Abstract
Acute myeloid leukemia (AML) is a common and lethal hematopoietic malignancy that is highly dependent on the bone marrow (BM) microenvironment. Infiltrating immune and stromal cells are important components of the BM microenvironment and significantly influence the progression of AML. This study aimed to elucidate the value of immune/stromal cell-associated genes for AML prognosis by integrated bioinformatics analysis. We obtained expression profiles from The Cancer Genome Atlas (TCGA) database and used the ESTIMATE algorithm to calculate immune scores and stromal scores; we then identified differentially expressed genes (DEGs) based on these scores. Overall survival analysis was applied to reveal common DEGs of prognostic value. Subsequently, we conducted a functional enrichment analysis, generated a protein–protein interaction (PPI) network and performed an interrelation analysis of immune system processes, showing that these genes are mainly associated with the immune/inflammatory response. Finally, eight genes (CD163, CYP27A1, KCNA5, PPM1J, FOLR1, IL1R2, MYOF, VSIG2) were verified to be significantly associated with AML prognosis in the Gene Expression Omnibus (GEO) database. In summary, we identified key microenvironment-related genes that affect the outcomes of AML patients and might serve as therapeutic targets.
Electronic supplementary material
The online version of this article (10.1007/s00262-019-02408-7) contains supplementary material, which is available to authorized users.
Keywords: AML, Immune microenvironment, Bioinformatics analysis, Overall survival
Introduction
Acute myeloid leukemia (AML) is one of the most common hematological cancers and is caused by clonal expansion of undifferentiated myeloid progenitor cells [1]. AML is characterized by impaired hematopoiesis and bone marrow (BM) failure, resulting in fatal outcomes [2]. Although many patients with AML achieve remission with chemotherapy, relapse is common and leads to treatment failure, which is caused by minimal residual disease in the protective BM microenvironment [3–5]. Accordingly, an improved understanding of the pathogenesis of AML within the BM microenvironment is crucially important for early diagnosis, prevention and personalized therapy.
Cytogenetic and molecular aberrations are key factors that influence treatment response and long-term outcomes in AML [6, 7]. In addition, the BM microenvironment plays an important role in tumor cell homing and survival. Overall, the BM environment is a dynamic system of immune cells, endothelial progenitor cells (EPCs), stromal cells, extracellular matrix (ECM), growth factors and cytokines [8]. Among them, immune cells and stromal cells are the major components necessary for leukemogenesis and progression [9, 10]. In recent years, novel immunotherapeutic strategies for AML have been developed [11–13].
Estimation of STromal and Immune cells in Malignant Tumours using Expression data (ESTIMATE) algorithm is based on single sample Gene Set Enrichment Analysis and generates stromal and immune scores to predict the infiltration of stromal and immune cells in tumors [14]. Various studies have employed the ESTIMATE algorithm to explore the microenvironment of prostate cancer [15], colon cancer [16] and glioblastoma [17]; however, evaluation of immune/stromal infiltration in AML has not been conducted.
In the current study, we obtained complete gene expression profiles for AML patients from The Cancer Genome Atlas (TCGA) database and calculated immune/stromal scores based on ESTIMATE. A series of microenvironment-related genes were identified as being associated with the overall survival of AML patients. Moreover, we verified the prognostic value of the genes identified in the Gene Expression Omnibus (GEO) database.
Materials and methods
Database
The RNA-Seq dataset of adult AML patients and corresponding clinical profiles were obtained from TCGA (https://gdc.nci.nih.gov/). Immune scores and stromal scores were calculated by the ESTIMATE algorithm of the downloaded database. We adopted two datasets (GSE12417 and GSE5122) from the GEO database. The data of GSE12417 were based on GPL570 platforms (Affymetrix Human Genome U133 Plus 2.0 Array, 79 AML patients), GPL96 platforms (Affymetrix Human Genome U133A Array, 163 AML patients) and GPL97 platforms (Affymetrix Human Genome U133B Array, 163 AML patients). The GSE5122 data were based on GPL96 platforms and included 58 AML patients.
DEGs identification based on immune scores and stromal scores
All AML patients were classified into high- and low-score groups according to their immune/stromal scores. Data analysis was conducted using the package edgeR. In this study, genes with a p value < 0.05 and |fold change| > 1.5 were defined as DEGs. The heatmap of the DEGs was drawn using the Morpheus website (https://software.broadinstitute.org/morpheus).
GO and pathway enrichment analyses
Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david-d.ncifcrf.gov/) was applied to analyze DEG functions and KEGG pathway enrichment. GO term analysis consists of BP, CC, and MF terms. Pathway enrichment was also performed based on the REACTOME online database (http://www.reactome.org). The ClueGO plug-in in Cytoscape software was used to perform interrelation analysis between pathways. A p value < 0.05 was set as the cutoff.
PPI network construction
The STRING database (http://string-db.org) was utilized to assess DEG-encoded proteins and PPI information. The PPI network was subsequently established using Cytoscape software. The MCODE plug-in in Cytoscape was applied to perform modular analysis, and the most significant module was identified based on the MCODE score and node number.
Survival analysis
Kaplan-Meier plots were constructed to illuminate correlations between expression of DEGs and the overall survival of AML patients. The statistical significance of the correlation was tested by the log-rank test. The online tool PrognoScan (http://dna00.bio.kyutech.ac.jp/PrognoScan/) was used to verify the prognostic values of the genes identified. A short step-by-step bioinformatics protocol was listed in supplementary materials.
Results
Immune conditions are associated with AML clinical characters
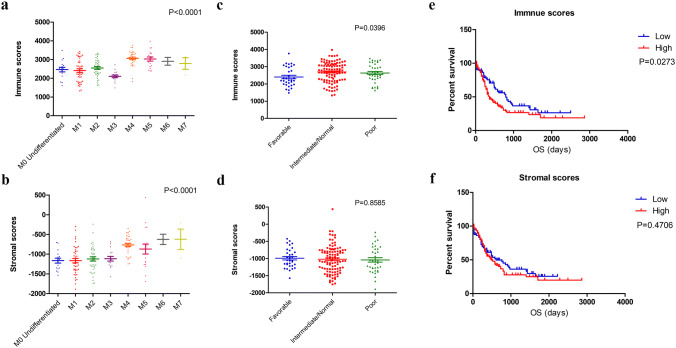
We obtained the complete gene expression profiles and clinical information for 173 AML patients from TCGA (Supplementary Table 1); 93 (53.8%) patients were male and 80 (46.2%) female. The age at initial pathological diagnosis ranged from 18 to 88 years, with a median age of 58 years. The eight subtypes of these patients included M0 undifferentiated (16, 9.2%), M1 (42, 24.3%), M2 (39, 22.5%), M3 (16, 9.2%), M4 (35, 20.2%), M5 (18, 10.4%), M6 (2, 1.2%), and M7 (3, 1.7%) [18]; 2 patients were not classified. Employing the ESTIMATE algorithm, we calculated immune scores and stromal scores for all these patients, ranging from 1329.53 to 3971.97 for the former and from −1888.81 to 435.75 for the latter. In addition, the immune and stromal scores were significantly associated with the subtype classification (Fig. 1a, b).
Fig. 1.
Immune conditions are associated with AML clinical features. a, b Distribution of immune scores and stromal scores for AML subtypes. c The significant correlation between immune scores and AML cytogenetic risk (p = 0.0396). d The stromal scores show no significant difference in cytogenetic risk (p = 0.8585). e Kaplan-Meier survival curve reveals that higher immune scores are associated with significantly shorter overall survival (log-rank test, p = 0.0273). f The low stromal score group showed a longer median overall survival than high stromal score group, with no significant difference (log-rank test, p = 0.4706)
The cytogenetic risk of AML patients was classified into three groups: favorable, intermediate/normal and poor [19]. We plotted the distribution of immune scores and stromal scores according to the degree of cytogenetic risk. As shown in Fig. 1c, the immune scores were meaningful in the correlation of cytogenetic risk (p = 0.0396), though no statistically significant differences in cytogenetic risk were found for the stromal scores (Fig. 1d, p = 0.8585).
To explore the potential association of overall survival with immune scores and stromal scores, we classified the 173 AML patients into high- and low-score groups. Kaplan-Meier survival analysis (Fig. 1e) revealed that the median overall survival of patients with low immune scores was longer than that of patients with high scores (792 vs. 365 days, p = 0.0273). In addition, the median overall survival of the patients in the low stromal score group was longer than that of the patients in the high stromal score group, with no significant difference (Fig. 1f, 608 vs. 489 days, p = 0.4706).
Identification of differentially expressed genes (DEGs) based on immune scores and stromal scores in AML
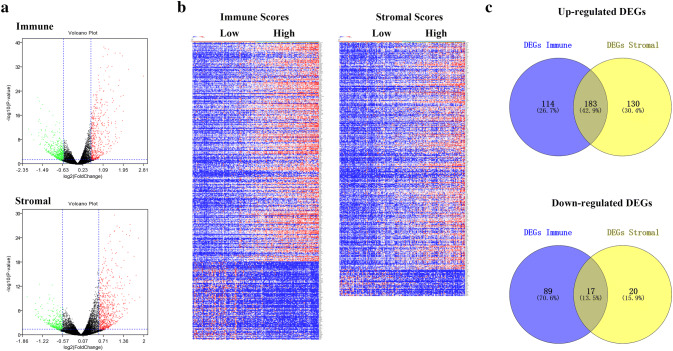
To determine the association of gene expression profiles with immune scores and/or stromal scores, we analyzed the RNA-Seq data for all 173 AML patients obtained from TCGA. Setting the cut-off criteria as p < 0.05 and |fold change| > 1.5, we identified 403 and 350 DEGs based on immune scores and stromal scores, respectively (Fig. 2a). The DEGs of the low vs. high immune score/stromal score groups are also illustrated in the heatmap shown in Fig. 2b. Through integrated bioinformatics analysis, we identified 183 commonly upregulated genes and 17 commonly downregulated genes from the immune score/stromal score groups (Fig. 2c). Our subsequent analysis focused on these common DEGs.
Fig. 2.
Identification of DEGs based on immune scores and stromal scores. a Volcano plot of DEGs from the low vs. high immune score/stromal score groups. Genes with p < 0.05 are shown in red (fold change > 1.5) and green (fold change < −1.5). Black plots represent the remaining genes (those with no significant difference). b Heatmap of DEGs for the immune and stromal score groups. c Commonly changed DEGs in the stromal and immune score groups (183 up- and 17 downregulated genes)
Gene ontology (GO) term and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of DEGs
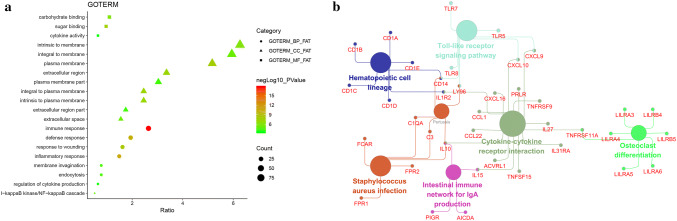
Based on the DAVID gene annotation tool, we performed GO analysis of the DEGs. As shown in Fig. 3a, the DEGs were analyzed for three subontologies: biological processes (BP), cellular component (CC), and molecular function (MF). For BP, DEGs were mainly enriched in the immune response, inflammatory response, defense response and response to wounding. With regard to CC, DEGs primarily clustered in the plasma membrane, integral to plasma membrane, extracellular region and intrinsic to plasma membrane. DEGs in the MF category are mainly associated with sugar binding, carbohydrate binding and cytokine activity. In addition, we conducted interrelation analysis by assessing BP (Supplementary Fig. 1), CC (Supplementary Fig. 2), and MF (Supplementary Fig. 3) for the DEGs in ClueGO and found that most of the genes are involved in more than two processes. Subsequently, we performed KEGG pathway enrichment and interrelation analysis. As shown in Fig. 3b, enrichment of DEGs was mainly observed for the cytokine–cytokine receptor interaction, osteoclast differentiation, the Toll-like receptor signaling pathway, hematopoietic cell lineage and the intestinal immune network for IgA production.
Fig. 3.
Gene ontology (GO) term enrichment analysis of common DEGs. a The top 30 significantly enriched GO terms, including three subontologies, biological process, molecular function and cellular component, are shown. b Interrelation analysis of KEGG pathways of common DEGs
Survival analysis of DEGs
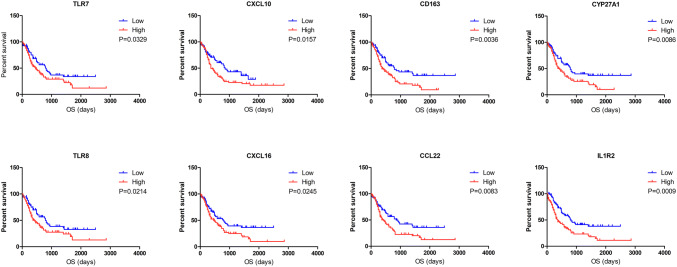
To determine the potential value of the DEGs in predicting the overall survival of AML patients, we constructed Kaplan-Meier survival curves. Among the 183 commonly upregulated DEGs, 55 (Supplementary Table 2) were negatively associated with overall survival according to the log-rank test (p < 0.05). Representative genes are shown in Fig. 4.
Fig. 4.
Correlation between expression of individual DEGs and AML overall survival in TCGA. Kaplan-Meier survival curves with the log-rank test were performed for the representative DEGs
Protein–protein interaction (PPI) network construction and functional enrichment of genes of prognostic value
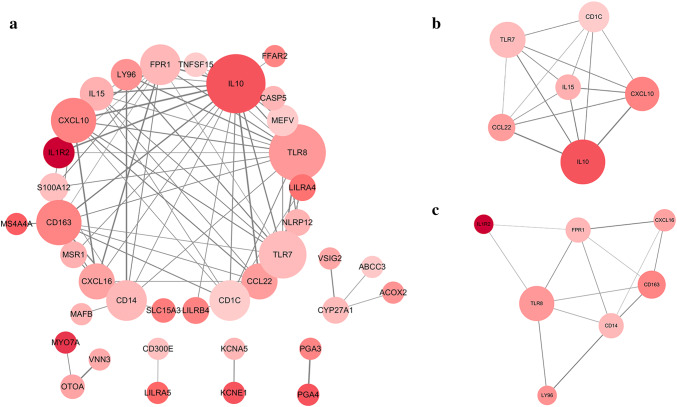
To further explore the interplay among the 55 genes with prognostic value, we constructed a PPI network based on the STRING online database and Cytoscape software. As shown in Fig. 5a, the network contains 38 nodes and 85 edges. Clustering analysis of the PPI network was then carried out using Cytotype MCODE, and the top two significant modules were selected based on the degree of importance. As shown in Fig. 5b, module 1 contains 6 nodes and 15 edges; module 2 contains 7 nodes and 13 edges (Fig. 5c). GO term analysis (Supplementary Fig. 4a) revealed the 55 genes of prognostic value to be mainly enriched in the immune response and the inflammatory response (BP), intrinsic to membrane and extracellular region (CC), and cytokine activity (MF). In addition, KEGG and REACTOME pathway enrichment analyses (Supplementary Fig. 4b) demonstrated that these genes are associated with IL-10 signaling, the immune system and the cytokine–cytokine receptor interaction. Interrelation analysis was also conducted using ClueGO to assess the immune system process. As depicted in Supplementary Fig. 5, we found enrichment of the genes primarily in the MyD88-dependent Toll-like receptor signaling pathway and negative regulation of myeloid leukocyte differentiation.
Fig. 5.
PPI network of DEGs of prognostic value and module identification. a Based on the STRING database and Cytoscape software, a PPI network containing 38 nodes and 85 edges was constructed. The size of the node represents the degree, and the color of the node represents the p value for prognosis. b Two significant modules were identified based on the degree of importance. Module 1 contains 6 nodes and 15 edges. c Module 2 contains 7 nodes and 13 edges
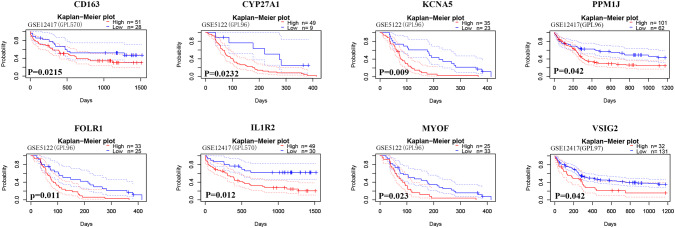
Validation in the GEO database
We further identified the prognostic values of the 55 genes described above using the PrognoScan online tool. Based on the GSE12417 and GSE5122 datasets, a total of 8 genes were verified (Fig. 6, Supplementary Table 3) to be significantly associated with a poor prognosis in AML. Among them, CD163 and IL1R2 are associated with the immune/inflammatory response. GO term analysis (CC) showed that IL1R2, KCNA5, MYOF, CD163 and VSIG2 clustered in the intrinsic to membrane.
Fig. 6.
Verification of genes with prognostic value in the GEO database. Kaplan-Meier survival curves with the log-rank test were performed for genes with prognostic value. Genes with statistical significance (p < 0.05) are shown
Discussion
AML is a rapidly progressive disease with a poor prognosis that is highly dependent on the BM microenvironment [20]. In this study, we analyzed BM microenvironment-associated genes of prognostic value in AML based on TCGA. Common DEGs were identified from low vs. high immune score/stromal score groups and subjected to overall survival analysis. We also utilized bioinformatics methods to deeply explore the DEGs, including GO term analysis, signaling pathway enrichment analysis and PPI network construction. Importantly, the genes identified as having prognostic value were also validated in the GEO database (Supplementary Fig. 6).
First, we calculated the immune scores and stromal scores of AML patients based on the ESTIMATE algorithm and found these scores to be significantly associated with the classification of AML subtype. In addition, the immune scores were meaningful in correlating cytogenetic risk and overall survival. Previous studies have indicated that immune cells and stromal cells are important components of the BM environment that influence AML cell survival, proliferation and therapeutic resistance [21]. Moreover, AML cells actively shape the BM environment and immune cells to promote disease progression through cellular, structural, and functional changes [20, 22, 23]. It is important to integrate and reanalyze genomic profiles from public databases to better understand correlations between AML cells and the BM environment [24, 25].
Furthermore, we identified common DEGs from the low vs. high immune score/stromal score groups. GO term analysis revealed these DEGs to be mainly enriched in the immune response and the inflammatory response (BP), the plasma membrane and integral to plasma membrane (CC), and cytokine activity (MF). Moreover, according to KEGG pathway enrichment analysis, the DEGs mainly clustered in cytokine–cytokine receptor interaction, the Toll-like receptor signaling pathway and hematopoietic cell lineage categories. Consistent with these results, previous studies have demonstrated that the biology of the immune system is crucial for the formation of a complex BM microenvironment [8, 21]. In recent years, knowledge of the immunological features of AML has increased, and the development of effective immunotherapeutic strategies for AML has attracted much attention [26–28].
Overall survival analysis of the commonly upregulated DEGs revealed that 55 genes correlate with unfavorable outcomes of AML patients. In addition, the PPI network of these genes consisted of two modules significantly associated with the immune/inflammatory response. Several genes in the two modules, such as IL-10, IL-15 and TLR8, have been indicated as being involved in the survival, proliferation and differentiation of AML cells [29–32]. Importantly, we verified the prognostic value of these 55 genes based on the GEO database. Eight genes were validated as unfavorable prognostic biomarkers for AML patients, a finding that needs to be further tested in the clinic. Among them, CD163 is expressed on M4/M5 AML cells but not on other subtypes and on normal hematopoietic progenitor cells [33, 34]. Thus, CD163 has been identified as a potential target for therapy. The functions of CYP27A1, FOLR1, IL1R2, KCNA5, MYOF, PPM1J and VSIG2 in AML have not been previously reported, but these factors might serve as biomarkers.
In conclusion, integrated bioinformatics analysis of the AML dataset from TCGA was performed with a focus on the immune microenvironment. Common DEGs were identified, tested and validated to determine their prognostic value for AML patients. Further investigation of these genes in the clinic is required and may provide new insight into the pathogenesis of AML. This study increases our understanding of the complex interactions between AML tumor cells and the BM microenvironment and might provide novel therapeutic targets.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank the TCGA and GEO databases for the availability of the data.
Abbreviations
- AML
Acute myeloid leukemia
- BM
Bone marrow
- BP
Biological processes
- CC
Cellular component
- DAVID
Database for Annotation, Visualization and Integrated Discovery
- DEGs
Differentially expressed genes
- ECM
Extracellular matrix
- EPCs
Endothelial progenitor cells
- ESTIMATE
Estimation of STromal and Immune cells in Malignant Tumours using Expression data
- GEO
Gene Expression Omnibus
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MF
Molecular function
- PPI
Protein–protein interaction
- TCGA
The Cancer Genome Atlas
Author contributions
HY and WC conceived and designed the study. JQ obtained the datasets. YL and GZ conducted data analysis. HY wrote the manuscript. EZ and ZC revised the manuscript. All authors reviewed and approved the final manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China [Grant number 91742110) and Zhejiang Provincial Natural Science Foundation (Grant number LY19H080004).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval and ethical standards
The data used in our study were obtained from public databases TCGA and GEO, therefore, ethical approval was not required.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Haimeng Yan and Jianwei Qu contributed equally to this work.
References
- 1.Rowe JM, Tallman MS. How I treat acute myeloid leukemia. Blood. 2010;116:3147–3156. doi: 10.1182/blood-2010-05-260117. [DOI] [PubMed] [Google Scholar]
- 2.Weinberg OK, Sohani AR. Diagnostic work-up of acute myeloid leukemia. 2017;92:317–321. doi: 10.1002/ajh.24648. [DOI] [PubMed] [Google Scholar]
- 3.Buckley SA, Kirtane K, Walter RB, Lee SJ, Lyman GH. Patient-reported outcomes in acute myeloid leukemia: Where are we now? Blood Rev. 2018;32:81–87. doi: 10.1016/j.blre.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 4.Zeng Z, Shi YX, Samudio IJ, Wang RY, Ling X, Frolova O, Levis M, Rubin JB, Negrin RR, Estey EH, Konoplev S, Andreeff M, Konopleva M. Targeting the leukemia microenvironment by CXCR4 inhibition overcomes resistance to kinase inhibitors and chemotherapy in AML. Blood. 2009;113:6215–6224. doi: 10.1182/blood-2008-05-158311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abdul-Aziz AM, Shafat MS, Mehta TK, Di Palma F, Lawes MJ, Rushworth SA, Bowles KM. MIF-induced stromal PKCbeta/IL8 is essential in human acute myeloid leukemia. Can Res. 2017;77:303–311. doi: 10.1158/0008-5472.can-16-1095. [DOI] [PubMed] [Google Scholar]
- 6.Dohner H, Estey EH, Amadori S, Appelbaum FR, Buchner T, Burnett AK, Dombret H, Fenaux P, Grimwade D, Larson RA, Lo-Coco F, Naoe T, Niederwieser D, Ossenkoppele GJ, Sanz MA, Sierra J, Tallman MS, Lowenberg B, Bloomfield CD. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115:453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 7.Kayser S, Levis MJ. Advances in targeted therapy for acute myeloid leukaemia. 2018;180:484–500. doi: 10.1111/bjh.15032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ayala F, Dewar R, Kieran M, Kalluri R. Contribution of bone microenvironment to leukemogenesis and leukemia progression. Leukemia. 2009;23:2233–2241. doi: 10.1038/leu.2009.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Austin R, Smyth MJ, Lane SW. Harnessing the immune system in acute myeloid leukaemia. Critical reviews in oncology/hematology. 2016;103:62–77. doi: 10.1016/j.critrevonc.2016.04.020. [DOI] [PubMed] [Google Scholar]
- 10.Yehudai-Resheff S, Attias-Turgeman S, Sabbah R, Gabay T, Musallam R, Fridman-Dror A, Zuckerman T. Abnormal morphological and functional nature of bone marrow stromal cells provides preferential support for survival of acute myeloid leukemia cells. Int J Cancer. 2019;144:2279–2289. doi: 10.1002/ijc.32063. [DOI] [PubMed] [Google Scholar]
- 11.Beyar-Katz O, Gill S. Novel approaches to acute myeloid leukemia immunotherapy. Clin Cancer Res. 2018;24:5502–5515. doi: 10.1158/1078-0432.ccr-17-3016. [DOI] [PubMed] [Google Scholar]
- 12.Vandsemb EN, Kim TK, Zeidan AM. Will deeper characterization of the landscape of immune checkpoint molecules in acute myeloid leukemia bone marrow lead to improved therapeutic targeting? Cancer. 2019;125:1410–1413. doi: 10.1002/cncr.32042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrmann M, Krupka C, Deiser K, Brauchle B, Marcinek A, Ogrinc Wagner A, Rataj F, Mocikat R, Metzeler KH, Spiekermann K, Kobold S, Fenn NC, Hopfner KP, Subklewe M. Bifunctional PD-1 x alphaCD3 x alphaCD33 fusion protein reverses adaptive immune escape in acute myeloid leukemia. Blood. 2018;132:2484–2494. doi: 10.1182/blood-2018-05-849802. [DOI] [PubMed] [Google Scholar]
- 14.Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, Trevino V, Shen H, Laird PW, Levine DA, Carter SL, Getz G, Stemke-Hale K, Mills GB, Verhaak RG. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shah N, Wang P, Wongvipat J, Karthaus WR, Abida W, Armenia J, Rockowitz S, Drier Y, Bernstein BE, Long HW (2017) Regulation of the glucocorticoid receptor via a BET-dependent enhancer drives antiandrogen resistance in prostate cancer. eLife 6. 10.7554/eLife.27861 [DOI] [PMC free article] [PubMed]
- 16.Alonso MH, Aussó S, Lopez-Doriga A, Cordero D, Guinó E, Solé X, Barenys M, de Oca J, Capella G, Salazar R, Sanz-Pamplona R, Moreno V. Comprehensive analysis of copy number aberrations in microsatellite stable colon cancer in view of stromal component. Br J Cancer. 2017;117:421–431. doi: 10.1038/bjc.2017.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia D, Li S, Li D, Xue H, Yang D, Liu Y. Mining TCGA database for genes of prognostic value in glioblastoma microenvironment. Aging. 2018;10:592–605. doi: 10.18632/aging.101415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lowenberg B, Downing JR, Burnett A. Acute myeloid leukemia. New Engl J Med. 1999;341:1051–1062. doi: 10.1056/nejm199909303411407. [DOI] [PubMed] [Google Scholar]
- 19.Slovak ML, Kopecky KJ, Cassileth PA, Harrington DH, Theil KS, Mohamed A, Paietta E, Willman CL, Head DR, Rowe JM, Forman SJ, Appelbaum FR. Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: a Southwest Oncology Group/Eastern Cooperative Oncology Group Study. Blood. 2000;96:4075–4083. doi: 10.1182/blood.V96.13.4075. [DOI] [PubMed] [Google Scholar]
- 20.Abdul-Aziz AM, Sun Y, Hellmich C, Marlein CR, Mistry J, Forde E, Piddock RE, Shafat MS, Morfakis A, Mehta T, Di Palma F, Macaulay I, Ingham CJ, Haestier A, Collins A, Campisi J, Bowles KM, Rushworth SA. Acute myeloid leukemia induces protumoral p16INK4a-driven senescence in the bone marrow microenvironment. Blood. 2019;133:446–456. doi: 10.1182/blood-2018-04-845420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tabe Y, Konopleva M. Role of Microenvironment in Resistance to Therapy in AML. Curr Hematol Malig Rep. 2015;10:96–103. doi: 10.1007/s11899-015-0253-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kogan AA, Lapidus RG, Baer MR, Rassool FV. Exploiting epigenetically mediated changes: Acute myeloid leukemia, leukemia stem cells and the bone marrow microenvironment. Adv Cancer Res. 2019;141:213–253. doi: 10.1016/bs.acr.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 23.Shafat MS, Oellerich T, Mohr S, Robinson SD, Edwards DR, Marlein CR, Piddock RE, Fenech M, Zaitseva L, Abdul-Aziz A, Turner J, Watkins JA, Lawes M, Bowles KM, Rushworth SA. Leukemic blasts program bone marrow adipocytes to generate a protumoral microenvironment. Blood. 2017;129:1320–1332. doi: 10.1182/blood-2016-08-734798. [DOI] [PubMed] [Google Scholar]
- 24.Huang R, Liao X, Li Q. Identification of key pathways and genes in TP53 mutation acute myeloid leukemia: evidence from bioinformatics analysis. OncoTargets Ther. 2018;11:163–173. doi: 10.2147/ott.s156003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng Y, Shen Y, Chen H, Wang X, Zhang R, Peng Y, Lei X, Liu T, Liu J, Gu L, Wang F, Yang Y, Bai J, Wang J, Zhao W, He A. Expression profile analysis of long non-coding RNA in acute myeloid leukemia by microarray and bioinformatics. . Cancer Sci. 2018;109:340–353. doi: 10.1111/cas.13465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nahas MR, Stroopinsky D. Hypomethylating agent alters the immune microenvironment in acute myeloid leukaemia (AML) and enhances the immunogenicity of a dendritic cell/AML vaccine. 2019;185(4):679–690. doi: 10.1111/bjh.15818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang D, Zhang X, Zhang X, Xu Y. The progress and current status of immunotherapy in acute myeloid leukemia. Ann Hematol. 2017;96:1965–1982. doi: 10.1007/s00277-017-3148-x. [DOI] [PubMed] [Google Scholar]
- 28.Rotiroti MC, Arcangeli S, Casucci M, Perriello V, Bondanza A, Biondi A, Tettamanti S, Biagi E. Acute myeloid leukemia targeting by chimeric antigen receptor T cells: bridging the gap from preclinical modeling to human studies. Hum Gene Ther. 2017;28:231–241. doi: 10.1089/hum.2016.092. [DOI] [PubMed] [Google Scholar]
- 29.Nishioka C, Ikezoe T, Yang J, Nobumoto A, Kataoka S, Tsuda M, Udaka K, Yokoyama A. CD82 regulates STAT5/IL-10 and supports survival of acute myelogenous leukemia cells. Int J Cancer. 2014;134:55–64. doi: 10.1002/ijc.28348. [DOI] [PubMed] [Google Scholar]
- 30.Meazza R, Basso S, Gaggero A, Detotero D, Trentin L, Pereno R, Azzarone B, Ferrini S. Interleukin (IL)-15 induces survival and proliferation of the growth factor-dependent acute myeloid leukemia M-07e through the IL-2 receptor beta/gamma. Int J Cancer. 1998;78:189–195. doi: 10.1002/(SICI)1097-0215(19981005)78:2<189::AID-IJC12>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 31.Ignatz-Hoover JJ, Wang H, Moreton SA, Chakrabarti A, Agarwal MK, Sun K, Gupta K, Wald DN. The role of TLR8 signaling in acute myeloid leukemia differentiation. Leukemia. 2015;29:918–926. doi: 10.1038/leu.2014.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smits EL, Cools N, Lion E, Van Camp K, Ponsaerts P, Berneman ZN, Van Tendeloo VF. The Toll-like receptor 7/8 agonist resiquimod greatly increases the immunostimulatory capacity of human acute myeloid leukemia cells. Cancer Immunol Immunother. 2010;59:35–46. doi: 10.1007/s00262-009-0721-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walter RB, Bachli EB, Schaer DJ, Ruegg R, Schoedon G. Expression of the hemoglobin scavenger receptor (CD163/HbSR) as immunophenotypic marker of monocytic lineage in acute myeloid leukemia. Blood. 2003;101:3755–3756. doi: 10.1182/blood-2002-11-3414. [DOI] [PubMed] [Google Scholar]
- 34.Bachli EB, Schaer DJ, Walter RB, Fehr J, Schoedon G. Functional expression of the CD163 scavenger receptor on acute myeloid leukemia cells of monocytic lineage. J Leukoc Biol. 2006;79:312–318. doi: 10.1189/jlb.0605309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.