Abstract
PD-L1 is a member of the B7 family co-inhibitory molecules and plays a critical role in tumor immune escape. In this study, we found a polymorphism rs10815225 in the PD-L1 promoter region was significantly associated with the occurrence of gastric cancer. The GG homozygous frequency was higher in the cancer patients than that in the precancerous lesions, which was higher than that in the health controls. This polymorphism locates in the binding-site of Sp1 transcription factor (SP1). The expression level of PD-L1 mRNA in the GG homozygous cancer patients was apparently higher than that in the GC heterozygotes. Luciferase reporter results showed that SP1 bonded to rs10815225 G-allelic PD-L1 promoter instead of C-allelic. Upregulation and knockdown of SP1 resulted in elevation and attenuation of PD-L1 in SGC-7901 cells, respectively. The chromatin immunoprecipitation results further confirmed the binding of SP1 to the promoter of PD-L1. Additionally, rs10815225 was found to be in disequilibrium with a functional polymorphism rs4143815 in the PD-L1 3′-UTR, and the haplotypes of these two polymorphisms were also markedly related to gastric cancer risk. These results revealed a novel mechanism underlying genetic polymorphisms influencing PD-L1 expression modify gastric cancer susceptibility.
Electronic supplementary material
The online version of this article (doi:10.1007/s00262-016-1936-0) contains supplementary material, which is available to authorized users.
Keywords: Gastric cancer, PD-L1, Polymorphism, SP1
Introduction
Gastric cancer is one of the most common malignant tumors in digestive tract, with estimated 951,600 new cancer cases and 723,100 deaths occurred in 2012 in the whole world, as well as 679,100 new cancer cases and 498,000 deaths in China [1, 2]. Gastric cancer development is a multi-step and multi-stage process, including chronic atrophic gastritis, high-grade intraepithelial neoplasia, malignant adenocarcinoma, and metastasis. Presently, the main treatment for gastric cancer is still surgery, which is benefit for the onset cancer patients but not for the advanced cancer patients. Although chemotherapy and radiation therapy can partly improve the survival rate of advanced cancer patients, the prognosis is still very poor [3]. Therefore, researches on the molecular mechanisms underlying the development of gastric cancer are essential to discover new therapy targets and/or biomarkers for early diagnosis.
PD-L1 (B7-H1, CD274), a key member of the B7 co-inhibitory molecules generally expresses on activated cells including T cells, B cells, monocytes/macrophages, DCs, and the cells derived from APC [4]. PD-L1 has been confirmed to negatively regulate immune response by interacting with its receptor PD-1 on T cells and consequently inhibiting T cell activation and proliferation [5]. PD-L1 was also found to contribute to cancer immune evasion such that numerous human cancer types have shown overexpression of PD-L1 [5], such as gastric cancer, esophageal cancer, pancreatic cancer, and other human gastrointestinal tumors [4]. The abnormal expression of PD-L1 firmly links to the development of cancers [6, 7] and the prognosis of patients [8]. Reorganizing the regulatory mechanism for PD-L1 overexpression in cancer microenvironment may provide novel insights to cancer immune evasion and potential new strategies for treating cancers [9–11].
We previously confirmed a wide expression of PD-L1 in gastric cancer, which was significantly related to the clinicopathological features including tumor size, depth of invasion, lymph node metastasis, and prognosis of patients [12–14]. A somatic mutation at a naturally occurring polymorphism locus rs4143815 in the 3′-UTR of PD-L1 gene contributed to the elevated PD-L1 protein expression in gastric cancer by disrupting the interaction between PD-L1 mRNA and miR-570 [14, 15]. The disruption of PD-L1 3′-UTR was further confirmed to invariably lead to a marked elevation of aberrant PD-L1 transcripts [16].
To elucidate the regulatory role of genetic variations of PD-L1 gene in its expression in gastric cancer, as well as the clinical significance of the genetic variations, we detected the genetic variations in the promoter of PD-L1 gene in gastric cancer patients and health controls. Statistical analysis results showed that a polymorphism rs10815225 was significantly related to the occurrence and development of gastric cancer. This polymorphism offered a binding-site for transcriptional factor SP1 on the promoter of PD-L1 gene, leading to upregulation of PD-L1 mRNA and protein in gastric cancer, and consequently cancer immune escaping. These findings revealed a novel mechanism for PD-L1 expression mediated by a polymorphism and a transcriptional factor, which will likely contribute to the immune escape of gastric cancer via co-inhibitor molecule PD-L1, and may offer a potential new strategy for treating gastric cancer.
Materials and methods
Subjects
This study was approved by the Institutional Review Board of Soochow University. A total of 350 gastric cancer patients, 156 precancerous patients, and 500 health controls were recruited from the First Affiliated Hospital of Soochow University between May 2012 and December 2014. The written informed consents and blood samples were collected from the participants according to the standard procedures. All participants were genetically unrelated ethnic Han Chinese in Suzhou. The gastric cancer patients and precancerous patients were pathologically diagnosed by two pathologists. The cancer patients did not receive preoperative chemotherapy or radiotherapy before blood collection and surgery. Forty-eight pairs of fresh gastric cancer tissues and distant normal tissues (>5 cm away from the edge of cancer) were collected from the cancer patients. The patients with previous cancer or metastasizing cancer from other origins were not included in this study. The clinicopathological features including tumor location, tumor diameter, tumor area, histological type, differentiation grade, depth of infiltration, tumor markers, lymph node metastasis, distant metastasis, and tumor-node-metastases (TNM) stage were obtained from the medical records of gastric cancer patients. Histological subtypes were classified according to the American Joint Commission for Cancer Staging in 2002. The precancerous patients include 16 gastritis patients, 88 gastric polyp patients, 9 gastric ulcer patients, 35 intraepithelial neoplasia patients, and 8 adenoma patients.
Extraction of genomic DNA
The whole blood samples were collected in EDTA-Vacutainer® tubes (BD Franklin Lakes, NJ). The total genomic DNAs were extracted from the whole blood using the phenol/chloroform method. The purity and concentration of the extracted DNA were determined by NanoDrop 2000 UV–VIS Spectrophotometer.
Selection and genotyping of polymorphisms in the PD-L1 gene
Firstly, the genetic variations in the PD-L1 gene promoter were searched from the NCBI dbSNP BUILED 129 (http://www.ncbi.nlm.nih.gov/SNP) and ENSEMBL v58 (http://www.ensembl.org/). The polymorphisms with minor allele frequency (MAF) of not less than 10% were investigated in this study. TFSEARCH (http://mbs.cbrc.jp/research/db/TFSEARCH.html) and MOTIF (http://www.genome.jp/tools/motif/) were used to predict potential binding-sites for transcription factors in the PD-L1 gene promoter. Finally, the DNA fragments containing the polymorphisms were amplified using the primers listed in Supplementary Table 1. The genotypes were determined by using sequencing technology (GeneWiz, Suzhou, China). The short tandem repeat (STR) analyses of the genomic DNAs were performed on 3500 DNA Genetic Analyzer (ABI).
Table 1.
Association of genotypes with the occurrence of gastric cancer
| SNP | Model | Genotype | Patient (n, %) | Control (n, %) | OR (95% CI) | P value | HWE |
|---|---|---|---|---|---|---|---|
| rs10815225 | Codominant | G/G | 310 (88.6) | 411 (82.2) | 1 | 0.0033 | 0.09 |
| G/C | 38 (10.9) | 89 (17.8) | 1.77 (1.18–2.66) | ||||
| C/C | 2 (0.6) | 0 (0) | 0.00 (0.00-NA) | ||||
| Dominant | G/G | 310 (88.6) | 411 (82.2) | 1 | 0.0098 | ||
| G/C–C/C | 40 (11.4) | 89 (17.8) | 1.68 (1.12–2.51) | ||||
| Recessive | G/G–G/C | 348 (99.4) | 500 (100) | 1 | 0.059 | ||
| C/C | 2 (0.6) | 0 (0) | 0.00 (0.00-NA) | ||||
| Overdominant | G/G–C/C | 312 (89.1) | 411 (82.2) | 1 | 0.0045 | ||
| G/C | 38 (10.9) | 89 (17.8) | 1.78 (1.18–2.67) | ||||
| rs4143815 | Codominant | G/G | 123 (35.5) | 117 (23.4) | 1 | <0.0001 | 0.08 |
| G/C | 153 (44.2) | 223 (44.6) | 1.53 (1.11–2.12) | ||||
| C/C | 70 (20.2) | 160 (32) | 2.40 (1.65–3.51) | ||||
| Dominant | G/G | 123 (35.5) | 117 (23.4) | 1 | 1.00E−04 | ||
| G/C–C/C | 223 (64.5) | 383 (76.6) | 1.81 (1.33–2.44) | ||||
| Recessive | G/G–G/C | 276 (79.8) | 340 (68) | 1 | 1.00E−04 | ||
| C/C | 70 (20.2) | 160 (32) | 1.86 (1.34–2.56) | ||||
| Overdominant | G/G–C/C | 193 (55.8) | 277 (55.4) | 1 | 0.91 | ||
| G/C | 153 (44.2) | 223 (44.6) | 1.02 (0.77–1.34) |
Significant results are shown in bold (P < 0.05)
Quantitative real-time polymerase chain reaction
Total RNAs from gastric cancer tissues and normal tissues were extracted using Trizol methods and subjected to reverse transcription with NxGen M-MuLV reverse transcriptase (MBI) and scramble primers (GeneWiz, Suzhou, China) for PD-L1, SP1, and GAPDH. Quantitative PCR (qPCR) was conducted in 20-μl reaction contained 400 nM of each primer (Supplementary Table 1) and quantitative RT-PCR master mix (Bio-Rad). The qPCR was conducted on the CFX96 TouchTM real-time PCR system (Bio-Rad). The relative expression was calculated by using the comparative threshold cycle (Ct) method. A ≥30 Ct value was interpreted as amplification too low to quantify.
Western blotting
The total proteins were prepared using RIPA buffer (Beyotime, Shanghai, China) supplemented with the complete protease inhibitor cocktail on ice for 30 min. Then, the cell lysates were centrifuged at 4 °C, 16,000 g for 15 min. The concentration of protein in the supernatant was measured by using the Pierce BCA Protein Assay Kit (Thermo). Twenty micrograms of protein was separated on 10% SDS-PAGE gel and electro-transferred onto PVDF membranes. The membranes were incubated with PD-L1 or GAPDH rabbit polyclonal antibodies (Santa Cruz Biotech.) and subsequently with a peroxidase goat anti-rabbit IgG (Santa Cruz Biotech.). The membranes were then incubated with Clarity Western ECL substrates (Merck Millipore) and visualized with the ChemiDocTM MP Imaging System (Bio-Rad).
Luciferase reporter assays
The construction of PD-L1/promoter/pGL-3 plasmids and luciferase reporter assays was performed as before [17]. Briefly, the DNA fragment containing rs10815225 G-allelic PD-L1 promoter was amplified from human genomic DNA using primers listed in Supplementary Table 1. The PCR products were cloned into the pGL-3 Basic vector (Promega) by using endonucleases KpnI and Xho1 (MBI). Positive clones were confirmed by PCR, restriction enzymes digestion, and DNA sequencing methods (GeneWiz, Suzhou, China). Then, the rs10815225 C-allelic PD-L1/promoter/pGL-3 plasmid was constructed using site-mutation reaction with the primers listed in Supplementary Table 1. For luciferase reporter analysis, the pGL3 constructs only or with SP1/CMV expression vector (kindly offered by professor Robert Tjian from University of California) were co-transfected into SGC-7901 cells (Type Culture Collection of Chinese Academy of Sciences, Shanghai, China) by using lipofectamine 2000 (Invitrogen). A pRL-TK plasmid (Promega) was used as a normalizing control. Transfection of pGL3-control vector or lipofectamine 2000 alone was used as negative control. 24 h later, the cells were collected and lysed for luciferase activity analysis using the dual-luciferase reporter assay system (Promega).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed using a ChIP kit for cell line samples (Millipore). Briefly, the cross-linked chromatin DNA was sonicated into 200–500-bp fragments. Then, the chromatin was immunoprecipitated using an anti-SP1 antibody (Santa Cruz Biotech). Normal mouse IgG was used as the negative control. Quantification of the immunoprecipitated DNA was performed using qPCR with SYBR Green Mix (TaKaRa) and the primers listed in Supplementary Table 1. The ChIP data were calculated as a ratio relative to the input DNA.
Statistical analysis
The association of alleles and genotypes of the polymorphisms with the occurrence and the clinicopathological features of diseases were assessed by using Chi-square test. From that, the crude and adjusted odds ratios (ORs) and 95% confidence intervals (CIs) were obtained using unconditional univariate and multivariate logistic regression models, respectively. Hardy–Weinberg equilibrium (HWE) of the genotype distribution among controls was analyzed using a goodness-of-fit Chi-square test. All of the statistical analyses were carried out by two analysts independently in a blind fashion using online bioinformatics software SNPStats (http://bioinfo.iconcologia.net/snpstats/start.htm). P < 0.05 was considered statistically significant.
Results
Rs10815225 and rs4143815 in the PD-L1 gene
Previously, we have found a polymorphism rs4143815 in the 3′-UTR of PD-L1 gene contributed to PD-L1 overexpression in gastric cancer through disrupting the interaction between PD-L1 mRNA and miR-570 [14, 15]. To further investigate the function of genetic variations of PD-L1 gene in gastric cancer, we analyzed the polymorphisms in the promoter of PD-L1 gene. From the online databases NCBI dbSNP BUILED 129 and ENSEMBL v58, we found a polymorphism rs10815225 had a MAF between 4% in Peru and 32% in Nigeria populations. To test whether these polymorphisms are associated with the occurrence and development of gastric cancer, we performed case–control studies on 350 gastric cancer patients, 156 precancerous patients, and 500 health controls using sequencing method.
The association of rs10815225 and rs4143815 with gastric adenocarcinoma risk
The genotype frequencies of rs10815225 and rs4143815 are presented in Table 1. Both of them are in HWE among the controls. Statistical results revealed that the genotype frequencies of rs10815225 and rs4143815 were significantly different between the cancer patients and the health controls (P = 0.0033 and P < 0.0001, respectively; Table 1). The individuals with rs10815225 GG genotype had a significantly higher risk of gastric adenocarcinoma than those with GC genotype (adjusted OR 1.77, 95% CI 1.18–2.66; Table 1). Moreover, compared to the C allele carriers, the G allele homozygotes also had a significantly higher risk of gastric adenocarcinoma (adjusted OR 1.68, 95% CI 1.12–2.51; Table 1).
The previously reported close relationship between rs4143815 genotypes and the occurrence of gastric cancer was further confirmed in this study [14, 15]. When compared to rs4143815 CC homozygotes, both GG homozygotes (adjusted OR 2.40, 95% CI 1.65–3.51) and G allele carriers (adjusted OR 1.86, 95% CI 1.34–2.56) were markedly more inclined to occur gastric cancer (Table 1). In addition, we found that rs10815225 and rs4143815 were in near-complete linkage disequilibrium with the haplotypes G–G and G–C of the frequency of 92.3% in the control group (Table 2). As compared to the dominant haplotype of G–C, both the G–G (adjusted OR 1.59, 95% CI 1.30–1.95) and C–C (adjusted OR 1.95, 95% CI 1.09–3.51) haplotypes were significantly more prevalent in the cancer patients (Table 2).
Table 2.
Association of haplotypes with the occurrence of gastric cancer
| Rs10815225 | Rs4143815 | Frequency (%) | OR (95% CI) | P value | |
|---|---|---|---|---|---|
| 1 | G | G | 47.27 | 1 | – |
| 2 | G | C | 45.03 | 1.59 (1.30–1.95) | <0.0001 |
| 3 | C | C | 4.35 | 1.95 (1.09–3.51) | 0.025 |
| 4 | C | G | 3.36 | 1.92 (0.97–3.77) | 0.06 |
| Global haplotype association | <0.0001 | ||||
Significant results are shown in bold (P < 0.05)
Somatic mutations at the rs10815225 locus in gastric cancer
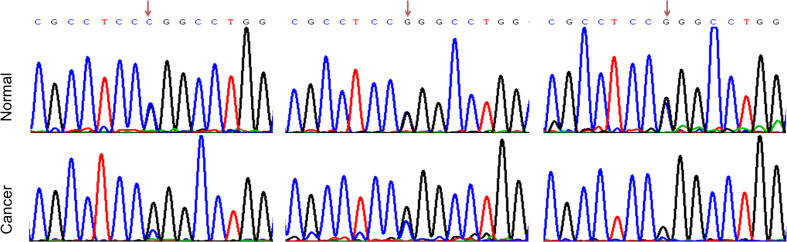
Previously, we found somatic mutations at the rs4143815 locus [14, 15]. Here, we also observed somatic mutations at the rs10815225 locus by sequencing the genomic DNA of PD-L1 promoter in 48 pairs of cancer tissues and distant tissues, as well as peripheral blood. We found that rs10815225 underwent G-to-C somatic mutations in three cancer tissues, as compared to blood (Fig. 1). Furthermore, we performed STR analysis and confirmed that each pair of tissue and blood samples was from the same individual (Supplementary Fig. 1).
Fig. 1.
Somatic mutations at the rs10815225 locus. The arrows indicate the mutation sites
The association of rs10815225 and rs4143815 with precancerous lesions risk
The associations of the genotype frequencies of rs10815225 and rs4143815 with the risk of precancerous lesions are presented in Table 3. Statistical results showed that the genotype frequencies of rs4143815 were significantly different between the precancerous patients and the health controls (P = 0.0013). The individuals with rs4143815 GG genotype were more likely to get gastric precancerous lesions compared to those with CC genotype (adjusted OR 2.51, 95% CI 1.51–4.15; Table 3). Moreover, the G allele carriers also had a significantly higher risk of gastric precancerous lesions compared to the C allele homozygotes (adjusted OR 1.95, 95% CI 1.25–3.02; Table 3). No significant difference in the genotype frequencies of rs10815225 was found between the precancerous patients and health controls (P = 0.11). Besides, as compared to the dominant haplotype G–C of rs10815225 and rs4143815, the G–G haplotypes were significantly more prevalent in the precancerous patients than those in the health controls (adjusted OR 1.65, 95% CI 1.26–2.17; Table 4).
Table 3.
Association of genotypes with the occurrence of precancerous lesions
| SNP | Model | Genotype | Patient (n, %) | Control (n, %) | OR (95% CI) | P value |
|---|---|---|---|---|---|---|
| rs10815225 | Codominant | G/G | 133 (85.8) | 411 (82.2) | 1 | 0.11 |
| G/C | 21 (13.6) | 89 (17.8) | 0.73 (0.44–1.22) | |||
| C/C | 1 (0.6) | 0 (0) | NA (0.00-NA) | |||
| Dominant | G/G | 133 (85.8) | 411 (82.2) | 1 | 0.29 | |
| G/C–C/C | 22 (14.2) | 89 (17.8) | 0.76 (0.46–1.27) | |||
| Recessive | G/G–G/C | 154 (99.3) | 500 (100) | 1 | 0.089 | |
| C/C | 1 (0.6) | 0 (0) | NA (0.00-NA) | |||
| Overdominant | G/G–C/C | 134 (86.5) | 411 (82.2) | 1 | 0.21 | |
| G/C | 21 (13.6) | 89 (17.8) | 0.72 (0.43–1.21) | |||
| rs4143815 | Codominant | C/C | 30 (19.5) | 160 (32) | 1 | 0.0013 |
| G/C | 69 (44.8) | 223 (44.6) | 1.65 (1.03–2.65) | |||
| G/G | 55 (35.7) | 117 (23.4) | 2.51 (1.51–4.15) | |||
| Dominant | C/C | 30 (19.5) | 160 (32) | 1 | 0.0021 | |
| G/C-G/G | 124 (80.5) | 340 (68) | 1.95 (1.25–3.02) | |||
| Recessive | C/C-G/C | 99 (64.3) | 383 (76.6) | 1 | 0.003 | |
| G/G | 55 (35.7) | 117 (23.4) | 1.82 (1.23–2.68) | |||
| Overdominant | C/C-G/G | 85 (55.2) | 277 (55.4) | 1 | 0.96 | |
| G/C | 69 (44.8) | 223 (44.6) | 1.01 (0.70–1.45) |
Significant results are shown in bold (P < 0.05)
Table 4.
Association of haplotypes with the occurrence of precancerous lesions
| Rs10815225 | Rs4143815 | Frequency (%) | OR (95% CI) | P value | |
|---|---|---|---|---|---|
| 1 | G | C | 46.48 | 1 | – |
| 2 | G | G | 44.96 | 1.65 (1.26–2.17) | 3.00E−04 |
| 3 | C | C | 4.86 | 1.13 (0.52–2.46) | 0.76 |
| 4 | C | G | 3.69 | 0.97 (0.40–2.35) | 0.95 |
| Global haplotype association | 0.0021 |
Significant results are shown in bold (P < 0.05)
The association of rs10815225 and rs4143815 with clinicopathological features
The relationships between the genotypes of rs10815225 or rs4143815 and the clinicopathological features of gastric cancer were also analyzed in this study. Strong associations between the rs10815225 genotypes and differentiation grade (adjusted OR 2.21, 95% CI 0.96–5.09 for the C carriers; Supplementary Table 3), as well as between the rs4143815 genotypes and tumor diameter (adjusted OR 1.93, 95% CI 1.10–3.38 for the GC heterozygotes; adjusted OR 1.73, 95% CI 1.02–2.95 for the C carriers; Supplementary Table 4), histological type (adjusted OR 0.50, 95% CI 0.26–0.95 for the CC homozygotes; Supplementary Table 4), and tumor markers CEA and CA199 were observed. In the advanced gastric cancer patients, the rs10815225 genotypes were found to be related to the clinicopathological features including tumor location (P = 0.08 for the GC heterozygotes; Supplementary Table 5), differentiation grade (P = 0.07 for the C carriers; Supplementary Table 5), and distant metastasis (P = 0.09 for the GC heterozygotes and P = 0.03 for the C carriers; Supplementary Table 5). The rs4143815 genotypes were found to be associated with the clinicopathological features including tumor diameter (P = 0.06 for the GC heterozygotes; Supplementary Table 6), histological type (P = 0.05 for the CC homozygotes; Supplementary Table 6), and the tumor markers specially CA199 (P = 0.003 for the CC homozygotes; Supplementary Table 6).
The effect of rs10815225 on the interaction between SP1 and PD-L1 promoter
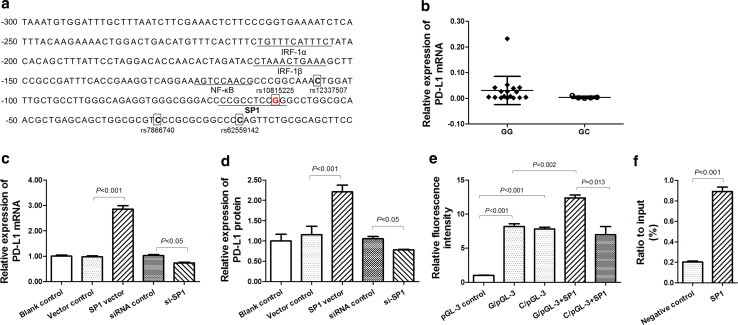
By using the online softwares TFSEARCH and MOTIF, we found four transcription factors interferon regulatory factor 1 (IRF-1), interferon regulatory factor 2 (IRF-2), NF-κB, and SP1 had possible binding-sites in the promoter of the PD-L1 gene. These results demonstrated that the polymorphism rs10815225 was located in the binding-site of SP1 (Fig. 2a). To explore the effect of rs10815225 on the PD-L1 gene regulation mediated by SP1, we first determined the expression level of PD-L1 and SP1 mRNA in 22 cancer tissues containing 17 GG homozygotes and 5 GC heterozygotes. We found that the PD-L1 expression in the GG homozygotes was apparently higher than that in the GC heterozygotes (Fig. 2b), but no difference in SP1 expression between these two corps was observed (Supplementary Fig. 3a). Based on the data from 1000 Genomes (http://www.1000genomes.org/), we found that the average expression levels of PD-L1 were the highest for the GG homozygotes, with more than 1.9 and 3.7 times of those of GC heterozygotes and CC homozygotes, respectively (Supplementary Fig. 3b). Then, we reinforced or attenuated SP1 expression in SGC-7910 cells by using SP1/CMV vector or SP1 siRNA, respectively, and found that both PD-L1 mRNA and protein were elevated by SP1/CMV vector and were diminished by SP1 siRNA (Fig. 2c and Supplementary Fig. 4). To further investigate the effect of rs10815225 on the interaction between SP1 and PD-L1 promoter, we constructed the PD-L1 promoter luciferase reporter plasmids (rs10815225 G or C-allelic) and transfected them alone or with SP1/CMV vector in SGC-7901 cells. For both constructs, the expression of luciferase was significantly enhanced (Fig. 2d), indicating that transcription factors binding with PD-L1 promoter led to increased luciferase expression in SGC-7901 cells. However, since the expression level of SP1 was low in SGC-7901 cells, no difference in the expression level of luciferase between G-allelic and C-allelic PD-L1/promoter/pGL-3 constructs was observed (Fig. 2e). While in the presence of SP1/CMV vector, the expression level of G-allelic PD-L1/promoter/pGL-3 constructs was significantly higher than that of C-allelic PD-L1/promoter/pGL-3 constructs (Fig. 2e), confirming the functional potential of the G-allelic PD-L1 mRNA-SP1 duplex. These findings indicate that SP1 can bind with G-allelic but not C-allelic PD-L1 promoter and promoting transcription of PD-L1 mRNA and consequent overexpression of PD-L1 protein. Furthermore, the results from ChIP assays confirmed direct binding of SP1 to PD-L1 promoter (Fig. 2f).
Fig. 2.
Impact of rs10815225 on the binding of SP1 in the promoter of the PD-L1 gene. a The schematic of PD-L1 promoter harbors the polymorphisms and putative binding-sites of transcription factors. The polymorphism loci are squared, and rs10815225 is located within the binding-site of SP1. The sequences of the binding-sites are underlined. b The expression level of PD-L1 mRNA in the gastric cancer tissues with rs10815225 GG genotype and GC genotype. c The impact of SP1 on the expression of PD-L1 mRNA in SGC-7901 cells. d The impact of SP1 on the expression of PD-L1 protein in SGC-7901 cells. e Luciferase reporter assays to show the impact of rs10815225 alleles on the transcription role of SP1. f Chromatin immunoprecipitation assays to show the binding of SP1 to the promoter of the PD-L1 gene
Discussion
In this study, we found a polymorphism rs10815225 in the PD-L1 promoter region was significantly associated with the occurrence of gastric cancer. G-allelic PD-L1 promoter offered a binding-site for a transcriptional factor SP1, resulting in upregulation of PD-L1 mRNA and protein in gastric cancer and consequently cancer immune escaping. Notably, somatic mutations were observed in both the promoter region (rs10815225 locus) and the 3′-UTR region (rs4143815 locus) of PD-L1. However, we did not find any cancer-related polymorphisms and mutations in the coding region of PD-L1 (data not shown). Furthermore, we found that rs10815225 was associated with the clinicopathological features including tumor location, differentiation grade, and distant metastasis in the advanced gastric cancer patients; while rs4143815 was associated with the clinicopathological features including tumor diameter, histological type, and the tumor markers specially CA199. In addition, we found that rs4143815, instead of rs10815225, was related to the risk of precancerous lesions. Rs10815225 and rs4143815 were in near-complete linkage disequilibrium. However, the genotype distribution of rs10815225 was different from that of rs4143815. The rs10815225 GG were the most prevalent in cancer patients, then in precancerous lesions, and the least in health controls; but the frequency of rs4143815 GG in cancer patients was similar to that in precancerous lesions, and was higher than that in health controls (Supplementary Fig. 2). These results revealed a novel mechanism underlying genetic polymorphisms influencing PD-L1 expression modify gastric cancer susceptibility.
It has been reported that the upstream fragment of 0 to −350 bp in the PD-L1 promoter was essential for PD-L1 transcription [18]. A number of transcription factors hypoxia inducible factor 1 alpha subunit (HIF-1α), forkhead box A1 (FoxA1), STAT3, IRF-1, and NF-κB have been found to bind to this fragment and to promote the transcription of PD-L1 gene [18–21]. In this study, we provided evidences to support the conclusion that a novel transcription factor SP1 bond to PD-L1 promoter to promote PD-L1 transcription. Firstly, the binding-sites for the transcription factors IRF-1, IRF-2, NF-κB, and SP1 were predicted in the promoter (0 to −300 bp) of PD-L1 gene. Secondly, overexpression or silence of SP1 in SGC-7901 cells directly increased or decreased the expression of both PD-L1 mRNA and protein. Thirdly, the expression of PD-L1/promoter/pGL3 constructs was obviously higher than that of empty pGL3 vectors in GSC-7901 cells. Fourthly, the expression of rs10815225 G-allelic PD-L1/promoter/pGL3 constructs was further enhanced when they were co-transfected with SP1/CMV expression plasmids in SGC-7901 cells. Fifthly, in the cases that the binding-site of SP1 in PD-L1 promoter was mutated from G-to-C at rs10815225 locus, the expression of C-allelic PD-L1/promoter/pGL3 constructs was not impacted by the overexpression of SP1. Finally, ChIP results further confirmed that SP1 could directly bind to the promoter of PD-L1.
Numerous studies have reported that genetic polymorphisms through disrupting the binding of transcription factors with the promoters of the target genes resulted in genes dysregulation and altered gastric cancer susceptibility [22–26]. For instance, a polymorphism −181A >G (rs11568818) in the matrix metallopeptidase 7 (MMP7) promoter is associated with gastric cancer risk in an eastern Indian population. Phosphorylated cAMP responsive element binding protein 1 (CREB1) through binding to the G-allelic promoter enhances MMP7 gene expression and thereby increases the risk of gastric cancer [22]. In this study, we revealed that a polymorphism rs10815225 in the promoter of PD-L1 gene offered a binding-site for a transcription factor SP1 to elevate PD-L1 expression and to increase the risk of gastric cancer. This conclusion is supported by the following findings: (1) rs10815225 locates in the predicted binding-site of SP1 in the PD-L1 promoter; (2) rs10815225 GG homozygotes and G allele carriers are significantly more prevalent in gastric cancer patients than those in the health controls; (3) the expression of PD-L1 mRNA is markedly higher in the rs10815225 GG homozygous patients than that in the GC heterozygous patients; (4) the expression level of rs10815225 G-allelic PD-L1/promoter/pGL3 constructs is apparently higher than that of C-allelic PD-L1/promoter/pGL3 constructs in SGC-7901 cells with overexpressed SP1.
SP1 is a well-known transcription factor. SP1 both promotes and suppresses the expression of numerous oncogenes and tumor suppressors, as well as genes involved in cell growth, differentiation, apoptosis, and angiogenesis, such as v-myc avian myelocytomatosis viral oncogene homolog (MYC), Jun proto-oncogene (JUN), Cyclin D1 (CCND1), platelet-derived growth factor subunit A (PDGFA), platelet-derived growth factor receptor alpha (PDGFRA), EGFR, VEGF, BCL2, and TGF-β. [27]. Abnormal SP1 expression and activation has been detected in human gastric cancer and was inversely correlated with patient survival, suggesting that SP1 may be a potential molecular marker for poor prognosis and directly contributes to gastric cancer development and progression [28, 29]. Interference of SP1 function has been shown to inhibit gastric cancer growth in vitro and in vivo [30, 31]. Several anticancer agents in clinical use and a number of experimental compounds have also been reported to be active for anticancer though interfering SP1 functions [32]. Since PD-L1 has been widely proved to be an effective anticancer therapy target [9–11], the compounds disrupting the DNA binding of SP1 or inhibiting the expression of SP1 might be able to reduce PD-L1 gene expression and to exert antitumor effects.
Multiple steps including transcription, posttranscriptional regulation, translation, and posttranslational modification participate in gene expression. At transcription, we and others have discovered that the transcription factors SP1, IRF-1, and NF-κB promoted the transcription of PD-L1 gene by binding to the PD-L1 promoter. On the other side, we previously identified a microRNA miR-570 posttranscriptionally inhibited the expression of PD-L1 through binding to the 3′-UTR of PD-L1 gene in gastric cancer [12, 13]. Before that, another microRNA miR-513 was also revealed to posttranscriptionally repress PD-L1 expression by binding with PD-L1 3′-UTR in human cholangiocytes [33]. Taken together with previous reports that PD-L1 overexpression is related to IFN-γ stimulation in vitro [5, 34–36] or loss of phosphatase and tensin homolog (PTEN) gene in human glioma [37], these results imply that multiple regulatory factors may contribute to PD-L1 expression. However, the regulatory mechanisms underlying the translation and posttranslational modification of PD-L1 molecules are still needed to be further investigated.
Multiple factors, including Helicobacter pylori infection, smoking, diet, gastroesophageal reflux disease, obesity, and certain dietary components, have been proposed to contribute to the risk of gastric carcinogenesis [38–40]. Unfortunately, information of these factors is not available in this study. It would be interesting to investigate the coaction of the genotypes of rs10815225 and/or rs4143815 and these factors on risk of gastric cancer. Also, since the survival information of gastric cancer patients is not available, our further analysis of the role of rs10815225 and/or rs4143815 in cancer prognosis is constrained. Another limitation of this study is the relative small sample sizes. However, our study provides the first evidence that the polymorphism rs10815225 is a genetic factor for the occurrence of gastric cancer, with the GG genotype being associated with increased risk of cancer in a Chinese population. These findings further support that PD-L1 plays an important role in gastric carcinogenesis. Studies on the impact of PD-L1 polymorphisms in cancer susceptibility in other populations and/or other cancer types will be of considerable value.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Nos. 81270031, 81272737, and 81372375), Science and Technology Special Project of Clinical Medicine in Jiangsu Province (BL2014046), Science and Technology Project in Suzhou (SYS201524), and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Abbreviations
- ChIP
Chromatin immunoprecipitation
- CI
Confidence interval
- Ct
Threshold cycle
- HWE
Hardy–Weinberg equilibrium
- MAF
Minor allele frequency
- OR
Odds ratio
- qPCR
Quantitative PCR
- SP1
Sp1 transcription factor
- STR
Short tandem repeat
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Li-Hua Tao and Xin-Ru Zhou have contributed equally to this work.
Contributor Information
Wei-Peng Wang, Email: wangweipeng@suda.edu.cn.
Wei-Chang Chen, Email: weichangchen@126.com.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Wesolowski R, Lee C, Kim R. Is there a role for second-line chemotherapy in advanced gastric cancer? Lancet Oncol. 2009;10:903–912. doi: 10.1016/S1470-2045(09)70136-6. [DOI] [PubMed] [Google Scholar]
- 4.Zou W, Chen L. Inhibitory B7-family molecules in the tumour microenvironment. Nat Rev Immunol. 2008;8:467–477. doi: 10.1038/nri2326. [DOI] [PubMed] [Google Scholar]
- 5.Dong H, Strome SE, Salomao DR, et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med. 2002;8:793–800. doi: 10.1038/nm0902-1039c. [DOI] [PubMed] [Google Scholar]
- 6.Cao Y, Zhang L, Ritprajak P, Tsushima F, Youngnak-Piboonratanakit P, Kamimura Y, Hashiguchi M, Azuma M. Immunoregulatory molecule B7-H1 (CD274) contributes to skin carcinogenesis. Cancer Res. 2011;71:4737–4741. doi: 10.1158/0008-5472.CAN-11-0527. [DOI] [PubMed] [Google Scholar]
- 7.Cao Y, Zhang L, Kamimura Y, Ritprajak P, Hashiguchi M, Hirose S, Azuma M. B7-H1 overexpression regulates epithelial-mesenchymal transition and accelerates carcinogenesis in skin. Cancer Res. 2011;71:1235–1243. doi: 10.1158/0008-5472.CAN-10-2217. [DOI] [PubMed] [Google Scholar]
- 8.Seliger B, Marincola FM, Ferrone S, Abken H. The complex role of B7 molecules in tumor immunology. Trends Mol Med. 2008;14:550–559. doi: 10.1016/j.molmed.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herbst RS, Soria JC, Kowanetz M, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Powles T, Eder JP, Fine GD, et al. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature. 2014;515:558–562. doi: 10.1038/nature13904. [DOI] [PubMed] [Google Scholar]
- 11.Brahmer JR, Tykodi SS, Chow LQ, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu C, Zhu Y, Jiang J, Zhao J, Zhang XG, Xu N. Immunohistochemical localization of programmed death-1 ligand-1 (PD-L1) in gastric carcinoma and its clinical significance. Acta Histochem. 2006;108:19–24. doi: 10.1016/j.acthis.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 13.Mao Y, Sun J, Wang WP, Zhang XG, Hua D. Clinical significance of costimulatory molecule B7-H3 expression on CD3(+) T cells in colorectal carcinoma. Chin Med J (Engl) 2013;126:3035–3038. [PubMed] [Google Scholar]
- 14.Wang W, Sun J, Li F, et al. A frequent somatic mutation in CD274 3′-UTR leads to protein over-expression in gastric cancer by disrupting miR-570 binding. Hum Mutat. 2012;33:480–484. doi: 10.1002/humu.22014. [DOI] [PubMed] [Google Scholar]
- 15.Wang W, Li F, Mao Y, et al. A miR-570 binding site polymorphism in the B7-H1 gene is associated with the risk of gastric adenocarcinoma. Hum Genet. 2013;132:641–648. doi: 10.1007/s00439-013-1275-6. [DOI] [PubMed] [Google Scholar]
- 16.Kataoka K, Shiraishi Y, Takeda Y, et al. Aberrant PD-L1 expression through 3′-UTR disruption in multiple cancers. Nature. 2016;534:402–406. doi: 10.1038/nature18294. [DOI] [PubMed] [Google Scholar]
- 17.Tang R, Qi Q, Wu R, et al. The polymorphic terminal-loop of pre-miR-1307 binding with MBNL1 contributes to colorectal carcinogenesis via interference with Dicer1 recruitment. Carcinogenesis. 2015;36:867–875. doi: 10.1093/carcin/bgv066. [DOI] [PubMed] [Google Scholar]
- 18.Lee SJ, Jang BC, Lee SW, et al. Interferon regulatory factor-1 is prerequisite to the constitutive expression and IFN-gamma-induced upregulation of B7-H1 (CD274) FEBS Lett. 2006;580:755–762. doi: 10.1016/j.febslet.2005.12.093. [DOI] [PubMed] [Google Scholar]
- 19.Noman MZ, Desantis G, Janji B, Hasmim M, Karray S, Dessen P, Bronte V, Chouaib S. PD-L1 is a novel direct target of HIF-1alpha, and its blockade under hypoxia enhanced MDSC-mediated T cell activation. J Exp Med. 2014;211:781–790. doi: 10.1084/jem.20131916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Carlsson R, Comabella M, et al. FoxA1 directs the lineage and immunosuppressive properties of a novel regulatory T cell population in EAE and MS. Nat Med. 2014;20:272–282. doi: 10.1038/nm.3485. [DOI] [PubMed] [Google Scholar]
- 21.Marzec M, Zhang Q, Goradia A, et al. Oncogenic kinase NPM/ALK induces through STAT3 expression of immunosuppressive protein CD274 (PD-L1, B7-H1) Proc Natl Acad Sci USA. 2008;105:20852–20857. doi: 10.1073/pnas.0810958105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kesh K, Subramanian L, Ghosh N, Gupta V, Gupta A, Bhattacharya S, Mahapatra NR, Swarnakar S. Association of MMP7 −181A–> G promoter polymorphism with gastric cancer risk: influence of nicotine in differential allele-specific transcription via increased phosphorylation of cAMP-RESPONSE ELEMENT-BINDING PROTEIN (CREB) J Biol Chem. 2015;290:14391–14406. doi: 10.1074/jbc.M114.630129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li C, Tong W, Liu B, Zhang A, Li F. The −1082A >G polymorphism in promoter region of interleukin-10 and risk of digestive cancer: a meta-analysis. Sci Rep. 2014;4:5335. doi: 10.1038/srep05335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang DS, Wang Z, He XJ, et al. Recurrent TERT promoter mutations identified in a large-scale study of multiple tumour types are associated with increased TERT expression and telomerase activation. Eur J Cancer. 2015;51:969–976. doi: 10.1016/j.ejca.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dey S, Stalin S, Gupta A, Saha D, Kesh K, Swarnakar S. Matrix metalloproteinase3 gene promoter polymorphisms and their haplotypes are associated with gastric cancer risk in eastern Indian population. Mol Carcinog. 2012;51(Suppl 1):E42–E53. doi: 10.1002/mc.21837. [DOI] [PubMed] [Google Scholar]
- 26.Kim DH, Park SE, Kim M, et al. A functional single nucleotide polymorphism at the promoter region of cyclin A2 is associated with increased risk of colon, liver, and lung cancers. Cancer. 2011;117:4080–4091. doi: 10.1002/cncr.25930. [DOI] [PubMed] [Google Scholar]
- 27.Beishline K, Azizkhan-Clifford J. Sp1 and the ‘hallmarks of cancer’. FEBS J. 2015;282:224–258. doi: 10.1111/febs.13148. [DOI] [PubMed] [Google Scholar]
- 28.Wang L, Wei D, Huang S, Peng Z, Le X, Wu TT, Yao J, Ajani J, Xie K. Transcription factor Sp1 expression is a significant predictor of survival in human gastric cancer. Clin Cancer Res. 2003;9:6371–6380. [PubMed] [Google Scholar]
- 29.Yao JC, Wang L, Wei D, Gong W, Hassan M, Wu TT, Mansfield P, Ajani J, Xie K. Association between expression of transcription factor Sp1 and increased vascular endothelial growth factor expression, advanced stage, and poor survival in patients with resected gastric cancer. Clin Cancer Res. 2004;10:4109–4117. doi: 10.1158/1078-0432.CCR-03-0628. [DOI] [PubMed] [Google Scholar]
- 30.Qiu T, Zhou X, Wang J, Du Y, Xu J, Huang Z, Zhu W, Shu Y, Liu P. MiR-145, miR-133a and miR-133b inhibit proliferation, migration, invasion and cell cycle progression via targeting transcription factor Sp1 in gastric cancer. FEBS Lett. 2014;588:1168–1177. doi: 10.1016/j.febslet.2014.02.054. [DOI] [PubMed] [Google Scholar]
- 31.Wang L, Guan X, Zhang J, Jia Z, Wei D, Li Q, Yao J, Xie K. Targeted inhibition of Sp1-mediated transcription for antiangiogenic therapy of metastatic human gastric cancer in orthotopic nude mouse models. Int J Oncol. 2008;33:161–167. [PubMed] [Google Scholar]
- 32.Vizcaino C, Mansilla S, Portugal J. Sp1 transcription factor: a long-standing target in cancer chemotherapy. Pharmacol Ther. 2015;152:111–124. doi: 10.1016/j.pharmthera.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 33.Gong AY, Zhou R, Hu G, et al. MicroRNA-513 regulates B7-H1 translation and is involved in IFN-gamma-induced B7-H1 expression in cholangiocytes. J Immunol. 2009;182:1325–1333. doi: 10.4049/jimmunol.182.3.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dong H, Zhu G, Tamada K, Chen L. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat Med. 1999;5:1365–1369. doi: 10.1038/70932. [DOI] [PubMed] [Google Scholar]
- 35.Gong AY, Zhou R, Hu G, et al. MicroRNA-513 regulates B7-H1 translation and is involved in IFN-gamma-induced B7-H1 expression in cholangiocytes. J. Immunol. 2009;182:1325–1333. doi: 10.4049/jimmunol.182.3.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kondo A, Yamashita T, Tamura H, et al. Interferon-gamma and tumor necrosis factor-alpha induce an immunoinhibitory molecule, B7-H1, via nuclear factor-kappaB activation in blasts in myelodysplastic syndromes. Blood. 2010;116:1124–1131. doi: 10.1182/blood-2009-12-255125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parsa AT, Waldron JS, Panner A, et al. Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nat Med. 2007;13:84–88. doi: 10.1038/nm1517. [DOI] [PubMed] [Google Scholar]
- 38.Parsonnet J, Friedman GD, Vandersteen DP, Chang Y, Vogelman JH, Orentreich N, Sibley RK. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325:1127–1131. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- 39.Correa P. Human gastric carcinogenesis: a multistep and multifactorial process–First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res. 1992;52:6735–6740. [PubMed] [Google Scholar]
- 40.Mayne ST, Navarro SA. Diet, obesity and reflux in the etiology of adenocarcinomas of the esophagus and gastric cardia in humans. J Nutr. 2002;132:3467S–3470S. doi: 10.1093/jn/132.11.3467S. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.