INT-HA-induced M2-like macrophages but miR-935 inhibitors caused an up-regulated expression of C/EBPβ (Fig. 5b). In conclusion, our results indicated that miR-935 inhibited the expression of C/EBPβ in INT-HA-induced M2-like macrophages.
Fig. 5.
miR-935 inhibits the expression of C/EBPβ. RT-qPCR (a) and Western blot (b) were used to determine the expression level of C/EBPβ in THP-1 cells transfected with miR-935 mimics, miR-935 inhibitors, and their controls. GAPDH served as the internal control. The representative histograms and quantification of the fold changes are shown (mean ± SD, n = 3 independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001)
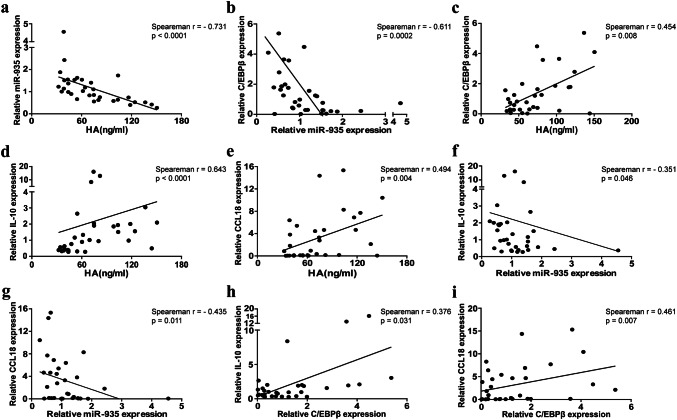
Correlation among HA, miR-935 expression, C/EBPβ, and M2-like monocytes in the peripheral blood of patients with solid tumors
Table 1 summarizes the clinical characteristics of solid tumor patients. A total of 33 patients with a median age of 43.5 years (range 12–75 years) were recruited into the study, and 32 healthy controls were also participants in our study. The age and sex distributions were not significantly different between the cancer and healthy groups. In the 33 cancer patients investigated, osteosarcoma (80.2%) was the main common solid tumor type, followed by lung cancer (12.1%) and nasopharyngeal cancer (12.1%). In the present study, miR-935 expression in the monocytes from the cancer group was negatively correlated with plasma HA and C/EBPβ (Fig. 6a, b). Furthermore, there was also a negative correlation between miR-935 in monocytes and M2-related markers in the cancer group (Fig. 6f, g). However, there was a positive correlation among plasma HA, C/EBPβ, and M2-like monocyte-related markers (IL10 and CCL18) in the cancer group (Fig. 6c–e, h, i). These results seem to be consistent with our research showing that INT-HA-induced M2-like differentiation in macrophages via miR-935/C/EBPβ.
Table 1.
The clinical characteristics of all subjects
| Cancer group (n = 33) | Healthy control (n = 32) | |
|---|---|---|
| Sex | ||
| Male | 18 (54.5%) | 12 (37.5%) |
| Female | 15 (45.5%) | 20 (62.5%) |
| Median age, years (range) | 43.50 (29.00-59.75) | 47.00 (37.75–59.25) |
| Plasma HA (ng/ml) | 69.43 (43.64–100.9) * | 44.48 (37.61–56.91) |
| Monocyte miR-935 (2− ΔΔCT) | 1.005 (0.630–1.541) | 0.944 (0.663-1.300) |
| Monocyte IL-10 (2− ΔΔCT) | 0.975 (0.423–2.020) | 1.022 (0.449–2.046) |
| Monocyte CCL18 (2− ΔΔCT) | 0.938 (0.120–5.035) | 2.175 (0.160–3.985) |
| Monocyte C/EBPβ (2− ΔΔCT) | 0.816 (0.277–1.826) | 0.917 (0.448–1.624) |
| Primary cancer | ||
| Osteosarcoma | 13 (39.4%) | |
| Lung cancer | 4 (12.1%) | |
| Nasopharyngeal cancer | 4 (12.1%) | |
| Osteoclastoma | 3 (9.1%) | |
| Cervical cancer | 2 (6.1%) | |
| Prostate cancer | 2 (6.1%) | |
| Others | 5 (15.1%) | |
*p = 0.0015
Fig. 6.
Correlations among plasma HA, miR-935 expression in monocytes, M2-like monocytes in and C/EBPβ in the peripheral blood from patients with solid tumors. Plasma HA levels were detected using a chemiluminescence method. MiR-935, CCL18, IL10, and C/EBPβ expression levels in the monocytes from peripheral blood of 33 patients with solid tumors were determined by RT-qPCR. a Correlations between HA and miR-935. b Correlations between miR-935 and C/EBPβ. c Correlations between HA and C/EBPβ. d, e Correlations between HA and M2-like monocytes (CCL18 and IL10). f, g Correlations between miR-935 and M2-like monocytes (CCL18 and IL10). h, i Correlations between C/EBPβ and M2-like monocytes (CCL18 and IL10). Proposed model of INT-HA-induced macrophage: M2-like polarization INT-HA potently polarizes M0 macrophages into M2-like macrophages through the TLR4/miR-935 pathway, which stimulates the up-regulation of C/EBPβ and M2 macrophage markers, such as ARG1, CCL18 and IL10
Discussion
In most solid tumors, tumor-associated macrophage (TAM) subtypes exhibit a distinct phenotype and perform specific functions [2, 13]. Previous studies have suggested that hyaluronan (HA) fragments (especially INT-HA, 50-1000 KD) are potent activators of macrophages [14, 30]. However, the mechanism by which HA fragments drives macrophages from the M0-like phenotype to the M2-like phenotype remains to be elucidated. In the current study, we used different molecular weights and different concentrations of HA to treat primary human monocyte-derived macrophages; we found that 50 µg/ml INT-HA could promote monocytes polarization to M2-like macrophages after 72 h. However, this finding is contrary to previous studies that have suggested that INT-HA induces the polarization of human macrophages toward the M1 phenotype [14, 17]. A possible explanation for this might be that the induction of macrophage differentiation by INT-HA appears to be time-dependent. In a previous study, macrophage differentiation to the M1-like phenotype was promoted 6–24 h after exposure to HA [17]. Furthermore, we also demonstrated that THP-1-derived macrophages were polarized toward the M2 phenotype by adding 50 µg/ml INT-HA for 72 h.
Accumulating evidence has revealed the importance of the potential contribution of miRNAs,such as miR-155, miR-142, miR-130a, and miR-let-7c,in altering macrophage phenotype and function [25, 26, 28, 31]. In this study, to clarify the mechanism by which HA activated M2-like macrophage polarization, we employed miRNA microarray analysis and several miRNA prediction databases and found that miR-935 may play a role in macrophage polarization. STAT1/CEBPβ are the potential target genes of miR-935 in macrophage polarization according to the online analysis software. However, there may be two opposite tendencies involved in miR-935-induced macrophage polarization. The transcriptional factors STAT1 and STAT3 control macrophage polarization from M0 to M1, but C/EBPβ drives macrophages toward M2 polarization [13, 32]. In our study, INT-HA treatment and transfection of the miR-935 mimics or miR-935 inhibitors did not change the gene expression of STAT1. Our finding by RT-qPCR and Western blot indicated that miR-935 may induce M2 macrophage differentiation through down-regulating C/EBPβ expression. It has been demonstrated that in activated macrophages the expression of C/EBPβ was required for gene characteristic of M2 macrophages (Arg1, IL10, IL13ra) but dispensable for induction of M1 genes (IL6, IL1β, TNFα) [20], which may explain our findings that the expression of M1 genes was not altered in HA-induced M2 macrophage activation.
Moreover, previous studies indicated that miR-935 play a role in regulating tumor cell proliferation, migration and invasion of tumor cells [33–35]. Up to now, very little information regarding the role of miR-935 in immune cell differentiation has been published in the literature. To further explore the pathway underlying miR-935-HA-mediated macrophage polarization, we first searched for receptors mediating the HA interaction in TAM cells. It is well-known that TLR4 and CD44 are the main receptors for HA in macrophages [18]. More importantly, several reports have shown that HA fragments play an important role in macrophage differentiation via CD44 or TLR4 [14–17]. In this study, we used anti-CD44 mAb and anti-TLR4 mAb to antagonize the interactions between HA and its receptors, and the results showed that anti-TLR4 mAb markedly inhibited the HA-mediated monocyte polarization, and miR-935 expression was down-regulated in M2-like macrophages. Therefore, HA promotes M2 macrophage polarization and acts in an anti-inflammatory manner through TLR4/miR-935.
M2-like TAMs are abundant in many solid tumors and play a vital role in the progression of tumors. Aberrant miRNA expressions levels have been observed in the serum and plasma from patients with different types of solid tumors, and their expression signatures can be used as diagnostic markers for cancer [36–38]. The above results indicate that miR-935 is an essential factor in HA-induced M2 modulation, which raises the question of whether the correlations exist among plasma HA, circulating M2-like monocytes, and miR-935 expression in monocytes. Therefore, we carried out a tentative observation on the levels of miR-935, C/EBPβ, M2-like macrophages, and HA concentrations in patients with solid tumors. Our results showed that there was a significant correlation between M2-like monocytes markers (Arg1, IL10 and CCL18) and plasma HA in the cancer group. However, miR-935 expression in monocytes in the cancer group was found to be negatively correlated with plasma HA. Furthermore, C/EBPβ expression in monocytes was negatively related to miR-935 but positively correlated with plasma HA and M2-like monocytes markers. These findings, while preliminary, may provide further support for the biological phenomenon that INT-HA induces M2-like macrophage differentiation through miR-935/C/EBPβ. Due to the limited sample size and the selection of tumor types, more and detailed studies are needed to confirm our results. Hence, it could be hypothesized that miR-935 may be a novel tumor‑suppressor target, because its regulation may contribute to cancer progression and metastasis through activating macrophage polarization to the M2-like phenotype.
In summary, our study demonstrates that INT-HA modulates the polarization of macrophages to an M2-like phenotype through TLR4. In addition, the subsequent intracellular regulator following TLR4 activation seems to be miR-935/C/EBPβ. This study provides a novel understanding of HA fragment involvement in M2-like macrophages polarization, and reveals a molecular mechanism that could be used in cancer targeting. However, the detailed mechanism by which miRNAs regulate macrophage polarization needs further investigation. Furthermore, our clinical data are limited and future studies are warranted to further support our study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Abbreviations
- Arg1
Arginase 1
- BC
Blank control
- ECM
Extracellular matrix
- GAPDH
Glyceraldehyde 3-phosphate dehydrogenase
- HA
Hyaluronan
- HMW-HA
High molecular weight hyaluronan
- HYALs
Hyaluronidases
- INT-HA
Intermediate-sized hyaluronan
- LMW-HA
Low molecular weight hyaluronan
- MiRNAs
MicroRNAs
- NC
Negative control
- RT-qPCR
Quantitative PCR with reverse transcription
- TAMs
Tumor-associated macrophages
- TLR4
Toll-like receptor 4
Authors’ contributions
BZ, YD, and FG contributed to the conception, design, and final approval of the submitted version. YH and YL performed the research and collected data. BZ, CY, and YD analyzed the data. BZ, GZ, and FG wrote and revised the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (81702852, 81572821, 81502491, 81502490, and 81672843), Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant Support (20171924), the Program of Shanghai Leading Talents (2013-038), and the Yuyan Program of Shanghai Jiao Tong University Affiliated Sixth People’s Hospital (LYZY-0245).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval and ethical standards
This study was performed according to the revised Declaration of Helsinki, 2013. All related human subject procedures were approved by the Ethics Committee of Shanghai Jiao Tong University (approval number: YS-2017-41). Peripheral blood samples were obtained from the Shanghai Jiao Tong University Affiliated Sixth People’s Hospital.
Informed consent
Informed consents were obtained from all participants in the study.
Footnotes
Boke Zhang and Yan Du are co-first authors.
References
- 1.Italiani P, Boraschi D. From monocytes to M1/M2 macrophages: phenotypical vs. functional differentiation. Front Immunol. 2014;5:514. doi: 10.3389/fimmu.2014.00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruffell B, Affara NI, Coussens LM. Differential macrophage programming in the tumor microenvironment. Trends Immunol. 2012;33:119–126. doi: 10.1016/j.it.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cook J, Hagemann T. Tumour-associated macrophages and cancer. Curr Opin Pharmacol. 2013;13:595–601. doi: 10.1016/j.coph.2013.05.017. [DOI] [PubMed] [Google Scholar]
- 4.Spinelli FM, Vitale DL, Demarchi G, et al. The immunological effect of hyaluronan in tumor angiogenesis. Clin Transl Immunol. 2015;4:e52. doi: 10.1038/cti.2015.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stern R, Asari AA, Sugahara KN. Hyaluronan fragments: an information-rich system. Eur J Cell Biol. 2006;85:699–715. doi: 10.1016/j.ejcb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 6.Misra S, Hascall VC, Markwald RR, et al. Interactions between hyaluronan and its receptors (CD44, RHAMM) regulate the activities of inflammation and cancer. Front Immunol. 2015;6:201. doi: 10.3389/fimmu.2015.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laurent TC, Laurent UB, Fraser JR. Functions of hyaluronan. Ann Rheum Dis. 1995;54:429–432. doi: 10.1136/ard.54.5.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian X, Azpurua J, Hine C, et al. High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature. 2013;499:346–349. doi: 10.1038/nature12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Termeer CC, Hennies J, Voith U, et al. Oligosaccharides of hyaluronan are potent activators of dendritic cells. J Immunol. 2000;165:1863–1870. doi: 10.4049/jimmunol.165.4.1863. [DOI] [PubMed] [Google Scholar]
- 10.Horton MR, Shapiro S, Bao C, et al. Induction and regulation of macrophage metalloelastase by hyaluronan fragments in mouse macrophages. J Immunol. 1999;162:4171–4176. [PubMed] [Google Scholar]
- 11.Termeer C, Benedix F, Sleeman J, et al. Oligosaccharides of Hyaluronan activate dendritic cells via toll-like receptor 4. J Exp Med. 2002;195:99–111. doi: 10.1084/jem.20001858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ponta H, Sherman L, Herrlich PA. CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol. 2003;4:33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- 13.Lawrence T, Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat Rev Immunol. 2011;11:750–761. doi: 10.1038/nri3088. [DOI] [PubMed] [Google Scholar]
- 14.Kuang DM, Wu Y, Chen N, et al. Tumor-derived hyaluronan induces formation of immunosuppressive macrophages through transient early activation of monocytes. Blood. 2007;110:587–595. doi: 10.1182/blood-2007-01-068031. [DOI] [PubMed] [Google Scholar]
- 15.Rayahin JE, Buhrman JS, Zhang Y, et al. High and low molecular weight hyaluronic acid differentially influence macrophage activation. ACS Biomater Sci Eng. 2015;1:481–493. doi: 10.1021/acsbiomaterials.5b00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weigel PH, Baggenstoss BA. What is special about 200 kDa hyaluronan that activates hyaluronan receptor signaling? Glycobiology. 2017;27:868–877. doi: 10.1093/glycob/cwx039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sokolowska M, Chen LY, Eberlein M, et al. Low molecular weight hyaluronan activates cytosolic phospholipase A2alpha and eicosanoid production in monocytes and macrophages. J Biol Chem. 2014;289:4470–4488. doi: 10.1074/jbc.M113.515106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamawaki H, Hirohata S, Miyoshi T, et al. Hyaluronan receptors involved in cytokine induction in monocytes. Glycobiology. 2009;19:83–92. doi: 10.1093/glycob/cwn109. [DOI] [PubMed] [Google Scholar]
- 19.Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 20.Ruffell D, Mourkioti F, Gambardella A, et al. A CREB-C/EBPbeta cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proc Natl Acad Sci USA. 2009;106:17475–17480. doi: 10.1073/pnas.0908641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho IJ, Woo NR, Kim SG. The identification of C/EBPbeta as a transcription factor necessary for the induction of MAPK phosphatase-1 by toll-like receptor-4 ligand. Arch Biochem Biophys. 2008;479:88–96. doi: 10.1016/j.abb.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 22.Zhou D, Huang C, Lin Z, et al. Macrophage polarization and function with emphasis on the evolving roles of coordinated regulation of cellular signaling pathways. Cell Signal. 2014;26:192–197. doi: 10.1016/j.cellsig.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 23.Squadrito ML, Etzrodt M, De Palma M, et al. MicroRNA-mediated control of macrophages and its implications for cancer. Trends Immunol. 2013;34:350–359. doi: 10.1016/j.it.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graff JW, Dickson AM, Clay G, et al. Identifying functional microRNAs in macrophages with polarized phenotypes. J Biol Chem. 2012;287:21816–21825. doi: 10.1074/jbc.M111.327031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai X, Yin Y, Li N, et al. Re-polarization of tumor-associated macrophages to pro-inflammatory M1 macrophages by microRNA-155. J Mol Cell Biol. 2012;4:341–343. doi: 10.1093/jmcb/mjs044. [DOI] [PubMed] [Google Scholar]
- 26.Su S, Zhao Q, He C, et al. miR-142-5p and miR-130a-3p are regulated by IL-4 and IL-13 and control profibrogenic macrophage program. Nat Commun. 2015;6:8523. doi: 10.1038/ncomms9523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Squadrito ML, Pucci F, Magri L, et al. miR-511-3p modulates genetic programs of tumor-associated macrophages. Cell Rep. 2012;1:141–154. doi: 10.1016/j.celrep.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 28.Banerjee S, Xie N, Cui H, et al. MicroRNA let-7c regulates macrophage polarization. J Immunol. 2013;190:6542–6549. doi: 10.4049/jimmunol.1202496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maharjan AS, Pilling D, Gomer RH. High and low molecular weight hyaluronic acid differentially regulate human fibrocyte differentiation. PLoS One. 2011;6:e26078. doi: 10.1371/journal.pone.0026078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang G, Guo L, Yang C, et al. A novel role of breast cancer-derived hyaluronan on inducement of M2-like tumor-associated macrophages formation. Oncoimmunology. 2016;5:e1172154. doi: 10.1080/2162402X.2016.1172154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eigsti RL, Sudan B, Wilson ME, et al. Regulation of activation-associated microRNA accumulation rates during monocyte-to-macrophage differentiation. J Biol Chem. 2014;289:28433–28447. doi: 10.1074/jbc.M114.599316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baer C, Squadrito ML, Laoui D, et al. Suppression of microRNA activity amplifies IFN-gamma-induced macrophage activation and promotes anti-tumour immunity. Nat Cell Biol. 2016 doi: 10.1038/ncb3371. [DOI] [PubMed] [Google Scholar]
- 33.Yan C, Yu J, Kang W, et al. miR-935 suppresses gastric signet ring cell carcinoma tumorigenesis by targeting Notch1 expression. Biochem Biophys Res Commun. 2016;470:68–74. doi: 10.1016/j.bbrc.2015.12.116. [DOI] [PubMed] [Google Scholar]
- 34.Yang M, Cui G, Ding M, et al. miR-935 promotes gastric cancer cell proliferation by targeting SOX7. Biomed Pharmacother. 2016;79:153–158. doi: 10.1016/j.biopha.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 35.Liu X, Li J, Yu Z, et al. miR-935 Promotes Liver Cancer Cell Proliferation and Migration by Targeting SOX7. Oncol Res. 2017;25:427–435. doi: 10.3727/096504016X14747300207374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carter JV, Galbraith NJ, Yang D, et al. Blood-based microRNAs as biomarkers for the diagnosis of colorectal cancer: a systematic review and meta-analysis. Br J Cancer. 2017;116:762–774. doi: 10.1038/bjc.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chauhan R, Lahiri N. Tissue- and serum-associated biomarkers of hepatocellular carcinoma. Biomark Cancer. 2016;8:37–55. doi: 10.4137/BIC.S34413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakamura K, Sawada K, Yoshimura A, et al. Clinical relevance of circulating cell-free microRNAs in ovarian cancer. Mol Cancer. 2016;15:48. doi: 10.1186/s12943-016-0536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.