Abstract
Molecular characterization of cytopathogenic (cp) bovine viral diarrhea virus (BVDV) strain CP Rit, a temperature-sensitive strain widely used for vaccination, revealed that the viral genomic RNA is about 15.2 kb long, which is about 2.9 kb longer than the one of noncytopathogenic (noncp) BVDV strains. Molecular cloning and nucleotide sequencing of parts of the genome resulted in the identification of a duplication of the genomic region encoding nonstructural proteins NS3, NS4A, and part of NS4B. In addition, a nonviral sequence was found directly upstream of the second copy of the NS3 gene. The 3′ part of this inserted sequence encodes an N-terminally truncated ubiquitin monomer. This is remarkable since all described cp BVDV strains with ubiquitin coding sequences contain at least one complete ubiquitin monomer. The 5′ region of the nonviral sequence did not show any homology to cellular sequences identified thus far in cp BVDV strains. Databank searches revealed that this second cellular insertion encodes part of ribosomal protein S27a. Further analyses included molecular cloning and nucleotide sequencing of the cellular recombination partner. Sequence comparisons strongly suggest that the S27a and the ubiquitin coding sequences found in the genome of CP Rit were both derived from a bovine mRNA encoding a hybrid protein with the structure NH2-ubiquitin-S27a-COOH. Polyprotein processing in the genomic region encoding the N-terminal part of NS4B, the two cellular insertions, and NS3 was studied by a transient-expression assay. The respective analyses showed that the S27a-derived polypeptide, together with the truncated ubiquitin, served as processing signal to yield NS3, whereas the truncated ubiquitin alone was not capable of mediating the cleavage. Since the expression of NS3 is strictly correlated with the cp phenotype of BVDV, the altered genome organization leading to expression of NS3 most probably represents the genetic basis of cytopathogenicity of CP Rit.
The genera Pestivirus, Flavivirus, and hepatitis C virus group constitute the family Flaviviridae (55). The genus Pestivirus currently comprises three members, bovine viral diarrhea virus (BVDV), classical swine fever virus, and border disease virus. The presence of a fourth separate group of pestiviruses comprising isolates from cattle and sheep has been recently described (3, 5, 40, 41, 46), and it is now generally accepted to refer to this additional species as BVDV-2; consequently, classical BVDV strains are frequently named BVDV-1. Pestivirus virions contain a positive-strand RNA genome of approximately 12.3 kb. Viral gene expression is believed to occur via synthesis of a polyprotein which is co- and posttranslationally processed by both viral and cellular proteases (6, 7, 11, 12, 14, 15, 31, 33, 38, 47). In the polyprotein, the mature viral proteins are arranged in the following order (from the N to the C terminus): Npro, C, Erns, E1, E2, p7, NS2-3, (NS2), (NS3), NS4A, NS4B, NS5A, and NS5B (see references 36 and 53 for reviews); the abbreviations Npro and Erns refer to an N-terminal autoprotease and a structural glycoprotein with RNase activity, respectively. The structural proteins are represented by C, Erns, E1, and E2, whereas the remaining proteins are presumably nonstructural (NS). In tissue culture, replication of pestiviruses can be accompanied by a cytopathic effect (20, 26). Accordingly, two biotypes of pestiviruses are distinguished, namely, cytopathogenic (cp) and noncytopathogenic (noncp).
BVDV is distributed worldwide, and it represents one of the most important bovine pathogens. BVDV infection can have quite different consequences, such as abortion, diarrhea, hemorrhagic syndrome, and, most frequently, inapparent courses (2, 53). Both cp and noncp BVDV strains are involved in the pathogenesis of mucosal disease (MD), a very severe clinical manifestation of BVDV infection (8, 9, 36). A prerequisite for the development of MD is an intrauterine infection with noncp BVDV during the first trimester of gestation, resulting in the birth of persistently infected animals with an acquired immunotolerance to the original BVDV strain. Interestingly, development of MD coincides with the appearance of cp BVDV.
For BVDV, cytopathogenicity is always correlated with expression of NS3, which is colinear with the carboxy-terminal part of NS2-3. While NS2-3 is expressed in both cp and noncp BVDV-infected cells, NS3 is found exclusively after infection with cp BVDV (11, 13, 17, 21, 34, 35, 42, 43). Accordingly, NS3 is regarded as the marker protein for cp BVDV strains and is supposed to be required for the induction of cytopathic effect.
Molecular analyses of several cp BVDV strains isolated from field cases of MD strongly suggested that each cp virus evolved from the respective persisting noncp virus by RNA recombination. The mutations identified in the genomes of cp BVDV strains include insertions of cellular sequences, frequently together with large duplications, and genomic rearrangements with large duplications and deletions (see reference 36 for a review). For pestiviruses, two different kinds of cellular insertions have been found which encode either (poly)ubiquitin or part of a cellular polypeptide of unknown function (4, 30, 32, 34, 44, 51). In this paper, we report the identification of a novel cellular insertion in the genome of a cp BVDV vaccine strain and its effect on polyprotein processing as well as the molecular characterization of the putative cellular recombination partner.
MATERIALS AND METHODS
Cells and viruses.
MDBK and BHK-21 cells were obtained from the American Type Culture Collection (Rockville, Md.). Cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum (FCS). Isolation of BVDV strain CP7 (13, 33, 50) and generation of the temperature-sensitive, cp BVDV vaccine strain Rit 4350 (CP Rit) (27) have been described previously. BVDV CP Rit was obtained from Pfizer (Karlsruher, Germany). BVDV CP Rit and BVDV CP7 represent BVDV-1 strains. The modified vaccinia virus Ankara expressing the T7 polymerase (MVA-T7pol) was kindly provided by G. Sutter (Institute of Molecular Virology, GSF-Centre for Environmental and Health Research, Oberschleissheim, Germany) (49).
Infection of cells.
Supernatants and lysates of infected cells were combined and used for infection of MDBK cells. Material for infection was prepared by freezing and thawing cultures 48 h postinfection and stored at −70°C. A multiplicity of infection of about 0.1 was used for infections. The proportion of virus-infected cells was assessed by indirect immunofluorescence with monoclonal antibody (MAb) 8.12.7 (directed against NS3), kindly provided by E. J. Dubovi (Cornell University, Ithaca, N.Y.). Cells and FCS were tested regularly for the absence of pestiviruses by reverse transcription-PCR (RT-PCR) and immunofluorescence. For FCS, the absence of anti-pestivirus antibodies was shown by lack of virus neutralization.
Oligonucleotides.
Oligonucleotides were purchased from MWG Biotech GmbH (Ebersberg, Germany). Sequences of oligonucleotides, their positions in the genomic sequence of BVDV SD-1 (15), and their polarities are as follows: Ol BVDV7100, 5′ AGACTAGARGAYACMACCCACCT 3′ (positions 7313 to 7335, sense); Ol NS3R, 5′ ATSCCGCCTTGGTGTGTGTA 3′ (5325 to 5343, antisense). Both primers were designed from published sequences of BVDV-1 strains NADL (12), Osloss (14), SD-1 (15), and CP7 (33). The following primers were derived from the BVDV Rit sequence: Ol Rit4AR (5′ GACTCTAGAAATCCGAGATAGATTCCATG 3′, antisense), Ol Rit-ubi4 (5′ GCATCCATGGTGAAGACCCTGACGGGGAAG 3′, sense), Ol RitNS3R (5′ CCTCACCTTTAGCAATGCTG 3′, antisense), Ol Rit4B1 (5′ GCATCCATGGCAGTGGGTGACCTGGAC 3′, sense), Ol S27a* (5′ GCATCCATGGAGAATGGCAAAATCAGTCG 3′, sense), and Ol S27a-del (5′ CTTCCCGGGGGACTCCGAAACCCAACAGTTCGTGAAGACCCTGACGG 3′, sense).
For cloning of the bovine mRNA encoding ubiquitin and S27a, oligonucleotides Ol UeR (5′ TTACTTGTCTTCTGGTTTGTTG 3′, antisense), and Ol Ue (5′ CGCCRCCRMRATGCAGAT 3′, sense) were deduced from published ubiquitin-S27a sequences of humans (GenBank accession no. AA627893), rats (X81839), and guinea pigs (D83209).
RNA preparation, gel electrophoresis, and Northern (RNA) hybridization.
RNA from pestivirus-infected cells was prepared by using either the RNeasy total-RNA kit (Qiagen GmbH, Hilden, Germany) or the RNA extraction kit (Pharmacia Biotech) as recommended by the supplier. Glyoxylated RNA (5 μg) (28) was separated in a phosphate-buffered 1.0% agarose gel containing 5.5% formaldehyde and transferred to Duralon-UV membranes (Stratagene, Heidelberg, Germany). An RNA ladder (Bethesda Research Laboratories) served as a size standard. Radioactive labelling of the probe, hybridization, and posthybridization washes were done as described previously (4). A 2.5-kb NotI-NsiI fragment from the cDNA clone pA/BVDV was used as a probe (33).
RT-PCR.
Reverse transcription (RT) of approximately 500 ng of heat-denatured RNA was done as described previously (5). Following amplification, the PCR products were characterized in agarose-ethidium bromide gels in Tris-acetate buffer.
Molecular cloning, nucleotide sequencing, and sequence analysis.
The cDNA fragments obtained after RT-PCR were separated by agarose gel electrophoresis and purified with the Qiaex DNA purification kit (Qiagen). The respective cDNA fragments were cloned with the TA cloning kit (Invitrogen, De Schelp, The Netherlands). Nucleotide sequences were determined by cycle sequencing with the Thermo Sequenase kit (Amersham Buchler, Braunschweig, Germany) and the DNA sequencer Li-Cor 4000 (MWG Biotech). All sequences were determined by sequencing both complementary strands of at least three independent cDNA clones. Computer analysis of sequence data was performed with HUSAR (DKFZ, Heidelberg, Germany), which provides the Genetics Computer Group software package (16).
Transient expression with the T7 vaccinia virus system.
BHK-21 cells (5 × 105 per 3.5-cm-diameter dish) were infected with the recombinant T7 vaccinia virus MVA-T7pol at a multiplicity of infection of 10 (49). After 1 h of incubation at 37°C, the cells were washed twice with medium lacking FCS. Subsequently, they were transfected with 2.0 μg of plasmid DNA by using Superfect reagent (Qiagen). After 3 h of incubation at 37°C, the supernatant was replaced with medium containing 10% FCS and the cells were incubated overnight at 37°C. Finally, the cells were washed with phosphate-buffered saline (PBS) and stored at −20°C.
Construction of T7 expression plasmids.
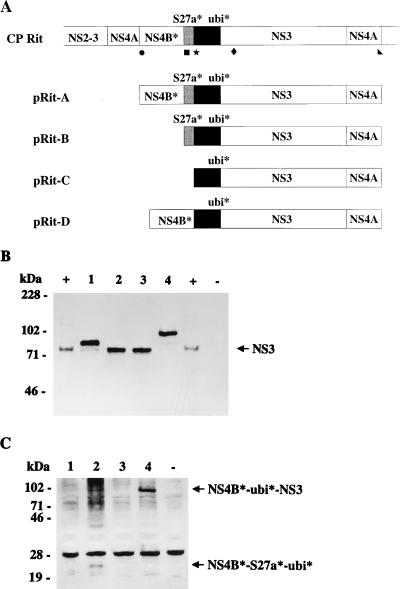
All T7 expression plasmids were based on the vector pCITE (Invitrogen). To establish a construct for the expression of ubiquitin* (truncated ubiquitin lacking the N-terminal 3 amino acids [aa]), NS3, and NS4A of BVDV Rit, the cDNA encoding the respective region of the CP Rit polyprotein was obtained by RT-PCR with primer Ol Rit4AR (including a XbaI site) and primer Ol Rit-ubi4 (including a NcoI site), and cloned into pCR2.1 (Invitrogen). The respective NcoI-XbaI fragment was cloned into pCITE (precut with NcoI and XbaI). The resulting plasmid is termed pRit-C. For construction of pCRRit-A, the genomic region encoding NS4B* (N-terminal 132 aa of NS4B), S27a* (the portion of S27a encoded by the genome of BVDV Rit), ubiquitin*, and part of NS3 was cloned into pCR2.1 after RT-PCR with primer Ol RitNS3R and primer Ol Rit4B1 (including a NcoI site); for generation of pCRRit-B, the genomic region encoding S27a*, ubiquitin*, and part of NS3 was cloned after RT-PCR with primer Ol RitNS3R and primer Ol S27a* (including a NcoI site). To obtain constructs pRit-A and pRit-B, NcoI-XhoI fragments from plasmids pCRRit-A and pCRRit-B were cloned into pRit-C (precut with NcoI and XhoI), respectively. Accordingly, pRit-A encompasses the genomic region encoding NS4B*, S27a*, ubiquitin*, NS3, and NS4A, while the fusion protein encoded by pRit-B starts with S27a*. A schematic representation of the different constructs together with the positions of primers used for cloning is shown in Fig. 5A.
FIG. 5.
(A) Schematic representation of part of the CP Rit genome organization and the fusion proteins encoded by constructs pRit-A, pRit-B, pRit-C, and pRit-D. For initiation of translation, each construct starts with an additional methionine. The positions of primers Ol Rit4B1 (•), Ol S27a* (■), Ol Rit-ubi4 (★), Ol RitNS3R (⧫), and Ol Rit4AR (◣), used for cloning of the respective constructs, are indicated below the bar on the top. (B and C) Immunoblot analysis of CP Rit nonstructural proteins. After infection with vaccinia virus MVA-T7pol, BHK-21 cells were transfected with pRit-C (lanes 1), pRit-A (lanes 2), pRit-B (lanes 3), and pRit-D (lanes 4). Cells were lysed 16 h posttransfection or postinfection, and the samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (8% polyacrylamide) under reducing conditions, transferred to nitrocellulose, and incubated with anti-NS3 monoclonal antibody (B) or anti-P1 serum directed against NS4A and NS4B (C). +, MDBK cells infected with CP Rit. −, Negative control (BHK-21 cells infected with MVA-T7pol but not transfected). The sizes (in kilodaltons) of marker proteins are indicated on the left. The positions of NS3 (B) and NS4B-specific fusion proteins (C) are marked with arrows. Additional bands visible in panel C represent nonspecific reactions due to the background level of the rabbit antiserum. With respect to pRit-D, it has actually not been demonstrated that NS4A is cleaved off.
Furthermore, the construct pRit-D, which lacks the region encoding S27a*, was generated. As a first step toward generating pRit-D, a cDNA fragment encompassing part of NS4B* directly fused to the N-terminal part of ubiquitin* was obtained after RT-PCR with primer Ol RitNS3R and primer Ol S27a-del (including a SmaI site) and subsequently cloned into pCR2.1. The resulting plasmid was termed pCRRITdel. The remaining part of NS4B* was obtained after digestion of pCRRit-A with XbaI and SmaI and subsequently cloned into pCRRitdel (precut with XbaI and SmaI). From the resulting construct, a 0.45-kb NcoI-XhoI fragment was cloned into pRit-C (precut with NcoI and XhoI), resulting in plasmid pRit-D. Construct pRit-D encodes a fusion protein with the structure NS4B*-ubiquitin*-NS3-NS4A.
Immunoblotting.
Infected MDBK cells were lysed 48 h postinfection in loading buffer containing 6 M urea, 2% sodium dodecyl sulfate, 10% glycerol, and 5% β-mercaptoethanol. BHK-21 cells were lysed 16 h posttransfection. Samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis under reducing conditions (48) and transferred to a nitrocellulose filter (Schleicher & Schuell, Dassel, Germany). The filters were blocked with 5% nonfat dry milk–0.05% Tween in PBS for 16 h. After being washed with PBS–0.05% Tween, the filters were incubated with either MAb 8.12.7 (directed against NS3) or anti-P1 antiserum (directed against NS4A and NS4B) (35). After several washes, the filters were incubated with the substrates of the ECL kit (Amersham) as specified by the manufacturer. They were then exposed to Kodak BioMax MR films. The prestained molecular weight standard was obtained from Gibco-BRL.
Nucleotide sequence accession numbers.
Sequence data from this article have been deposited in the EMBL and GenBank data libraries and assigned accession no. AF058699 (partial sequence of CP Rit) and AF058700 (bovine mRNA encoding ubiquitin and S27a).
RESULTS
Characterization of the BVDV vaccine strain CP Rit by hybridization and RT-PCR.
For noncp and also some cp BVDV strains, the RNA genomes are about 12.3 kb long. In contrast, the genomes of several cp BVDV strains contain either large duplications or deletions leading to viral RNAs significantly longer or shorter than the ones of noncp BVDV (25, 34, 35, 52). As a first step toward molecular characterization of the BVDV vaccine strain CP Rit, a Northern blot analysis with total RNA from MDBK cells infected with CP Rit was performed. Hybridization of RNA from cells infected with BVDV Rit with a BVDV CP7-derived cDNA probe showed that the genomic RNA is about 15 kb (Fig. 1). Similar sizes have been reported for other cp BVDV strains, and analyses of these isolates led to the identification of duplications of the NS3 gene (34, 35, 44). It was therefore likely that the genome of CP Rit also contains a large duplication.
FIG. 1.
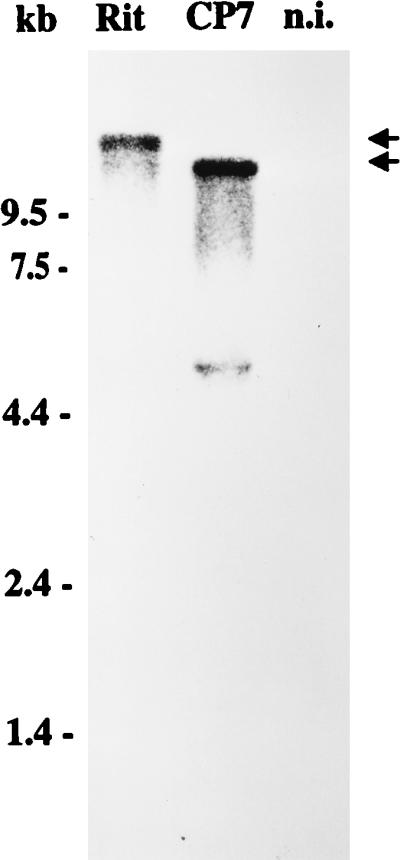
Northern blot analysis of total RNA from MDBK cells infected with BVDV strain CP Rit or CP7 and noninfected cells (n.i.). Before transfer and hybridization, RNA was separated on a 1.0% agarose gel under denaturing conditions. The blot was hybridized with a 2.5-kb NotI-NsiI fragment from the cDNA clone pA/BVDV (33). RNA ladder sizes (in kilobases) are indicated. Migration positions of the viral genomic RNAs are marked with arrows. The additional band visible below the viral RNA of CP7 represents a gel artifact resulting from large amounts of rRNA.
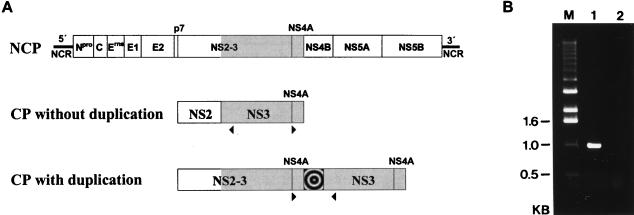
As the next step toward molecular characterization of CP Rit, we performed an RT-PCR analysis in which specific amplification of cDNA is possible only in the case of a duplicated NS3 gene (Fig. 2). This RT-PCR assay generated a fragment of about 950 bp that was subsequently cloned into pCR2.1. To determine the consensus sequence, the complementary strands of three independent clones were sequenced.
FIG. 2.
(A) Genome organization of a noncp pestivirus (NCP) and RT-PCR strategy for identification of a duplication of the NS3 gene. The positions and orientations of primers Ol NS3R and Ol BVDV7100 are indicated by arrowheads below the bars. RT-PCR with this primer pair allows a specific amplification only for a duplicated NS3 gene. The bull’s-eye icon indicates a putative insertion. (B) RT-PCR product obtained from RNA of BVDV CP Rit-infected cells (lane 1). RNA from cells infected with noncp BVDV strain 519-NCP (2a) served as negative control (lane 2). The cDNA fragment was separated on a 1.0% agarose gel and stained with ethidium bromide. M, size standard; KB, kilobases.
Genome organization of CP Rit.
To determine the genome organization of CP Rit, the obtained nucleotide sequence was first compared with the genomic sequence of BVDV SD-1, which represents the first completely sequenced noncp strain (15). With regard to numbering of nucleotides, the SD-1 sequence has been widely used for comparison of pestivirus genomes. The 5′ part of the determined Rit sequence corresponds to positions 7312 to 7787 of the SD-1 sequence. This part of the genome encodes the C-terminal 27 aa of NS4A and the N-terminal 132 aa of NS4B. Downstream of this part, a nonviral insertion comprising 300 nucleotides was found. The sequence located downstream of this insertion is colinear with the pestivirus NS3 gene, starting at position 5153 of the SD-1 sequence (Fig. 3). This position corresponds to the N terminus of NS3.
FIG. 3.
(A) Schematic representation of the genome organization of a noncp pestivirus (NCP) and CP Rit. For CP Rit, the genomic region encoding NS3, NS4A, and the N-terminal 132 aa of NS4B (NS4B*) is duplicated. In addition, two cellular insertions are located directly upstream of the NS3 gene. These cellular sequences encode part of ribosomal protein S27a (S27a*, shaded box) and an N-terminally truncated ubiquitin (ubi*, solid box), respectively. (B) Deduced amino acid sequence of part of the CP Rit sequence. The positions of NS4B*, S27a*, ubiquitin*, and NS3 are indicated. The two cellular insertions are underlined.
The genome organization of CP Rit is reminiscent of that described for other cp BVDV isolates with large duplications. For all these viruses, an insertion encoding either viral Npro or cellular (poly)ubiquitin was found directly upstream of a duplicated NS3 gene (36). All ubiquitin insertions described so far for cp BVDV strains encode at least one complete ubiquitin monomer consisting of 76 aa (32, 34, 44, 51). Interestingly, the 3′-terminal 219 nucleotides of the insertion identified within the genome of CP Rit encodes ubiquitin*, an N-terminally truncated ubiquitin lacking the first 3 amino acids. In contrast, the 5′-terminal 81 nucleotides of the CP Rit insertion do not show any homology to (poly)ubiquitin-specific sequences or cINS, the other cellular sequence identified in the genome of cp pestiviruses (4, 30). It was therefore interesting to investigate the nature and origin of this second insertion found within CP Rit.
Identification of S27a coding sequences.
A databank search revealed that the 5′-terminal 81 nucleotides of the nonviral sequences within the genome of CP Rit show similarities to cellular sequences encoding part of ribosomal protein S27a. The nucleotide sequence identities between the respective genomic region of CP Rit and cellular S27a coding sequences from humans, rats, and guinea pigs were about 90%. To our knowledge, this is the first identification of cellular S27a coding sequences within the genome of any virus including cp pestivirus strains. An alignment of the respective deduced amino acid sequences showed that the inserted sequence of CP Rit encodes aa 34 to 60 of cellular S27a; in the following, this part of S27a is designated S27a*. Cellular S27a is a highly conserved, very basic protein consisting of 80 aa. S27a has been demonstrated to represent the carboxy-terminal part of a ubiquitin fusion protein which is processed to ubiquitin and S27a (18, 45). In contrast, the S27a-derived sequences within CP Rit are located upstream of the ubiquitin coding sequence (Fig. 3). With regard to this unusual organization of S27a* and ubiquitin* coding sequences within the genome of CP Rit, it was important to identify the putative cellular recombination partner(s), in particular to look for a cellular mRNA with a similar arrangement of S27a and ubiquitin coding sequences.
Search for the putative cellular recombination partner.
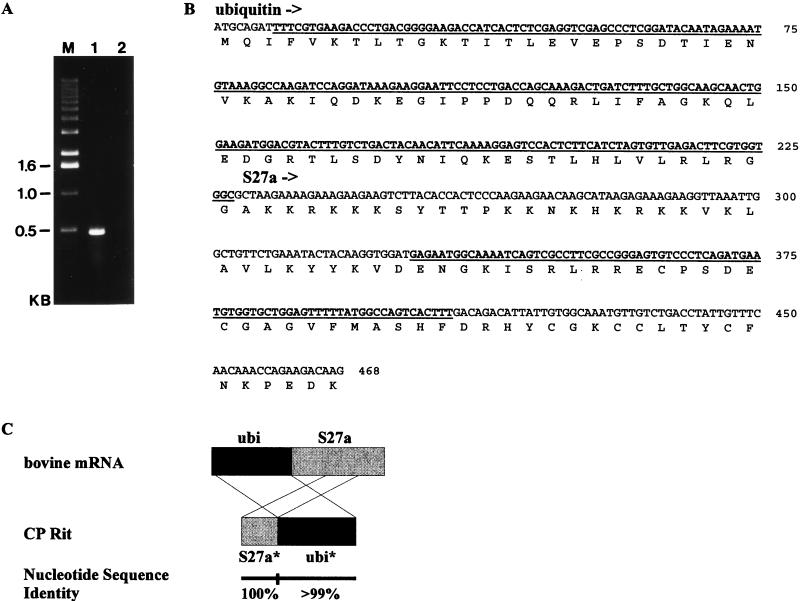
BVDV CP Rit has been isolated from cattle, and bovine cells were used for propagation of this virus strain (27). It was therefore likely that the two cellular insertions of CP Rit originated from bovine cells. To identify the putative cellular recombination partner of the two CP Rit insertions, oligonucleotides Ol UeR and Ol Ue were deduced from published sequences encoding ubiquitin-S27a from other species. By using total RNA of MDBK cells, RT-PCR with these primers resulted in amplification of a cDNA fragment of about 500 bp (Fig. 4A). This fragment was cloned into a PCR cloning vector and subjected to sequence analysis. The obtained nucleotide sequence of the bovine mRNA encodes the complete ubiquitin-S27a fusion protein (Fig. 4B). A comparative analysis revealed that both the S27a* and the ubiquitin* coding sequences identified within the viral genome of CP Rit are more than 99% identical to the respective sequences from the bovine mRNA (Fig. 4C).
FIG. 4.
Identification of the bovine mRNA encoding ubiquitin and S27a. (A) RT-PCR product obtained from RNA of noninfected MDBK cells with primers Ol UeR and Ol Ue (lane 1). The cDNA fragment was separated on a 1.0% agarose gel and stained with ethidium bromide. Negative control, double-distilled H2O (lane 2); M, size standard; KB, kilobases. (B) Nucleotide and deduced amino acid sequence of the coding region of the bovine mRNA. The positions of ubiquitin and S27a are indicated. Regions which correspond to the two cellular insertions found within the genome of CP Rit are underlined. (C) Comparison of the structure of the bovine mRNA sequence and the respective part of the CP Rit genome encompassing the two cellular insertions, S27a* and ubi*, respectively. Nucleotide sequence identities between the bovine mRNA and the two cellular insertions of CP Rit are indicated.
The two cellular insertions of CP Rit are arranged in the order 5′-S27a*-ubiquitin*-3′, while the identified bovine mRNA encodes a fusion protein with the structure 5′-ubiquitin-S27a-3′ (Fig. 4C). With regard to this remarkable difference, it was tempting to speculate about the existence of a bovine mRNA with an arrangement of S27a* and ubiquitin* coding sequence as was found for CP Rit. RT-PCR with several appropriate primers, involving total RNA of uninfected MDBK cells, did not, however, result in identification of cellular sequences encoding a polypeptide with such a primary structure (data not shown). Taking into account the high nucleotide sequence identity between the two cellular insertions of CP Rit and the identified bovine mRNA sequence encoding the ubiquitin-S27a fusion protein, it appears highly likely that both cellular insertions of CP Rit were derived from this mRNA.
S27a*-ubiquitin* as processing signal.
The molecular analysis of several cp BVDV strains led to the identification of various genomic rearrangements (36). Interestingly, all these mutations are located in the genomic region encoding NS2-3 and lead to expression of NS3, the marker protein of cp BVDV. This nonstructural protein is not present in cells infected with noncp BVDV strains. As expected, infection of cells with CP Rit allowed the detection of NS3 in addition to NS2-3. NS3 of CP Rit has the same apparent molecular weight as NS3 found in cells after infection with other cp BVDV strains (data not shown).
In a previous study, it was suggested that at least one complete ubiquitin monomer is required for processing of ubiquitin-NS3 fusion proteins to yield NS3; the lack of 8 aa at the N terminus of the ubiquitin monomer abolished cleavage (51). It was not known whether the ubiquitin of CP Rit which lacks the N-terminal 3 aa was capable of mediating the respective processing event. To investigate the assumed role of the cellular insertions identified within the genome of CP Rit for generation of NS3, the genomic region encoding the N-terminal part of NS4B (NS4B* with aa 1 to 132), the two cellular insertions, and NS3 was transiently expressed in the MVA-T7pol virus system. Expression of NS3-specific proteins was monitored by immunoblotting with a monoclonal antibody (MAb) directed against NS3. As a first step, the construct pRit-A, which comprises the genomic region encoding NS4B*-S27a*-ubiquitin*-NS3, was generated. Expression of pRit-A led to detection of an 80-kDa protein that comigrated with NS3 from CP Rit-infected MDBK cells (Fig. 5A and B, lane 2). A protein with the same apparent molecular mass was also detected after expression of pRit-B, which encodes S27a*-ubiquitin*-NS3. In contrast, after expression of construct pRit-C encoding ubiquitin*-NS3, the anti-NS3 MAb reacted with a protein with an apparent molecular mass of 87 kDa. The predicted molecular mass of ubiquitin* is 7 kDa. Accordingly, the 87-kDa protein detected after expression of pRit-C most probably represents a ubiquitin*-NS3 fusion protein. Taken together, our results demonstrate that ubiquitin* lacking the N-terminal 3 aa is not sufficient to serve as processing signal to yield NS3 whereas addition of S27a* to the N terminus of ubiquitin* results in processing at the C terminus of ubiquitin*.
To identify additional cleavage product(s), immunoblotting with an antiserum directed against NS4B (35) was performed. After expression of pRit-A, this serum recognized a protein with an apparent molecular mass of about 24 kDa (Fig. 5C, lane 2). This size fits with the predicted molecular mass of a putative NS4B*-S27a*-ubiquitin* fusion protein. For pRit-A, a protein with the same apparent molecular mass was also detected by using an antiserum directed against ubiquitin (data not shown). These results show that the two cellular insertions are apparently expressed as part of a fusion protein with the structure NS4B*-S27a*-ubiquitin*.
It was interesting to investigate whether replacement of S27a* by another sequence fused to the N terminus of ubiquitin* would allow processing after Gly76 to yield NS3. An additional construct, pRit-D, which encodes a fusion protein with the structure NS4B*-ubiquitin*-NS3, was generated. Expression of pRit-D and subsequent immunoblot analysis with the anti-NS3 MAb resulted in identification of a protein with an apparent molecular mass of about 105 kDa (Fig. 5A and B, lane 4). Detection of a protein of the same size by the antiserum directed against NS4B (Fig. 5C, lane 4) demonstrates that the fusion protein with the structure NS4B*-ubiquitin*-NS3 is not further processed. To generate NS3, S27a* can apparently not be replaced by any sequence directly upstream of ubiquitin*.
DISCUSSION
Molecular characterization of pestiviruses led to the detection of different alterations present only in the genomes of cp viruses; these include insertions of cellular sequences with or without duplications of viral sequences, as well as deletions, duplications, and rearrangements of viral sequences (for a comprehensive review, see reference 36). Two types of cellular insertions which encode either ubiquitin (32, 34, 44, 51) or part of a cellular protein of unknown function, termed cINS (4, 30), have been identified. For the BVDV strain CP Rit described here, a duplication of the genomic region encoding NS3, NS4A, and part of NS4B and two cellular insertions were found; the latter encode part of ribosomal protein S27a and a ubiquitin fragment (Fig. 3). To our knowledge, this is the first report on the presence of S27a coding sequences in a viral genome. Ribosomal protein S27a is a highly conserved, very basic protein consisting of 80 amino acids. It has been reported that S27a is expressed as the C-terminal part of a ubiquitin fusion protein, which is cleaved to ubiquitin and S27a. Cellular S27a is incorporated into nascent ribosomes and is required for efficient ribosome biogenesis (18, 45).
Further analysis of the genome structure of CP Rit revealed three major differences from other cp BVDV strains with ubiquitin coding sequences: (i) the ubiquitin monomer encoded by CP Rit is N-terminally truncated; (ii) a different mRNA served as the source of the ubiquitin coding sequences; and (iii) ribosomal S27a coding sequences are present. All cp pestiviruses with ubiquitin insertions described so far encode at least one complete ubiquitin monomer, and it was therefore surprising that the ubiquitin-specific insertion identified within the genome of CP Rit encodes a truncated ubiquitin lacking the N-terminal 3 aa (14, 32, 34, 44, 51). Interestingly, the ubiquitin insertions of two cp BVDV strains carry mutations. cp BVDV Osloss carries two point mutations, both of which lead to amino acid changes (T55→S, G76→S), while BVDV Ill-C carries a duplication of codons 48 to 51 (32, 44). It remains to be investigated whether such mutations lead to functionally significant effects. Comparison of ubiquitin coding sequences of several cp BVDV strains including Osloss, CP1, CP14, 190, Ill-C, and TGAC with bovine polyubiquitin coding sequences strongly suggests that a bovine mRNA encoding polyubiquitin was the source of the cellular insertions for all these virus isolates (Table 1). In contrast, the ubiquitin insertion of CP Rit is less than 80% identical to either of the two available bovine polyubiquitin gene sequences. This difference in codon usage was unexpected, since the respective deduced amino acid sequences differ at only one residue; we observed a replacement of valine17 by alanine in the ubiquitin* encoded by CP Rit. For eukaryotic cells, two types of ubiquitin genes have been described. These encode either polyubiquitin, consisting of multiple, exact head-to-tail repeats of ubiquitin, or a single ubiquitin monomer fused to a ribosomal protein (18, 19, 45). To elucidate the nature and origin of the CP Rit insertions, the bovine mRNA encoding the ubiquitin-S27a hybrid protein was identified and the coding region of this mRNA was cloned and sequenced. Comparative sequence analysis revealed that the identity between this bovine mRNA sequence and the ubiquitin coding sequence of CP Rit is greater than 99% (Fig. 4; Table 1). It can be concluded that the ubiquitin coding insertion of CP Rit was derived not from a polyubiquitin gene but most probably from a bovine mRNA encoding ubiquitin together with ribosomal protein S27a. Comparison of the second cellular insertion of CP Rit encoding part of S27a with this bovine mRNA sequence revealed that the viral and cellular sequences are again identical. This strongly suggests that both cellular insertions of CP Rit were derived from the same bovine mRNA. It should be emphasized that the S27a* coding sequences within the genome of CP Rit are located directly upstream of the ubiquitin* coding insertion whereas the respective bovine mRNA encodes a fusion protein with the structure NH2-ubiquitin-S27a-COOH.
TABLE 1.
Nucleotide sequence identity between the ubiquitin coding insertions of different BVDV strains and cellular ubiquitin coding sequences
| BVDV strain | Nucleotide sequence identity (%)
|
||
|---|---|---|---|
| Bovine polyubiquitina
|
Bovine ubiquitin-S27a | ||
| rpub1 | rpub2 | ||
| CP Rit | 78.6 | 79.5 | 99.5 |
| Osloss | 89.0 | 98.2 | 78.5 |
| CP1 | 89.9 | 100 | 80.2 |
| CP14 | 98.7 | 90.8 | 79.3 |
| 190 | 88.7 | 99.4 | 79.8 |
| TGAC | 88.6 | 99.3 | 79.8 |
| Ill-C | 98.3 | 91.3 | 79.8 |
The two polyubiquitin sequences (rpub1 and rpub2) have been reported previously (34).
For BVDV, cytopathogenicity is correlated with expression of NS3, which is not found after infection of cells with noncp BVDV (11, 13, 17, 21, 34, 35, 42, 43, 52, 53). In the case of ubiquitin insertions, it has been reported that ubiquitin functions as a processing signal leading to an additional cleavage of the viral polyprotein by cellular ubiquitin C-terminal hydrolases; at least one entire ubiquitin monomer is required for processing at the C terminus of ubiquitin (51). Furthermore, it has been demonstrated that replacement of the N-terminal 2 aa MQ by the tripeptide MEL did not affect the cleavage (51). With regard to CP Rit, our analysis of polyprotein processing revealed that engineered fusion proteins with the structure ubiquitin*-NS3 or NS4B*-ubiquitin*-NS3 were not cleaved whereas release of NS3 could be observed after expression of (NS4B*-)S27a*-ubiquitin*-NS3 polypeptides. These results show that (NS4B*-)S27a*-ubiquitin* serves as processing signal to yield NS3 whereas the N-terminally truncated ubiquitin alone is not sufficient to allow the cleavage. The mutations responsible for expression of NS3 most probably also represent the genetic basis for cytopathogenicity of BVDV CP Rit.
For all cp pestiviruses with ubiquitin coding sequences, including CP Rit, the 3′ crossing-over site is conserved; this results in fusion of a given N terminus of NS3 to the C terminus of ubiquitin. Specific sequences which might serve as signals for recombination have not been identified within the genomes of pestiviruses including CP Rit. The observed conservation of the 3′ recombination site is probably the result of a functional selection allowing the expression of NS3 with a defined N-terminus. In contrast, the 5′ recombination sites between the viral and cellular sequences either vary between nucleotides 7456 (located in the NS4B gene) and 8788 (located in the NS5A gene) or are located in the NS2 gene. The respective fusion of viral and cellular sequences results in the expression of fusion proteins which have been described for several cp pestiviruses (4, 34, 35). Our data demonstrate that both cellular insertions of CP Rit are expressed as parts of one fusion protein with the structure NS4B*-S27a*-ubiquitin*.
It is assumed that recombination of pestiviruses occurs at the RNA level. The most widely accepted model of RNA recombination is the replicase-driven template-switching model, although it is not possible to favor a particular mechanism of recombination on the basis of sequences of the recombination end products (39). RNA recombination may occur during the synthesis of either positive or negative RNA strands (1, 22, 24). The frequency of recombination is presumed to depend heavily on the availability of acceptor templates. Since the concentration of positive-strand RNAs is much higher than that of negative-strand RNAs, it has been suggested that recombination during negative-strand synthesis occurs more frequently. The preferred occurrence of recombination during negative-strand RNA synthesis is also supported by the assumption that the negative-strand RNAs exist predominantly as part of replicative intermediates in a double-stranded form and that in this form they are not available as a template (1). With respect to cp pestiviruses, all cellular sequences including the two insertions within the genome of CP Rit are present in coding orientation. Accordingly, recombination must have occurred during negative-strand synthesis, since the corresponding cellular mRNAs are present only as positive strands. The majority of RNA virus recombinants can be explained by a single template switch, while pestiviruses with cellular sequences are considered to be the result of at least two template switches (36). CP Rit represents the first pestivirus with two cellular insertions; for generation of its genome, at least three template switches are required. Our finding that both insertions were derived from the same bovine mRNA might be significant for the interpretation of the respective recombination. For integration of both cellular insertions during one step, an intramolecular template switch on the bovine mRNA appears to be more likely than an intermolecular one. Alternatively, the genome of CP Rit might have evolved by two separate recombination events. Accordingly, an intermediate genome with integration of either S27a or ubiquitin coding sequences was first generated. In a second step, recombination of this hypothetical intermediate with the same bovine mRNA generated the CP Rit genome analyzed here. An interaction between the insertion integrated within the intermediate genome and the bovine mRNA may have promoted the second recombination. However, on the basis of the determined nucleotide sequences, it is not possible to favor one of these models.
With the exception of transduction of cellular proto-oncogene sequences in the genomes of retroviruses, recombinations between host cellular RNAs and viral genomic RNAs represent rare events and have been described for only a few other RNA viruses including influenza virus (23), poliovirus (10, 29), and Sindbis virus (37, 54). For influenza virus and poliovirus, 28S rRNA-derived sequences were found, while in the case of several defective interfering particles of Sindbis virus, cellular tRNA as well as 26S RNA sequences were detected. As a unique feature of pestiviruses, all insertions of cellular sequences identified so far in their genomes were derived from protein coding sequences. Remarkably, the insertions are directly or indirectly responsible for an additional processing of the pestiviral polyprotein and thereby for expression of NS3; occurrence of the latter is strictly correlated with the cp phenotype of BVDV. Future studies on cp pestiviruses are expected to result in identification of additional genomic alterations including the detection of novel cellular insertions. The respective analyses will help to understand the different mechanisms for generation of NS3, in particular with respect to the introduction of processing signals into a viral genome.
ACKNOWLEDGMENTS
We thank Norbert Tautz for critical reading of the manuscript.
This study was supported by Intervet International BV (project 75/73,1808.720) and SFB 535 “Invasionsmechanismen und Replikationsstrategien von Krankheitserregern” from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Agol V I. Recombination and other genomic rearrangements in picornaviruses. Semin Virol. 1997;8:77–84. [Google Scholar]
- 2.Baker J C. Bovine viral diarrhea virus: a review. J Am Vet Med Assoc. 1987;190:1449–1458. [PubMed] [Google Scholar]
- 2a.Becher, P. Unpublished data.
- 3.Becher P, König M, Paton D, Thiel H-J. Further characterization of border disease virus isolates: evidence for the presence of more than three species within the genus pestivirus. Virology. 1995;209:200–206. doi: 10.1006/viro.1995.1243. [DOI] [PubMed] [Google Scholar]
- 4.Becher P, Meyers G, Shannon A D, Thiel H-J. Cytopathogenicity of border disease virus is correlated with integration of cellular sequences into the viral genome. J Virol. 1996;70:2992–2998. doi: 10.1128/jvi.70.5.2992-2998.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becher P, Orlich M, Shannon A D, Horner G, König M, Thiel H-J. Phylogenetic analysis of pestiviruses from domestic and wild ruminants. J Gen Virol. 1997;78:1357–1366. doi: 10.1099/0022-1317-78-6-1357. [DOI] [PubMed] [Google Scholar]
- 6.Becher P, Orlich M, Thiel H-J. Complete genomic sequence of border disease virus, a pestivirus from sheep. J Virol. 1998;72:5165–5173. doi: 10.1128/jvi.72.6.5165-5173.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becher P, Shannon A D, Tautz N, Thiel H-J. Molecular characterization of border disease virus, a pestivirus from sheep. Virology. 1994;198:542–551. doi: 10.1006/viro.1994.1065. [DOI] [PubMed] [Google Scholar]
- 8.Bolin S R, McClurkin A W, Cutlip R C, Coria M F. Severe clinical disease induced in cattle persistently infected with noncytopathogenic bovine viral diarrhea virus by superinfection with cytopathogenic bovine viral diarrhea virus. Am J Vet Res. 1985;46:573–576. [PubMed] [Google Scholar]
- 9.Brownlie J, Clarke M C, Howard C J. Experimental production of fatal mucosal disease in cattle. Vet Rec. 1984;114:535–536. doi: 10.1136/vr.114.22.535. [DOI] [PubMed] [Google Scholar]
- 10.Charini W A, Todd S, Gutman G A, Semler B L. Transduction of a human RNA sequence by poliovirus. J Virol. 1994;68:6547–6552. doi: 10.1128/jvi.68.10.6547-6552.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collett M S, Larson R, Belzer S, Retzel E. Proteins encoded by bovine viral diarrhea virus: the genome organization of a pestivirus. Virology. 1988;165:200–208. doi: 10.1016/0042-6822(88)90673-3. [DOI] [PubMed] [Google Scholar]
- 12.Collett M S, Larson R, Gold C, Strick D, Anderson D K, Purchio A F. Molecular cloning and nucleotide sequence of the pestivirus bovine viral diarrhea virus. Virology. 1988;165:191–199. doi: 10.1016/0042-6822(88)90672-1. [DOI] [PubMed] [Google Scholar]
- 13.Corapi W V, Donis R O, Dubovi E J. Monoclonal antibody analyses of cytopathic and noncytopathic viruses from fatal bovine viral diarrhea virus infections. J Virol. 1988;62:2823–2827. doi: 10.1128/jvi.62.8.2823-2827.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Moerlooze L, Lecomte C, Brown-Shimmer S, Schmetz D, Guiot C, Vandenbergh D, Allaer D, Rossius M, Chappuis G, Dina D, Renard A, Martial J A. Nucleotide sequence of the bovine viral diarrhoea virus Osloss strain: comparison with related viruses and identification of specific DNA probes in the 5′ untranslated region. J Gen Virol. 1993;74:1433–1438. doi: 10.1099/0022-1317-74-7-1433. [DOI] [PubMed] [Google Scholar]
- 15.Deng R, Brock K V. Molecular cloning and nucleotide sequence of a pestivirus genome, noncytopathogenic bovine viral diarrhea virus strain SD-1. Virology. 1992;191:867–879. doi: 10.1016/0042-6822(92)90262-n. [DOI] [PubMed] [Google Scholar]
- 16.Devereux J, Haeberli P, Smithies O A. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donis R O, Dubovi E J. Characterization of bovine diarrhoea-mucosal disease virus-specific proteins in bovine cells. J Gen Virol. 1987;68:1597–1605. doi: 10.1099/0022-1317-68-6-1597. [DOI] [PubMed] [Google Scholar]
- 18.Finley D, Bartel B, Varshavsky A. The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature. 1989;338:394–401. doi: 10.1038/338394a0. [DOI] [PubMed] [Google Scholar]
- 19.Finley D, Özkaynak E, Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation and other stresses. Cell. 1987;48:1035–1046. doi: 10.1016/0092-8674(87)90711-2. [DOI] [PubMed] [Google Scholar]
- 20.Gillespie J H, Baker J A, McEntee K. A cytopathogenic strain of virus diarrhea virus. Cornell Vet. 1960;50:73–79. [PubMed] [Google Scholar]
- 21.Greiser-Wilke I, Dittmar K E, Liess B, Moennig V. Heterogeneous expression of the non-structural protein p80/p125 in cells infected with different pestiviruses. J Gen Virol. 1992;73:47–52. doi: 10.1099/0022-1317-73-1-47. [DOI] [PubMed] [Google Scholar]
- 22.Jarvis T C, Kirkegaard K. Poliovirus RNA recombination: mechanistic studies in the absence of selection. EMBO J. 1992;11:3135–3145. doi: 10.1002/j.1460-2075.1992.tb05386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khatchikian D, Orlich M, Rott R. Increased viral pathogenicity after insertion of a 28S ribosomal RNA sequence into the haemagglutinin gene of an influenza virus. Nature. 1989;340:156–157. doi: 10.1038/340156a0. [DOI] [PubMed] [Google Scholar]
- 24.Kirkegaard K, Baltimore D. The mechanism of RNA recombination in poliovirus. Cell. 1986;47:433–443. doi: 10.1016/0092-8674(86)90600-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kupfermann H, Thiel H-J, Dubovi E J, Meyers G. Bovine viral diarrhea virus: characterization of a cytopathogenic defective interfering particle with two internal deletions. J Virol. 1996;70:8175–8181. doi: 10.1128/jvi.70.11.8175-8181.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee K M, Gillespie J H. Propagation of virus diarrhea virus of cattle in tissue culture. Am J Vet Res. 1957;18:953. [PubMed] [Google Scholar]
- 27.Lobmann M, Charlier P, Florent G, Zygraich N. Clinical evaluation of a temperature-sensitive bovine viral diarrhea vaccine strain. Am J Vet Res. 1984;45:2498–2503. [PubMed] [Google Scholar]
- 28.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 29.McClure M, Perrault J. Poliovirus genome RNA hybridizes specifically to higher eukaryotic rRNAs. Nucleic Acids Res. 1985;13:6797–6816. doi: 10.1093/nar/13.19.6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyers G, Rümenapf T, Thiel H-J. Insertion of ubiquitin-coding sequence identified in the RNA genome of a Togavirus. In: Brinton M A, Heinz F X, editors. New aspects of positive-strand RNA viruses. Washington, D.C: American Society for Microbiology; 1990. pp. 25–29. [Google Scholar]
- 31.Meyers G, Rümenapf T, Thiel H-J. Molecular cloning and nucleotide sequence of the genome of hog cholera virus. Virology. 1989;171:555–567. doi: 10.1016/0042-6822(89)90625-9. [DOI] [PubMed] [Google Scholar]
- 32.Meyers G, Rümenapf T, Thiel H-J. Ubiquitin in a togavirus. Nature. 1989;341:491. doi: 10.1038/341491a0. [DOI] [PubMed] [Google Scholar]
- 33.Meyers G, Tautz N, Becher P, Thiel H-J, Kümmerer B. Recovery of cytopathogenic and noncytopathogenic bovine viral diarrhea viruses from cDNA constructs. J Virol. 1996;70:8606–8613. doi: 10.1128/jvi.70.12.8606-8613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyers G, Tautz N, Dubovi E J, Thiel H-J. Viral cytopathogenicity correlated with integration of ubiquitin-coding sequences. Virology. 1991;180:602–616. doi: 10.1016/0042-6822(91)90074-L. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyers G, Tautz N, Stark R, Brownlie J, Dubovi E J, Collett M S, Thiel H-J. Rearrangement of viral sequences in cytopathogenic pestiviruses. Virology. 1992;191:368–386. doi: 10.1016/0042-6822(92)90199-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meyers G, Thiel H-J. Molecular characterization of pestiviruses. Adv Virus Res. 1996;47:53–118. doi: 10.1016/s0065-3527(08)60734-4. [DOI] [PubMed] [Google Scholar]
- 37.Monroe S S, Schlesinger S. RNAs from two independently isolated defective interfering particles of Sindbis virus contain a cellular tRNA sequence at their 5′ ends. Proc Natl Acad Sci USA. 1983;80:3279–3283. doi: 10.1073/pnas.80.11.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moormann R J M, Warmerdam P A M, Van der Meer B, Schaaper W M M, Wensvoort G, Hulst M M. Molecular cloning and nucleotide sequence of hog cholera virus strain brescia and mapping of the genomic region encoding envelope glycoprotein E1. Virology. 1990;177:184–198. doi: 10.1016/0042-6822(90)90472-4. [DOI] [PubMed] [Google Scholar]
- 39.Nagy P D, Simon A E. New insights into the mechanisms of RNA recombination. Virology. 1997;235:1–9. doi: 10.1006/viro.1997.8681. [DOI] [PubMed] [Google Scholar]
- 40.Paton D J. Pestivirus diversity. J Comp Pathol. 1995;112:215–236. doi: 10.1016/s0021-9975(05)80076-3. [DOI] [PubMed] [Google Scholar]
- 41.Pellerin C, Van Den Hurk J, Lecomte J, Tijssen P. Identification of a new group of bovine viral diarrhea virus strains associated with severe outbreaks and high mortalities. Virology. 1994;203:260–268. doi: 10.1006/viro.1994.1483. [DOI] [PubMed] [Google Scholar]
- 42.Pocock D H, Howard C J, Clarke M C, Brownlie J. Variation in the intracellular polypeptide profiles from different isolates of bovine viral diarrhea virus. Arch Virol. 1987;94:43–53. doi: 10.1007/BF01313724. [DOI] [PubMed] [Google Scholar]
- 43.Purchio A F, Larson R, Collett M S. Characterization of bovine viral diarrhea viral proteins. J Virol. 1984;50:666–669. doi: 10.1128/jvi.50.2.666-669.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qi F, Ridpath J F, Lewis T, Bolin S R, Berry E S. Analysis of the bovine viral diarrhea virus genome for possible cellular insertions. Virology. 1992;189:285–292. doi: 10.1016/0042-6822(92)90704-s. [DOI] [PubMed] [Google Scholar]
- 45.Redmann K L, Rechsteiner M. Identification of the long ubiquitin extension as ribosomal protein S27a. Nature. 1989;338:438–440. doi: 10.1038/338438a0. [DOI] [PubMed] [Google Scholar]
- 46.Ridpath F F, Bolin S R, Dubovi E J. Segregation of bovine viral diarrhea virus into genotypes. Virology. 1994;205:66–74. doi: 10.1006/viro.1994.1620. [DOI] [PubMed] [Google Scholar]
- 47.Ridpath J F, Bolin S R. The genomic sequence of a virulent bovine viral diarrhea virus (BVDV) from the type 2 genotype: detection of a large genomic insertion in a noncytopathic BVDV. Virology. 1995;212:39–46. doi: 10.1006/viro.1995.1451. [DOI] [PubMed] [Google Scholar]
- 48.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 49.Sutter G, Ohlmann M, Erfle V. Non-replicating vaccinia vector efficiently expresses bacteriophage T7 RNA polymerase. FEBS Lett. 1995;371:9–12. doi: 10.1016/0014-5793(95)00843-x. [DOI] [PubMed] [Google Scholar]
- 50.Tautz N, Meyers G, Stark R, Dubovi E J, Thiel H-J. Cytopathogenicity of a pestivirus correlated with a 27-nucleotide insertion. J Virol. 1996;70:7851–7858. doi: 10.1128/jvi.70.11.7851-7858.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tautz N, Meyers G, Thiel H-J. Processing of poly-ubiquitin in the polyprotein of an RNA virus. Virology. 1993;197:74–85. doi: 10.1006/viro.1993.1568. [DOI] [PubMed] [Google Scholar]
- 52.Tautz N, Thiel H-J, Dubovi E J, Meyers G. Pathogenesis of mucosal disease: a cytopathogenic pestivirus generated by internal deletion. J Virol. 1994;68:3289–3297. doi: 10.1128/jvi.68.5.3289-3297.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thiel H-J, Plagemann P G W, Moennig V. Pestiviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1059–1073. [Google Scholar]
- 54.Tsiang M, Monroe S S, Schlesinger S. Studies of defective interfering RNAs of Sindbis virus with and without tRNA-ASP sequences at their 5′ termini. J Virol. 1985;54:38–44. doi: 10.1128/jvi.54.1.38-44.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wengler G, Bradley D W, Collett M S, Heinz F X, Schlesinger R W, Strauss J H. Flaviviridae. In: Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D, editors. Virus taxonomy. Sixth report of the International Committee on Taxonomy of Viruses. Vienna, Austria: Springer Verlag; 1995. pp. 415–427. [Google Scholar]