Abstract
The effects of phosphorylation of histone H3 at serine 10 have been studied in the context of other posttranslational modifications such as lysine methylation. We set out to investigate the impact of phosphoserine-10 on arginine-8 methylation. We performed methylation reactions using peptides based on histone H3 that contain a phosphorylated serine and compared the extent of arginine methylation with unmodified peptides. Results obtained via fluorography indicate that peptides containing a phosphorylated serine-10 inhibit deposition of methyl groups to arginine-8 residues. To further explore the effects of phosphoserine on neighboring arginine residues, we physically characterized the non-covalent interactions between histone H3 phosphoserine-10 and arginine-8 using 31P NMR spectroscopy. A salt bridge was detected between the negatively charged phosphoserine-10 and the positively charged unmodified arginine-8 residue. This salt bridge was not detected when arginine-8 was symmetrically dimethylated. Finally, molecular simulations not only confirm the presence of a salt bridge but also identify a subset of electrostatic interactions present when arginine is replaced with alanine. Taken together, our work suggests that the negatively charged phosphoserine maximizes its interactions. By limiting its exposure and creating new contacts with neighboring residues, it will inhibit deposition of neighboring methyl groups, not through steric hindrance, but by forming intrapeptide interactions that may mask substrate recognition. Our work provides a mechanistic framework for understanding the role of phosphoserine on nearby amino acid residues and arginine methylation.
Keywords: post-translational modification (PTM), histone crosstalk, phosphoserine, phosphorylation, symmetric arginine dimethylation (SDMA), methylarginine, PRMT, methylation
INTRODUCTION
Histone proteins organize DNA, making it accessible in part through modifications on their N-terminal tails (Y. Wang et al., 2004). Histone posttranslational modifications (PTMs) can alter protein-protein interactions and are heavily implicated in gene expression (Taverna, Li, Ruthenburg, Allis, & Patel, 2007). Although single modifications on the N-terminus of histone proteins are established independent epigenetic marks such as the association of acetylation with gene activation (Allfrey, Faulkner, & Mirsky, 1964), information about how these modifications affect each other is now emerging. Multiple studies on the combinatorial crosstalk of histone proteins have revealed synergistic and/or antagonistic effects. The synergistic coupling of histone H3 phosphorylated serine-10 (H3S10p) and acetylated lysine-14 (H3K14ac) is a canonical example of one PTM enhancing the deposition of another (Cheung et al., 2000; Lo et al., 2000; Manning & Manning, 2018).
Protein phosphorylation introduces a negatively charged phosphate group onto a serine/threonine/tyrosine residue, and this residue, which significantly changes the interactions with other amino acid residues, causing a change in the conformation of this substrate. Its contributions to neighboring PTMs is unclear. For example, it is not clear whether a negatively charged phosphate will provoke a major structural change and mask substrate recognition by the enzymes that deposit neighboring PTMs, or whether it will alter the hydrogen-bonding local regime enough to physically block deposition of neighboring groups. At the molecular level, several investigations have reported that phosphorylation in general reorganizes a salt-bridge network to bring about a localized conformational change and thus effects an exchange of signaling partners (Skinner et al., 2017). Of these, the phosphoserine arginine salt bridge has been identified (Papamokos et al., 2012). Histone H3 harbors an arginine-lysine-serine (RKS) motif that can become phosphorylated at serine-10 (Ibuki, Toyooka, Zhao, & Yoshida, 2014), and is symmetrically dimethylated at arginine-8 (Hyun, Lee, Han, & Yu, 2011; Pal, Vishwanath, Erdjument-Bromage, Tempst, & Sif, 2004).
To understand the role of phosphoserine-10 on arginine-8 methylation, we performed in vitro methylation reactions using synthetic phosphoserine-10 peptides based on the N-terminal tail of histone H3. Arginine methylation of H3R8 was detected through fluorography and thin layer chromatography. We show that a pre-existing phosphate group on serine 10 (S10p) will inhibit both asymmetric and symmetric dimethylated arginine products (ADMA and SDMA) in neighboring arginine 8 (R8) within an RXS motif of histone H3. We then measured the non-covalent interactions by 31P NMR spectroscopy and confirm a salt bridge between phosphoserine-10 and unmodified arginine-8 of histone H3. Briefly, we estimated the free energy contributions of salt bridges by measuring the change in pKa of ionizable groups caused by altered electrostatic interactions. This salt bridge was absent in phosphoserine-10 peptides containing symmetrically dimethylated arginine-8 residues. Finally, computational modeling was carried out to measure the interactions in these peptides. We conclude that the presence of a phosphate group on serine-10 does not physically prevent methylation of neighboring arginine-8 within the RKS motif of histone H3. Instead, the negatively charged phosphoserine creates interactions that most likely mask PRMT enzyme recognition.
MATERIALS AND METHODS
PRMT Expression and Purification of GST fusion proteins of PRMT1 and PRMT4.
All methyltransferase constructs were purified from E. coli and were obtained from Dr. Steven Clarke (UCLA). Plasmids expressing rat GST-PRMT1 or GST-PRMT4 were expressed in BL21 Star (DE3) cells. Briefly, cells containing GST-PRMT1 or GST-PRMT4 were induced for 4 h at 37°C with 0.4 mM isopropyl ß-D-thiogalactopyranoside (IPTG). They were lysed with seven 20-s sonicator pulses (Fisherbrand Model 120 Sonic Dismembrator). The cell lysates were centrifuged for 50 min at 23,000 × g at 4°C. The proteins were then purified from extracts by incubating them with glutathione-sepharose 4B beads (GE Health) according to the manufacturer’s instructions. They were next eluted with 30 mM glutathione, 50 mM Tris-HCl, 120 mM NaCl, and 4% glycerol at pH 7.5. Purified protein preparations were quantified by a Lowry assay after trichloroacetic acid precipitation (Lowry, Rosebrough, Farr, & Randall, 1951).
PRMT5 expressed in bacteria yields inactive enzyme. In addition, and for optimal activity, human PRMT5 acts as part of a larger complex that requires another protein known as methylosome protein 50 (MEP50) (Antonysamy et al., 2012). Thus, PRMT5 methylation reactions were carried out using commercially available human recombinant PRMT5 in complex with human recombinant MEP50, produced by co-expression in an insect cell/baculovirus expression system, PRMT5 2–637(C-term), MEP50 2–342(C-term) (HMT-22-148, Reaction Biology Corp.).
Methylation Reactions.
Full-length GST-PRMT1 (1–2 μg) or GST-PRMT4 (1–2 μg) was incubated with 1–30 μg of either full-length histone H3 (positive control), unmodified peptide 1 or phosphorylated peptide 2, and [3H]AdoMet at 37°C in a 30 μl – 90 μl total reaction volume. See figure legends for actual amounts. Negative controls such as an enzyme alone or peptide alone control were carried out in the presence of [3H]AdoMet and all other reagents. Methylation reactions were carried out as outlined in the literature (Jain, Jin, & Clarke, 2017) using full-length histone proteins (New England Biolabs) and unmodified or phosphorylated peptide substrates (VCP Bio, Inc.) corresponding to the histone H3 tail (amino acids 4–16; ~1.4 kDa): NH2-TKQTARKSTGGKAP-COOH (unmodified) and NH2-TKQTARKSpTGGKAP-COOH (phosphoserine). The accuracy of the sequence for the peptide substrates were verified by mass spectrometry by the manufacturer. To the best of our abilities, careful handling and preparation of peptides 1 and 2 was taken to ensure that equal amounts were used in subsequent methylation reactions and analyses by fluorography and TLC. For example, both peptides were dissolved in water to an equal concentration of 1μg/μl.
For SDS-PAGE analysis, methylation reactions were stopped with the addition of an equal volume of SDS gel sample buffer. Briefly, methylation reactions containing peptide substrates were quenched using 2X sample buffer (0.05 M Tris, 20% glycerol, 0.1% bromophenol blue, pH 6.8), heated at 100 °C for 3 min, and loaded on a 20% acrylamide and 0.43% N,N-methylene bisacrylamide gel. Electrophoresis was performed at 80 V at room temperature. The gel was stained with Coomassie Brilliant Blue R-250 for 1 h and destained with 10% methanol and 5% acetic acid for 5 h. The destained gel was prepared for fluorography by incubation in EN3HANCE (PerkinElmer Life Sciences) for 1 h and washed with water for 20 min. The gel was dried at 70 °C in vacuo and exposed to Kodak BIOMAX XAR scientific imaging film at −80 °C for different lengths of time ranging from seven days to several weeks using molecular mass standards 1.7–42 kDa (Spectra Multicolor Low Range Protein Ladder, ThermoFisher).
For samples analyzed by thin layer chromatography (TLC), 10 μmol of either monomethylated arginine (ω-MMA), asymmetric dimethylarginine (ADMA), or symmetric dimethylarginine (SDMA) were analyzed separately and also added to approximately 1/3 of the methylation reactions for acid hydrolysis (Debler et al., 2016). The samples were resuspended in approximately 5 μL of water and immediately spotted onto 20 × 20 cm silica-coated plates (Sigma-Aldrich Z122807–25EA) in 1-μL aliquots. After pre-equilibrating the chamber with the mobile phase: 30% ammonium hydroxide/chloroform/methanol/water (2:0.5:4.5:1), samples were spotted on the plate and run for approximately 5 h. After air drying overnight, the plate was sprayed with a ninhydrin solution [15 mg/mL ninhydrin dissolved in 97% (vol/vol) butanol, 3% (vol/vol) glacial acetic acid] and allowed to air dry. To visualize the ninhydrin spots indicating arginine residues, the plate was heated at 100 °C for 10 min and sectioned into 3-mm-wide slices. The silica from each slice was mixed with 950 μL of water and vortexed. This material was added to 5 mL of scintillation fluid (Research Products International). The samples were loaded on to a Beckmann LS6500 Liquid Scintillation Counter where three 5-min counting cycles were averaged to determine the corresponding counts per minute of each fraction.
Peptides used for 31P NMR.
Six peptides (14 amino acids long) corresponding to the histone H3 tail were identical except for specific changes in the amino acid sequences underlined. Note: all peptides used for NMR were phosphorylated. Peptide 1-RKS(PO3H2): Thr-Lys-Gln-Thr-Ala-Arg-Lys-Ser(PO3H2)-Thr-Gly-Gly-Lys-Ala-Pro; peptide 2-RAS(PO3H2): Thr-Lys-Gln-Thr-Ala-Arg-Ala-Ser(PO3H2)-Thr-Gly-Gly-Lys-Ala-Pro; peptide 3-AKS(PO3H2): Thr-Lys-Gln-Thr-Ala-Ala-Lys-Ser(PO3H2)-Thr-Gly-Gly-Lys-Ala-Pro; peptide 4-AAS(PO3H2): Thr-Lys-Gln-Thr-Ala-Ala-Ala-Ser(PO3H2)-Thr-Gly-Gly-Lys-Ala-Pro; peptide 5-R(SDMA)KS(PO3H2): Thr-Lys-Gln-Thr-Ala-Arg(Symmetrically Dimethylated)-Lys-Ser(PO3H2)-Thr-Gly-Gly-Lys-Ala-Pro; peptide 6-R(SDMA)AS(PO3H2):: Thr-Lys-Gln-Thr-Ala-Arg(Symmetrically Dimethylated)-Ala-Ser(PO3H2)-Thr-Gly-Gly-Lys-Ala-Pro (1 mg; CPS Scientific, Sunnyvale, CA). In addition, all precautions were taken to ensure the peptides were treated equally and preserved to prevent degradation. The peptides came lyophilized and were stored at −20 °C.
NMR Sample Preparation.
A modified protocol optimized for these peptides was based on the work of Theillet et al., 2012 (Theillet et al., 2012). Briefly, approximately 1 mg of commercially purchased customized peptide was dissolved in 520–550 μL of buffer, final concentration 1 mM. The buffer was made as a stock solution of 25 mL consisting of 30 mM HEPES, 5 mM MgCl2, 5 mM KCl, 10 % glycerol and 8 % D2O at 22 °C. The peptide-buffer solution was then transferred to a 5 mm diameter Wilmad NMR tube. The pH was adjusted in small increments with 1 M HCl and 1 M NaOH to obtain at least 10 NMR spectra. The pH is first lowered to a value less than 3.00 and then raised to a value above 8.00.
NMR Spectroscopy.
31P NMR data were acquired on a Bruker 400 MHz spectrometer using TopSpin software (version 3.2.6). Final sample volumes were approximately 600 μL. The parameters used for the NMR spectrometer include 1000 scans, 22 – 25°C, degree of substitution of 4, and a relaxation delay of 3 seconds (Supplemental Materials Figures 2–6). 31P spectra referenced indirectly to 85 % H3PO3.
pKa Determination.
Data analysis for NMR titration was done under the assumption that the titration sites are noninteracting. Starting with the Henderson-Hasselbalch equation:
| #(1) |
we replaced protonated and deprotonated concentrations with the chemical shifts using the following relationships:
| #(2) |
| #(3) |
to arrive at the modified version of the Henderson-Hasselbalch equation:
| #(4) |
where is the observed chemical shift and the and are the chemical shifts of the protonated and deprotonated forms (determined by the fit), respectively. To fit the experimental data of pH versus observed chemical shifts, the following equation has been rewritten as .
| #(5) |
We can arrive at the Hill equation or four-parameter logistic equation by introducing the Hill Slope coefficient and algebraically manipulating equation 4, which results in
| #(6) |
MATLAB (Release 2019b, Math Works Inc.) scripts and functions were developed in-house to carry out nonlinear regression to model the experimental results. The MATLAB functions were used to determine the values of the , and Hill Slope parameters of the Hill equation. The nlinfit MATLAB function used in this step estimates the coefficients using iterative least squares estimation. Confidence intervals at 95% were calculated to determine the uncertainty of the curve fitting.
Molecular dynamics simulations.
Starting structures for the six peptides used in the experiments described above were built by extracting the appropriate residues from PDB structure 1KX5 (Davey, Sargent, Luger, Maeder, & Richmond, 2002). These sequences were then edited to include a phosphate group at serine-10 (all peptides, in keeping with the peptides used for NMR), symmetric dimethylation at arginine-8 (peptides 5-RSDMAKSp and 6-RSDMAASp), and/or an alanine substitution for lysine-9 (peptides 2-RASp, 4-AASp, and 6-RSDMAASp).
Solvated peptide systems were built using the program tleap in AmberTools 18 (Case et al., 2018). Each system contained the peptide, approximately 4400 water molecules, and the number of chloride (Cl−) ions necessary to neutralize the system. The peptide was modeled using the AMBER-FB15 force field (L. P. Wang et al., 2017) with new parameters for phosphorylated residues (Stoppelman, Ng, Nerenberg, & Wang, 2020). The water and ions were modeled using TIP3P-FB water model (L. P. Wang, Martinez, & Pande, 2014) and Joung-Cheatham ion parameters for SPC/E, a water model with similar overall parameters to TIP3P-FB (Joung & Cheatham, 2008).
Molecular dynamics simulations were performed using pmemd and pmemd.cuda (Salomon-Ferrer, Götz, Poole, Le Grand, & Walker, 2013) of Amber 18 and all the heavy atom-hydrogen covalent bonds were constrained with the SHAKE algorithm. The real-space cutoff for nonbonded interactions was set at 9.0 Å. Before running dynamics, each system was subjected tox a two-step energy minimization procedure. In the first step, 1000 steps of minimization were performed with the peptide atoms restrained to their initial positions using a harmonic potential of 10 kcal mol−1 Å−2. The second round of minimization for 1000 steps was performed without any restraints on the peptide.
After minimization was complete, each system was heated at constant volume from a starting temperature of 100 K to a final temperature of 295.15 K (22 °C) over 30 ps with the peptide atoms again restrained using a harmonic potential of 10 kcal mol−1 Å−2. Next, the density of the system was equilibrated by simulating in the NPT ensemble at 1.0 atm and 295.15 K using a Berendsen barostat with coupling constant 2.0 ps−1 and a Langevin thermostat with a collision frequency of 1.0 ps−1 for 50 ps. The final structure from this equilibration was used as the starting structure for the replica exchange MD simulations, which were performed in the NVT ensemble.
The replica exchange MD simulations (REMD) were performed at 24 exponentially spaced temperatures ranging from 295.15 K to 370 K. To decorrelate the starting structures, the replicas were first simulated for 10 ns before any exchanges were performed. We then performed 200 or 400 ns (depending on convergence of the conformational ensemble, see below) of replica exchange MD simulation, with exchange attempts every 0.20 ps. Structures were saved every 5.0 or 10 ps, yielding 40,000 structures for each of the six peptides.
MD simulation data analysis.
To check the convergence of each simulation, we split each simulation into equal halves, discarded the first 10% of each half, and then computed the Kullback-Leibler (K-L) divergence of the end-to-end distance for the peptide, defined as the distance between the Cα atoms of the first and last residues of the peptide. We observed rapid exponential decay of the K-L divergence toward zero for all systems, as would be expected for independent simulations converging to the same ensemble average. Close intramolecular contacts to the phosphate group of phosphorylated Ser10 were detected using the ‘hbond’ function of cpptraj in AmberTools 18 (Roe & Cheatham, 2013). We defined the “acceptors” as the oxygen atoms of the phosphate group; the “donors” were N and O atoms with bonded H atoms (including, for example, Lys and Arg side chains, Thr side chain, the peptide backbone, etc.). We used an O-N/O interatomic distance cutoff of 3.5 Å and an O-H-N/O angle cutoff of 135°; in other words, interactions had to simultaneously have an interatomic distance of <3.5 Å and an angle of >135° in order to be counted. Interactions that occurred in fewer than 1% of the conformational ensemble were ignored.
RESULTS
Detection of methylated arginine via fluorography and TLC.
To determine whether a phosphorylated serine-10 residue would inhibit methylation of neighboring arginine-8 in histone H3, we carried out in vitro methylation reactions using unmodified and phosphorylated peptides corresponding to histone H3. Protein arginine methylation can form three different products: mono-, symmetric- or asymmetric- arginine methylation (MMA, SDMA, ADMA) catalyzed by a family of nine protein arginine methyltransferases (PRMT1–9) (Bedford & Clarke, 2009). Histone H3 is physiologically asymmetrically dimethylated by PRMT4/CARM1 at arginine-17 (Jacques et al., 2016), at arginine-26 (Schurter et al., 2001), and symmetrically dimethylated by PRMT5 at arginine-8 (Pal et al., 2004; Southall, Wong, Odho, Roe, & Wilson, 2009). PRMT4 does not methylate histone H3 at arginine-8; however, since H3 is a physiological substrate, it was used as a control. In addition, Tang et al have shown that PRMT1, which primarily forms ADMA, catalyzes 85% of the total protein arginine methylation in rat fibroblast and in mouse liver (Tang et al., 2000). Thus, in addition to in vitro reactions with PRMT4 and PRMT5, we also performed methylation reactions with PRMT1, the most active member of the PRMT family. We expected that only PRMT5 would methylate peptides corresponding to arginine-8 and serine-10 of histone H3.
Figure 1, panel A shows that PRMT5 will methylate both unmodified peptide 1 (lane 1) and full-length histone H3 (lane 3) but not phosphorylated peptide 2 (lane 2). PRMT5 is the physiological catalyst, responsible for producing symmetric dimethylated arginine-8 in histone H3 (Pal et al., 2004). Figure 1 panel B shows that both PRMT4 (lane 1) and PRMT5 (lane 3) catalyze the methylation of full-length histone H3. While PRMT4 does methylate histone H3, it does not methylate it in this region. Thus, PRMT4 was not capable of methylating either peptide 1 or 2 (see supplemental information, Figure 1). To further explore whether the inhibition is unique to PRMT5, which forms SDMA, or whether the neighboring phosphate could inhibit other members of the PRMT family, we performed in vitro methylation reactions with PRMT1, the most active and predominant member of the PRMT family (Figure 1, panels C and D and Figure 2) (Tang et al., 2000). PRMT1 forms ADMA, thus this reaction serves to determine whether the phosphorylated serine only inhibits SDMA, catalyzed by PRMT5 or both ADMA and SDMA. Here we show that PRMT1 will methylate full-length histone H3 (lane 1, Figure 1C) and unmodified peptide 1 (lane 4, Figure 1C and lane 4, Figure 1D), but not phosphorylated peptide 2 (lane 2, Figure 1D) (Figure 2). The two PRMT1 alone controls show that although one enzyme preparation was more active than the other, both preparations were inhibited by a neighboring phosphate group. The data is representative of several methylation reactions detected by fluorography. Thus, regardless of the methyltransferase used or the methylated mark deposited (ADMA or SDMA), detection of radioactivity (methylation) is greater for the unmodified peptide when compared to the phosphorylated peptide.
Figure 1.
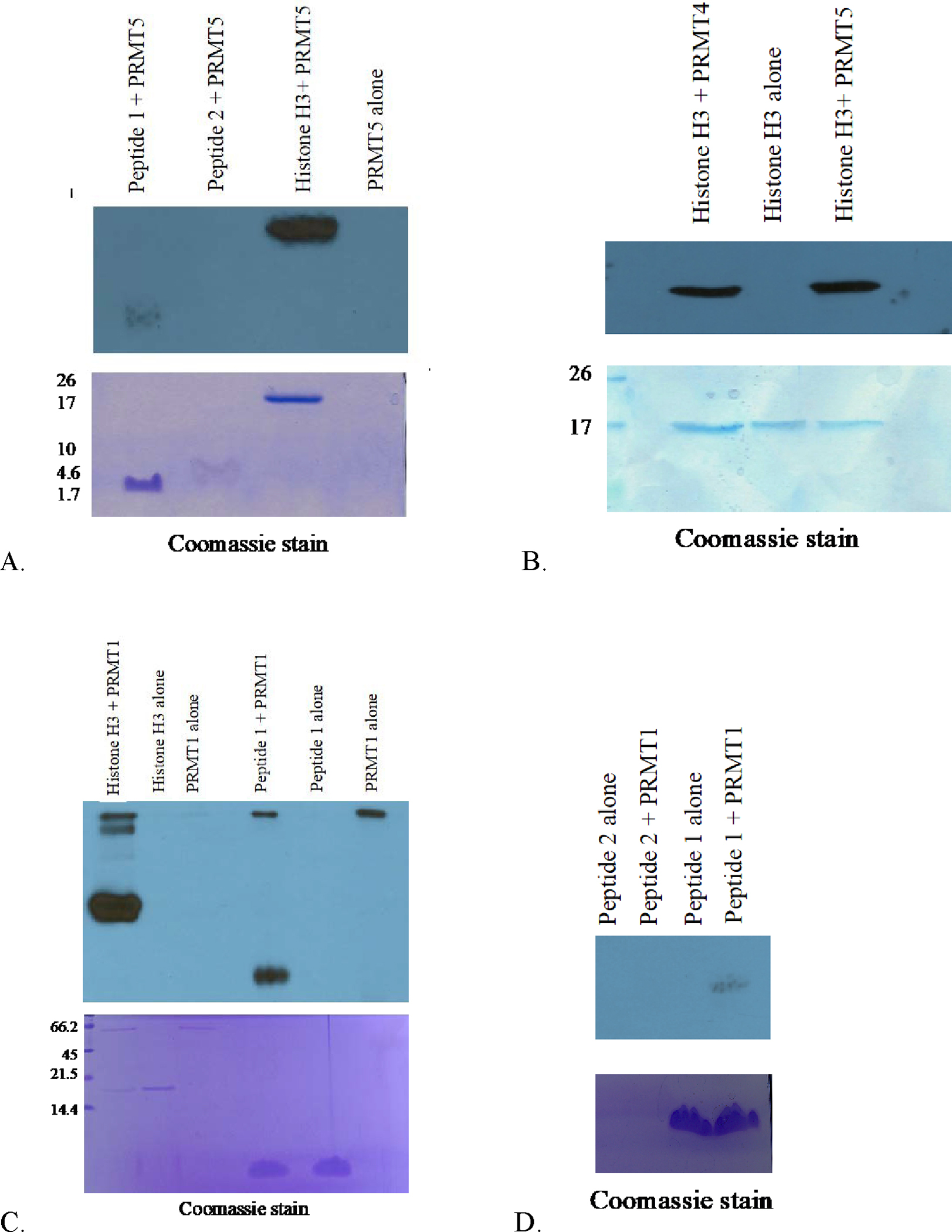
Methylation of peptide 1 (TKQTARKSTGGKAP) or peptide 2 (TKQTARKSpTGGKAP) corresponding to histone H3 detected via fluorography. A) GST-PRMT5 (1μg), B) GST-PRMT4 (1μg), C and D) GST-PRMT1 (1μg), were incubated with substrates histone H3, peptide 1 (1μg) or peptide 2 (1 μg) in the presence of 0.5 μM S-adenosyl-L-[methyl-3H]methionine for 1 hr at 37 °C in a final volume of 30 μl of HEPES buffer as described in Materials and Methods. The samples were then resolved via 20% SDS-PAGE. Full-length histone H3 was used as a positive control, PRMT, and peptide alone reactions lacking enzyme or substrates were used as negative controls. Negative controls such as PRMT1 alone or Peptide 1 alone were carried out in the presence of all reagents, including [3H]AdoMet. The radioactive methylation reactions were exposed on film as described Materials and Methods for A) 8 days, B) 7 days, C) 3 weeks and D) 9 days.
Figure 2.

Hydrolysates of methylation reactions of PRMT1 incubated with either unmodified peptide 1 or phosphorylated peptide 2 and resolved by TLC. TLC for hydrolysates of the reaction mixture and individual and mixed standards of ADMA, MMA, and SDMA. The upper portion shows the ninhydrin staining of the TLC plate containing the arginine derivative standards; the lower portion shows the radioactivity corresponding to the TLC slices of the reaction mixture lane. A) TLC of hydrolysates for PRMT1, 5001 cpms; mix lane contains reaction mixture and standards; B) TLC of hydrolysates for PRMT1 incubated with phosphorylated peptide 2, 1596 cpms; C) TLC of hydrolysates for PRMT1 alone (control), 1702 cpms, and D) TLC of hydrolysates for peptide 1 and peptide 2 alone controls. All controls contained [3H]AdoMet.
Analysis of peptides by fluorography requires optimization because low molecular weight proteins and peptides (<10 kDa) are difficult to resolve by polyacrylamide gel electrophoresis (Sarfo, Moorhead, & Turner, 2003). The SDS-PAGE gels in Figure 1A and 1D of Coomassie blue-stained peptides reveals that peptide 1 stains darker than phosphorylated peptide 2. Coomassie blue is a negatively charged dye that binds to positively charged residues, specifically arginine, lysine, and histidine residues (de Moreno, Smith, & Smith, 1986). We hypothesize that the presence of a negatively charged phosphate group is preventing adherence of the dye, especially to neighboring arginine-8 and lysine-9. Unmodified peptide 1 always stained darker than phosphorylated peptide 2. Thus, to further confirm the difference in production of methylated arginine species between peptides 1 and 2, TLC was performed with acid hydrolysates of methylation reactions catalyzed by PRMT1, the most active member of the methyltransferase family (Figure 2). Three additional methylation reactions served as controls: PRMT1 alone, peptide 1 alone and peptide 2 alone. These controls were carried out in parallel and in the presence of [3H]AdoMet. Radioactivity from TLC slices show that when PRMT1 is incubated with unmodified peptide 1, the reaction produces over 5001 cpms (Figure 2A). However, when PRMT1 is incubated with phosphorylated peptide 2, 1597 cpms are produced (Figure 2B). Since this amount is not greater than the PRMT1 alone control (1703 cpms) (Figure 2C) or the peptide alone controls (Figure 2D), we conclude that a phosphate group on serine-10 inhibits methylation of neighboring arginine-8, even in the presence of the most active member of the PRMT family, PRMT1.
NMR.
Methylation does not change the overall positive charge of the arginine guanidinium group; however, depending on the number of methyl groups added can change the number of hydrogen bond donor sites (Cannizzaro & Houk, 2002; Fuhrmann, Clancy, & Thompson, 2015; Yesselman, Horowitz, Brooks, & Trievel, 2015). The basic nature of arginine and lysine mean that they add a positive charge to the peptide. Table 1 shows a list of the peptides as well as their net charge at a neutral pH (7.0). To investigate this interaction at a molecular level, we focused on detecting non-covalent interactions between phosphoserine and arginine within the RKS motif of histone H3. This type of experiment is studied by NMR in two ways. The first method involves measuring one or two of the pKa values of the residues involved and then substituting those potential residues to remove any interactions. The second method involves creating changes in pKa values by studying the residues involved and comparing changes upon protein unfolding (Bosshard, Marti, & Jelesarov, 2004). Since the histone H3 N-terminal tail is inherently unstructured and not folded, the first method (i.e. residue substitution) was used for the detection of the salt bridge between phosphoserine-10 and unmodified arginine-8. Phosphorylated peptides corresponding to the first 14 amino acid residues of histone H3 were purchased. We compared the changes in pKa value of the original sequence against the pKa of the peptides with altered amino acids. We used 31P NMR for the observation of a salt bridge because the natural abundance of phosphorus (100 %) and gyromagnetic ratio (17.235 MHz T−1) is high enough that we can observe a peak without the need to isotopically label our peptide substrates.
Table 1.
31P NMR titration pKa derived for the 6 peptides based on the N-terminal sequence of histone H3 amino acid residues 3–16.
| # | Peptide Sequence | Net charge (pH 7) | Modification | pKa Values | Mean pKa |
Std. Dev. | Delta pKa (AASp) |
|---|---|---|---|---|---|---|---|
| 1 | TKQTARKSpTGGKAP | +2 | None | 5.55 5.60 5.51 |
5.55 | 0.05 | 0.37 |
| 2 | TKQTARASpTGGKAP | +1 | K9 to A | 5.78 5.69 5.68 |
5.72 | 0.06 | 0.20 |
| 3 | TKQTAAKSpTGGKAP | +1 | R8 to A | 5.68 5.90 5.91 |
5.83 | 0.13 | 0.09 |
| 4 | TKQTAAASpTGGKAP | 0 | R8 to A and K9 to A | 6.04 5.84 5.87 |
5.92 | 0.17 | - |
| 5 | TKQTARSDMAKSpTGGKAP | +2 | R8 SDMA |
5.70 5.77 |
5.74 | 0.05 | 0.18 |
| 6 | TKQTARSDMAASpTGGKAP | +1 | K9 to A R8 SDMA |
6.37 5.76 |
6.07 | 0.43 | −0.15 |
A salt bridge or network of salt bridges form between oppositely charged residues. Two types of salt bridges exist: the surface-exposed salt bridge and the buried salt bridge that can interact in both an inter- and intramolecular manner (Williamson et al., 2013). Salt bridges buried within the same protein are expected to have stronger electrostatic interactions. For instance, the largest salt bridge observed to date, was detected in T4 lysozyme, whose buried salt bridge between Asp-70 and His-30 contributes favorable stability at about 3–5 kcal/mol, and a pKa shift of 3 units for Asp-70 (Anderson, Becktel, & Dahlquist, 1990). In contrast, a single salt bridge within two oppositely charged residues, an arginine and phosphoserine salt bridge (i, i+4) was calculated as 0.45 kcal/mol using synthetic peptides (Liehr & Chenault, 1999). Although the pKa was not reported, it is expected that this value is small. Indeed, a study using 31P NMR reported a downshifted pKa value of about 0.7 units in an arginine-phosphoserine salt bridge interaction with a small peptide of 24 amino acid residues (Kumar et al., 2012). Thus, we developed NMR methods for a 14 amino acid long peptide expecting small changes in pKa (0.3–0.7 units) as an indication of a salt bridge and performed a 31P NMR titration by varying the pH which allowed for the calculation of the pKa of the side chain. A list of the experimentally derived pKa values is shown in Table 1.
A Salt Bridge Interaction in Peptide 1-RKSp (TKQTARKSpTGGKAP).
Titrations of peptide 1-RKSp were carried out using the phosphoserine for 31P NMR detection. This allowed for the determination of the acid dissociation constant of the monophosphorylated peptides. Figure 3 shows representative sigmoidal Henderson-Hasselbalch fitting of the chemical shifts versus pH of all peptides. See supplemental information for additional NMR values and corresponding spectra. As shown in Table 1, the average pKa value of three trials for peptide 1-RKSp was 5.55. A salt bridge for NMR spectroscopy is detected at a pKa shift below 0.7 units, as was detected for peptides based on CLASP2 protein (Kumar et al., 2012). Our value falls within this range when compared to peptide 4-AASp which lacks a positively charged arginine or lysine residue, thus served as a control, and has an average pKa value of 5.92. The value of the experimentally obtained control was compared to the published pKa value of solvent exposed phosphoserine model peptide glycylglycylserylalanine (GGSpA) which has a pKa of 6.1 (Hoffmann, Reichert, Wachs, Zeppezauer, & Kalbitzer, 1994). Hence, the pKa value of peptide 1-RKSp shifted 0.37 units (5.92–5.55), indicative of a salt bridge between the neighboring arginine-8 and phosphorylated serine-10. This result also confirms computational simulations described in this study as well as those previously described in the literature (Papamokos et al., 2012).
Figure 3.

Dependence of the 31P NMR chemical shifts of 1 mg of A) Peptide 1 (TKQTARKS(PO3H2)TGGKAP), one of three trials, B) Peptide 2 (TKQTARAS(PO3H2)TGGKAP), one of three trials, C) Peptide 3 (TKQTAAKS(PO3H2)TGGKAP), one of three trials, D) Peptide 4 (TKQTAAAS(PO3H2)TGGKAP), one of three trials, E) Peptide 5 (TKQTAR(SDMA)KS(PO3H2)TGGKAP), one of two trials, and F) Peptide 6 (TKQTAR(SDMA)AS(PO3H2)TGGKAP), one of two trials. All peptides were dissolved in approximately 0.5 ml of 30 mM HEPES, 5 mM MgCl2, 5 mM KCl, 10 % glycerol and 8 % D2O at 22 °C. The titration curves were calculated with the parameters described in Materials and Methods. The x-axis is the experimental pH measured and the y-axis is the chemical shift observed at the respective pH value.
To determine the contribution of the lysine residue, we carried out similar titrations using peptide 2-RASp and peptide 3-AKSp which contains an alanine in place of lysine-9 or arginine-8 respectively. The average pKa values were 5.72 and 5.83. Since these values fall between both the pKa of the control peptide 4-AASp and peptide 1-RKSp, with small shifts in delta pKa (< 0.2, Table 1), we postulate that in the absence of an arginine residue, an interaction between the lysine residue and the phosphoserine becomes prominent.
To determine whether methylation of arginine residues affects salt bridge formation between positively charged arginine and negatively charged phosphoserine, we performed titrations using peptide 5-RSDMAKSp and peptide 6-RSDMAASp where arginine is modified with a symmetrically dimethylated group. The delta pKa values of peptide 5-RSDMAKSp and peptide 6-RSDMAASp shifted 0.18 and −0.15 units respectively, indicating an absence in salt-bridge upon methylation, and underscoring the contribution of the lysine residue when the arginine residue is either methylated or absent.
Molecular Dynamics Simulations.
MD simulations of these peptides allow us to visualize and quantify the interactions of phosphoserine-10 in the presence or absence of positively charged residues such as arginine and lysine. In our simulations of peptide 1-RKSp, as well as peptide 2-RASp, we observed a consistently strong interaction (salt bridge) between phosphoserine-10 and the neighboring arginine-8 (Figure 4A). When both arginine-8 and lysine-9 are replaced by alanine residues, however, other interactions are formed with phosphoserine-10 that at least partially make up for the elimination (Figure 4B).
Figure 4.
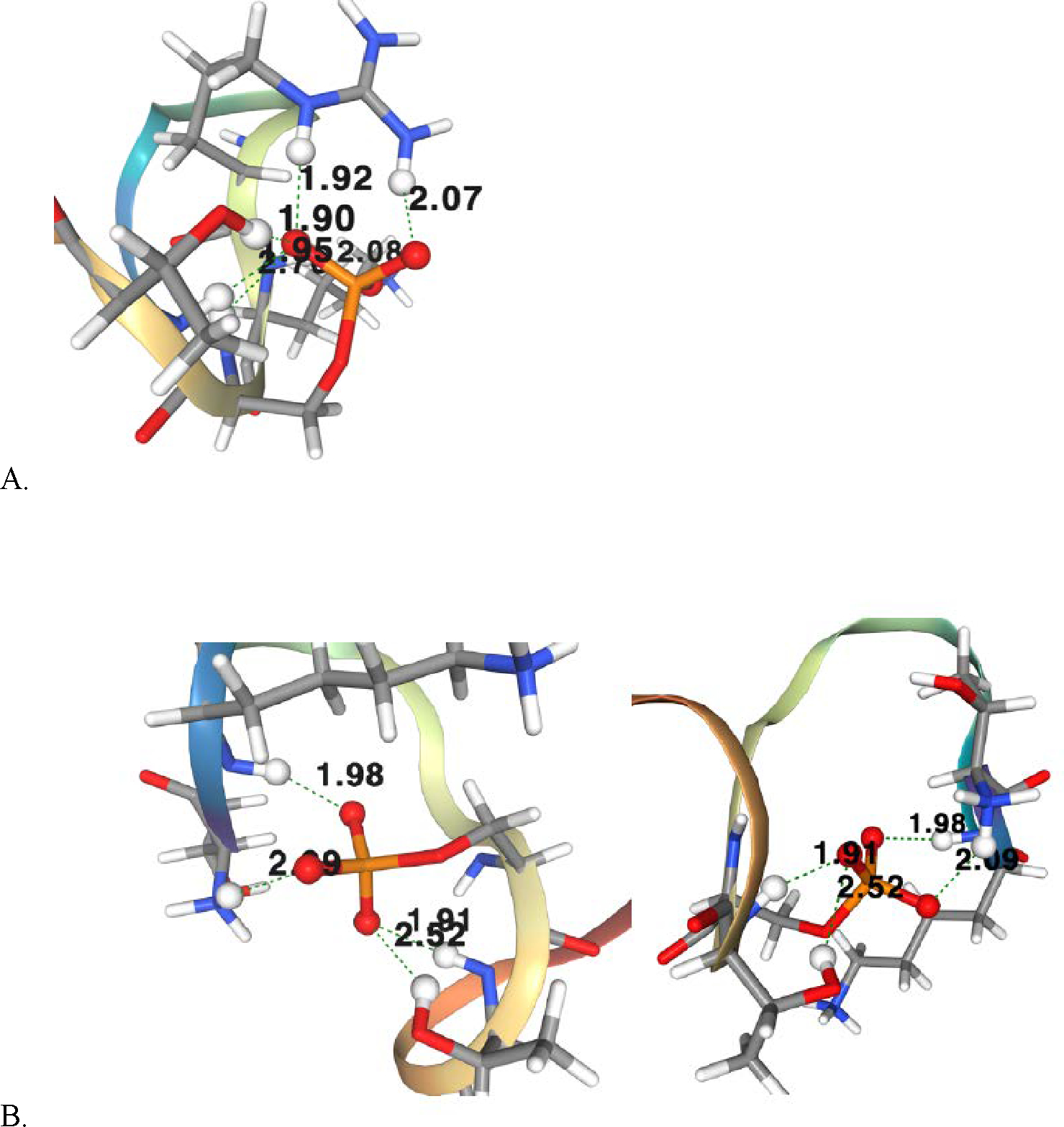
Comparision of histone H3 peptides 1-RKS(PO3H2), and 4-AAS(PO3H2), (PDB file 1kx5, Python, chain; ribbon). A) Phosphoserine-10 (red/orange), creates a salt bridge with arginine-8 (blue/white). The salt-bridge is denoted by dashed lines and by the the proximity of the atoms (1.92 Å and 2.07 Å , respectively). B) In the absence of positively charged arginine-8 and lysine-9, phosphoserine-10 (red/orange), maintains or creates interactions with other atoms. These include the threonine-11 residue, other lysine residues, and backbone amide protons found both nearby and more distant to phosphoserine-10.
To quantify these interactions in a comprehensive way, we computed the presence of intramolecular interactions with the phosphate group of phosphoserine-10 and all of the possible positively charged or partially-positively charged moieties in the peptide. Table 2 lists the phosphoserine-10 interactions (close contacts) that can take place with each constituent group. To more readily understand what is happening, we have separated these interactions into several categories: interactions with the positively charged guanidino group of (unmodified or symmetrically dimethylated) arginine-8, interactions with the positively charged ammonium groups of lysine residues and the N-terminus of the peptide, interactions with the hydroxyl groups of threonine residues, and interactions with the amide protons of the peptide backbone.
Table 2.
Probabilities of close contacts* between various residues/moieties and the phosphate group of serine-10 from REMD simulations.
| Peptide | Arg8/Arg(SDMA)8 | Lys9 | Other Lys/NH3+ | Thr11 | Local backbone | Distant backbone |
|---|---|---|---|---|---|---|
| 1 | 1.53 | 0.25 | 0.20 | 0.88 | 1.79 | 0.06 |
| 2 | 1.74 | n/a | 0.36 | 0.78 | 1.64 | 0.18 |
| 3 | n/a | 0.55 | 0.47 | 0.80 | 1.22 | 0.18 |
| 4 | n/a | n/a | 0.75 | 0.93 | 1.02 | 1.02 |
| 5 | 0.26 | 0.55 | 0.27 | 0.84 | 1.48 | 0.26 |
| 6 | 0.24 | n/a | 0.72 | 0.77 | 1.26 | 0.40 |
Close contacts are defined as those for which the heavy atom distance between an oxygen atom of the phosphate group and the heavy atom “donor” of a hydrogen atom on another residue/moiety are within 3.5 Å and have a bond angle between these two atoms of >135°. Values above 1.00 (100%) can be interpreted as more than one interaction occurring simultaneously. “Arg8/Arg(SDMA)8” represents the charged arginine-8 or symmetrically dimethylated arginine-8 side chain. “Lys9” represents the lysine-9 side chain, while “Other Lys/NH3+” counts interactions with the other Lys residue side chains and the N-terminus of the peptide. “Thr11” represents interactions with the threonine-11 side chain, specifically the hydroxyl group. Finally, “Local backbone” counts interactions with the backbone amide protons of residues 9–11, while “Distant backbone” encompasses the backbone amide protons of the other residues.
At a glance our results demonstrate that there are many intrapeptide interactions formed with the phosphate group of phosphoserine-10, but Table 2 indicates that a few trends stand out. Perhaps most noticeably, we find that the positively charged guanidino group of arginine-8 of peptide 1 is able to form more than one interaction at a time with the phosphate group of phosphoserine-10 (i.e., interactions that are sometimes bidentate), with the average interaction count of 1.53. When lysine-9 is replaced by an alanine in peptide 2-RASp, the average interaction count between these two residues increases further to 1.74. Conversely, when arginine-8 is symmetrically dimethylated, the average interaction count decreases to 0.24–0.26 in peptides 5-RSDMAKSp and 6-RSDMAASp. We also observe some interaction with lysine-9, but this is relatively infrequent (0.25) when arginine-8 is present (peptide 1-RKSp) and increases in frequency (0.55) only when arginine-8 is either replaced by an alanine (peptide 3-AKSp) or symmetrically dimethylated (peptide 5-RSDMAKSp). We see that the interaction with lysine-9 can be partially compensated for by interactions with other lysine residues or the N-terminus of the peptide when lysine-9 is replaced by an alanine residue. One interaction that remains relatively constant across all peptides, however, is that with the hydroxyl group of the threonine-11 sidechain (0.77–0.93). Finally, we observed considerable interaction between the phosphate group and the amide protons of the nearby backbone (residues 9–11), as well as amide protons that are more distant (all other residues). Most notably, the phosphate group of serine-10 has the greatest interaction with the distant backbone in the absence of arginine or lysine residues. In general, these distant moieties contribute a total of 1.40–2.04 interactions to the phosphate group. Together these data suggest that the phosphate group of phosphoserine-10 is not fully solvent-exposed and engages in a variety of intrapeptide interactions both when arginine-8 is present (and forms a salt bridge with this residue) and when arginine-8 is not present or is symmetrically demethylated (thereby preventing or hindering the formation of a salt bridge). These intrapeptide interactions may explain why the change in pKa of the phosphate group is not as large when the salt bridge with arginine-8 is present as compared to other studies (e.g., Kumar et al.).
CONCLUSION
How one PTM can alter another is the focus of intense investigation (Chen et al., 2020; Khoury, Baliban, & Floudas, 2011; Kirsch, Jensen, & Schwämmle, 2020). Moreover, the interplay between arginine methylation and serine phosphorylation has been identified in several proteins (Basso & Pennuto, 2015; Hsu et al., 2011; Smith et al., 2020). Using peptide substrates based on histone H3, we set out to understand the role of phosphoserine-10 on neighboring arginine-8. We focus on the RKS motif because interactions between unmodified arginine-8 and phosphorylated serine-10 affect protein binding (Papamokos et al., 2012). Building on this evidence, we carried out in vitro methylation reactions using peptides based on the RKS motif of histone H3. The fluorography and TLC data show that phosphoserine-10 will inhibit deposition of methylated groups (both ADMA and SDMA) to neighboring arginine-8.
We further investigated why methylation was inhibited in the presence of a phosphoserine through NMR and computational studies. We conclude that a salt bridge forms between phosphoserine-10 and arginine-8 within the RKS motif of histone H3, limiting interactions with other neighboring atoms. This was validated by comparing the pKa values of several peptides. For example, we expected peptide 2-RASp, which is lacking a lysine residue, to have a similar pKa value to that of peptide 1-RKSp. However, the average pKa values for these peptides were 5.55 and 5.72 respectively. This interaction is further confirmed when the arginine residue is completely eliminated (peptide 3-AKSp), with a pKa value of 5.83. The symmetrically dimethylated arginine peptides 5 and 6 represent the possible interaction between the two post-translational modifications: arginine methylation and serine phosphorylation. The average pKa values for peptide 5-RSDMAKSp and 6-RSDMAASp were determined to be 5.74 and 6.07, respectively. When we compare these values to each other, we conclude that the lysine residue is contributing to protein stability. We reason that when the arginine residue is either absent or symmetrically dimethylated, the contribution of the lysine residue becomes significant.
Our molecular dynamic simulation results suggest that the largest observed differences in pKa of the phosphate group would occur when comparing peptide 1-RKSp, to peptides 3-AKSp, and 4-AASp, which have no arginine residues, or to peptides 5-RSDMAKSp, and 6-RSDMAASp,which have symmetrically dimethylated arginine residues. We would expect to see smaller differences between peptides 1-RKSp and 2-RASp, peptides 3-AKSp, and 4-AASp, or peptides 5-RSDMAKSp, and 6-RSDMAASp. Indeed, the experimental data bear these predictions out quite well, with the sole exception of peptide 6-RSDMAASp which has a large difference in pKa from peptide 5-RSDMAKSp. We note, however, that the experimental pKa for this peptide has the largest uncertainty of all of the peptides examined. Additional computational studies may generate greater insights into the nature of these interactions.
Phosphorylation plays a central role in cellular regulation (Hunter, 2012). With respect to histone H3 serine 10, phosphorylation is associated with cell cycle progression and also increases in response to environmental stimuli (Bode & Dong, 2005; Duan, Chen, Costa, & Dai, 2008). Symmetric dimethylation of arginine-8 at histone H3 (H3R8(SDMA)) represses expression of tumor suppressor genes (Pal et al., 2004) (Dong et al., 2018) and is implicated in germ cell development in zebrafish (Zhu et al., 2019). We show that the addition of a charged phosphate group onto a serine residue will significantly change interactions with neighboring amino acid residues and cause a change in the conformation of the dynamic histone tail (Figure 4). By limiting its exposure and creating new contacts with neighboring residues, it will inhibit deposition of neighboring methyl groups, not through steric hindrance, but by forming intrapeptide interactions that most likely result in masking enzymatic recognition. We thus conclude that deposition of a methyl group in proximity of a phosphoserine is physically possible, but that the substrate landscape may be altered such that it is unrecognizable by the methyltransferase enzyme. Further work should be done to highlight the interplay between arginine methylation and serine phosphorylation and investigate whether the active site of PRMT5 and PRMTs in general, can accommodate an adjacent phosphate group so that they may recognize and methylate arginine substrates in the presence of phosphoserine.
Supplementary Material
Highlights.
Methylation of H3R8 is inhibited in the presence of phosphoserine-10.
A salt-bridge occurs between phosphoserine-10 and unmodified arginine-8.
Intrapeptide interactions with phosphoserine-10 were computationally modeled.
H3R8sdma and H3S10ph are not likely to coexist under physiological conditions.
Acknowledgements:
The authors thank S. G. Clarke at UCLA for providing the cDNA encoding the E. coli PRMT constructs and for providing insightful comments on this manuscript. Funding: This work was supported by the National Institutes of Health NIGMS SC2GM118202-01 Award. NIH MBRS-RISE M.S. to Ph.D. (GM61331) for J. A. Leal, M. Mendoza, A. Meza, and A. Mendiola and LSAMP Bridge to the Doctorate Grant (HRD-1602210) for Z. M. Estrada-Tobar.
ABBREVIATIONS
- H3K8(SDMA)
- PRMT1
- PRMT5
- ADMA
- MMA
- SDMA
- NMR
- TLC
- REMD
References
- Allfrey VG, Faulkner R, & Mirsky AE (1964). Acetylation and Methylation of Histones and their Possible Role in the Regulation of RNA Synthesis. Proc Natl Acad Sci U S A, 51, 786–794. doi: 10.1073/pnas.51.5.786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DE, Becktel WJ, & Dahlquist FW (1990). pH-induced denaturation of proteins: a single salt bridge contributes 3–5 kcal/mol to the free energy of folding of T4 lysozyme. Biochemistry, 29(9), 2403–2408. [DOI] [PubMed] [Google Scholar]
- Antonysamy S, Bonday Z, Campbell RM, Doyle B, Druzina Z, Gheyi T, . . . Emtage S. (2012). Crystal structure of the human PRMT5:MEP50 complex. Proc Natl Acad Sci U S A, 109(44), 17960–17965. doi: 10.1073/pnas.1209814109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso M, & Pennuto M (2015). Serine phosphorylation and arginine methylation at the crossroads to neurodegeneration. Exp Neurol, 271, 77–83. doi: 10.1016/j.expneurol.2015.05.003 [DOI] [PubMed] [Google Scholar]
- Bedford MT, & Clarke SG (2009). Protein arginine methylation in mammals: who, what, and why. Mol Cell, 33(1), 1–13. doi: 10.1016/j.molcel.2008.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode AM, & Dong Z (2005). Inducible covalent posttranslational modification of histone H3. Sci STKE, 2005(281), re4. doi: 10.1126/stke.2812005re4 [DOI] [PubMed] [Google Scholar]
- Bosshard HR, Marti DN, & Jelesarov I (2004). Protein stabilization by salt bridges: concepts, experimental approaches and clarification of some misunderstandings. J Mol Recognit, 17(1), 1–16. doi: 10.1002/jmr.657 [DOI] [PubMed] [Google Scholar]
- Cannizzaro CE, & Houk KN (2002). Magnitudes and chemical consequences of R(3)N(+)-C-H...O[double bond]C hydrogen bonding. J Am Chem Soc, 124(24), 7163–7169. doi: 10.1021/ja012417q [DOI] [PubMed] [Google Scholar]
- Case DA, Ben-Shalom IY, Brozell SR, Cerutti DS, Cheatham I, T.E., V. W. D Cruzeiro, . . . P. A Kollman. (2018). AMBER2018, University of California, San Francisco. [Google Scholar]
- Chen P, Zhuo Y, Tian S, Zhang T, Zhai G, Fan E, . . . Zhang, K. (2020). An Integrated Approach for Combinatorial Readout of Dual Histone Modifications by Epigenetic Tandem Domains. Analytical chemistry, 92(9), 6218–6223. doi: 10.1021/acs.analchem.9b05394 [DOI] [PubMed] [Google Scholar]
- Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, & Allis CD (2000). Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell, 5(6), 905–915. [DOI] [PubMed] [Google Scholar]
- Davey CA, Sargent DF, Luger K, Maeder AW, & Richmond TJ (2002). Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J Mol Biol, 319(5), 1097–1113. doi: 10.1016/S0022-2836(02)00386-8 [DOI] [PubMed] [Google Scholar]
- de Moreno MR, Smith JF, & Smith RV (1986). Mechanism studies of coomassie blue and silver staining of proteins. J Pharm Sci, 75(9), 907–911. [DOI] [PubMed] [Google Scholar]
- Debler EW, Jain K, Warmack RA, Feng Y, Clarke SG, Blobel G, & Stavropoulos P (2016). A glutamate/aspartate switch controls product specificity in a protein arginine methyltransferase. Proc Natl Acad Sci U S A, 113(8), 2068–2073. doi: 10.1073/pnas.1525783113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald JE, Kulp DW, & DeGrado WF (2011). Salt bridges: geometrically specific, designable interactions. Proteins, 79(3), 898–915. doi: 10.1002/prot.22927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F, Li Q, Yang C, Huo D, Wang X, Ai C, . . . Wu, X. (2018). PRMT2 links histone H3R8 asymmetric dimethylation to oncogenic activation and tumorigenesis of glioblastoma. Nat Commun, 9(1), 4552. doi: 10.1038/s41467-018-06968-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Q, Chen H, Costa M, & Dai W (2008). Phosphorylation of H3S10 blocks the access of H3K9 by specific antibodies and histone methyltransferase. Implication in regulating chromatin dynamics and epigenetic inheritance during mitosis. J Biol Chem, 283(48), 33585–33590. doi: 10.1074/jbc.M803312200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira de Freitas R, Eram MS, Szewczyk MM, Steuber H, Smil D, Wu H, . . . Schapira M. (2016). Discovery of a Potent Class I Protein Arginine Methyltransferase Fragment Inhibitor. J Med Chem, 59(3), 1176–1183. doi: 10.1021/acs.jmedchem.5b01772 [DOI] [PubMed] [Google Scholar]
- Fuhrmann J, Clancy KW, & Thompson PR (2015). Chemical biology of protein arginine modifications in epigenetic regulation. Chem Rev, 115(11), 5413–5461. doi: 10.1021/acs.chemrev.5b00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann R, Reichert I, Wachs WO, Zeppezauer M, & Kalbitzer HR (1994). 1H and 31P NMR spectroscopy of phosphorylated model peptides. Int J Pept Protein Res, 44(3), 193–198. doi: 10.1111/j.1399-3011.1994.tb00160.x [DOI] [PubMed] [Google Scholar]
- Hsu JM, Chen CT, Chou CK, Kuo HP, Li LY, Lin CY, . . . Hung, M. C. (2011). Crosstalk between Arg 1175 methylation and Tyr 1173 phosphorylation negatively modulates EGFR-mediated ERK activation. Nat Cell Biol, 13(2), 174–181. doi: 10.1038/ncb2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T (2012). Why nature chose phosphate to modify proteins. Philos Trans R Soc Lond B Biol Sci, 367(1602), 2513–2516. doi: 10.1098/rstb.2012.0013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun S, Lee KH, Han A, & Yu J (2011). An RNA aptamer that selectively recognizes symmetric dimethylation of arginine 8 in the histone H3 N-terminal peptide. Nucleic Acid Ther, 21(3), 157–163. doi: 10.1089/nat.2011.0300 [DOI] [PubMed] [Google Scholar]
- Ibuki Y, Toyooka T, Zhao X, & Yoshida I (2014). Cigarette sidestream smoke induces histone H3 phosphorylation via JNK and PI3K/Akt pathways, leading to the expression of proto-oncogenes. Carcinogenesis, 35(6), 1228–1237. doi: 10.1093/carcin/bgt492 [DOI] [PubMed] [Google Scholar]
- Jacques SL, Aquino KP, Gureasko J, Boriack-Sjodin PA, Porter Scott M, Copeland RA, & Riera TV (2016). CARM1 Preferentially Methylates H3R17 over H3R26 through a Random Kinetic Mechanism. Biochemistry, 55(11), 1635–1644. doi: 10.1021/acs.biochem.5b01071 [DOI] [PubMed] [Google Scholar]
- Jain K, Jin CY, & Clarke SG (2017). Epigenetic control via allosteric regulation of mammalian protein arginine methyltransferases. Proc Natl Acad Sci U S A, 114(38), 10101–10106. doi: 10.1073/pnas.1706978114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung IS, & Cheatham TE (2008). Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J Phys Chem B, 112(30), 9020–9041. doi: 10.1021/jp8001614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury GA, Baliban RC, & Floudas CA (2011). Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Sci Rep, 1. doi: 10.1038/srep00090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch R, Jensen ON, & Schwämmle V (2020). Visualization of the dynamics of histone modifications and their crosstalk using PTM-CrossTalkMapper. Methods. doi: 10.1016/j.ymeth.2020.01.012 [DOI] [PubMed] [Google Scholar]
- Kumar P, Chimenti MS, Pemble H, Schonichen A, Thompson O, Jacobson MP, & Wittmann T (2012). Multisite phosphorylation disrupts arginine-glutamate salt bridge networks required for binding of cytoplasmic linker-associated protein 2 (CLASP2) to end-binding protein 1 (EB1). J Biol Chem, 287(21), 17050–17064. doi: 10.1074/jbc.M111.316661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liehr S, & Chenault HK (1999). A comparison of the alpha-helix forming propensities and hydrogen bonding properties of serine phosphate and alpha-amino-gamma-phosphonobutyric acid. Bioorg Med Chem Lett, 9(18), 2759–2762. doi: 10.1016/s0960-894x(99)00469-2 [DOI] [PubMed] [Google Scholar]
- Lo WS, Trievel RC, Rojas JR, Duggan L, Hsu JY, Allis CD, . . . Berger, S. L. (2000). Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol Cell, 5(6), 917–926. doi: 10.1016/s1097-2765(00)80257-9 [DOI] [PubMed] [Google Scholar]
- Lobanov M. I. u., Bogatyreva NS, & Galzitskaia OV (2008). [Radius of gyration is indicator of compactness of protein structure]. Mol Biol (Mosk), 42(4), 701–706. [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, & Randall RJ (1951). Protein measurement with the Folin phenol reagent. J Biol Chem, 193(1), 265–275. [PubMed] [Google Scholar]
- Manning LR, & Manning JM (2018). Phosphorylation of Serine Induces Lysine p Ka Increases in Histone N-Termini and Signaling for Acetylation. Transcription Implications. Biochemistry, 57(50), 6816–6821. doi: 10.1021/acs.biochem.8b01040 [DOI] [PubMed] [Google Scholar]
- Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, & Sif S (2004). Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol, 24(21), 9630–9645. doi: 10.1128/MCB.24.21.9630-9645.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papamokos GV, Tziatzos G, Papageorgiou DG, Georgatos SD, Politou AS, & Kaxiras E (2012). Structural role of RKS motifs in chromatin interactions: a molecular dynamics study of HP1 bound to a variably modified histone tail. Biophys J, 102(8), 1926–1933. doi: 10.1016/j.bpj.2012.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z, Mizianty MJ, Xue B, Kurgan L, & Uversky VN (2012). More than just tails: intrinsic disorder in histone proteins. Mol Biosyst, 8(7), 1886–1901. doi: 10.1039/c2mb25102g [DOI] [PubMed] [Google Scholar]
- Roe DR, & Cheatham TE (2013). PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J Chem Theory Comput, 9(7), 3084–3095. doi: 10.1021/ct400341p [DOI] [PubMed] [Google Scholar]
- Salomon-Ferrer R, Götz AW, Poole D, Le Grand S, & Walker RC (2013). Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. J Chem Theory Comput, 9(9), 3878–3888. doi: 10.1021/ct400314y [DOI] [PubMed] [Google Scholar]
- Sarfo K, Moorhead GBG, & Turner RJ (2003). A novel procedure for separating small peptides on polyacrylamide gels. Letters in Peptide Science, 10(2), 127–133. doi:DOI 10.1023/B:LIPS.0000032383.40916.e2 [DOI] [Google Scholar]
- Schurter BT, Koh SS, Chen D, Bunick GJ, Harp JM, Hanson BL, . . . Aswad DW. (2001). Methylation of histone H3 by coactivator-associated arginine methyltransferase 1. Biochemistry, 40(19), 5747–5756. [DOI] [PubMed] [Google Scholar]
- Skinner JJ, Wang S, Lee J, Ong C, Sommese R, Sivaramakrishnan S, . . . Rosner MR. (2017). Conserved salt-bridge competition triggered by phosphorylation regulates the protein interactome. Proc Natl Acad Sci U S A, 114(51), 13453–13458. doi: 10.1073/pnas.1711543114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DL, Erce MA, Lai YW, Tomasetig F, Hart-Smith G, Hamey JJ, & Wilkins MR (2020). Crosstalk of Phosphorylation and Arginine Methylation in Disordered SRGG Repeats of Saccharomycescerevisiae Fibrillarin and Its Association with Nucleolar Localization. J Mol Biol, 432(2), 448–466. doi: 10.1016/j.jmb.2019.11.006 [DOI] [PubMed] [Google Scholar]
- Southall SM, Wong PS, Odho Z, Roe SM, & Wilson JR (2009). Structural basis for the requirement of additional factors for MLL1 SET domain activity and recognition of epigenetic marks. Mol Cell, 33(2), 181–191. doi: 10.1016/j.molcel.2008.12.029 [DOI] [PubMed] [Google Scholar]
- Stoppelman JP, Ng TT, Nerenberg PS, & Wang LP (2020). FB18: AMBER FB15-compatible parameters for phosphorylated amino acids, in preparation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Frankel A, Cook RJ, Kim S, Paik WK, Williams KR, . . . Herschman HR. (2000). PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J Biol Chem, 275(11), 7723–7730. [DOI] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, & Patel DJ (2007). How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol, 14(11), 1025–1040. doi: 10.1038/nsmb1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theillet FX, Smet-Nocca C, Liokatis S, Thongwichian R, Kosten J, Yoon MK, . . . Selenko, P. (2012). Cell signaling, post-translational protein modifications and NMR spectroscopy. Journal of Biomolecular Nmr, 54(3), 217–236. doi: 10.1007/s10858-012-9674-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson JH, Ullah S, Hansen PE, & Williamson MP (2009). Characterization of salt bridges to lysines in the protein G B1 domain. J Am Chem Soc, 131(13), 4674–4684. doi: 10.1021/ja808223p [DOI] [PubMed] [Google Scholar]
- Wang LP, Martinez TJ, & Pande VS (2014). Building Force Fields: An Automatic, Systematic, and Reproducible Approach. J Phys Chem Lett, 5(11), 1885–1891. doi: 10.1021/jz500737m [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LP, McKiernan KA, Gomes J, Beauchamp KA, Head-Gordon T, Rice JE, . . . Pande VS. (2017). Building a More Predictive Protein Force Field: A Systematic and Reproducible Route to AMBER-FB15. J Phys Chem B, 121(16), 4023–4039. doi: 10.1021/acs.jpcb.7b02320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wysocka J, Perlin JR, Leonelli L, Allis CD, & Coonrod SA (2004). Linking covalent histone modifications to epigenetics: the rigidity and plasticity of the marks. Cold Spring Harb Symp Quant Biol, 69, 161–169. doi: 10.1101/sqb.2004.69.161 [DOI] [PubMed] [Google Scholar]
- Williamson MP, Hounslow AM, Ford J, Fowler K, Hebditch M, & Hansen PE (2013). Detection of salt bridges to lysines in solution in barnase. Chem Commun (Camb), 49(84), 9824–9826. doi: 10.1039/c3cc45602a [DOI] [PubMed] [Google Scholar]
- Yesselman JD, Horowitz S, Brooks CL, & Trievel RC (2015). Frequent side chain methyl carbon-oxygen hydrogen bonding in proteins revealed by computational and stereochemical analysis of neutron structures. Proteins, 83(3), 403–410. doi: 10.1002/prot.24724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Zhang D, Liu X, Yu G, Cai X, Xu C, . . . Xiao W. (2019). Zebrafish prmt5 arginine methyltransferase is essential for germ cell development. Development, 146(20). doi: 10.1242/dev.179572 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


