Abstract
Chordoma is difficult to eradicate due to high local recurrence rates. The immune microenvironment is closely associated with tumor prognosis; however, its role in skull base chordoma is unknown. The expression of Galectin-9 (Gal9) and tumor-infiltrating lymphocyte (TIL) markers was assessed by immunohistochemistry. Kaplan–Meier and multivariate Cox analyses were used to assessing local recurrence-free survival (LRFS) and overall survival (OS) of patients. MiR-455-5p was identified as a regulator of Gal9 expression. Immunopositivity for Gal9 was associated with tumor invasion (p = 0.019), Karnofsky performance status (KPS) score (p = 0.017), and total TIL count (p < 0.001); downregulation of miR-455-5p was correlated with tumor invasion (p = 0.017) and poor prognosis; and the T-cell immunoglobulin and mucin-domain 3 (TIM3)+ TIL count was associated with chordoma invasion (p = 0.010) and KPS score (p = 0.037). Furthermore, multivariate analysis indicated that only TIM3+ TIL density was an independent prognostic factor for LRFS (p = 0.010) and OS (p = 0.016). These results can be used to predict clinical outcome and provide a basis for immune therapy in skull base chordoma patients.
Electronic supplementary material
The online version of this article (10.1007/s00262-019-02349-1) contains supplementary material, which is available to authorized users.
Keywords: Skull base chordoma, TIM3, Galectin–9, CD8, FOXp3, miR-455-5p
Introduction
Chordoma is a rare malignant neoplasm with an incidence of 0.08 per 100,000 persons that arises from embryonic notochord remnants, with as many as 32% of tumors located in the clivus region [1–3]. Chordomas can invade surrounding neurovascular tissues, making total resection difficult to achieve, and there are no effective radiotherapeutic or chemotherapeutic options for treatment, which poses a significant challenge for clinical treatment and patient prognosis [4–8]. Several studies have shown that immunosuppression affects the proliferation and invasion of tumors in gastric and breast cancer and glioma. Novel immune checkpoint proteins such as PD-1, PD-L1, and cytolytic T lymphocyte-associated Ag-4 (CTLA-4) modulate the apoptosis and immunosuppression of tumor-infiltrating lymphocytes (TILs) to enable tumors to escape immune surveillance [9–13]. However, little is known about the efficacy of immunotherapy for chordomas [14, 15]. The identification of chordoma-specific immune checkpoint proteins could improve therapeutic efficacy and patient prognosis.
As the major subtypes of TILs, the cluster of differentiation 8 (CD8)+ TILs and FOXp3+ TILs have been shown to correlate with the clinical prognosis of different cancers [16, 17]. Most CD8+ TILs can recognize particular tumor-associated antigens presented on MHC class I molecules at the cancer cell surface and possess the ability to destroy cancer cells directly [18]. FOXp3+ TILs have been known to disrupt anti-tumor activity by inhibiting activation of various immune cells and are implicated in the immune escape of cancer cells [17]. T-cell immunoglobulin and mucin-domain 3 (TIM3)+ TIL density was associated with the accumulation of TILs in the microenvironment. TIM3, a transmembrane protein that contains an immunoglobulin and a mucin-like domain, was originally identified as a specific molecule expressed on Th1 cells and cytotoxic T cells [19–21]. Galectin-9 (Gal9), a member of the S-type lectins, is one of the previously identified TIM3 ligands [22–24]. When binding to TIM3 on T cells, it can generate an inhibitory signal to induce the apoptosis of T cells [25]. In addition, other studies have shown that the expression of TIM3 is associated with dysfunctional or exhausted lymphocytes, which depend on the ability of TIM3 to modulate TCR signaling [26]. Previous clinical trials have already shown immunosurveillance by TIM3/Gal9 pathway expression in gliomas, osteosarcomas and gastrointestinal stromal tumors [27–29].
MicroRNAs (miRNAs) are a class of small, non-coding, single-stranded RNAs that are known to regulate the immune response [30, 31]. Several studies have shown that miR-21 and miR-124 are downregulated in the tumor microenvironment as compared to normal tissue and target relevant immune signaling pathways to modulate the immunosuppressive effects of tumors [32, 33]. A recent study showed that high miR-22 expression in liver cancer disrupted the interaction between Tim-3 and Gal9, prevented apoptosis of lymphocytes, partially restored effector T-cell function, and enhanced the tumor immune response, thereby reducing tumor cell proliferation and immune escape [34]. In the present study, we investigated the relationship between the expression of Gal9, miR-455-5p, and TIM3+ TIL densities and clinicopathological features and prognosis of skull base chordoma patients. Gal9 overexpression in chordoma was associated with TIL infiltration, whereas low miR-455-5p expression was related to increased tumor invasion. In addition, patients with both negative Gal9 and high miR-455-5p expression had longer survival times. Finally, TIM3+ TILs were independent predictors of LRFS and OS.
Methods and materials
Patients and specimens
We retrospectively examined 93 chordoma tissue specimens from skull base chordoma patients who underwent surgery at the First Affiliated Hospital of China Medical University between January 2010 and January 2013. Clinicopathological features, including age, sex, tumor size, invasion condition, preoperative recurrence, the extent of resection, Karnofsky performance status (KPS) score and histopathology were retrospectively reviewed from patients’ medical records (Table 1). The exclusion criteria for this study included patients who received any type of tumor-specific therapy and patients who suffered from other diseases that affect genetic changes or prognosis.
Table 1.
The correlations between miR-455-5p, galectin-9, TILs and clinicopathological features of skull base chordoma patients
| Clinicopathological factors | TIM3+ TILs | Tumor galectin-9 expression | Tumor miR-455-5p expression | CD8+ TILs | FOXp3+ TILs | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of patients (n = 93) | Mean ± SD | p values | Negative (n = 23) | Positive (n = 70) | p values | No. of patients (n = 93) | Mean ± SD | p values | No. of patients (n = 93) | Mean ± SD | p values | No. of patients (n = 93) | Mean ± SD | p values | |
| Age (years) | |||||||||||||||
| ≤ 50 | 45 | 231.5 ± 51.6 | 0.283 | 9 | 36 | 0.433 | 45 | 4.38 ± 2.39 | 0.679 | 45 | 367.3 ± 44.2 | 0.908 | 45 | 168.5 ± 46.2 | 0.751 |
| > 50 | 48 | 245.5 ± 54.2 | 14 | 34 | 48 | 4.16 ± 2.64 | 48 | 366.2 ± 47.1 | 48 | 165.8 ± 45.9 | |||||
| Sex | |||||||||||||||
| Male | 53 | 232.3 ± 53.9 | 0.278 | 13 | 40 | 1.000 | 53 | 4.04 ± 2.57 | 0.297 | 53 | 369.9 ± 47.2 | 0.439 | 53 | 170.1 ± 46.1 | 0.475 |
| Female | 40 | 247.2 ± 51.5 | 10 | 30 | 40 | 4.59 ± 2.39 | 40 | 362.5 ± 43.4 | 40 | 163.2 ± 45.8 | |||||
| Tumor size (cm) | |||||||||||||||
| ≤ 4 | 37 | 232.4 ± 53.1 | 0.947 | 11 | 26 | 0.508 | 37 | 3.88 ± 2.51 | 0.059 | 37 | 366.8 ± 42.1 | 0.988 | 37 | 168.4 ± 48.2 | 0.827 |
| > 4 | 56 | 243 ± 53.2 | 12 | 44 | 56 | 4.88 ± 2.38 | 56 | 366.6 ± 47.9 | 56 | 166.3 ± 44.6 | |||||
| Location | |||||||||||||||
| Superior clivus | 45 | 243.2 ± 51.8 | 0.641 | 12 | 33 | 0.677 | 45 | 4.20 ± 2.55 | 0.934 | 45 | 407.6 ± 45.5 | 0.206 | 45 | 178.8 ± 31.8 | 0.070 |
| Middle clivus | 30 | 254.3 ± 46.9 | 8 | 22 | 30 | 4.11 ± 2.49 | 30 | 417.4 ± 41.3 | 30 | 172.5 ± 29.7 | |||||
| Inferior clivus | 18 | 255.4 ± 56.0 | 3 | 15 | 18 | 4.74 ± 2.46 | 18 | 393.8 ± 44.9 | 18 | 166.5 ± 20.2 | |||||
| Invasion condition | |||||||||||||||
| Yes | 68 | 247.3 ± 51. | 0.010 | 12 | 56 | 0.019 | 68 | 3.91 ± 2.57 | 0.017 | 68 | 364.9 ± 44.3 | 0.534 | 68 | 165.5 ± 47.2 | 0.814 |
| No | 25 | 215.6 ± 52.1 | 11 | 14 | 25 | 5.29 ± 1.99 | 25 | 371.6 ± 49.2 | 25 | 169.0 ± 42.7 | |||||
| Preoperative recurrence | |||||||||||||||
| Yes | 76 | 239.1 ± 52.4 | 0.893 | 16 | 60 | 0.153 | 76 | 4.37 ± 2.43 | 0.517 | 76 | 367.8 ± 44.8 | 0.616 | 76 | 167.5 ± 45.8 | 0.891 |
| No | 17 | 237.2 ± 57.9 | 7 | 10 | 17 | 3.98 ± 2.72 | 17 | 361.7 ± 49.4 | 17 | 165.7 ± 47.2 | |||||
| Rate of resection | |||||||||||||||
| > 90% | 50 | 227.9 ± 50.8 | 0.683 | 10 | 40 | 0.368 | 50 | 4.19 ± 2.20 | 0.615 | 50 | 362.4 ± 46.4 | 0.326 | 50 | 165.5 ± 48.4 | 0.720 |
| ≤ 90% | 43 | 251.5 ± 53.6 | 13 | 30 | 43 | 4.47 ± 2.27 | 43 | 372.7 ± 44.4 | 43 | 168.9 ± 43.1 | |||||
| KPS | |||||||||||||||
| < 80 | 38 | 252.5 ± 54.9 | 0.037 | 2 | 36 | 0.001 | 38 | 4.41 ± 2.59 | 0.538 | 38 | 363.9 ± 43.6 | 0.629 | 38 | 162.7 ± 44.0 | 0.439 |
| ≥ 80 | 55 | 229.3 ± 50.2 | 21 | 34 | 55 | 4.08 ± 2.37 | 55 | 368.6 ± 47.1 | 55 | 170.2 ± 47.2 | |||||
| Histopathology | |||||||||||||||
| Conventional | 74 | 238.8 ± 52.1 | 0.996 | 18 | 56 | 1.000 | 74 | 4.51 ± 2.53 | 0.072 | 74 | 367.9 ± 45.5 | 0.628 | 74 | 168.3 ± 45.9 | 0.619 |
| Chondroid | 19 | 237.9 ± 58.4 | 5 | 14 | 19 | 3.36 ± 2.18 | 19 | 362.1 ± 46.4 | 19 | 162.4 ± 46.6 | |||||
As gross resection of skull base chordoma is difficult to achieve, tumor progression was defined as recurrence or re-growth of the residual tumor in this study. Chordoma diagnosis was made from the histological examination of hematoxylin and eosin (HE)-stained tumor tissue sections by two pathologists as previously described [35]. As a result, this group only included conventional and chondroid types. To facilitate comparison of resection extent, tumor resection was defined as follows: (> 90%) gross total resection and subtotal resection; (≤ 90%) partial removal and biopsy. Tumor invasion was defined as chordoma invading into adjacent bone or the dura structure, which was detected by radiographical examinations such as preoperative MRI and computed tomography (CT) images [36]. All patients underwent pre-operative MRI or CT to determine whether the invasion was present.
Patient characteristics
This study included 93 patients with skull base chordoma who underwent radical resection in our hospital. There were 53 males and 40 females with an average age of 45.8 years (range 9–74 years). Seventy-four patients had the conventional chordoma subtype and 19 patients had the chondroid subtype. The largest tumor diameter was 4.6 cm. All skull base chordomas were located in the clival region. The clivus was divided into three parts (superior, middle and inferior) by anatomical structure [37]. There were 45 chordomas in the superior clivus, 30 chordomas in the middle clivus and 18 chordomas in the inferior clivus. Fifty patients (53.8%) underwent postoperative image-guided radiation therapy. The radiotherapy dose-fractionation schedule was approximately 66–78 Gy at 2 Gy per day. Other patient characteristics are summarized in Supplementary Table 1.
Follow-up
The median follow-up period was 36.9 months (range 14–66 months), during which 35 patients died. Local recurrence-free survival (LRFS) was defined as the time interval from the date of surgery to the diagnosis of the first local recurrence. Similarly, OS was defined as the interval between surgery and death from any cause. No patient was lost to follow-up in this study. For surviving patients, data were censored at the last follow-up at the time of the analysis.
Cell culture
The human chordoma cell line UM-Chor1 was maintained in a 4:1 mixture of Iscove’s modified Dulbecco’s medium (HyClone, Logan, UT, USA) and RPMI-1640 medium (HyClone), supplemented with 10% fetal bovine serum (FBS, Gibco, Carlsbad, CA, USA) and 1% penicillin/strepto-mycin (Gibco) at 37 °C with 5% CO2.
Quantitative real-time PCR (qPCR)
Total miRNA was extracted from the frozen skull base chordoma tissues via RNAiso for Small RNA (TaKaRa, Kyoto, Japan) according to the manufacturer’s instructions. RNA quantity and quality were determined using the NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA). The isolated RNA was then reverse transcribed using the TianScript RT kit (Tiangen Biotech, Beijing, China) and real-time PCR was performed using SYBR FAST qPCR Master Mix (Kapa Biosystems, Inc., Wilmington, MA, USA) according to the manufacturer’s protocol. U6 RNA was chosen as an internal control for normalization. The primer sequences were designed by TaKaRa and the sequences were as follows: miR-455-5p stem–loop: 5ʹ-GTCGTATCGAGTGGAGCGTCGGAGCTATACGCACTCGATACGACACAAA-3ʹ, miR-455-5p forward: 5′-CGAGCTTCCTTCTGCAGGT-3′, miR-455-5p reverse: 5′-CACCACTGCCATCCCACA-3′, U6 stem–loop: 5ʹ-GTCCTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACAAAATATGGAAC-3ʹ, U6 forward: 5ʹ-TGCGGGTGCTCGCT TCGCAGC-3ʹ, U6 reverse: 5ʹ-CCAGTGCAGGGTCCGAGGT-3ʹ.
Immunohistochemistry
Immunohistochemistry staining of chordoma specimens was performed as previously described [38]. Paraffin-embedded sections were labeled with primary antibody against Gal9, TIM3, CD8, and FOXp3 (1:200, Abcam, Cambridge, UK) and samples were imaged under a BX-51 light microscope (Olympus).
Semiquantitative analysis
Immunoreactivity was evaluated and scored semi-quantitatively by two pathologists who were blinded to the patients’ clinical data. As previously described, the overall degree of TILs was evaluated to be: absent (0), rare/few (1), moderate (2) or prominent (3); the tissue samples were classified into negative expression (score 0–1) and positive (score 2–3) [39, 40]. Gal9 expression was evaluated by staining intensity and percentage of positive cells. The staining intensity was determined as follows: absent—0; weak—1; moderate—2; and strong—3. The percentage of positive cells was scored as follows: 0%, 0; 1–10%, 1; 11–50%, 2; 51–80%, 3; and 81–100%, 4. The immunohistochemical score was defined as the multiplication of both grading results (percentage of positive cells × staining intensities) and the positive expression was defined as a score ≥ 4 [35, 41, 42].
Quantitative evaluation
Quantitative evaluation was performed by examining each section using at least three different high-power fields with the most abundant TILs. The number of CD8+, FOXp3+ and TIM3+ TILs was manually counted five times for each photograph and the score was re-evaluated when an obvious difference occurred. Finally, the numbers of positively stained cells per unit area (mm2) were calculated and the mean densities were obtained [43].
Bioinformatic analysis
Three online miRNA databases—miRanda (www.microrna.org), miRDB (http://mirdb.org) and TargetScan (www. targetscan.org)—were used to predict the possible miRNAs that target Gal9 by examining the Gal9 3′-UTR with bioinformatics algorithms [44–46]. We summarized the results from these three databases and chose candidate miRNAs to validate our experiments.
Lentivirus vector, plasmid construction, and transduction
The mature miR-455-5p sequence was obtained from the miRBase database. The lentivirus-based vector for miR-445-5p was constructed by Gene-Chem (Shanghai, China) and transfected into UM-Chor1 cells as previously described [38]. Then, 10 μg/ml puromycin (Sigma, Santa Clara, CA, USA) was used to screen the transfected cells for 2 weeks and the effectiveness of miR-445-5p was assessed using qPCR.
Dual luciferase reporter assay
The dual luciferase reporter assay was performed as previously described [38]. In brief, UM-Chor1 cells were seeded in 96-well plates at a density of 1 × 104 cells per well, following 3′-UTR plasmids co-transfection for 48 h. The Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) was used to harvest cell lysates and detect firefly and renilla luciferase activities.
Statistical analysis
All statistical analyses were performed using GraphPad Prism software version 6.0. The data were analyzed using the independent samples t test for continuous variables (presented as mean ± standard deviation) and the Chi-square test for categorical variables. Pearson’s correlation test was used to examine the relationship between two continuous variables. Cutoff Finder Web Application (http://molpath.charite.de/cutof) was used to determine the threshold value for prognosis analysis [47]. Patient survival curves of OS and LRFS were generated using the Kaplan–Meier method and log-rank test. The prognostic factors associated with LRFS and OS were identified by multivariate Cox analysis. A two-tailed p value of 0.05 was regarded as significant.
Results
Identification of miR-455-5p as a potential regulator of Gal9 expression in skull base chordoma
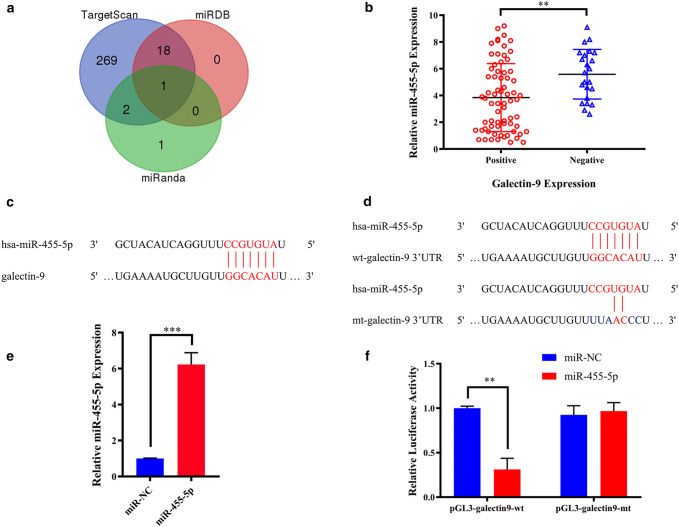
We predicted possible miRNAs that target Gal9 using the miRNA databases, miRanda, miRDB, and TargetScan. Only miR-455-5p was predicted in all three databases (Fig. 1a). We, therefore, evaluated miR-455-5p levels by qPCR and Gal9 expression by immunohistochemistry in the 93 chordoma cases. There was a negative correlation between miR-455-5p and Gal9 expression levels (t = 3.548, p = 0.001, r = − 0.629, p < 0.001; Fig. 1b and Supplementary Fig. 1a). Potential miR-455-5p binding sites in the 3′-UTR of Gal9 transcript were predicted with miRanda (Fig. 1c). We performed a luciferase reporter assay to determine whether miR-455-5p regulated Gal9 expression (Fig. 1d). The transfection efficiency of miR-445-5p in UM-Chor1 cells was confirmed by qPCR (Fig. 1e). Relative luciferase activity was lower in UM-Chor1 cells co-transfected with pGL3-Gal9-wt and miR-455-5p than in those co-transfected with pGL3-Gal9-wt and miR-negative control (NC) (p < 0.01). Additionally, there was no difference in luciferase activity between UM-Chor1 cells co-transfected with pGL3-Gal9-mt and miR-455-5p as compared to pGL3-Gal9-mt and miR-NC co-transfection (Fig. 1f). These results indicated that miR-455-5p was a regulator of Gal9 expression in skull base chordoma.
Fig. 1.
a Identification of a miRNA that potentially regulates Gal9 expression. b Independent t test results for the association between miR-455-5p and Gal9 expression. c Putative miR-455-5p binding sites in the 3′-untranslated region of Gal9 mRNA predicted by miRanda. d Luciferase reporter assays for evaluating Gal9 regulation by miR-455-5p. e Transfection efficiency of miR-455-5p in UM-Chor1 cells. f UM-Chor1 cells co-transfected with pGL3-Gal9-wt and miR-455-5p vs. cells co-transfected with pGL3-Gal9-wt and miR-NC
Relationship between miR-455-5p and Gal9 expression, TIL expression, and clinicopathological factors
The level of miR-455-5p in chordoma tissue was significantly associated with reduced invasion (t = 2.430, p = 0.017, Table 1). Positive Gal9 expression in tumor cells was observed in 70/93 patients (75.3%: Supplementary Fig. 2a–c). We also found that positive Gal9 expression was more common in patients with tumor invasion and lower KPS scores (Z = 5.477, p = 0.019 and Z = 11,374, p = 0.001, respectively; Table 1).
TILs were present in all 93 patient samples, as determined by HE staining. TILs were scored as rare or few in 30 cases (32.3%), moderate in 37 cases (39.7%), and prominent in 26 cases (28%) (Supplementary Fig. 2d–f). Thus, the extent of overall TIL expression was negative in 30 (32.3%) and positive in 63 (67.7%) cases. TIM3 was expressed in TILs in all 93 specimens (Supplementary Table 2 and Supplementary Fig. 2g–i). Average TIM3+ TIL density was 249.1 cells/mm2 (Supplementary Table 1). TIM3+ TILs densities were associated with higher invasion into surrounding bone structures and lower KPS score (t = 2.627, p = 0.010, and t = − 2.112, p = 0.037, respectively; Table 1). The pattern of CD8+ or FOXp3+ TILs in chordoma tissues were classified into three categories and these TIL populations were also detected in all specimens (Supplementary Fig. 2j–l and Supplementary Fig. 2m–o). The mean CD8+ and FOXp3+ TILs densities were 408.1 and 174.4 cells/mm2, respectively (Supplementary Table 1). However, there was no statistically significant association between CD8+ or FOXp3+ TIL density and clinicopathological features (Table 1).
Correlation between TIL subtypes in the chordoma microenvironment and miR-455-5p/Gal9 axis
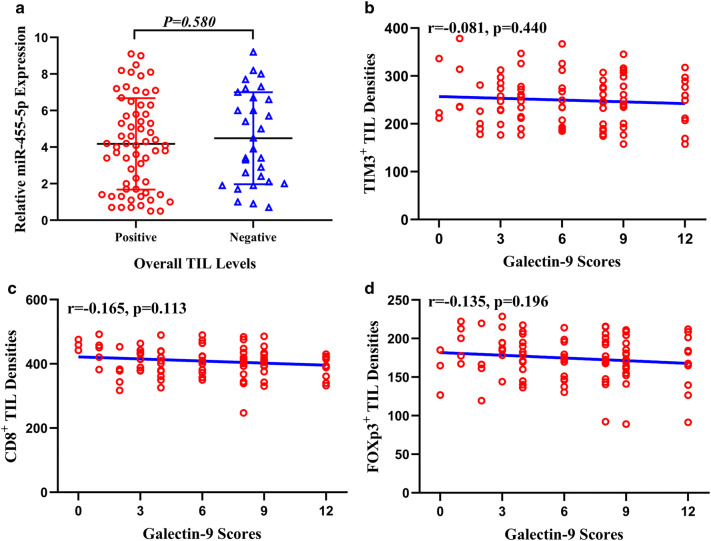
Tumors positive for Gal9 expression were more likely to exhibit total infiltration of TILs (χ2 = 15.19, p < 0.001, Supplementary Table 3 and Supplementary Fig. 3a). However, miR-455-5p expression was not correlated with TIL infiltration into the chordoma microenvironment (Fig. 2a). However, the density of TIM3+, CD8+, and FOXp3+ TILs were not associated with Gal9 expression (Fig. 2b–d). Other correlations/associations presented in Supplemental Table 3 are shown in Supplementary Figs. 1 and 3.
Fig. 2.
a Association between miR-455-5p expression and total TIL counts. b–d Association between Gal9 expression and TIM3+ TIL densities (b), CD8+ TIL densities (c), FOXp3+ TIL densities (d)
Prognostic value of miR-455-5p and Gal9 expression and TIL subtypes in skull base chordoma patients
During the follow-up period, tumor progression was observed in 45 patients (48.4%) and estimated 1- and 3-year LRFS rates were 84.7% and 34.6%, respectively. 41 patients (44.1%) died and estimated 1-, 3-, and 5-year OS rates were 100, 79.2, and 44.3%, respectively. The median survival time of LRFS and OS was 23.0 months (range 3–40 months) and 42.0 months (range 14–66 months), respectively.
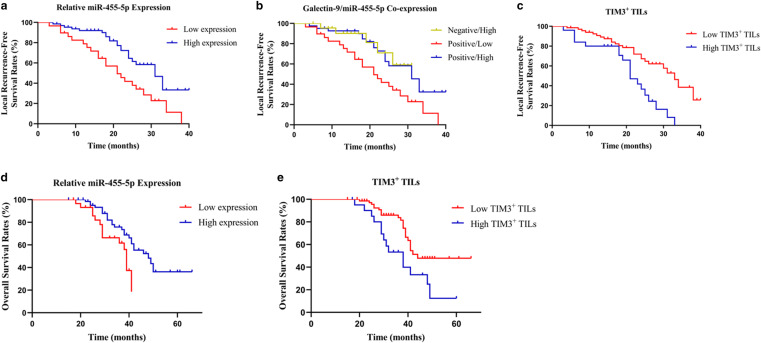
We predicted the cutoff values for miR-455-5p expression level and TIL densities associated with LRFS and OS (Supplementary Table 4). According to the cutoff value, patients were divided into high and low expression groups. We found that high tumor miR-455-5p expression was associated with better LRFS (p = 0.002, Table 2 and Fig. 3a). Furthermore, Kaplan–Meier analysis showed that coexpression of Gal9 and miR-455-5p in tumor cells was related to survival time (p = 0.010, Table 2 and Fig. 3b). When compared to positive Gal9 and high miR-455-5p levels, positive Gal9 expression and low miR-455-5p levels were correlated with worse survival (p = 0.006). In addition, patients with low TIM3+ TIL counts had longer LRFS (p < 0.001, Table 2 and Fig. 3c). Multivariate Cox analysis revealed that TIM3+ TIL count was an independent predictor of LRFS (p = 0.010, Table 2).
Table 2.
Univariate and multivariate analyses of different prognostic parameters for local recurrence-free survival of 93 skull base chordoma patients
| Factors | Categories | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| χ 2 | p values | p values | HR (95%CI) | ||
| Sex | Male/female | 0.706 | 0.401 | ||
| Age | ≤ 50/> 50 | 0.005 | 0.943 | ||
| Tumor size | ≤ 4 cm/> 4 cm | 0.442 | 0.506 | ||
| Location | Superior/middle /inferior | 0.426 | 0.808 | 0.099 | 0.363 (0.113–1.166) |
| Invasion condition | Yes/no | 15.591 | 0.001 | 0.079 | 1.899 (0.929–3.883) |
| Preoperative recurrence | Yes/no | 2.540 | 0.111 | ||
| Extent of resection | > 90%/≤ 90% | 9.307 | 0.002 | 0.237 | 0.655 (0.325–1.321) |
| KPS | ≥ 80/< 80 | 0.358 | 0.549 | ||
| Histopathology | Conventional/chondroid | 0.094 | 0.759 | ||
| Expression of overall TILs | Positive/negative | 0.326 | 0.568 | ||
| Tumor galectin-9 expression | Positive/negative | 1.098 | 0.295 | ||
| Tumor miR-455-5p expression | High/low | 9.251 | 0.002 | 0.357 | 1.379 (0.696–2.733) |
| Galectin-9/miR-455-5p coexpression | Negative/low | 9.269 | 0.010 | 0.826 | 1.126 (0.391–3.239) |
| Negative/high | |||||
| Positive/low | |||||
| Positive/high | |||||
| TIM3+ TILs | High/low | 12.606 | < 0.001 | 0.010 | 0.425 (0.221–0.815) |
Fig. 3.
Kaplan–Meier plots of local recurrence-free survival rates. a–c Stratification by miR-455-5p expression (a), Gal9 and miR-455-5p co-expression (b), TIM3+ TIL densities (c). Kaplan–Meier plots of overall survival rates. d–e Stratification by miR-455-5p expression (d), TIM3+ TIL densities (e)
Survival analysis revealed that high tumor miR-455-5p expression was related to longer OS (p = 0.018, Table 3 and Fig. 3d). Furthermore, coexpression of Gal9 and miR-455-5p in chordoma was associated with OS (p = 0.049, Table 3). Positive Gal9 expression and low miR-455-5p levels were related to worse survival compared with positive Gal9 expression and high miR-455-5p levels (p = 0.016). We also found that there was a significant difference in OS between patients with high vs. low TIM3+ TIL counts (p = 0.009, Table 3 and Fig. 3e). Multivariate Cox analysis showed that TIM3+ TIL counts were associated with an increased risk of death (p = 0.016, Table 3).
Table 3.
Univariate and multivariate analyses of different prognostic parameters for overall survival of 93 skull base chordoma patients
| Factors | Categories | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| χ 2 | p values | p values | HR (95%CI) | ||
| Sex | Male/female | 0.014 | 0.905 | ||
| Age | ≤ 50/> 50 | 0.149 | 0.699 | ||
| Tumor size | ≤ 4 cm/> 4 cm | 0.059 | 0.807 | ||
| Location | Superior/middle /inferior | 3.858 | 0.146 | 0.467 | 1.454 (0.574–3.681) |
| Invasion condition | Yes/no | 5.237 | 0.026 | 0.351 | 0.665 (0.282–1.569) |
| Preoperative recurrence | Yes/no | 0.600 | 0.439 | ||
| Extent of resection | >90%/≤ 90% | 0.662 | 0.416 | ||
| KPS | ≥ 80/< 80 | 9.924 | 0.002 | 0.192 | 0.604 (0.283–1.289) |
| Histopathology | Conventional/chondroid | 1.695 | 0.193 | ||
| Expression of overall TILs | Positive/negative | 1.609 | 0.205 | ||
| Tumor galectin-9 expression | Positive/negative | 1.741 | 0.187 | ||
| Tumor miR-455-5p expression | High/low | 5.634 | 0.018 | 0.372 | 1.680 (0.292–3.586) |
| Galectin-9/miR-455-5p coexpression | Negative/low | 7.840 | 0.049 | 0.106 | 1.942 (0.537–7.026) |
| Negative/high | |||||
| Positive/low | |||||
| Positive/high | |||||
| TIM3+ TILs | High/low | 4.879 | 0.009 | 0.016 | 1.453 (0.921–1.607) |
Discussion
In the present study, we found that miR-455-5p expression was associated with patient survival. In addition, patients with both negative Gal9 and high miR-455-5p expression had longer survival times. The results showed that TIM3+ TILs were independent predictors of LRFS and OS. These findings may be useful for predicting the prognosis of chordoma patients and provide a basis for individualized immunotherapy.
Although the brain was previously believed to be an immune privileged organ, there is an evidence suggesting that it serves as an immune site with a microenvironment that provides opportunities for immunotherapy of central nervous system tumors [13]. TIM3 protein is mainly localized on the cell surface and its expression on T cells could modulate the immune response [48]. TIM3 also has been investigated in myeloid cells, such as monocytes, macrophages, and dendritic cells. However, the function of TIM-3 in myeloid cells and the underlying mechanisms are not fully understood [49]. Gal9 is a ligand of TIM3 that is expressed on numerous tumor cells and whose expression may be induced by interferon-γ secreted by multiple cell types [50, 51]. Gal9 binding to TIM3 on T cells can lead to T cell apoptosis, inhibition of T cell responses, and immune escape of tumor cells [52]. Thus, the TIM3/Gal9 pathway negatively regulates T cell-mediated immune responses [53]. However, in our study, we found that tumors positive for Gal9 expression were more likely to exhibit total infiltration of TILs by immunohistochemistry and semiquantitative analysis. We speculated that TILs contained not only T cells, but also other cells such as NK cells and dendritic cells and that the binding of Gal9 and TIM3 resulted in the apoptosis of T cells, which might increase the infiltration of other types of lymphocyte, thereby increasing overall TIL infiltration. There was no direct relationship between TIM3+ TILs and overall TIL infiltration. We also found that according to our previous inference, Gal9 and TIM3+ TIL expression should be negatively correlated. However, our results showed that there was no correlation between Gal9 and TIM3+ TIL expression. This may be because of the small sample size in our study. We will expand the sample size in future studies to confirm our inference. In addition, recent studies have shown that low TIM3+ TIL counts are associated with better prognosis in hepatocellular carcinoma and gastric cancer patients, which is consistent with our findings [54, 55].
Recent studies showed that miR-1, miR-16-5p, miR-219-5p, miR-574-3p, and miR-1237-3p were downregulated in chordoma tissues. MiR-1, miR-16-5p, and miR-219-5p were shown to inhibit chordoma cell proliferation or invasion. Moreover, miR-574-3p was investigated to promote immune escape by regulating the expression of PD-L1 in the PD-1/PD-L1 axis [56–61]. In this study, we predicted and confirmed that miR-455-5p is downregulated and can negatively regulate Gal9 expression. It has been suggested that miR-455-5p expression is downregulated in gastric cancer tissue and overexpression of miR-455-5p can inhibit the proliferation, invasion, and metastasis of gastric cancer cells [62], which is consistent with our findings. However, another study showed that miR-455-5p expression was upregulated in colon cancer and promoted colon cancer cell proliferation and inhibited apoptosis by suppressing Gal9 expression [63]. Thus, the expression level of miR-455-5p varies depending on the cancer type. Our results also showed that there was a positive correlation between TIM3 and miR-455-5p expression. Therefore, we inferred that the lower expression of miR-455-5p could upregulate the expression of Gal9 in chordoma and then induce the apoptosis of TIM3+ lymphocytes and lower TIM3+ TIL densities in chordomas. It remains unclear whether miR-455-5p can prevent the binding between TIM3 and Gal9. Furthermore, patients with high miR-455-5p and negative Gal9 levels showed prolonged survival. These results indicate that miR-455-5p may provide a therapeutic strategy for patients with chordoma by targeting the TIM3/Gal9 pathway.
Several studies have reported that oncogenes such as sex determining region Y (SRY)-box 9 (SOX9) or c-Casitas B lineage lymphoma (c-Cbl) are associated with the prognosis of skull base chordoma patients [35, 64]. However, a single biomarker cannot predict clinical outcome and drug treatment response in chordoma due to intratumoral heterogeneity. It has been suggested that the tumor immune microenvironment is minimally affected by tumor heterogeneity [65]. In our study, we found that TIM3+ TILs in the tumor microenvironment were associated with LRFS or OS of skull base chordoma patients. These findings may allow the prediction of patient survival and provide a basis for personalized immune therapy. However, additional studies are required to elucidate the detailed mechanisms of miR-455-5p and Gal9 signaling and to explore the influence of other immune factors on the prognosis of skull base chordoma patients.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Abbreviations
- ATCC
American Type Culture Collection
- C-Cbl
C-Casitas B lineage lymphoma
- CD8
Cluster of differentiation 8
- CT
Computed tomography
- CTLA-4
Cytolytic T lymphocyte-associated Ag-4
- Gal9
Galectin-9
- HE
Hematoxylin and eosin
- KPS
Karnofsky performance status
- LRFS
Local recurrence-free survival
- MiRNAs
MicroRNAs
- NC
Negative control
- qPCR
Quantitative real-time polymerase chain reaction
- SOX9
Sex-determining region Y (SRY)-box 9
- TIM3
T-cell immunoglobulin and mucin-domain 3
Author contributions
ZJ and YZ conceived and designed the study; JZ and YJ performed the experiments and collected the data; HZ, JZ, LC, PL, LL, JZ, and YJ produced the figures and tables; all authors performed the analysis and analyzed the data. JZ, YJ, YZ, and ZJ interpreted results and wrote the manuscript. FL, DZ, and HZ modified the manuscript. All authors read and approved the final version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 81101917, 81270036, 30901736), the Liaoning Province Natural Science Foundation (No. 20170541022), the Plan to Focus on Research and Development from Science and Technology project of Liaoning Province (No. 2017225029), the Science and Technology Plan Project of Shenyang City (No. 18-014-4-11), and the Fund for Scientific Research of The First Hospital of China Medical University (No. FHCMU-FSR).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interests.
Ethical approval
The study was approved by the Research Ethics Committee of the First Hospital of China Medical University and was in accordance with the ethical standards of the institutional committees and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The study approval number is AF-SOP-07-01.
Informed consent
Informed consent was obtained from all individual participants included in the study. With approval from the National Science Foundation of China (81101917), we designed informed consent forms that were signed by eligible patients before recruitment into the study and admission to the hospital. The patients agreed to the use of their specimens and clinical data for research purposes only.
Cell line authentication
The human chordoma cell line UM-Chor1 was obtained as a gift from Professor Yazhuo Zhang, Department of Neurosurgery, Beijing Tiantan Hospital. The origin of UM-Chor1 was human clivus chordoma tissue. The American Type Culture Collection (ATCC) performed authentication of cell line UM-Chor1 via cytochrome C oxidase I assay and short tandem repeat analysis. We obtained a cell line authentication certificate from the ATCC.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jinpeng Zhou and Yang Jiang contributed equally to this work.
Contributor Information
Ye Zhang, Email: yzhang21@cmu.edu.cn.
Zhitao Jing, Email: jingzhitao@hotmail.com.
References
- 1.McMaster ML, Goldstein AM, Bromley CM, Ishibe N, Parry DM. Chordoma: incidence and survival patterns in the United States, 1973–1995. Cancer Causes Control. 2001;12(1):1–11. doi: 10.1023/A:1008947301735. [DOI] [PubMed] [Google Scholar]
- 2.Heffelfinger MJ, Dahlin DC, MacCarty CS, Beabout JW. Chordomas and cartilaginous tumors at the skull base. Cancer. 1973;32(2):410–420. doi: 10.1002/1097-0142(197308)32:2<410::AID-CNCR2820320219>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 3.Salisbury JR. The pathology of the human notochord. J Pathol. 1993;171(4):253–255. doi: 10.1002/path.1711710404. [DOI] [PubMed] [Google Scholar]
- 4.Stacchiotti S, Sommer J. Building a global consensus approach to chordoma: a position paper from the medical and patient community. Lancet Oncol. 2015;16(2):e71–e83. doi: 10.1016/s1470-2045(14)71190-8. [DOI] [PubMed] [Google Scholar]
- 5.Chibbaro S, Cornelius JF, Froelich S, Tigan L, Kehrli P, Debry C, Romano A, Herman P, George B, Bresson D. Endoscopic endonasal approach in the management of skull base chordomas—clinical experience on a large series, technique, outcome, and pitfalls. Neurosurg Rev. 2014;37(2):217–224. doi: 10.1007/s10143-013-0503-9. [DOI] [PubMed] [Google Scholar]
- 6.Di Maio S, Rostomily R, Sekhar LN. Current surgical outcomes for cranial base chordomas: cohort study of 95 patients. Neurosurgery. 2012;70(6):1355–1360. doi: 10.1227/neu.0b013e3182446783. [DOI] [PubMed] [Google Scholar]
- 7.Hines JP, Ashmead MG, Stringer SP. Clival chordoma of the nasal septum secondary to surgical pathway seeding. Am J Otolaryngol. 2014;35(3):431–434. doi: 10.1016/j.amjoto.2013.12.018. [DOI] [PubMed] [Google Scholar]
- 8.Koutourousiou M, Gardner PA, Tormenti MJ, Henry SL, Stefko ST, Kassam AB, Fernandez-Miranda JC, Snyderman CH. Endoscopic endonasal approach for resection of cranial base chordomas: outcomes and learning curve. Neurosurgery. 2012;71(3):614–624. doi: 10.1227/neu.0b013e31825ea3e0. [DOI] [PubMed] [Google Scholar]
- 9.Jackson CM, Lim M, Drake CG. Immunotherapy for brain cancer: recent progress and future promise. Clin Cancer Res. 2014;20(14):3651–3659. doi: 10.1158/1078-0432.CCR-13-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dougan M, Dranoff G. Immune therapy for cancer. Annu Rev Immunol. 2009;27:83–117. doi: 10.1146/annurev.immunol.021908.132544. [DOI] [PubMed] [Google Scholar]
- 11.Sharma P, Allison JP. Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell. 2015;161(2):205–214. doi: 10.1016/j.cell.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garber ST, Hashimoto Y, Weathers SP, Xiu J, Gatalica Z, Verhaak RG, Zhou S, Fuller GN, Khasraw M, de Groot J, Reddy SK, Spetzler D, Heimberger AB. Immune checkpoint blockade as a potential therapeutic target: surveying CNS malignancies. Neurooncology. 2016;18(10):1357–1366. doi: 10.1093/neuonc/now132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim M, Xia Y, Bettegowda C, Weller M. Current state of immunotherapy for glioblastoma. Nat Rev Clin Oncol. 2018 doi: 10.1038/s41571-018-0003-5. [DOI] [PubMed] [Google Scholar]
- 14.Sounni NE, Noel A. Targeting the tumor microenvironment for cancer therapy. Clin Chem. 2013;59(1):85–93. doi: 10.1373/clinchem.2012.185363. [DOI] [PubMed] [Google Scholar]
- 15.Nirschl CJ, Drake CG. Molecular pathways: coexpression of immune checkpoint molecules: signaling pathways and implications for cancer immunotherapy. Clin Cancer Res. 2013;19(18):4917–4924. doi: 10.1158/1078-0432.CCR-12-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiraoka K, Miyamoto M, Cho Y, Suzuoki M, Oshikiri T, Nakakubo Y, Itoh T, Ohbuchi T, Kondo S, Katoh H. Concurrent infiltration by CD8+ T cells and CD4+ T cells is a favourable prognostic factor in non-small-cell lung carcinoma. Br J Cancer. 2006;94(2):275–280. doi: 10.1038/sj.bjc.6602934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shang B, Liu Y, Jiang SJ, Liu Y. Prognostic value of tumor-infiltrating FOXp3+ regulatory T cells in cancers: a systematic review and meta-analysis. Sci Rep. 2015;5:15179. doi: 10.1038/srep15179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seo AN, Lee HJ, Kim EJ, Kim HJ, Jang MH, Lee HE, Kim YJ, Kim JH, Park SY. Tumour-infiltrating CD8+ lymphocytes as an independent predictive factor for pathological complete response to primary systemic therapy in breast cancer. Br J Cancer. 2013;109(10):2705–2713. doi: 10.1038/bjc.2013.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li G, Wang Z, Zhang C, Liu X, Cai J, Wang Z, Hu H, Wu F, Bao Z, Liu Y, Zhao L, Liang T, Yang F, Huang R, Zhang W, Jiang T. Molecular and clinical characterization of TIM-3 in glioma through 1,024 samples. Oncoimmunology. 2017;6(8):e1328339. doi: 10.1080/2162402X.2017.1328339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu S, Lin J, Qiao G, Wang X, Xu Y. Tim-3 identifies exhausted follicular helper T cells in breast cancer patients. Immunobiology. 2016;221(9):986–993. doi: 10.1016/j.imbio.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 21.Tan Y, Trent JC, Wilky BA, Kerr DA, Rosenberg AE. Current status of immunotherapy for gastrointestinal stromal tumor. Cancer Gene Ther. 2017;24(3):130–133. doi: 10.1038/cgt.2016.58. [DOI] [PubMed] [Google Scholar]
- 22.Monney L, Sabatos CA, Gaglia JL, Ryu A, Waldner H, Chernova T, Manning S, Greenfield EA, Coyle AJ, Sobel RA, Freeman GJ, Kuchroo VK. Th1-specific cell surface protein Tim-3 regulates macrophage activation and severity of an autoimmune disease. Nature. 2002;415(6871):536–541. doi: 10.1038/415536a. [DOI] [PubMed] [Google Scholar]
- 23.Zhu C, Anderson AC, Schubart A, Xiong H, Imitola J, Khoury SJ, Zheng XX, Strom TB, Kuchroo VK. The Tim-3 ligand galectin-9 negatively regulates T helper type 1 immunity. Nat Immunol. 2005;6(12):1245–1252. doi: 10.1038/ni1271. [DOI] [PubMed] [Google Scholar]
- 24.Sharma P, Allison JP. Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell. 2015;161(2):205–214. doi: 10.1016/j.cell.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ngiow SF, Teng MW, Smyth MJ. Prospects for TIM3-targeted antitumor immunotherapy. Can Res. 2011;71(21):6567–6571. doi: 10.1158/0008-5472.CAN-11-1. [DOI] [PubMed] [Google Scholar]
- 26.Ferris RL, Lu B, Kane LP. Too much of a good thing? Tim-3 and TCR signaling in T cell exhaustion. J Immunol. 2014;193(4):1525–1530. doi: 10.4049/jimmunol.1400557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Z, Han H, He X, Li S, Wu C, Yu C, Wang S. Expression of the galectin-9-Tim-3 pathway in glioma tissues is associated with the clinical manifestations of glioma. Oncol Lett. 2016;11(3):1829–1834. doi: 10.3892/ol.2016.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X, Chen Y, Liu X, Zhang J, He X, Teng G, Yu D. Tim3/Gal9 interactions between T cells and monocytes result in an immunosuppressive feedback loop that inhibits Th1 responses in osteosarcoma patients. Int Immunopharmacol. 2017;44:153–159. doi: 10.1016/j.intimp.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 29.Komita H, Koido S, Hayashi K, Kan S, Ito M, Kamata Y, Suzuki M, Homma S. Expression of immune checkpoint molecules of T cell immunoglobulin and mucin protein 3/galectin-9 for NK cell suppression in human gastrointestinal stromal tumors. Oncol Rep. 2015;34(4):2099–2105. doi: 10.3892/or.2015.4149. [DOI] [PubMed] [Google Scholar]
- 30.Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223(2):102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le XF, Merchant O, Bast RC, Calin GA. The roles of MicroRNAs in the cancer invasion-metastasis cascade. Cancer Microenviron. 2010;3(1):137–147. doi: 10.1007/s12307-010-0037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z, Han J, Cui Y, Zhou X, Fan K. miRNA-21 inhibition enhances RANTES and IP-10 release in MCF-7 via PIAS3 and STAT3 signalling and causes increased lymphocyte migration. Biochem Biophys Res Commun. 2013;439(3):384–389. doi: 10.1016/j.bbrc.2013.08.072. [DOI] [PubMed] [Google Scholar]
- 33.Jiang S, Zhang HW, Lu MH, He XH, Li Y, Gu H, Liu MF, Wang ED. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Can Res. 2010;70(8):3119–3127. doi: 10.1158/0008-5472.CAN-09-4250. [DOI] [PubMed] [Google Scholar]
- 34.Yang Q, Jiang W, Zhuang C, Geng Z, Hou C, Huang D, Hu L, Wang X. MicroRNA-22 downregulation of galectin-9 influences lymphocyte apoptosis and tumor cell proliferation in liver cancer. Oncol Rep. 2015;34(4):1771–1778. doi: 10.3892/or.2015.4167. [DOI] [PubMed] [Google Scholar]
- 35.Luo P, Wang X, Zhou J, Li L, Jing Z. C-Cbl and Cbl-b expression in skull base chordomas is associated with tumor progression and poor prognosis. Hum Pathol. 2018;74:129–134. doi: 10.1016/j.humpath.2017.12.019. [DOI] [PubMed] [Google Scholar]
- 36.Wang L, Wu Z, Tian K, Wang K, Li D, Ma J, Jia G, Zhang L, Zhang J. Clinical features and surgical outcomes of patients with skull base chordoma: a retrospective analysis of 238 patients. J Neurosurg. 2017;127(6):1257–1267. doi: 10.3171/2016.9.JNS16559. [DOI] [PubMed] [Google Scholar]
- 37.Fernandez-Miranda JC, Gardner PA, Snyderman CH, Devaney KO, Mendenhall WM, Suarez C, Rinaldo A, Ferlito A. Clival chordomas: A pathological, surgical, and radiotherapeutic review. Head Neck. 2014;36(6):892–906. doi: 10.1002/hed.23415. [DOI] [PubMed] [Google Scholar]
- 38.Jiang Y, Han S, Cheng W, Wang Z, Wu A. NFAT1-regulated IL6 signalling contributes to aggressive phenotypes of glioma. Cell Commun Signal. 2017;15(1):54. doi: 10.1186/s12964-017-0210-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zou MX, Peng AB, Lv GH, Wang XB, Li J, She XL, Jiang Y. Expression of programmed death-1 ligand (PD-L1) in tumor-infiltrating lymphocytes is associated with favorable spinal chordoma prognosis. Am J Transl Res. 2016;8(7):3274–3287. [PMC free article] [PubMed] [Google Scholar]
- 40.Bellmunt J, Mullane SA, Werner L, Fay AP, Callea M, Leow JJ, Taplin ME, Choueiri TK, Hodi FS, Freeman GJ, Signoretti S. Association of PD-L1 expression on tumor-infiltrating mononuclear cells and overall survival in patients with urothelial carcinoma. Ann Oncol. 2015;26(4):812–817. doi: 10.1093/annonc/mdv009. [DOI] [PubMed] [Google Scholar]
- 41.Hutterer M, Knyazev P, Abate A, Reschke M, Maier H, Stefanova N, Knyazeva T, Barbieri V, Reindl M, Muigg A, Kostron H, Stockhammer G, Ullrich A. Axl and growth arrest specific gene 6 are frequently overexpressed in human gliomas and predict poor prognosis in patients with glioblastoma multiforme. Clin Cancer Res. 2008;14(1):130–138. doi: 10.1158/1078-0432.ccr-07-0862. [DOI] [PubMed] [Google Scholar]
- 42.Remmele W, Schicketanz KH. Immunohistochemical determination of estrogen and progesterone receptor content in human breast cancer. Pathol Res Pract. 1993;189(8):862–866. doi: 10.1016/s0344-0338(11)81095-2. [DOI] [PubMed] [Google Scholar]
- 43.Sato E, Olson SH, Ahn J, Bundy B, Nishikawa H, Qian F, Jungbluth AA, Frosina D, Gnjatic S, Ambrosone C, Kepner J, Odunsi T, Ritter G, Lele S, Chen YT, Ohtani H, Old LJ, Odunsi K. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc Natl Acad Sci USA. 2005;102(51):18538–18543. doi: 10.1073/pnas.0509182102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11(8):R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43(Database issue):D146–D152. doi: 10.1093/nar/gku1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 47.Budczies J, Klauschen F, Sinn BV, Gyorffy B, Schmitt WD, Darb-Esfahani S, Denkert C. Cutoff Finder: a comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS One. 2012;7(12):e51862. doi: 10.1371/journal.pone.0051862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han G, Chen G, Shen B, Li Y. Tim-3: an activation marker and activation limiter of innate immune cells. Front Immunol. 2013;4:449. doi: 10.3389/fimmu.2013.00449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ocana-Guzman R, Torre-Bouscoulet L, Sada-Ovalle I. TIM-3 regulates distinct functions in macrophages. Front Immunol. 2016;7:229. doi: 10.3389/fimmu.2016.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Imaizumi T, Kumagai M, Sasaki N, Kurotaki H, Mori F, Seki M, Nishi N, Fujimoto K, Tanji K, Shibata T, Tamo W, Matsumiya T, Yoshida H, Cui XF, Takanashi S, Hanada K, Okumura K, Yagihashi S, Wakabayashi K, Nakamura T, Hirashima M, Satoh K. Interferon-gamma stimulates the expression of galectin-9 in cultured human endothelial cells. J Leukoc Biol. 2002;72(3):486–491. [PubMed] [Google Scholar]
- 51.Yoshida H, Imaizumi T, Kumagai M, Kimura K, Satoh C, Hanada N, Fujimoto K, Nishi N, Tanji K, Matsumiya T, Mori F, Cui XF, Tamo W, Shibata T, Takanashi S, Okumura K, Nakamura T, Wakabayashi K, Hirashima M, Sato Y, Satoh K. Interleukin-1beta stimulates galectin-9 expression in human astrocytes. Neuroreport. 2001;12(17):3755–3758. doi: 10.1097/00001756-200112040-00030. [DOI] [PubMed] [Google Scholar]
- 52.Sabatos CA, Chakravarti S, Cha E, Schubart A, Sanchez-Fueyo A, Zheng XX, Coyle AJ, Strom TB, Freeman GJ, Kuchroo VK. Interaction of Tim-3 and Tim-3 ligand regulates T helper type 1 responses and induction of peripheral tolerance. Nat Immunol. 2003;4(11):1102–1110. doi: 10.1038/ni988. [DOI] [PubMed] [Google Scholar]
- 53.Sanchez-Fueyo A, Tian J, Picarella D, Domenig C, Zheng XX, Sabatos CA, Manlongat N, Bender O, Kamradt T, Kuchroo VK, Gutierrez-Ramos JC, Coyle AJ, Strom TB. Tim-3 inhibits T helper type 1-mediated auto- and alloimmune responses and promotes immunological tolerance. Nat Immunol. 2003;4(11):1093–1101. doi: 10.1038/ni987. [DOI] [PubMed] [Google Scholar]
- 54.Cheng G, Li M, Wu J, Ji M, Fang C, Shi H, Zhu D, Chen L, Zhao J, Shi L, Xu B, Zheng X, Wu C, Jiang J. Expression of Tim-3 in gastric cancer tissue and its relationship with prognosis. Int J Clin Exp Pathol. 2015;8(8):9452–9457. [PMC free article] [PubMed] [Google Scholar]
- 55.Li H, Wu K, Tao K, Chen L, Zheng Q, Lu X, Liu J, Shi L, Liu C, Wang G, Zou W. Tim-3/galectin-9 signaling pathway mediates T-cell dysfunction and predicts poor prognosis in patients with hepatitis B virus-associated hepatocellular carcinoma. Hepatology. 2012;56(4):1342–1351. doi: 10.1002/hep.25777. [DOI] [PubMed] [Google Scholar]
- 56.Duan Z, Shen J, Yang X, Yang P, Osaka E, Choy E, Cote G, Harmon D, Zhang Y, Nielsen GP, Spentzos D, Mankin H, Hornicek F. Prognostic significance of miRNA-1 (miR-1) expression in patients with chordoma. J Orthop Res. 2014;32(5):695–701. doi: 10.1002/jor.22589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang H, Yang K, Ren T, Huang Y, Tang X, Guo W. miR-16-5p inhibits chordoma cell proliferation, invasion and metastasis by targeting Smad3. Cell Death Dis. 2018;9(6):680. doi: 10.1038/s41419-018-0738-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wei W, Zhang Q, Wang Z, Yan B, Feng Y, Li P. miR-219-5p inhibits proliferation and clonogenicity in chordoma cells and is associated with tumor recurrence. Oncol Lett. 2016;12(6):4568–4576. doi: 10.3892/ol.2016.5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zou MX, Guo KM, Lv GH, Huang W, Li J, Wang XB, Jiang Y, She XL. Clinicopathologic implications of CD8(+)/FOXp3(+) ratio and miR-574-3p/PD-L1 axis in spinal chordoma patients. Cancer Immunol Immunother. 2018;67(2):209–224. doi: 10.1007/s00262-017-2080-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zou MX, Huang W, Wang XB, Li J, Lv GH, Wang B, Deng YW. Reduced expression of miRNA-1237-3p associated with poor survival of spinal chordoma patients. Eur Spine J. 2015;24(8):1738–1746. doi: 10.1007/s00586-015-3927-9. [DOI] [PubMed] [Google Scholar]
- 61.Osaka E, Yang X, Shen JK, Yang P, Feng Y, Mankin HJ, Hornicek FJ, Duan Z. MicroRNA-1 (miR-1) inhibits chordoma cell migration and invasion by targeting slug. J Orthop Res. 2014;32(8):1075–1082. doi: 10.1002/jor.22632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu J, Zhang J, Li Y, Wang L, Sui B, Dai D. MiR-455-5p acts as a novel tumor suppressor in gastric cancer by down-regulating RAB18. Gene. 2016;592(2):308–315. doi: 10.1016/j.gene.2016.07.034. [DOI] [PubMed] [Google Scholar]
- 63.Yang Q, Hou C, Huang D, Zhuang C, Jiang W, Geng Z, Wang X, Hu L. miR-455-5p functions as a potential oncogene by targeting galectin-9 in colon cancer. Oncol Lett. 2017;13(3):1958–1964. doi: 10.3892/ol.2017.5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen H, Garbutt CC, Spentzos D, Choy E, Hornicek FJ, Duan Z. Expression and therapeutic potential of SOX9 in chordoma. Clin Cancer Res. 2017;23(17):5176–5186. doi: 10.1158/1078-0432.CCR-17-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fritzsching B, Fellenberg J, Moskovszky L, Sapi Z, Krenacs T, Machado I, Poeschl J, Lehner B, Szendroi M, Bosch AL, Bernd L, Csoka M, Mechtersheimer G, Ewerbeck V, Kinscherf R, Kunz P. CD8(+)/FOXP3(+)-ratio in osteosarcoma microenvironment separates survivors from non-survivors: a multicenter validated retrospective study. Oncoimmunology. 2015;4(3):e990800. doi: 10.4161/2162402X.2014.990800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.