Abstract
Calnexin is an endoplasmic reticulum (ER)-associated molecular chaperone proposed to promote folding and assembly of glycoproteins that traverse the secretory pathway in eukaryotic cells. In this study we examined if calnexin interacts with the ER-associated luminal (VP7) and transmembrane (NSP4) proteins of rotavirus. Only glycosylated NSP4 interacted with calnexin and did so in a time-dependent manner (half-life, 20 min). In vitro translation experiments programmed with gene 10 of rhesus rotavirus confirmed that calnexin recognizes only glycosylated NSP4. Castanospermine (a glucosidase I and II inhibitor) experiments established that calnexin associates only with partly deglucosylated (di- or monoglucosylated) NSP4. Furthermore, enzymatic removal of the remaining glucose residues on the N-linked glycan units was essential to disengage the NSP4-calnexin complex. Novel experiments with castanospermine revealed that glucose trimming and the calnexin-NSP4 interaction were not critical for the assembly of infectious virus.
Rotavirus, a segmented double-stranded RNA (dsRNA) virus, undergoes a unique maturation process in the endoplasmic reticulum (ER) (10, 11). The assembly process, which includes the translocation of subviral particles across the ER membrane and the retention of mature virus in the ER, has provided a system in which posttranslational events, such as folding, targeting, and retention, can be studied (2, 4, 24, 26–29, 35, 39–41). Of the 12 structural and nonstructural proteins of rotavirus, two have been in particular focus with regard to assembly and pathogenesis.
One of these is the VP7 outer capsid protein, which is a luminal and ER-associated polypeptide with only N-linked high-mannose oligosaccharide residues, for which the rhesus rotavirus (RRV) strain used in this study contains only a single glycosylation site (10, 23). Biochemical and morphological studies have established that calcium, an oxidizing milieu, and N-linked glycosylation are critical for the correct folding of VP7 (8, 28, 36, 41). It has also been shown that VP7 becomes endo-β-N-acetylglucosaminidase H resistant after brefeldin A treatment, a finding which suggests modifications by Golgi apparatus-associated enzymes (29).
NSP4 is a nonstructural glycoprotein that has been given significant attention in recent years. It is a novel type of trans-ER resident glycoprotein that functions not only as a receptor for subviral particles in the cytoplasm (2, 4) but also purportedly as a toxin capable of inducing diarrhea in mice and reorganizing Ca2+ in cells (3, 30, 43). NSP4 contains two N-linked high-mannose oligosaccharide residues that appear to be critical for the assembly of rotavirus (10, 34).
The mechanisms and structural motifs involved in the retention of NSP4 and VP7 in the ER are not yet fully recognized, nor have late events in the assembly process been identified in more detail. While 3 amino acids in the N terminus have been proposed to function as a retrieval or retention signal for VP7 (24), no information is yet available on how NSP4 remains associated with the ER. A critical role in the retention and unique viral assembly process may be played by components of the quality control system of the ER (14, 15). This system includes foldases and chaperones responsible for the prevention of protein aggregation, the retention of incorrect and incompletely folded glycoproteins in the ER, and the promotion of correct folding of polypeptides (9, 14, 17, 19, 21, 37). A key chaperone in this quality control family is calnexin, a trans-ER-associated type I membrane protein of 64 kDa that preferentially, but not exclusively, interacts with monoglucosylated glycoproteins (12, 16, 33). Calnexin has been found associated with folding and assembly intermediates of a wide array of membrane-associated glycoproteins. While the role of calnexin in the folding of proteins that traverse the secretory pathway has been explored (15, 25, 33, 42, 45), information is not yet available concerning the role of this lectin-like chaperone in the maturation of ER-resident proteins and its participation in the assembly of ER-associated virus, such as rotavirus.
In a recent study, we found that protein disulfide isomerase (PDI), a chaperone that catalyzes disulfide bond formation, interacted with glycosylated VP7 of rotavirus and that the binding was glycosylation dependent and prevented VP7 from aggregation (28). In this study, we found that calnexin interacted with glycosylated NSP4 in vivo and in vitro. Biochemical studies revealed that calnexin only interacted with glucose-trimmed NSP4. Furthermore, we found that the trimming of glucose residues on the N-linked glycan was not essential for viral infectivity.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
MA-104 cells were grown in Dulbecco’s modified Eagle’s minimal essential medium (Eagle’s MEM) supplemented with 10% fetal calf serum. RRV was obtained from infected MA-104 cells by freezing and thawing. The antibodies used in this study included monoclonal antibody M60, which recognizes a cross-reactive nonneutralizing epitope on VP7 (38) and which is dependent on correct disulfide bond formation (41); a polyclonal rabbit anti-NSP4 antiserum which recognizes amino acids 114 to 134 of NSP4; and a rabbit polyclonal anticalnexin (C-terminal) antibody.
Reagents.
Tunicamycin (TM) and protease inhibitors were purchased from Boehringer GmbH, Mannheim, Germany. Castanospermine (CST), N-ethylmaleimide (NEM), cycloheximide, and 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) were purchased from Sigma.
Rotavirus infection.
RRV was activated with 10 μg of trypsin per ml for 30 min at 37°C before inoculation of MA-104 cells. After 1 h of infection, the inoculum was replaced with serum-free Eagle’s MEM. Virus titers were determined by peroxidase staining as previously described (28, 29, 41).
DNA constructs.
To synthesize cDNA encoding the NSP4 gene of simian group A rotavirus (SA11), dsRNA was extracted with phenol-chloroform and purified with silica. The dsRNA was subsequently denatured with methyl mercuric hydroxide and converted to cDNA at 37°C with Moloney murine leukemia virus reverse transcriptase (Life Technologies) and a 3′ primer (5′-AATGAATTCCCCGGGACGGCAGCTCAACCT-3′; the underlined sequence indicates restriction sites for EcoRI and SmaI). A full-length cDNA clone corresponding to the open reading frame of gene 10 was produced by PCR with primers (5′-AATGAATTCCCCGGGATGGAAAAGCTTACC-3′ and 5′-AATGAATTCCCCGGGACGGCAGCTCAACCT-3′) and Pfu polymerase (Stratagene). After electrophoresis in 1% agarose and gel purification (PCR purification kit; Qiagen, Hilden, Germany), the blunt-ended PCR product (gene 10) was immediately ligated into SrfI-digested pCRScript (Stratagene). Positive clones in the T7 orientation were selected and used in an in vitro transcription-translation assay (TNT lysate system; Promega).
In vitro translation.
Plasmid pCRScript carrying gene 10 (0.5 μg) was subjected to in vitro coupled transcription-translation with the TNT system. The construct was translated both in the absence and in the presence of canine pancreatic microsomes (Promega) in accordance with the manufacturer’s instructions. Briefly, 25 μl of rabbit reticulocyte lysate, 2.5 μl of nuclease-treated microsomes, 0.5 μl of amino acids minus methionine, 0.02 μCi of [35S]methionine, 0.5 μl of T7 polymerase, 0.5 μl of RNasin, and 0.5 μg of gene 10 DNA-carrying plasmid were mixed, and the total volume was adjusted to 25 μl with H2O. After incubation of the mixture for 90 min at 30°C, the samples were lysed in lysis buffer (ice-cold 150 mM NaCl, 2% CHAPS, 50 mM HEPES, and protease inhibitors [6 μg of leupeptin per ml, 3 μg of antipain per ml, 1 μg of aprotinin per ml, and 0.1 mg of Pefabloc per ml]) and analyzed by immunoprecipitation.
Metabolic labeling of viral proteins.
To produce metabolically labeled cell lysates, MA-104 cells were infected with trypsin-activated RRV at a multiplicity of infection (MOI) of 10 as described previously (28, 29, 41). At 7 h postinfection (hpi), infected cells were starved for 1 h in methionine- and cysteine-free medium before being labeled for 5 min with 250 μCi of [35S]methionine-cysteine (Trans-label; Dupont). For chase experiments, cells were washed and incubated with Eagle’s MEM containing an excess of methionine (10 mM) and 1 mM cycloheximide. At the end of each radioactive pulse or after a chase period, cells were incubated with ice-cold phosphate-buffered saline (PBS) containing 40 mM NEM for 2 min to prevent disulfide bond rearrangement. Cells were then lysed in ice-cold lysis buffer, and the lysate was clarified of cell debris by centrifugation at 13,000 × g for 2 min in a microcentrifuge before use.
Treatment of cells with TM and CST.
To inhibit N-linked glycosylation, 2 μg of TM per ml was added to the media 3 h before pulse-labeling and maintained throughout the chase. To inhibit glucosidases I and II, 1 mM CST (13, 16, 45) was added to the media 1 h before or directly after pulse-labeling and maintained throughout the chase.
RIPA.
Radioimmunoprecipitation (RIPA) was performed essentially as described previously (29). Briefly, radiolabeled lysates (100 μl) were incubated with 10 μl of the desired antibody and 400 μl of RIPA buffer (150 mM NaCl, 50 mM HEPES, 0.5% CHAPS) overnight at 4°C. Fifty microliters of Staphylococcus aureus protein A–Sepharose CL-4B (Pharmacia, Uppsala, Sweden) was subsequently added to the mixture, which was then incubated for 2 h at 4°C. The immune complexes were washed three times with RIPA buffer, suspended in reducing sample buffer, and boiled for 5 min before separation by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE).
SDS-PAGE.
Polypeptide separation was performed by SDS-PAGE with a 4.5% stacking gel and a 10% separation gel as previously described (41). Electrophoresis was carried out at a constant voltage of 50 V at room temperature, followed by fixation with 10% glacial acetic acid and 35% methanol for 1 h at room temperature. Autoradiography was performed as previously described (41). Molecular mass standards included myosin (200 kDa), phosphorylase b (97 kDa), bovine serum albumin (69 kDa), ovalbumin (46 kDa), carbonic anhydrase (30 kDa), and lysozyme (14 kDa).
RESULTS AND DISCUSSION
A glycosylated 28-kDa protein of rotavirus interacts with calnexin in a time-dependent manner.
To analyze if calnexin interacts with rotavirus glycoproteins, mock- and RRV-infected cells were pulse-labeled for 5 min at 8 hpi and immediately harvested or chased in media containing an excess of methionine (10 mM) and 1 mM cycloheximide. At the end of each radioactive pulse-chase period, cells were briefly incubated with ice-cold PBS-NEM to prevent artificial disulfide bond formation (5, 13, 28), harvested in lysis buffer, and immunoprecipitated.
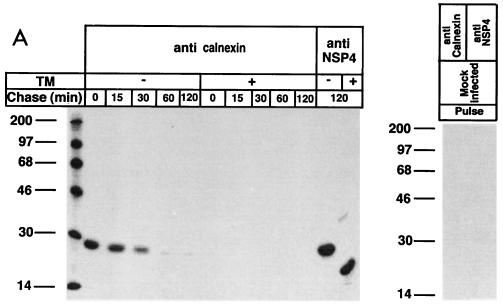
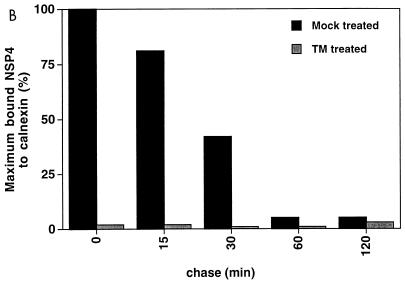
The results showed that a 28-kDa protein was coprecipitated with calnexin immediately following biosynthesis (Fig. 1A). Immunoprecipitation with an anti-NSP4-antibody suggested, based on migration similarities, that calnexin recognized NSP4 (Fig. 1A). The association was transient, and the 28-kDa protein was dissociated from calnexin over the course of 1 h (Fig. 1). Quantification by densitometry revealed that the half-life for the interaction was 20 min (Fig. 1B). These kinetics are thus similar to the observations made for several other glycoproteins: 5 min for influenza virus hemagglutinin (12), 10 min for G protein of vesicular stomatitis virus (12), 35 min for transferrin (32), and 15 min for gp160 of cytomegalovirus (45). It should be emphasized that most of these proteins are heavily glycosylated, in contrast to NSP4, which contains only two N-linked oligosaccharide units.
FIG. 1.
Association of calnexin with rotavirus proteins during protein maturation. Cells were mock infected or infected with RRV (MOI, 10). Cells infected with RRV were treated with TM or not (mock) treated. TM (2 μg/ml) was added to the medium of the monolayers at 5 hpi and was maintained throughout the experiment. At 7 hpi, cells were starved for 1 h in methionine- and cysteine-free medium and then metabolically labeled (250 μCi) for 5 min. To examine posttranslational processing, labeled proteins were chased with Eagle’s MEM supplemented with 1 mM cycloheximide and 10 mM methionine for the lengths of time indicated over the lanes. At the end of the chase, the monolayers were incubated with ice-cold PBS containing 40 mM NEM for 2 min. Cells were then harvested in lysis buffer. (A) The cell lysates from the pulse-chase experiments were immunoprecipitated with antibodies to calnexin and NSP4 and analyzed by reducing SDS-PAGE. Numbers at left are kilodaltons. (B) The amount of NSP4 bound to calnexin was measured by densitometry from the fluorograph shown in panel A, and the results are expressed as percentages of coprecipitated NSP4 and calnexin.
The fact that the calnexin-NSP4 interaction was transient and disappeared over time excludes nonspecific reactivity of an anticalnexin antibody. Furthermore, the fact that NSP4 could be immunoprecipitated at 120 min after biosynthesis by an anti-NSP4 antibody proves that NSP4 was not degraded during the chase (Fig. 1A). Therefore, the absence of a calnexin-NSP4 interaction at 60 min of chase (Fig. 1A) cannot be explained as protein degradation. The possibility that the precipitated protein was calnexin itself can also be ruled out, as calnexin has a molecular mass of 64 kDa (12). Furthermore, the anticalnexin antibody did not coprecipitate any 28-kDa protein from mock-infected cells. Thus, we conclude that calnexin interacted in a time-dependent manner with a virus-encoded protein of 28 kDa.
To investigate whether N-linked glycosylation was a prerequisite for binding of the 28-kDa protein to calnexin, TM (2 μg/ml) was used for treatment of RRV-infected cells at 5 hpi and included during the pulse and chase periods. Figure 1A shows that the inhibition of N-linked glycosylation by TM completely prevented the association of calnexin with the 28-kDa protein, suggesting that calnexin requires carbohydrates for interaction. This suggestion is also supported by other studies that reported a high specificity of calnexin for N-linked oligosaccharide units on newly synthesized proteins (12, 15, 45).
The most reasonable explanation for the observation that VP7, which possesses only a single N-linked glycan (23), was not recognized by calnexin (Fig. 1A) is that calnexin does not associate with proteins containing only a single N-linked glycan. This explanation is supported by a previous study reporting a requirement of two or more N-linked glycans for a calnexin interactions (6). Another possibility is that calnexin preferentially interacts with transmembrane glycoproteins (e.g., NSP4) rather then soluble ones (e.g., VP7), as suggested by Hebert and coworkers (18).
Calnexin recognizes in vitro-translated and glycosylated NSP4.
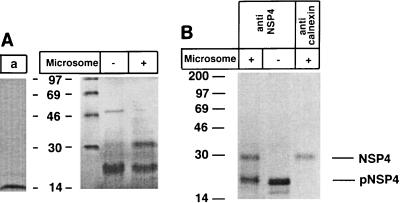
To firmly establish that calnexin recognized NSP4, an in vitro translation protocol with gene 10 encoding NSP4 of RRV was used. In the absence of canine microsomes, which do not support N-linked glycosylation, gene 10 was translated into a single protein with an estimated molecular mass of 20 kDa (Fig. 2A). However, in the presence of microsomes, a second, more slowly migrating polypeptide, corresponding in molecular mass to the glycosylated form of NSP4 (28 kDa), was produced. Immunoprecipitation with an anti-NSP4 antibody recognized both the glycosylated and the nonglycosylated forms of NSP4 (Fig. 2B). Immunoprecipitation experiments with an anticalnexin antibody revealed that only the glycosylated form of NSP4 interacted with calnexin (Fig. 2B). Taken together, these results (Fig. 1 and 2) indicate that glycosylated NSP4 interacts with calnexin in vivo and in vitro.
FIG. 2.
Calnexin recognizes in vitro-translated NSP4. (A) In vitro translation control with rabbit pancreatic microsomes (lane a) and with gene 10 in the presence or absence of rabbit pancreatic microsomes. (B) Immunoprecipitation of in vitro-translated NSP4 shown in panel A. Numbers at left are kilodaltons.
Association of calnexin with NSP4 is dependent on glucose trimming.
It was previously proposed that lectin-like chaperons, such as calnexin and calreticulin, have preferences for monoglycosylated N-linked oligosaccharides (16, 31, 44). Monoglycosylated oligosaccharides are generated by trimming of the two outermost glucose residues from the core oligosaccharide (22). Glucosidase I removes the outermost of the three glucose residues, and glucosidase II hydrolyzes the middle and the third glucose residues (1, 16). To determine whether glucose trimming was a prerequisite for the binding of NSP4 to calnexin, we used a glucosidase inhibitor, CST, that blocks the action of both glucosidases I and II (12, 13, 45). RRV-infected cells were mock or CST (1 mM) treated 1 h before a metabolic pulse and throughout the pulse-chase (Fig. 3). Immunoprecipitation experiments revealed that mock-treated NSP4 migrated faster than CST-treated NSP4 and that mock-treated NSP4 in cells chased for 60 min migrated faster than mock-treated NSP4 in cells lysed immediately after the pulse (Fig. 3), suggesting that glucose trimming of mock-treated NSP4 started cotranslationally and continued during the chase. Furthermore, inhibition of oligosaccharide processing of NSP4 by CST confirmed that glucosidases I and II are responsible for removal of the glucose residues on NSP4. These observations provide a biochemical explanation for a previously suggested model proposing that the N-linked glycan of NSP4 is trimmed within 60 to 90 min from Glc3Man9GlcNAc2 to Man8GlcNAc2 (20).
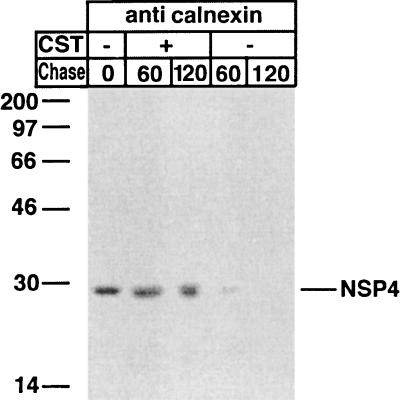
FIG. 3.
Effect of CST on the folding of NSP4. Cells were infected with RRV (MOI, 10), and at 7 hpi, monolayers were either not treated or treated with CST (1 mM) for 1 h before the pulse, during the pulse, and during the chase. Cells were starved for 1 h in methionine- and cysteine-free medium and then metabolically labeled (250 μCi) for 5 min. Labeled proteins were chased in the presence or absence of 1 mM CST in Eagle’s MEM supplemented with 1 mM cycloheximide and 10 mM methionine for the lengths of time indicated over the lanes. At the end of the chase, the monolayers were incubated with ice-cold PBS containing 40 mM NEM for 2 min. Cells were then harvested in lysis buffer. The cell lysates were immunoprecipitated with antibodies to calnexin and NSP4 and analyzed by reducing SDS-PAGE. Numbers at left are kilodaltons.
The results presented in Fig. 3 also show that inhibition of deglucosylation by CST prevented an association between calnexin and NSP4, suggesting that calnexin requires di- or monoglucosylated core glycans for an interaction. Others have also reported that treatment with CST prevents an interaction between secretory glycoproteins and calnexin (7, 13, 21, 45).
Dissociation of calnexin from NSP4 is dependent on glucose trimming by glucosidase II.
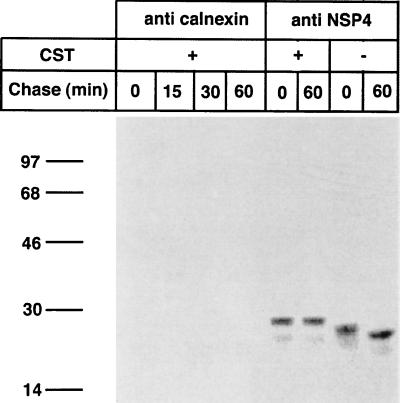
To further explore if the removal of the remaining (one or two) glucose residues on the N-linked glycans of NSP4 was required for dissociation from calnexin, RRV-infected cells were pulse-labeled without CST and chased for 60 and 120 min in the presence or absence of CST, followed by immunoprecipitation with an anticalnexin antibody (Fig. 4).
FIG. 4.
Dissociation of calnexin from NSP4. Cells were infected with RRV (MOI, 10). At 7 hpi, cells were starved for 1 h in methionine- and cysteine-free medium and then metabolically labeled (250 μCi) for 5 min. Labeled proteins were chased in the presence or absence of 1 mM CST in Eagle’s MEM supplemented with 1 mM cycloheximide and 10 mM methionine for the lengths of time indicated over the lanes. At the end of the chase, the monolayers were incubated with ice-cold PBS containing 40 mM NEM for 2 min. Cells were then harvested in lysis buffer. The cell lysates were immunoprecipitated with antibodies to calnexin and analyzed by reducing SDS-PAGE. Numbers at left are kilodaltons.
In contrast to mock-treated cells, in which >98% of initially bound NSP4 was dissociated from calnexin within 60 min (Fig. 4), no significant release of NSP4 from calnexin occurred within 60 min in CST-treated cells (Fig. 4). These observations led to the conclusion that enzymatic removal of the remaining second or/and third glucose residues of NSP4 by glucosidase II is essential to dissociate the NSP4-calnexin complex. Our observations support a previous proposal that glucosidase II serves a dual function: it removes the second glucose residue on N-linked glycans to allow glycoproteins to attach to calnexin, and it removes the innermost glucose residue to allow the dissociation of calnexin from its substrate (12, 16).
Glucose trimming and calnexin interaction are not critical for viral infectivity.
The fact that CST could prevent the association as well as the dissociation of calnexin from NSP4 led us to investigate if this interaction was indeed essential from a viral infectivity point of view. Furthermore, we were also interested in establishing whether glucose trimming of VP7 and NSP4 by glucosidases I and II was critical for viral infectivity. To address these questions, which to our knowledge have not yet been raised (13, 16, 45), RRV-infected cells were mock treated or treated with 2 mM CST at 1 hpi. To maintain a constant CST concentration, fresh CST was added to the medium every 4 h. At 8 or 18 hpi, the CST- and mock-treated infected cells were frozen and thawed twice, and progeny virus titers were determined. The results showed no significant reduction in progeny virus yield between CST-treated (2 × 106 and 3 × 109 PFU) and mock-treated (3 × 106 and 4 × 109 PFU) cells after 8 and 18 hpi, respectively. Results were obtained from three separate experiments. We conclude that glucosidase I and II trimming is not critical for the assembly of infectious virus. These results also show that the binding of calnexin to NSP4 is not required for the assembly process leading to infectious virus. A possible explanation for these observations is that glucose trimming has limited conformational effects on proteins with few glycans, such as NSP4, and more serious effects on export proteins, which usually are more heavily glycosylated.
In summary, this study, together with our recent report (28), shows that molecular chaperones such as PDI and calnexin interact in a time-dependent manner with rotavirus proteins during protein and virus maturation. We believe that this new information will contribute to a better understanding of the unique maturation process for rotavirus.
ACKNOWLEDGMENTS
This project received financial support from the Swedish Medical Council (grant K97-06X-10392-05A) and the European Community (grant ERBIC18CT960027).
We are grateful to Harry Greenberg for monoclonal antibody M60 and Ralf Pettersson for the anticalnexin antibody.
REFERENCES
- 1.Atkinson P H, Lee J T. Co-translational excision of alpha-glucose and alpha-mannose in nascent vesicular stomatitis virus G protein. J Cell Biol. 1984;98:2245–2249. doi: 10.1083/jcb.98.6.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Au K S, Chan W K, Burns J W, Estes M K. Receptor activity of rotavirus nonstructural glycoprotein NS28. J Virol. 1989;63:4553–4562. doi: 10.1128/jvi.63.11.4553-4562.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ball J M, Tian P, Zeng C Q, Morris A P, Estes M K. Age-dependent diarrhea induced by a rotaviral nonstructural glycoprotein. Science. 1996;272:101–104. doi: 10.1126/science.272.5258.101. [DOI] [PubMed] [Google Scholar]
- 4.Bergmann C C, Maass D, Poruchynsky M S, Atkinson P H, Bellamy A R. Topology of the non-structural rotavirus receptor glycoprotein NS28 in the rough endoplasmic reticulum. EMBO J. 1989;8:1695–1703. doi: 10.1002/j.1460-2075.1989.tb03561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braakman I, Helenius J, Helenius A. Manipulating disulfide bond formation and protein folding in the endoplasmic reticulum. EMBO J. 1992;11:1717–1722. doi: 10.1002/j.1460-2075.1992.tb05223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cannon K S, Hebert D N, Helenius A. Glycan-dependent and -independent association of vesicular stomatitis virus G protein with calnexin. J Biol Chem. 1996;271:14280–14284. doi: 10.1074/jbc.271.24.14280. [DOI] [PubMed] [Google Scholar]
- 7.Chen W, Helenius J, Braakman I, Helenius A. Cotranslational folding and calnexin binding during glycoprotein synthesis. Proc Natl Acad Sci USA. 1995;92:6229–6233. doi: 10.1073/pnas.92.14.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dormitzer P R, Greenberg H B. Calcium chelation induces a conformational change in recombinant herpes simplex virus-1-expressed rotavirus VP7. Virology. 1992;189:828–832. doi: 10.1016/0042-6822(92)90616-w. [DOI] [PubMed] [Google Scholar]
- 9.Earl P L, Moss B, Doms R W. Folding, interaction with GRP78-BiP, assembly, and transport of the human immunodeficiency virus type 1 envelope protein. J Virol. 1991;65:2047–2055. doi: 10.1128/jvi.65.4.2047-2055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Estes M K, Cohen J. Rotavirus gene structure and function. Microbiol Rev. 1989;53:410–449. doi: 10.1128/mr.53.4.410-449.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Estes M K, Palmer E L, Obijeski J F. Rotaviruses: a review. Curr Top Microbiol Immunol. 1983;105:123–184. doi: 10.1007/978-3-642-69159-1_3. [DOI] [PubMed] [Google Scholar]
- 12.Hammond C, Braakman I, Helenius A. Role of N-linked oligosaccharide recognition, glucose trimming, and calnexin in glycoprotein folding and quality control. Proc Natl Acad Sci USA. 1994;91:913–917. doi: 10.1073/pnas.91.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammond C, Helenius A. Folding of VSV G protein: sequential interaction with BiP and calnexin. Science. 1994;266:456–458. doi: 10.1126/science.7939687. [DOI] [PubMed] [Google Scholar]
- 14.Hammond C, Helenius A. Quality control in the secretory pathway: retention of a misfolded viral membrane glycoprotein involves cycling between the ER, intermediate compartment, and Golgi apparatus. J Cell Biol. 1994;126:41–52. doi: 10.1083/jcb.126.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hebert D N, Foellmer B, Helenius A. Calnexin and calreticulin promote folding, delay oligomerization and suppress degradation of influenza hemagglutinin in microsomes. EMBO J. 1996;15:2961–2968. [PMC free article] [PubMed] [Google Scholar]
- 16.Hebert D N, Foellmer B, Helenius A. Glucose trimming and reglucosylation determine glycoprotein association with calnexin in the endoplasmic reticulum. Cell. 1995;81:425–433. doi: 10.1016/0092-8674(95)90395-x. [DOI] [PubMed] [Google Scholar]
- 17.Hebert D N, Simons J F, Peterson J R, Helenius A. Calnexin, calreticulin, and Bip/Kar2p in protein folding. Cold Spring Harbor Symp Quant Biol. 1995;60:405–415. doi: 10.1101/sqb.1995.060.01.045. [DOI] [PubMed] [Google Scholar]
- 18.Hebert D N, Zhang J X, Chen W, Foellmer B, Helenius A. The number and location of glycans on influenza hemagglutinin determine folding and association with calnexin and calreticulin. J Cell Biol. 1997;139:613–623. doi: 10.1083/jcb.139.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helenius A. How N-linked oligosaccharides affect glycoprotein folding in the endoplasmic reticulum. Mol Biol Cell. 1994;5:253–265. doi: 10.1091/mbc.5.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kabcenell A K, Atkinson P H. Processing of the rough endoplasmic reticulum membrane glycoproteins of rotavirus SA11. J Cell Biol. 1985;101:1270–1280. doi: 10.1083/jcb.101.4.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim P S, Arvan P. Calnexin and BiP act as sequential molecular chaperones during thyroglobulin folding in the endoplasmic reticulum. J Cell Biol. 1995;128:29–38. doi: 10.1083/jcb.128.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 23.Mackow E R, Shaw R D, Matsui S M, Vo P T, Benfield D A, Greenberg H B. Characterization of homotypic and heterotypic VP7 neutralization sites of rhesus rotavirus. Virology. 1988;165:511–517. doi: 10.1016/0042-6822(88)90595-8. . (Erratum, 167:660.) [DOI] [PubMed] [Google Scholar]
- 24.Maass D R, Atkinson P H. Retention by the endoplasmic reticulum of rotavirus VP7 is controlled by three adjacent amino-terminal residues. J Virol. 1994;68:366–378. doi: 10.1128/jvi.68.1.366-378.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mathieu M E, Grigera P R, Helenius A, Wagner R R. Folding, unfolding, and refolding of the vesicular stomatitis virus glycoprotein. Biochemistry. 1996;35:4084–4093. doi: 10.1021/bi952924i. [DOI] [PubMed] [Google Scholar]
- 26.Meyer J C, Bergmann C C, Bellamy A R. Interaction of rotavirus cores with the nonstructural glycoprotein NS28. Virology. 1989;171:98–107. doi: 10.1016/0042-6822(89)90515-1. [DOI] [PubMed] [Google Scholar]
- 27.Michelangeli F, Liprandi F, Chemello M E, Ciarlet M, Ruiz M C. Selective depletion of stored calcium by thapsigargin blocks rotavirus maturation but not the cytopathic effect. J Virol. 1995;69:3838–3847. doi: 10.1128/jvi.69.6.3838-3847.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mirazimi A, Svensson L. Carbohydrates facilitate correct disulfide bond formation and folding of rotavirus VP7. J Virol. 1998;72:3887–3892. doi: 10.1128/jvi.72.5.3887-3892.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mirazimi A, von Bonsdorff C H, Svensson L. Effect of brefeldin A on rotavirus assembly and oligosaccharide processing. Virology. 1996;217:554–563. doi: 10.1006/viro.1996.0150. [DOI] [PubMed] [Google Scholar]
- 30.Newton K, Meyer J C, Bellamy A R, Taylor J A. Rotavirus nonstructural glycoprotein NSP4 alters plasma membrane permeability in mammalian cells. J Virol. 1997;71:9458–9465. doi: 10.1128/jvi.71.12.9458-9465.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ora A, Helenius A. Calnexin fails to associate with substrate proteins in glucosidase-deficient cell lines. J Biol Chem. 1995;270:26060–26062. doi: 10.1074/jbc.270.44.26060. [DOI] [PubMed] [Google Scholar]
- 32.Ou W J, Cameron P H, Thomas D Y, Bergeron J J. Association of folding intermediates of glycoproteins with calnexin during protein maturation. Nature. 1993;364:771–776. doi: 10.1038/364771a0. [DOI] [PubMed] [Google Scholar]
- 33.Peterson J R, Ora A, Van P N, Helenius A. Transient, lectin-like association of calreticulin with folding intermediates of cellular and viral glycoproteins. Mol Biol Cell. 1995;6:1173–1184. doi: 10.1091/mbc.6.9.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petrie B L. Biological activity of rotavirus particles lacking glycosylated proteins. In: Compans R W, Bishop D H L, editors. Double-stranded RNA virus. New York, N.Y: Elsevier Science Publishing, Inc.; 1983. pp. 145–156. [Google Scholar]
- 35.Poruchynsky M S, Tyndall C, Both G W, Sato F, Bellamy A R, Atkinson P H. Deletions into an NH2-terminal hydrophobic domain result in secretion of rotavirus VP7, a resident endoplasmic reticulum membrane glycoprotein. J Cell Biol. 1985;101:2199–2209. doi: 10.1083/jcb.101.6.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruiz M C, Charpilienne A, Liprandi F, Gajardo R, Michelangeli F, Cohen J. The concentration of Ca2+ that solubilizes outer capsid proteins from rotavirus particles is dependent on the strain. J Virol. 1996;70:4877–4883. doi: 10.1128/jvi.70.8.4877-4883.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruoppolo M, Freedman R B. Refolding by disulfide isomerization: the mixed disulfide between ribonuclease T1 and glutathione as a model refolding substrate. Biochemistry. 1995;34:9380–9388. doi: 10.1021/bi00029a014. [DOI] [PubMed] [Google Scholar]
- 38.Shaw R D, Vo P T, Offit P A, Coulson B S, Greenberg H B. Antigenic mapping of the surface proteins of rhesus rotavirus. Virology. 1986;155:434–451. doi: 10.1016/0042-6822(86)90205-9. [DOI] [PubMed] [Google Scholar]
- 39.Stirzaker S C, Both G W. The signal peptide of the rotavirus glycoprotein VP7 is essential for its retention in the ER as an integral membrane protein. Cell. 1989;56:741–747. doi: 10.1016/0092-8674(89)90677-6. [DOI] [PubMed] [Google Scholar]
- 40.Stirzaker S C, Whitfeld P L, Christie D L, Bellamy A R, Both G W. Processing of rotavirus glycoprotein VP7: implications for the retention of the protein in the endoplasmic reticulum. J Cell Biol. 1987;105:2897–2903. doi: 10.1083/jcb.105.6.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Svensson L, Dormitzer P R, von Bonsdorff C H, Maunula L, Greenberg H B. Intracellular manipulation of disulfide bond formation in rotavirus proteins during assembly. J Virol. 1994;68:5204–5215. doi: 10.1128/jvi.68.8.5204-5215.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tatu U, Hammond C, Helenius A. Folding and oligomerization of influenza hemagglutinin in the ER and the intermediate compartment. EMBO J. 1995;14:1340–1348. doi: 10.1002/j.1460-2075.1995.tb07120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tian P, Ball J M, Zeng C Q, Estes M K. The rotavirus nonstructural glycoprotein NSP4 possesses membrane destabilization activity. J Virol. 1996;70:6973–6981. doi: 10.1128/jvi.70.10.6973-6981.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ware F E, Vassilakos A, Peterson P A, Jackson M R, Lehrman M A, Williams D B. The molecular chaperone calnexin binds Glc1Man9GlcNAc2 oligosaccharide as an initial step in recognizing unfolded glycoproteins. J Biol Chem. 1995;270:4697–4704. doi: 10.1074/jbc.270.9.4697. [DOI] [PubMed] [Google Scholar]
- 45.Yamashita Y, Shimokata K, Mizuno S, Daikoku T, Tsurumi T, Nishiyama Y. Calnexin acts as a molecular chaperone during the folding of glycoprotein B of human cytomegalovirus. J Virol. 1996;70:2237–2246. doi: 10.1128/jvi.70.4.2237-2246.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]