Abstract
Double-stranded linear DNA is synthesized as a minor viral DNA species by all hepadnaviruses. In a previous study (W. Yang and J. Summers, J. Virol. 69:4029–4036, 1995) we showed that virus particles containing linear DNA of the duck hepatitis B virus (DHBV) could initiate an infection of primary duck hepatocytes. In cells infected by linear DNA containing viruses the transcriptional template, covalently closed circular DNA, was formed by circularization of linear DNA by nonhomologous recombination between the two ends. This process was shown to result in viral DNA replication through multiple generations of linear DNA intermediates, a process we called illegitimate replication. In this study we showed that viruses containing linear DHBV DNA produced by engineered insertions in the r sequence, which encodes the 5′ end of the pregenome, could infect hepatocytes in vivo, and these hepatocytes proceeded to carry out illegitimate replication. Nonhomologous recombination quickly produced revertants and partial revertants in which all or part of the insertion was deleted. One such partial revertant that replicated primarily through circular DNA intermediates, but which synthesized elevated levels of linear DNA, could be sustained for several days as the predominant genotype in vivo, but this mutant was eventually displaced by variants showing full reversion to legitimate replication and that synthesized normal low levels of linear DNA. Full revertants did not necessarily contain the wild-type r sequence. The results suggest that the linear DNA produced during DHBV infection initiates cycles of illegitimate replication by generating mutants with altered r sequences. Some r sequence mutants carry out a mixture of legitimate and illegitimate replication that can contribute to elevated production of linear DNA in individual cells.
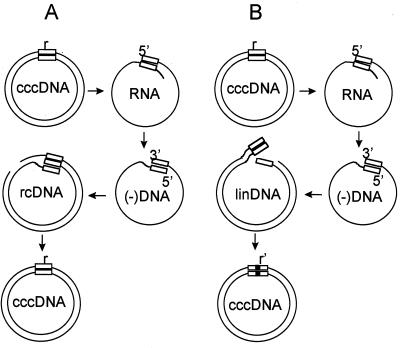
Hepadnaviruses are a family of small DNA-containing viruses that replicate their genomes through reverse transcription of RNA intermediates called pregenomes (5, 6, 17, 21, 23). The template for the transcription of pregenomic RNA is nuclear covalently closed circular DNA (cccDNA) (17, 26, 28, 30). Pregenomic RNA is encapsidated in the cytoplasm and is used as a template for the synthesis of a double-stranded circular form of the genome, produced through a reverse transcription step. Capsids containing double-stranded DNA, called relaxed circular DNA (rcDNA), can be assembled into an envelope and secreted from the infected cell as infectious virus or they can be transported to the nucleus, where rcDNA is released and converted to additional cccDNA molecules (10, 11, 25, 28, 30). An important feature of this conversion is that all genomic information stored in the rcDNA is precisely conserved in the resulting cccDNA molecule so that pregenomic RNA transcribed from this cccDNA can direct the synthesis of progeny rcDNAs that are genetically identical to their parental rcDNA molecule. We have called this process, which occurs in every cycle of hepadnavirus infection, legitimate replication (31) (Fig. 1A).
FIG. 1.
Legitimate and illegitimate pathways of DHBV DNA replication. (A) Legitimate replication occurs through transcription of viral cccDNA to produce the RNA pregenome (RNA). Transcription initiates at the 5′ boundary (left boundary) of the 9-bp r sequence (box), at nucleotide 2529, and after one circuit of the genome, terminates around nucleotide 2800, so that the pregenome contains a 5′ copy and a 3′ copy of r. Reverse transcription initiates at the 3′ boundary (right boundary) of the 3′ r and proceeds to the 5′ end of the pregenome, producing a cDNA, (−)DNA, with terminal duplications of r. Plus-strand DNA synthesis initiates at position 2491 on the (−)DNA, and elongation proceeds through r, causing circularization of the genome through a template switch (rcDNA). rcDNA is converted to a progeny copy of cccDNA that is identical to the parental copy. (B) Illegitimate replication occurs through the same pathway, except rcDNA is not formed. Instead, plus-strand synthesis is initiated near the 3′ end of (−)DNA, producing a linear double-stranded DNA (linDNA). linDNA is converted to cccDNA by nonhomologous recombination. The progeny cccDNA is not identical to the parental molecule, as indicated by the partially filled box. Please see reference 31 for details.
However, rcDNA is not the only double-stranded viral DNA synthesized in the cytoplasm of an infected cell. The production of genome-sized double-stranded viral linear DNA is observed in all natural hepadnavirus infections (24, 27, 29). The mechanism underlying the linear DNA synthesis has been elucidated and is due to the failure of a specific step in plus-strand synthesis, i.e., RNA primer translocation from DR1 to DR2 (12, 13), a step required for genome circularization. As a result of this failure, plus-strand DNA synthesis is initiated in situ 19 nucleotides from the 3′ end of the minus-strand DNA (15, 22). In situ priming produces a double-stranded linear copy of the viral DNA, with nine nucleotides of information repeated at each end (13). In situ priming of plus-strand DNA occurs during DNA replication of wild-type hepadnaviruses at a frequency of 5 to 10% of all plus strands synthesized (22, 29, 31).
We previously found that double-stranded linear DNA-containing duck hepatitis B virus (DHBV) (18) could infect primary duck hepatocyte cultures and deliver linear DNA molecules to the nucleus of the infected hepatocytes, where cccDNA was formed as efficiently as from rcDNA-containing virus (31). We found that cccDNA had been converted from the double-stranded linear DNA by means of nonhomologous recombination occurring preferentially around the two ends of the linear DNA (Fig. 1B). Most cccDNAs formed by this mechanism had lost one or more cis- or trans-acting elements required for DNA replication; however, more than 10% could participate in further DNA synthesis. In most of the molecules that retained sufficient genetic information to produce progeny DNA, alterations in the length or in the sequence of the r region of the pregenomes that were transcribed resulted in the synthesis of a higher-than-wild-type ratio of linear DNAs to rcDNAs. Thus, the formation of cccDNA from linear DNA initiated a process in which linear viral DNA production was favored, resulting in second and higher generations of cccDNAs with complex multiple recombination joints within the r region. We argued that, in theory, this process of DNA replication was capable of producing unlimited generations of DNA molecules, a subset of which contained r sequences that were able to revert to the wild-type r sequence through subsequent generations of nonhomologous recombination. Since the precise sequence in a parental cccDNA was not conserved in its progeny cccDNA due to the step of nonhomologous recombination, we called this mechanism for propagating DNA molecules “illegitimate replication.” Because the rate of illegitimate replication is very low, the presence of illegitimate replication is obscured by the relatively higher levels of legitimate replication in natural infections. We speculated that the low levels of replication inherent in illegitimate replication, however, might be favored in some in vivo situations, e.g., in evading antigen-specific cellular cytotoxicity.
In the livers of woodchucks chronically infected with the mammalian hepadnavirus woodchuck hepatitis virus and undergoing a chronic inflammatory reaction we found evidence that nonhomologous recombination of double-stranded linear DNA was responsible for the formation of a significant fraction of cccDNA. In one infected liver at least 10% of total cccDNA was apparently formed from linear precursors. Sequence analysis predicted that some of these cccDNAs would be functional for progeny linear DNA synthesis (32). Evidence for the occurrence of illegitimate replication through multiple generations of linear DNA, however, was not obtained. To date, the fate of hepatocytes infected in vivo with linear DNA-containing viruses has not been described due to the overwhelming level of legitimate replication that occurs in the liver.
In this study we have examined the fate of hepatocytes infected in vivo with linear DNA-containing viruses that mimicked the types of virus that can arise during a natural infection, i.e., viruses that could revert to wild type through nonhomologous recombination between the ends of the linear DNA. Our results directly demonstrated that linear DNA-containing viruses initiated an infection in vivo through the production of cccDNA by nonhomologous recombination. Some of these first-generation cccDNAs were used for subsequent generations of illegitimate replication. Illegitimate replication also produced a spectrum of cccDNAs that acquired the ability to carry out various levels of legitimate replication, through rcDNA, yet at the same time produced higher-than-wild-type levels of linear DNA in infected cells. Variants showing complete reversion for legitimate replication also occurred but these did not necessarily acquire the original wild-type r sequence.
MATERIALS AND METHODS
Ducks.
One-day-old Pekin common hybrid ducks were purchased from Metzer Farms (Redlands, Calif.). All ducks used for experimental infection were screened by dot blot hybridization of duck sera, and DHBV DNA-negative ducks at an age of 3 to 5 days posthatch were inoculated intravenously with virus particles containing DHBV wild-type or mutant genomes.
Plasmid and mutations.
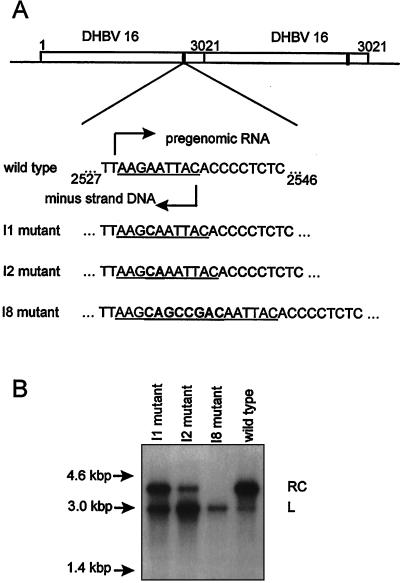
A plasmid expression vector, pSPDHBV5.1(2X), containing EcoRI-linearized DHBV16 DNA (16) cloned into the plasmid vector pSP65 as a head-to-tail dimer was previously described (20). This plasmid was used as the template in the site-directed mutagenesis described below. To introduce an insertion in the r sequence, a procedure involving PCR and a biotin-labeled primer was designed. The first PCR amplification included a 5′ end biotin-labeled plus-orientation (+) primer (DHBV nucleotides 2217 to 2240) and a mutagenic minus-orientation (−) primer (nucleotides 2544 to 2511) containing one of three specific insertions in the nine-nucleotide r sequence. A high-fidelity DNA polymerase Pwo (Boehringer Mannheim) was chosen for the amplification, in which about one error in a total of 5 kb of DNA could be detected by sequencing of individual clones (data not shown). The resulting biotinylated (+) PCR product then was used as a megaprimer and annealed with cloned DHBV DNA template cleaved with BamHI at DHBV nucleotide 1658. One cycle of elongation was carried out by Pwo DNA polymerase, and the 5′ end biotin-labeled single-stranded DNA containing the engineered mutation was isolated by absorbing the biotinylated DNA strand to streptavidin-coated M 280 Dynabeads (Dynal Corp.) and eluting nonbiotinylated DNA with alkali. The remaining DNA was then used as a template for a final amplification (DHBV 2217 to 2240 and DHBV 70 to 45). The resulting PCR products were cut with the restriction enzymes NcoI and NsiI, and the 500-bp fragment was gel purified and exchanged into both copies of the DHBV monomer in pSPDHBV5.1(2X). The region between NcoI and NsiI in the plasmid was sequenced to exclude possible secondary mutations. The plasmids containing the r sequence mutations are shown in Fig. 2A.
FIG. 2.
Insertion mutants of DHBV and the effects of the insertions on rcDNA synthesis. (A) Top: map of the DHBV dimer in pSPDHBV5.1(2×) showing two copies of the mutated r region. Bottom: the mutations I1, I2, and I8 within the r sequence (underlined). The site of initiation of pregenome transcription in cccDNA (top arrow) and the site of minus-strand initiation at the 3′ boundary of the downstream copy of r (bottom arrow) in the wild-type pregenome are indicated. (B) DHBV DNA extracted from enveloped virus particles in the culture medium of LMH cells transfected with DHBV dimer plasmids containing the indicated mutations. The migration positions of molecular size markers are shown on the left (arrows). The two bands are indicated by RC (rcDNA) and L (linear DNA).
Transfections.
Transfection of the chicken hepatoma cell line LMH (2, 8), isolation and assay of replicative intermediates by Southern blot hybridization, and concentration and assay of enveloped virus by the pronase-DNase I method were performed as previously described (10, 11).
Analysis of viral DNA in sera.
Duck serum (50 μl) was mixed with 150 μl of digestion solution containing a final concentration of 10 mM EDTA, 1% sodium dodecyl sulfate (SDS), 0.1 M NaCl, and 0.5 mg of Pronase/ml and incubated at 45°C for 2 h. After phenol extraction, viral DNA was precipitated with ethanol and analyzed by Southern blot hybridization.
Analysis of DHBV DNA replicative intermediates and cccDNAs in the infected livers.
For viral DNA purification, 60 mg of liver tissue was homogenized with 2 ml of ice-cold TE buffer (50 mM Tris-HCl [pH 8.0], 1 mM EDTA [pH 8.0]) with a loose-fitting plunger in a 7-ml Dounce homogenizer. The homogenate then was equally divided into two parts. For purification of viral replicative intermediates, one part of the homogenate was mixed with 50 μl of 10% Nonidet P-40 and incubated on ice for 30 min and the nuclei and other debris were removed by microcentrifugation. The supernatant was adjusted to a final concentration of 10 mM EDTA–1% SDS–0.1 M NaCl–0.5 mg of Pronase/ml and incubated at 37°C for 1 to 2 h. After phenol extraction, DNA was recovered by ethanol precipitation and dissolved in TE (10 mM Tris-HCl [pH 8.0], 1 mM EDTA [pH 8.0]), and the nucleic acid concentrations were normalized by absorbance at 260 μm.
For viral cccDNA purification, the second portion of homogenate was mixed with an equal volume of 4% SDS by vortexing. Cellular DNA, proteins, and viral protein-bound DNAs were precipitated by the addition of 0.5 ml of 2.5 M KCl. After centrifugation at 4°C for 10 min, the supernatant was removed and extracted with phenol. Viral DNA was collected by ethanol precipitation and dissolved in TE (10 mM Tris-HCl [pH 8.0], 1 mM EDTA [pH 8.0]). The total nucleic acid concentration of each sample was normalized by absorbance at 260 μm. Viral replicative intermediates and cccDNA from equal amounts of total nucleic acids (usually 20 μg) were analyzed by 1.7% agarose gel electrophoresis and Southern blot hybridization.
PCR amplification and sequencing.
A pair of primers corresponding to DHBV16 DNA sequence 70 to 45 [(−) primer] and 2217 to 2240 [(+) primer] was used in all PCR amplification experiments described in this study unless otherwise indicated (see Fig. 5C). Hot start (2 min at 80°C before adding Mg2+-containing buffer) was used in all PCR amplifications to decrease nonspecific template binding. Amplification reactions consisted of a denaturation step at 94°C for 1 min, annealing at 55°C for 2 min, and elongation at 72°C for 3 min for 30 cycles with Taq DNA polymerase. To construct a library, PCR products were diluted 100-fold in fresh reaction buffer and subjected to two additional cycles of amplification to eliminate heteroduplex molecules before being cloned. The reamplified PCR products were cloned in the TA cloning vector pCRII and transformed into Escherichia coli INVαF′. Colony screening and sequencing were performed as previously described (32).
FIG. 5.
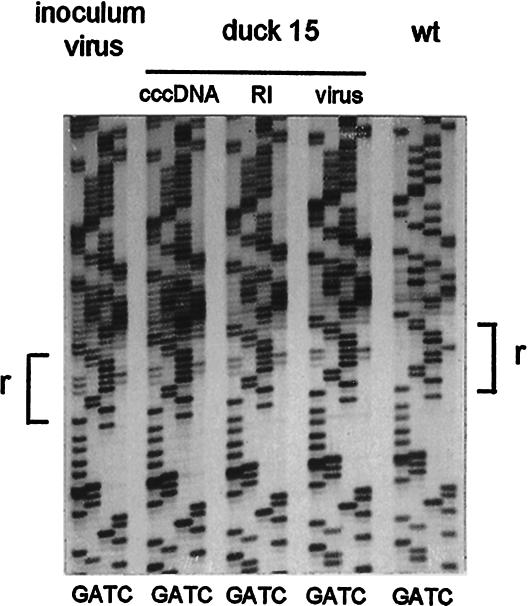
Infection of ducklings with the I2 mutant. Virus particles containing the mutant I2 or wild-type genome were prepared by transfection of LMH cells and used to infect four 4-day-old ducklings at a dose of 5 × 109 viral genomes per bird. (A) Virus present at the indicated times in 25 μl of serum was extracted and analyzed by agarose gel electrophoresis and blot hybridization. The top panel was exposed approximately four times as long as the bottom panel (compare hybridization marker signals). (B) Liver samples (equivalent to 5 mg) were analyzed for viral replicative intermediates present at day 6 postinfection by gel electrophoresis and blot hybridization. Each lane contained the DNA extracted from an equivalent of 5 mg of liver tissue. (C) rcDNA extracted from enveloped virus in the blood and cccDNA from the liver at day 6 postinfection was subjected to PCR amplification and direct sequencing of the PCR products through the r region. Primers corresponding to nucleotides 2492 to 2516 and 70 to 45 were used for amplification of the rcDNA. The sequencing gel shows the presence of the I2 mutation in the day 6 postinfection samples in comparison with the wild-type sequence.
RESULTS
We previously showed that linear DNA-containing viruses could initiate a cycle of replication through linear DNA intermediates in primary duck hepatocytes. Because this process, which we called illegitimate replication, has potential consequences for infected cells that differ from the consequences of legitimate replication, we wanted to know if illegitimate replication also occurs in vivo and what the fate of cells carrying out illegitimate replication in vivo would be. In vivo, illegitimate replication would be difficult to detect because it would contribute only a small fraction of the replicative intermediates in the DNA extracted from a piece of liver tissue. To circumvent this technical difficulty, we infected ducklings with high titers of virus particles that contained mutant DNAs that mimicked the type of DNAs that could be formed from linear DNA in a natural infection. To produce these mutant genomes we inserted 1, 2, or 8 nucleotides into the r regions of a standard wild-type DHBV plasmid expression vector to produce the mutants 1I, 2I, and 8I, respectively. These insertion mutants resembled those that would occur in natural infections in that they could be reverted to produce the wild-type sequence by deleting the inserted nucleotides during nonhomologous recombination. The structure of these mutant genomes is shown in Fig. 2A. As assayed by transfection of the plasmid expression vectors into LMH cells, the various insertions were found to produce corresponding defects in the production of rcDNA in favor of linear DNA in the enveloped virus particles released from the transfected cells (Fig. 2B). The virus produced by transfection of these insertion mutant plasmids was used for the infection of ducklings.
Infection initiated by linear DNA.
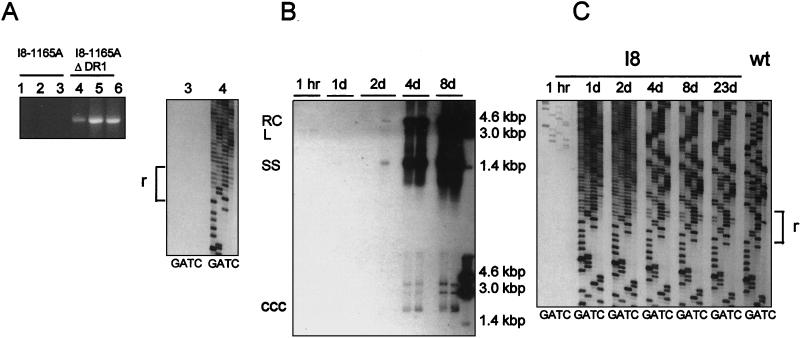
In order to determine if an infection could be initiated in vivo by linear DNA-containing virus particles, we assayed for the appearance of cccDNA in the livers of ducklings after inoculation with the I8 mutant virus. As a negative control for infection we tested whether the appearance of cccDNA was dependent on the presence of the preS envelope protein. Finally, by sequencing, we tested whether the structure of the cccDNA was consistent with its having been formed by nonhomologous recombination of the input linear DNA. To carry out this experiment, we modified the I8 genome by introducing a second mutation (1165A) that destroyed the ability of the expression vector to produce the large envelope protein, preS (26). The expression vector containing the double-mutant genome, I8/1165A, was transfected into LMH cells in the presence or absence of a preS expression vector, ΔDR1 (4). Particles were concentrated from the supernatants and inoculated into six ducklings at 4 days posthatch. After 2 days, cccDNA was selectively extracted from the livers and assayed by PCR (Fig. 3A, left). Only those three ducklings inoculated with enveloped virus particles contained cccDNA by this assay. To determine if the cccDNA detected was derived from linear DNA provided by the inoculated virus, direct sequencing through the r region was performed on two of the samples (Fig. 3A, right). Only the product from the duckling infected with enveloped virus particles produced a sequence ladder, as expected. More importantly, the sequence ladder showed a high degree of degeneracy beginning approximately at the boundary of the r region, consistent with a population of cccDNA molecules formed by nonhomologous recombinations at different sites. This result indicated that linear DNA-containing virus particles had initiated an infection through the production of cccDNA by nonhomologous recombination.
FIG. 3.
Infection of ducklings with the I8 mutant virus. (A) Specificity of the assay for infected hepatocytes. Supernatants from LMH cells transfected with the DHBV I8/1165A plasmid, defective in preS envelope protein, with and without an envelope expression vector (ΔDR1), were used to infect 4-day-old ducklings (1.3 × 109 and <106 enveloped viral genomes per bird, respectively). The ducklings were sacrificed at day 2 postinfection, and cccDNA was extracted from the livers and amplified by PCR (left panel). Two samples were subjected to direct sequencing. (B) In a parallel experiment, ducklings were infected with DHBV/I8 virus containing a wild-type preS gene. Two ducklings were sacrificed at the indicated times up to day 23 postinfection, and replicative intermediates and cccDNA were extracted from their livers. Fractions containing replicative intermediates (equivalent to 5 mg of liver tissue from ducklings sacrificed up to day 8 postinfection) were analyzed for viral DNA by agarose gel electrophoresis and blot hybridization with a 32P-labeled riboprobe specific for detection of the minus strand. (C) cccDNAs from the samples used for panel B were subjected to PCR and direct sequencing across the r region. Degeneracy in the sequence ladder indicates the presence of different sequences in the template population. wt, wild type.
Emergence of revertants following infection with linear DNA.
We next followed the progress of virus infection initiated by linear DNA. Twelve ducklings were infected with virus particles containing the linear I8 genome produced in LMH cells, and the livers were removed at various times postinfection. Viral replicative intermediates were extracted from the livers harvested through 8 days postinfection to determine the extent of replication and spread of infection. Southern blot analysis of these replicative intermediates (Fig. 3B) showed detectable viral DNA signals beginning at day 2 postinfection, increasing through day 8. All virus replication detectable by Southern blot hybridization was apparently due to infection with spontaneously occurring viral variants that carry out legitimate replication, since rcDNA was present in normal proportions (overexposure of the film makes this difficult to see). The data are consistent with a rapid emergence and spread of such phenotypically revertant viruses within and from cells originally infected with linear DNAs.
This interpretation was confirmed by analysis of the cccDNAs that were present in the livers of the ducklings infected with the I8 genome. cccDNAs extracted from these livers were subjected to PCR amplification, and the sequences around the r regions were determined by direct sequencing of the products. As shown in Fig. 3C, only a very weak sequence ladder was visible in these assays in the sample harvested at 1 h postinfection, but in samples taken at 1 day and beyond, strong sequence ladders were produced. A high degree of degeneracy of the sequence ladder at 1 and 2 days postinfection was seen in this experiment as in the previous experiment (Fig. 3A), and this degeneracy was substantially reduced at 4, 8, and 23 days, when high levels of virus replication were detectable by Southern blot analysis (Fig. 3B). As early as 4 days, the major species were resolved into two r sequence lengths, of eight and nine nucleotides (as indicated by doublet bands throughout the regions in the sequencing gel above the r region), apparently consisting of more than one sequence for each species (see below). This result is consistent with the conclusion that the genomes responsible for legitimate replication were derived from the input linear DNA rather than from contamination with wild-type virus.
Illegitimate replication following infection with linear DNA.
Evidence for illegitimate replication occurring in the livers of ducks infected with linear DNA-containing viruses was obtained by amplifying, cloning, and sequencing individual r regions from the cccDNA of a duck infected with the I8 mutant and harvested at day 2 postinfection. As indicated by the direct sequencing experiments shown in Fig. 3A and C, the cccDNA molecules present at day 2 were highly diverse with respect to the sequences around the r region. Of 67 individual clones sequenced in this experiment, no clone was found to contain a wild-type r sequence, which could only be derived by a single type of nonhomologous recombination. Sixty-four of 67 r sequences were apparently formed by nonhomologous recombination, since they differed from both the wild-type and the I8 sequences. The three r sequence clones that matched the I8 sequence could have been derived either by homologous recombination or by amplification of trace amounts of contaminating plasmid DNA from the I8 expression vector.
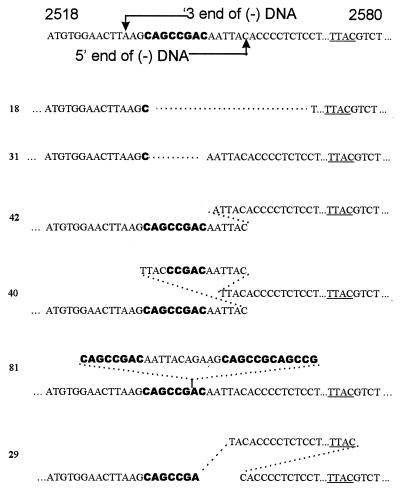
Of 64 clones with apparent nonhomologous recombination joints, 46 clones had sequences that could have been generated by a single recombination event between the ends of the linear I8 DNA. The sequences of three such clones are shown in Fig. 4. Clone 18 exemplifies one class of recombinants in which the loss of a cis-acting sequence (DR1 in this case) would render pregenomes produced by this cccDNA largely inactive for further DNA synthesis. Twelve of the 46 single-site recombinants were presumed to be inactive in further DNA synthesis because of deletions extending into DR1 or the pol open reading frame. Clone 31 is an example of a partial revertant clone, identical to the I1 mutant, which would be capable of supporting further DNA synthesis, with the production of excess levels of linear DNA and a second generation of cccDNAs by both legitimate and illegitimate replication. In clone 42 a nonhomologous recombination generated an additional insertion of 5 bp in the r region. In this clone, DNA synthesis would be expected to occur, but only linear DNA would be produced because of the excessive length of the r sequence. This type of recombinant would be likely to produce a second generation of cccDNAs through illegitimate replication.
FIG. 4.
Sequences of individual clones of r regions formed by nonhomologous recombination in I8-infected ducklings at 2 days postinfection. Nucleotides derived from the I8 insertion are shown in bold and are not included in the numbering system. A reference I8 sequence between nucleotides 2518 and 2580 is shown at the top, and the positions of the two ends of in situ-primed linear DNA, defined by the 5′ and 3′ ends of the minus strand, are indicated. Nucleotides 2549 to 2572 are omitted for brevity. The sequences show evidence of single (18, 32, and 42) or multiple (40, 81, and 30) nonhomologous recombinations. See the text for further explanation. The site of reverse transcription of the minus-strand primer in the 3′ ε is underlined.
More direct evidence for illegitimate replication was obtained from the sequences of 18 other clones, whose sequences around the r region showed evidence of more than one nonhomologous recombination event, consistent with more than one generation of cccDNA formed from linear DNA. Two such sequences, clones 40 and 81, are shown in Fig. 4. Fourteen of these 18 sequences required two (clone 40) or more recombination events to generate the observed sequence, while 4 sequences required up to 4 generations of cccDNA synthesis to be produced by nonhomologous recombination events, as seen in clone 81. These results support the conclusion that illegitimate replication through several generations of linear DNA occurred within an initial period of less than 2 days following infection of cells with linear DNA.
A third group of recombination joints, previously not seen by us, was observed in five different clones in this library, exemplified by clone 29. The sequence of each of the five clones required two nonhomologous recombination events, one of which occurred between sites near the ends of conventional in situ primed linear DNA. The second recombination event occurred between a site located near the conventional 3′ end of the minus strand and a site at or just upstream of nucleotide 2576 at the 5′ end of the minus strand in linear DNA. The structures of these joints indicated that a double-stranded linear DNA with a long direct repeat on each end might have been the precursor of these cccDNA molecules and that the 5′ end of the minus strand in this linear precursor mapped to the UUAC in the 3′ ɛ of pregenomic RNA. This model is consistent with previous reports (3) that a minor fraction of the minus-strand 5′ end maps to nucleotide 2576. The exact mechanism for generation of such minus strand ends is not known.
Partial revertants of illegitimate replication.
By day 4 postinfection with the I8 mutant, the infection was dominated by genomes carrying out legitimate replication, as judged by the preponderance of rcDNA in the liver. However, among the genomes produced by nonhomologous recombination, some genomes suffering short deletions or insertions would be predicted to carry out a mixture of legitimate and illegitimate DNA synthesis in individual cells. To investigate the course of an infection initiated by such partial revertants, we generated a massive infection with a mutant DHBV genome containing a 2-bp insertion in the r sequence (mutant I2). Virus prepared from this mutant was used to inoculate two 4-day-old ducklings at 5 × 109 viral genomes per duckling. For comparison, two ducklings were inoculated with a similar dose of wild-type virus particles. The ducklings were bled at days 1, 4, 5, and 6 postinfection, and they were sacrificed at day 6 and DNA was extracted from their livers. Results from this experiment are shown in Fig. 5. All ducklings were viremic at day 4 postinfection, after which titers of virus in the blood of both groups decreased (Fig. 5A). The decrease in circulating wild-type virus seen in this experiment is commonly observed in experimentally infected ducklings and occurs in conjunction with sustained levels of replicative intermediates in the liver, as shown in this experiment. We do not know the cause of this reduction in the titer of circulating virus.
The patterns of viral DNA observed in the I2-infected ducklings showed a substantial enrichment in linear double-stranded DNA compared with that of the wild type, suggesting that the I2 virus was propagated in vivo. This mutant phenotype was also seen in the replicative intermediates extracted from liver tissue at day 6 postinfection (Fig. 5B), demonstrating that the production of high levels of linear DNA in the liver, compared with levels seen in wild-type infection, could be obtained in fully infected livers. The presence of the original I2 mutation, detected by direct PCR sequencing of the cccDNA as well as the viral DNA (Fig. 5C), explains the enhanced production of linear DNA and indicates that the replication of partial revertants of insertion mutants such as I8, as represented by the mutant I2, could be sustained in the infected cells for at least 6 days. However, passage of the virus in the sera of these infected ducks through a second group of ducklings at an inoculation dose of 108 viral genomes resulted in a complete reversion to legitimate replication (data not shown). As in the I8 infection, however, the phenotypically revertant viruses were a mixture of different viruses containing r sequences of 8 or 9 bp (see below).
Phenotypically revertant r sequences that carry out legitimate replication.
To determine what r sequences supporting legitimate replication were selected in vivo, viral cccDNA was extracted from the liver of an I8-infected duckling at 23 days postinfection. The r region from the cccDNA was amplified and cloned to produce a library of plasmids, and individual clones were sequenced through the r region. The sequences and frequency of their occurrence among 38 clones analyzed are shown in Table 1. Only one example of a genotypically wild-type revertant was seen among the 38 clones. One r sequence of 10 bp, 26 r sequences of 9 bp, and 11 r sequences of 8 bp were found. All but 2 of 38 examples could have been generated by a single nonhomologous recombination event between the predicted ends of I8 linear DNA. The basis for the selection of this particular collection of non-wild-type r sequences is not understood. While r sequences containing 9 bp were more frequent than any other length, some 8-bp lengths were stable through a subsequent passage in ducklings. To test the stability of these variant r sequences during a second passage, serum (2 × 108 viral genomes) from a duckling infected with the I8 mutant, collected at day 8 postinfection (see Fig. 3B and C), was used to infect a second bird. At day 7 postinfection, viral DNA from the serum and replicative intermediates and cccDNA from the liver were all purified and amplified by PCR for direct sequencing (Fig. 6). The mixed sequence ladder characteristic of the inoculum was still present in virus particles, replicative intermediates, and cccDNA at day 7 postinfection. Because the wild-type sequence was shown to be present in the inoculum, we concluded that this sequence (shown in the ladder on the right) did not have a replication advantage over the mixed sequences under these outgrowth conditions.
TABLE 1.
Different r sequences and their frequencies in a library of PCR products of cccDNAs at day 23 postinfection with the I8 mutant
| r sequence (plus strand)a | Size of r sequence (bp) | Frequency |
|---|---|---|
| AAGCAATTAC | 10 | 1/38 |
| AACAATTAC | 9 | 16/38 |
| AAGCATTAC | 9 | 8/38 |
| AAGAATTAC | 9 | 1/38 |
| AGAATTTAC | 9 | 1/38 |
| AAGATTAC | 8 | 5/38 |
| AAAATTAC | 8 | 5/38 |
| AAGTTTAC | 8 | 1/38 |
Nucleotides derived from the inserted sequence are shown in bold.
FIG. 6.
Stability of non-wild-type r sequences in a second passage. Virus (2 × 108 viral genomes) from an I8-infected duck from the experiment described in the legend for Fig. 3B and C was used as the inoculum for a 4-day-old duckling, and viral DNA was extracted from serum and the liver at day 7 postinfection. DNA was subjected to PCR amplification, and direct sequencing of the PCR product was performed. The sequence of the virus particles used as the inoculum and that of the wild-type virus are shown. In all five samples, the primers used for amplification were outside the cohesive end region, as described in Materials and Methods. As has been previously reported (9), the amplification efficiency of an rcDNA template was greatly reduced but not eliminated with these primers.
DISCUSSION
Illegitimate replication was previously shown to occur in primary hepatocyte cultures (31). These cultures were infected with mutants of DHBV that were defective in rcDNA synthesis as a result of nucleotide substitutions in and around DR1. Illegitimate replication, which occurs at very low levels, could be detected in the experiments because of the suppression of legitimate replication by a stable defect in rcDNA synthesis. In natural infections, illegitimate replication is likely to be initiated by a different process, i.e., by infection of cells with viruses that contain naturally occurring linear double-stranded DNAs of wild-type sequence, produced by in situ priming. Linear DNAs were shown to be converted efficiently to cccDNA by nonhomologous recombination, and those cccDNAs suffered deletions or insertions of nucleotides within the r region, changing the length of the r region. Alterations in the length of the r sequence have been shown to increase in situ priming of plus-strand DNA synthesis, independent of any DR1-associated mutation (15).
As a result of changes in the r sequence, cccDNA synthesized during the infection of a hepatocyte by naturally occurring linear DNA-containing virus would frequently carry out only linear DNA synthesis and illegitimate replication. These types of mutations are easily reverted, however, in a subsequent generation of nonhomologous recombination by insertion or deletion of the exact number of nucleotides that were originally deleted or inserted, respectively, in the r region during the initial nonhomologous recombination event. Because of the high level of linear DNA synthesis (5 to 10%) in a wild-type infection, it is possible that cycles of illegitimate replication are repeatedly initiated. We speculated that in an infected liver, a small proportion of infected cells may be carrying out illegitimate replication at any one time, and that these cells might have a survival advantage under some circumstances, for example, in the presence of antigen-specific cytotoxic T cells. Such cells carrying out illegitimate replication in the liver would be very difficult to detect by biochemical means since illegitimate replication levels are so low in comparison with those of legitimate replication (31). In addition, microscopic methods might not distinguish between such cells and uninfected cells. Even though evidence for the existence of such a population of cells in natural infections is not available, we set out to characterize the fate of DHBV illegitimate replication in such cells.
By these experiments we demonstrated that hepatocytes infected with linear DNAs in vivo were able to carry out illegitimate replication and to give rise to variant viruses that replicate by legitimate replication. Illegitimate replication was readily detected by day 2 postinfection, as judged by the appearance of cccDNAs containing more than one nonhomologous recombination joint per r sequence. Assuming that each joint corresponded to at least one generation of illegitimate replication, we could detect up to four generations occurring within the 2-day postinfection period. As we expected, replication quickly was dominated by variants carrying out legitimate replication by day 4 postinfection. Since the outgrowth of phenotypic revertants resulted in an approximately 50-fold increase in viral replicative intermediates in the liver between days 2 and 4 postinfection it was not possible to follow the fate of illegitimate replication past day 2. We concluded that revertants arose from nonhomologous recombination since most revertants did not contain the wild-type r sequence.
Infection with viruses containing linear DNA also resulted in the production of partial phenotypic revertants containing less extensive changes in the r region. As judged by infection experiments with one such partial revertant, I2, partial revertants possessed the potential to establish an infection in individual hepatocytes and to produce elevated levels of linear DNA in infected cells. Such partial revertants also gave rise to full phenotypic revertants by nonhomologous recombination. Full revertants were able to displace partial revertants in multiple cycles of replication, presumably due to their replication advantage in the synthesis of functional cccDNA. Full revertants did not necessarily contain the wild-type r sequence, and these non-wild-type revertants did not appear to be at a disadvantage compared with wild-type virus in these short-term experiments. This result extends the previous finding of Loeb et al. (14) that some variations of the wild-type r sequence do not result in defects in genome circularization. In addition, this interpretation seems to predict that r sequence variation would be observed among and within the various DHBV strains; however, no such variation has been reported to our knowledge.
The experiments suggest that in natural infections linear DNA may be generated in three different populations of infected hepatocytes. In cells infected with virus particles that contain wild-type rcDNA, linear DNA production is only 5 to 10% that of rcDNA. Since the average cccDNA content of infected cells has been estimated to be between 3 and 20 copies per cell (7, 26), many of the cells in this population would contain cccDNA that was derived only from rcDNA, while others would contain only one or two copies of cccDNA derived from linear DNA. In cells infected with viruses that contained rcDNA with small insertions or deletions in the r region, such as the I2 mutant, the amount of linear DNA synthesized from the original cccDNA can exceed the amount of rcDNA produced. Thus, a large fraction of cccDNA produced by amplification in such cells may be formed from linear DNA, as previously reported (31), and may, therefore, preferentially produce linear DNA. In cells infected with virus particles containing linear DNA, as in the I8 mutant, the original cccDNA molecule formed by nonhomologous recombination would, in most cases, be unable to generate rcDNA, so that the entire pool of amplified cccDNA might be derived from linear DNAs. Such cells would be expected to carry out predominantly illegitimate replication. The three populations of cells that produce linear viral DNA would be predicted to show differences in their levels of virus replication according to the amount of their cccDNA that was derived from rcDNA. These differences could contribute to the ability of a chronic virus infection to survive antigen-specific cell-mediated cytotoxicity or other selective pressures that might be based on the levels of virus replication. In addition, the three populations of cells would be expected to differ, according to the level of linear DNA production, in their susceptibility to insertional mutagenesis by nonhomologous recombination of linear viral DNA with cellular DNA ends generated by DNA damaging agents (19). In the mammalian hepadnavirus woodchuck hepatitis virus, insertional mutagenesis is thought to be an important step in the malignant transformation of hepatocytes (reviewed in reference 1).
ACKNOWLEDGMENT
This work was supported by Public Health Service grant CA-42542 from the National Cancer Institute.
REFERENCES
- 1.Beundia M A. Hepatitis B viruses and hepatocellular carcinoma. Adv Cancer Res. 1992;59:167–226. doi: 10.1016/s0065-230x(08)60306-1. [DOI] [PubMed] [Google Scholar]
- 2.Condreay L, Aldrich C, Coates L, Mason W, Wu T-T. Efficient duck hepatitis B virus production by an avian tumor cell line. J Virol. 1990;64:3249–3258. doi: 10.1128/jvi.64.7.3249-3258.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Condreay L D, Wu T T, Aldrich C E, Delaney M A, Summers J, Seeger C, Mason W S. Replication of DHBV genomes with mutations at the sites of initiation of minus and plus strand synthesis. Virology. 1992;188:208–216. doi: 10.1016/0042-6822(92)90751-a. [DOI] [PubMed] [Google Scholar]
- 4.Horwich A L, Furtak K, Pugh J C, Summers J. Synthesis of hepadnavirus particles containing replication-defective duck hepatitis B virus genomes in cultured Huh-7 cells. J Virol. 1990;64:642–650. doi: 10.1128/jvi.64.2.642-650.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang M, Summers J. Infection initiated by the RNA pregenome of a DNA virus. J Virol. 1991;65:5435–5439. doi: 10.1128/jvi.65.10.5435-5439.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang M, Summers J. pet, a small sequence distal to the pregenome cap site, is required for transcription of the duck hepatitis B virus pregenome. J Virol. 1994;68:1564–1572. doi: 10.1128/jvi.68.3.1564-1572.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jilbert A R, Wu T-T, England J M, Hall P D L M, Carp N Z, O’Connell A P, Mason W S. Rapid resolution of duck hepatitis B virus infection occurs after massive hepatocellular involvement. J Virol. 1992;66:1377–1388. doi: 10.1128/jvi.66.3.1377-1388.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawaguchi T, Nomura K, Hirayama Y, Kitagawa T. Establishment and characterization of a chicken hepatocellular carcinoma cell line LMH. Cancer Res. 1987;47:4460–4464. [PubMed] [Google Scholar]
- 9.Koeck J, Schlicht H-J. Analysis of the earliest steps of hepadnaviral replication: genome repair after infectious entry into hepatocytes does not depend on viral polymerase activity. J Virol. 1993;67:4867–4874. doi: 10.1128/jvi.67.8.4867-4874.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lenhoff R, Summers J. Coordinate regulation of replication and virus assembly by the large envelope protein of an avian hepadnavirus. J Virol. 1994;68:4565–4571. doi: 10.1128/jvi.68.7.4565-4571.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lenhoff R, Summers J. Construction of avian hepadnavirus variants with enhanced replication and cytopathicity in primary hepatocytes. J Virol. 1994;68:5706–5713. doi: 10.1128/jvi.68.9.5706-5713.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lien J M, Aldrich C, Mason W S. Evidence that a capped oligonucleotide is the primer for duck hepatitis B virus plus-strand DNA synthesis. J Virol. 1986;57:229–236. doi: 10.1128/jvi.57.1.229-236.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lien J M, Petcu D, Aldrich C, Mason W S. Initiation and termination of duck hepatitis B virus DNA synthesis during virus maturation. J Virol. 1987;61:2286–2296. doi: 10.1128/jvi.61.12.3832-3840.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loeb D, Gulya K J, Tian R. Sequence identity of the terminal redundancies on the minus-strand DNA template is necessary but not sufficient for the template switch during hepadnavirus plus-strand DNA synthesis. J Virol. 1997;71:152–160. doi: 10.1128/jvi.71.1.152-160.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loeb D, Hirsch R, Ganem D. Sequence-independent RNA cleavages generate the primers for plus strand DNA synthesis in hepatitis B viruses: implications for other reverse transcribing elements. EMBO J. 1991;10:3533–3540. doi: 10.1002/j.1460-2075.1991.tb04917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mandart E, Kay A, Galibert F. Nucleotide sequence of a cloned duck hepatitis B virus genome: comparison with woodchuck and human hepatitis B virus sequences. J Virol. 1984;49:782–792. doi: 10.1128/jvi.49.3.782-792.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mason W S, Aldrich C, Summers J, Taylor J M. Asymmetric replication of duck hepatitis B virus DNA in liver cells (free minus strand DNA) Proc Natl Acad Sci USA. 1982;79:3997–4001. doi: 10.1073/pnas.79.13.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mason W S, Seal G, Summers J. A virus of Pekin ducks with structural and biological relatedness to human hepatitis B virus. J Virol. 1980;36:829–836. doi: 10.1128/jvi.36.3.829-836.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petersen J, Dandri M, Burkle A, Zhang L, Rogler C E. Increase in the frequency of hepadnavirus DNA integrations by oxidative DNA damage and inhibition of DNA repair. J Virol. 1997;71:5455–5463. doi: 10.1128/jvi.71.7.5455-5463.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pugh J C, Yaginuma K, Koike K, Summers J. Duck hepatitis B virus (DHBV) particles produced by transient expression of DHBV DNA in a human hepatoma cell line are infectious in vitro. J Virol. 1988;62:3513–3516. doi: 10.1128/jvi.62.9.3513-3516.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seeger C, Ganem D, Varmus H E. Biochemical and genetic evidence for the hepatitis B virus replication strategy. Science. 1986;232:477–484. doi: 10.1126/science.3961490. [DOI] [PubMed] [Google Scholar]
- 22.Staprans S, Loeb D, Ganem D. Mutations affecting hepadnavirus plus-strand synthesis dissociate primer cleavage from translocation and reveal the origin of linear viral DNA. J Virol. 1991;65:1255–1262. doi: 10.1128/jvi.65.3.1255-1262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Summers J, Mason W S. Replication of the genome of hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell. 1982;29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 24.Summers J, O’Connell A, Millman I. Genome of hepatitis B virus: restriction enzyme cleavage and structure of the DNA extracted from Dane particles. Proc Natl Acad Sci USA. 1975;72:4597–4601. doi: 10.1073/pnas.72.11.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Summers J, Smith P, Horwich A L. Hepadnaviral envelope proteins regulate amplification of covalently closed circular DNA. J Virol. 1990;64:2819–2824. doi: 10.1128/jvi.64.6.2819-2824.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Summers J, Smith P, Huang M, Yu M. Morphogenetic and regulatory effects of mutations in the envelope proteins of an avian hepadnavirus. J Virol. 1991;65:1310–1317. doi: 10.1128/jvi.65.3.1310-1317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Summers J, Smolec J M, Snyder R. A virus similar to hepatitis B virus associated with hepatitis and hepatoma in woodchucks. Proc Natl Acad Sci USA. 1978;75:4533–4537. doi: 10.1073/pnas.75.9.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tuttleman J, Pourcel C, Summers J. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell. 1986;47:451–460. doi: 10.1016/0092-8674(86)90602-1. [DOI] [PubMed] [Google Scholar]
- 29.Wei Y, Tavis J E, Ganem D. Relationship between viral DNA synthesis and virion envelopment in hepatitis B viruses. J Virol. 1996;70:6455–6458. doi: 10.1128/jvi.70.9.6455-6458.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu T-T, Coates L, Aldrich C E, Summers J, Mason W S. In hepatocytes infected with duck hepatitis B virus, the template for viral RNA synthesis is amplified by an intracellular pathway. Virology. 1990;175:255–261. doi: 10.1016/0042-6822(90)90206-7. [DOI] [PubMed] [Google Scholar]
- 31.Yang W, Summers J. Illegitimate replication of linear hepadnaviral DNA through nonhomologous recombination. J Virol. 1995;69:4029–4036. doi: 10.1128/jvi.69.7.4029-4036.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang W, Mason W S, Summers J. Covalently closed circular viral DNA formed from two types of linear DNA in woodchuck hepatitis virus-infected liver. J Virol. 1996;70:4567–4575. doi: 10.1128/jvi.70.7.4567-4575.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]