Abstract
Gastric cancer remains the second leading cause of cancer-related deaths worldwide. Although Helicobacter pylori (H. pylori) is considered to be a critical risk factor, the molecular mechanisms underlying H. pylori-induced gastric carcinogenesis are still poorly defined. Recently, accumulating studies have revealed that microRNAs play key roles in development, differentiation, immune regulation, and even carcinogenesis. This study was performed to explore the mechanism of microRNA-375 (miR-375) in H. pylori promotion of gastric carcinogenesis. It was shown that miR-375 was down-regulated in response to H. pylori infection in gastric epithelial cell lines; this finding was quite opposite to the expression patterns of pro-inflammatory cytokines observed in a co-culture cell model. Moreover, the ectopic expression of miR-375 aggravated cell proliferation and migration. It was further observed that Janus kinase 2 (JAK2) was a bona fide target of miR-375 and further activated signal transducer and activator of transcription 3 (STAT3) and other downstream target molecules. Both gain-of-function and loss-of-function experiments showed that decreased miR-375 expression could mimic the oncogenic effects of the JAK2–STAT3 pathway. In addition, pretreatment with siRNAs targeting JAK2 prevented gastric epithelial cells from increasing proliferation and migration even in response to H. pylori infection. For the first time, our results demonstrate that the JAK2–STAT3 pathway regulated by miR-375 is involved in H. pylori-induced inflammation; this pathway promotes neoplastic transformation by affecting the expression of BCL-2 and TWIST1, hence offering a potential therapeutic target for inflammation-related cancers, especially those related to H. pylori.
Electronic supplementary material
The online version of this article (doi:10.1007/s00262-014-1550-y) contains supplementary material, which is available to authorized users.
Keywords: miRNA-375, Helicobacter pylori, Gastric carcinogenesis, Inflammation, JAK2–STAT3
Introduction
Gastric cancer is one of the most common diseases worldwide and has an extremely high mortality that is only second to lung cancer. Helicobacter pylori (H. pylori) is considered to be a critical risk factor during the process of gastric carcinogenesis [1]. Infection with H. pylori causes strong and persistent immune responses, inducing changes in the expression of inflammatory molecules, such as cytokines and other immune mediators [2]. These immune mediators activated by H. pylori initiate the important step in inflammation-promoted neoplastic transformation. However, the precise molecular interactions between these pivotal mediators and the way in which they participate in H. pylori-induced gastric carcinogenesis remain to be elucidated.
MicroRNAs (miRNAs) are one such family of inflammatory mediators. They are typically 18–24 nucleotide noncoding RNAs with the ability to regulate messenger RNA (mRNA) expression at the posttranscriptional level by binding to the 3′ un-translated regions (UTR) of target mRNAs and further inducing mRNA degradation or translation suppression [3, 4]. Previous studies have indicated that miRNAs play key roles in development, differentiation, cell proliferation, tumor metastasis, immune regulation, and many other biological processes [5]. Indeed, miRNAs act as oncogenes or tumor suppressors by targeting the corresponding mRNAs.
Recent reports have revealed that H. pylori infection or chronic inflammation can cause an aberrant expression of miRNAs [6, 7]. Zhang et al. [8] found that miR-21 was significantly up-regulated in human gastric cancer tissues and in H. pylori-infected gastric mucosa and that this up-regulation enhanced cell proliferation and invasion in a gastric cancer cell line. Petrocca et al. [9] compared nontumor tissues with histological signs of chronic gastritis to normal mucosa and identified seven miRNAs that are associated with chronic inflammation. Statistical analyses indicated that miR-223, miR-155, and miR-200c are the most relevant miRNAs in the colonization density of H. pylori, which plays crucial roles in gastric cancer pathogenesis and progression [2, 10–12]. miR-375 was also previously reported to be involved in gastric cancer [13], but the role of miR-375 in H. pylori-induced gastritis or even gastric carcinogenesis is poorly defined. Recently, Isomoto et al. [2] found that 17 miRNAs (including miR-375) had significant correlations with H. pylori-associated gastritis based on endoscopic biopsies. Based on the complex role of miR-375 in different cancers [14–17], we finally focused on miR-375 and its involvement in H. pylori-induced gastric cancer onset and progression.
The JAK–STAT (Janus kinase–signal transducer and activator of transcription) signaling pathway is involved in numerous aspects of development, immune system regulation, and cell physiology [18]. The binding of ligands to their receptors leads to the activation of JAKs, which in turn phosphorylate and activate STATs. In particular, the aberrant activation of JAK2–STAT3 signaling is now considered to be closely linked to inflammation-associated tumorigenesis initiated by direct transcriptional regulation of target genes in malignant cells [19]. Activated STAT3 has been reported to induce cancer transformation by elevating anti-apoptotic genes and pro-metastatic genes, such as BCL-2 and TWIST1, respectively [20–23]. Nevertheless, whether this pathway is involved in H. pylori-induced gastric carcinogenesis has not yet been reported.
In this study, we confirmed that miR-375 was deregulated after H. pylori infection, and the potential mechanism underlying this effect was further investigated. This is the first report on the role of miR-375 in regulating the JAK2–STAT3 pathway and facilitating H. pylori-induced gastric carcinogenesis.
Materials and methods
Cell lines and H. pylori culture
The human embryonic kidney cell line HEK-293T was obtained from the ATCC and cultured in DMEM (Gibco) supplemented with 10 % fetal bovine serum (FBS, Gibco) in a humidified atmosphere at 37 °C in 5 % CO2. The human gastric cancer cell lines BGC-823, AGS, SGC-7901, and MKN-45 were purchased from the China Academia Sinica Cell Repository (Shanghai). The GES-1 immortalized gastric epithelial cell line was a gift from the Shanghai Institute of Digestive Disease. All of the gastric cancer cell lines and GES-1 cells were maintained in RPMI 1640 medium (Gibco) supplemented with 10 % FBS (Gibco) under the same conditions as the HEK-293T cells. The standard strain of H. pylori 26695 was obtained from the ATCC and grown on brain–heart infusion (BHI) plates containing 8 % goat blood, trimethoprim (5 μg/ml), polymyxin B (5 μg/ml), and vancomycin (10 μg/ml) in a microaerobic humidified atmosphere at 37 °C. After 48 h of culture, H. pylori was harvested and re-suspended in RPMI 1640 at a final concentration of 3 × 108 colony forming units (CFU)/ml. H. pylori was added to the cells at a multiplicity of infection (MOI) of 100:1 and co-cultured with cell lines for the indicated times [11].
Tissue specimens
Patients undergoing gastrointestinal endoscopy at the Anhui provincial hospital were enrolled. General information about these patients is provided in Supplementary Table 1. In total, tissues were harvested from ten patients with gastric cancer at the time of surgery, and another eight tissues from patients with chronic gastritis and eight matched normal tissues were collected. The diagnoses of all the specimens were histopathologically confirmed. The study was approved by the Ethics Committee of China Pharmaceutical University.
ELISA
After co-culture of GES-1 or BGC-823 cells with H. pylori at a MOI of 100:1, the release of the pro-inflammatory cytokines IL-8 and TNF-α in the culture supernatant was determined by corresponding ELISA kits (R&D system, USA) according to the manufacturer’s instructions.
Cell transfection
GES-1 and BGC-823 cells were seeded in six-well plates and transfected at approximately 50 % confluence (3 × 105 cells). Mimics, inhibitors, and scrambled negative control (NC) of miR-375 were purchased from Biomics Biotechnology. The BCL-2, TWIST1, and JAK2 siRNAs were also purchased from Biomics Biotechnology. Transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Total RNA or total protein was extracted at the designated time points after transfection.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted using Trizol reagent (Invitrogen, USA). Reverse transcription was performed with M-MLV reverse transcriptase (Promega, USA) following standard protocols. For quantitative PCR of miRNAs, the miR-375, U6 snRNA primer, and EzOmics SYBR qPCR kit were purchased from Biomics. The amplification procedure was as follows: 94 °C for 5 min, followed by 30 cycles at 94 °C for 30 s, 62 °C for 30 s, and finally 72 °C for 10 min. For the quantification of mRNAs, the primer sequences were provided in Supplementary Table 2. The expression of U6 snRNA or GAPDH was assayed for normalization. All reactions were performed in triplicate. The relative expression levels were determined using the 2−ΔΔCt analysis method.
Luciferase assay
Luciferase reporters were constructed as described previously [24]. Wild-type and mutant sequences containing fragments of the 3′-UTR of JAK2 mRNA, which contains binding sites for miR-375, were synthesized by Sangon Biotech (Shanghai, China) and annealed according to the manufacturer’s instructions. The annealed oligonucleotides were digested and ligated at HindIII and SpeI sites into the pMIR-Report Fluc vectors (Ambion, USA). The sequences were provided in Supplementary Table 3. HEK-293T cells were co-transfected with the recombinant pMIR-Report vectors, miR-375 mimics or NC, and pMIR-Report β-gal control plasmid (Ambion) using Lipofectamine 2000. Forty-eight hours after transfection, luciferase activity was measured and normalized to β-galactosidase activity.
Western blotting and immunohistochemistry (IHC)
The proteins (60 μg) were subjected to electrophoresis through 10 % SDS-polyacrylamide gels and transferred onto PVDF membranes (Millipore). The membranes were blocked overnight with 5 % nonfat milk at 4 °C and incubated with anti-JAK2 RabMab (1:1,000, Cell Signaling Technology, CST), anti-phospho-JAK2 (Tyr1007/1008) RabMab (1:1,000, CST), anti-STAT3 RabMab (1:1,000, CST), anti-phospho-STAT3 (Tyr705) RabMab (1:2,000, CST), anti-BCL-2 rabbit polyconal Ab (1: 750, Signalway Antibody, SAB), or anti-TWIST1 rabbit polyconal Ab (1:750, SAB) as indicated for 12–16 h at 4 °C or with anti-β-actin monoclonal antibody (1:500, ZSGB-Bio) as a control. After three washes with TBST, the secondary antibody was added according to standard procedures. The membrane was washed another three and developed using the ECL system (Amersham Pharmacia, GE Healthcare Bio-Sciences AB, Uppsala, Sweden). The levels of JAK2 in the tissue specimens were determined by IHC staining with primary antibody (JAK2, 1:100) incubation at 4 °C overnight. The experiments were conducted by Wuhan Google Biotechnology.
Cell proliferation assay and colony formation
Cells were seeded in 96-well plates at approximately 3,000 cells per well and incubated at 37 °C the day before transfection or H. pylori infection. The growth of the cells was determined using a Cell Counting Kit-8 assay (Beyotime) according to the manufacturer’s protocol. Cells transfected with miRNAs for 24 h were seeded in 6-well plates (300 cells per well) for approximately 10 days. The cells were stained with crystal violet, and the colonies containing more than 50 cells were counted.
Transwell migration assay
The migration assay was performed using transwell insert chambers (8-μm pore size, Millipore, USA). A total of 1 × 105 cells transfected with miRNAs for 24 h or infected with H. pylori for 12 h were placed into the upper chamber in serum-free medium. The lower chamber was filled with complete medium containing 10 % fetal bovine serum. After incubation for another 24 h, the migrated cells were fixed with methanol and stained with crystal violet. Five random fields from each of the triplicate migration assays were counted using phase-contrast microscopy. The dye was eluted with 33 % acetic acid, and the absorbance was measured at 570 nm.
Statistical analysis
All statistical analyses were performed using SPSS Statistical software (SPSS version 16.0, Chicago, USA). The values are presented as the mean ± SD of three independent experiments. The result’s comparisons among multiple groups were performed using analysis of variance (ANOVA) followed by Student’s t-tests. And the result’s comparisons between two groups were examined by Student’s t test. * P < 0.05 was considered statistically significant compared with negative control group (NC).
Results
The expression of pro-inflammatory cytokines is up-regulated in response to H. pylori infection in gastric epithelial cell lines
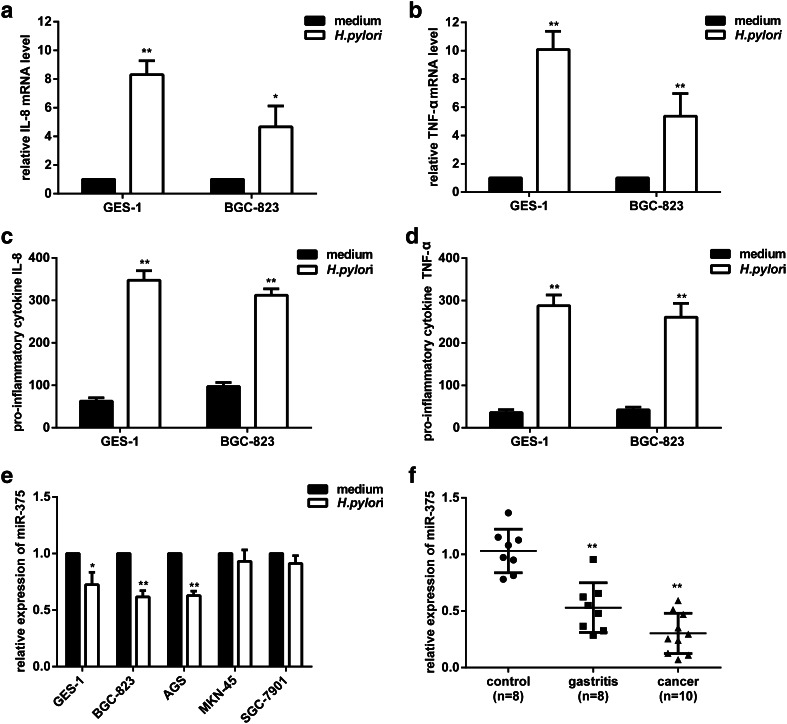
To determine whether H. pylori infection is associated with the inflammatory responses of gastric epithelial cells, we examined the release of pro-inflammatory cytokines. As previously reported [2], the immune response caused by H. pylori infection was characterized by a profile of cytokines, including IL-1β, IL-6, IL-8, and TNF-α. Here, we chose IL-8 and TNF-α as representatives of the humoral and cellular immune responses; these factors were tested after H. pylori infection in a human nonmalignant gastric epithelial cell line (GES-1) and a poorly differentiated gastric adenocarcinoma cell line (BGC-823). qRT-PCR results (Fig. 1a, b) showed significant increases in IL-8 and TNF-α mRNA levels after H. pylori infection. The results of ELISA assays further confirmed that IL-8 was obviously stimulated in GES-1 (5.5-fold) and BGC-823 (3.2-fold) cells (Fig. 1c). Meanwhile, TNF-α was also noted for their greatest changes of elevated expression in GES-1 (8.0-fold) and BGC-823 (6.2-fold) cells (Fig. 1d). These data suggested that H. pylori infection caused strong immune responses that may be implicated in the process of neoplastic transformation.
Fig. 1.
Pro-inflammatory cytokines are up-regulated, and miR-375 is down-regulated in response to H. pylori infection of gastric epithelial cell lines or tissue specimens of gastritis and gastric cancer. a, b GES-1 and BGC-823 cells were infected with H. pylori for 12 h; total RNA was extracted, reverse-transcribed, and the levels of IL-8 (a) and TNF-α (b) mRNA were quantified by qRT-PCR. The results were normalized to the noninfected cells. c, d GES-1 and BGC-823 cells were infected with H. pylori for 24 h; the culture supernatant was collected, and the levels of IL-8 (c) and TNF-α (d) protein were determined by ELISA assays. e The relative expression of miR-375 in GES-1, BGC-823, AGS, MKN-45, and SGC-7901 cells was quantified by qRT-PCR after 12 h of H. pylori infection. The results were normalized to the noninfected cells. f The relative expression of miR-375 in gastric tissue specimens. Each point represents one sample. The data are presented as the mean ± SD of triplicate samples, *P < 0.05, **P < 0.01 versus the control group
miR-375 expression is significantly decreased in response to H. pylori infection and in gastric tissue specimens
To explore the potential role of miR-375 in carcinogenesis related to H. pylori infection, we measured its expression level first. Compared with the negative controls, qRT-PCR revealed that miR-375 levels were reduced by 27.6, 38.3, and 37.2 % in GES-1, BGC-823, and AGS cells, respectively, while H. pylori infection did not significantly decrease miR-375 expression in the SGC-7901 and MKN-45 cell lines (Fig. 1e). However, we also found that miR-375 was markedly down-regulated in human gastric cancer specimens (n = 10) and chronic gastritis specimens (n = 8) relative to matched normal tissues (n = 8) (Fig. 1f). These consistent results suggested that H. pylori infection may contribute to a reduction in miR-375 expression in vitro and in vivo.
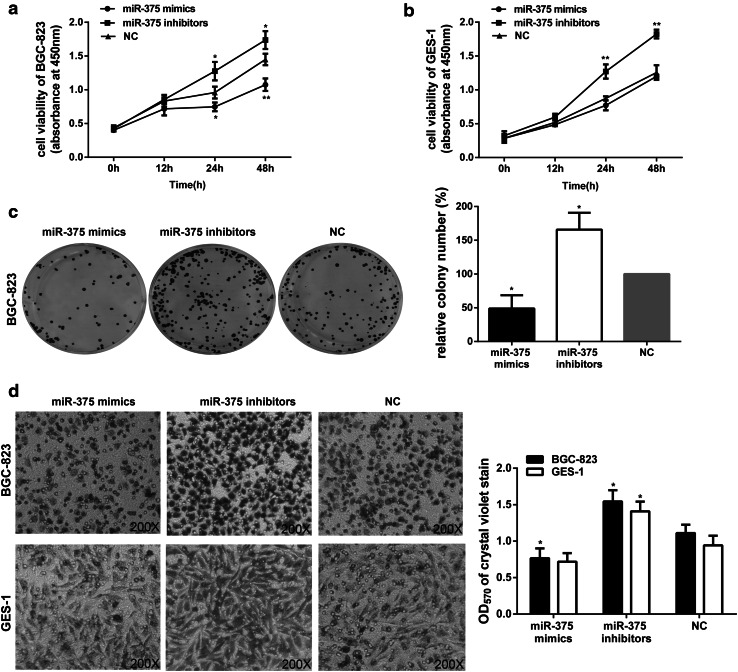
miR-375 expression is inversely correlated with cell proliferation and migration in gastric epithelial cell lines
To verify whether miR-375 functions as a tumor suppressor, the correlation between low-level miR-375 expression and gastric carcinogenesis was tested. First, we demonstrated the effects of miR-375 on cell proliferation. After transfection with miR-375 mimics, inhibitors, and NC, the relative expression of miR-375 in GES-1 and BGC-823 cell lines was measured by qRT-PCR (Supplementary Fig. 1). The cells transfected with the indicated oligonucleotides were then subjected to CCK-8 analysis at different time points. As expected, the miR-375 mimics showed a notable inhibition of proliferation in BGC-823 cells, and the miR-375 inhibitors promoted cell proliferation (Fig. 2a). Surprisingly, miR-375 mimics had almost no growth inhibitory effects in GES-1 cells, though the cells exhibited a much higher growth rate after the addition of miR-375 inhibitors (Fig. 2b). These unexpected results may be due in part to the fact that miR-375 expression level in nonmalignant gastric epithelial cell line (GES-1) was not as low as other cancer cell lines (Supplementary Fig. 2). Therefore, the proliferation rate of GES-1 cells was nearly unaffected by treatment with miR-375 mimics under basic conditions or without H. pylori infection. On the contrary, the overexpression of miR-375 in both GES-1 and BGC-823 cells could markedly suppress cell growth when the cells were infected with H. pylori (Supplementary Fig. 3).
Fig. 2.
The effects of miR-375 expression on cell proliferation and migration in GES-1 and BGC-823 cells. a, b BGC-823 (a) and GES-1 (b) cells treated with the indicated oligonucleotides (miR-375 mimics, inhibitors, and NC) at 100 nM were subjected to CCK-8 assays at the indicated time points. c The colony numbers of BGC-823 cells were detected after transfection of the indicated oligonucleotides. The relative percentage of the NC group is designated as 100 %. d The migratory ability of BGC-823 and GES-1 cells was evaluated by transwell assays after transfection of the indicated oligonucleotides. Representative image fields of the migratory cells on the membrane stained with crystal violet (×200), and OD570 of crystal violet stain was determined to quantify the transwell assays. The data are represented as the mean ± SD of triplicate samples, *P < 0.05, **P < 0.01 versus the control group
The results of BGC-823 cells were confirmed by colony formation assays; the results of which were consistent with the CCK-8 analysis. As shown in Fig. 2c, the colony number of BGC-823 cells transfected with miR-375 inhibitors was significantly higher than that of the control groups, and the overexpression of miR-375 resulted in far fewer colonies. Altogether, these results suggested that miR-375 may function as a tumor suppressor, at least by suppressing cell proliferation.
We next assessed the requirement of miR-375 for cell migration. As shown in Fig. 2d, transfecting both GES-1 and BGC-823 cells with miR-375 inhibitors increased cell migration to a large extent. On the contrary, treating BGC-823 cells with miR-375 mimics for 24 h resulted in a significantly lower rate of migration, but this effect was reduced in GES-1 cells. This difference may be due to the relatively high basal level of miR-375 in GES-1 cells.
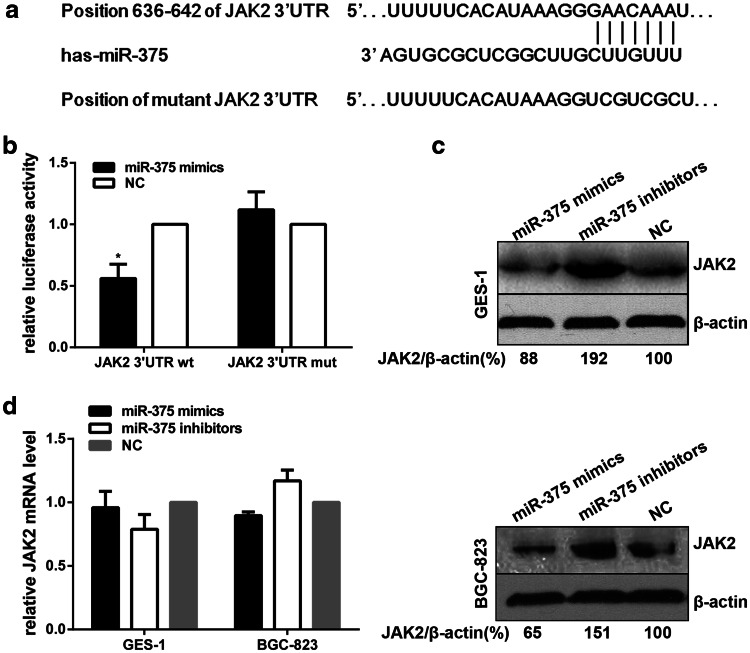
JAK2 is a bona fide target of miR-375
We used TargetScan (version6.2, http://www.targetscan.org/) to identify potential target genes of miR-375; found that JAK2, a known oncogene, might be a potential one with a putative miR-375 binding sites within its 3′UTR (Fig. 3a). To verify whether JAK2 is a bona fide target of miR-375, two luciferase reporter vectors containing the putative miR-375 binding sites in the wild-type JAK2 3′UTR (WT) and mutant 3′UTR (MUT) were constructed. As shown in Fig. 3b, co-transfection of HEK-293T cells with miR-375 mimics and the pMIR-Report-JAK2 3′UTR WT construct led to a significant reduction in luciferase activity compared with transfection with the NC. Conversely, no effect of luciferase activity in pMIR-Report-JAK2 3′UTR MUT was found by miR-375 up-regulation, suggesting that JAK2 is a direct target of miR-375. These results were further confirmed by western blotting, which demonstrated that the over-expression of miR-375 indeed reduced JAK2 protein expression in both GES-1 and BGC-823 cells (Fig. 3c). However, no significant difference in JAK2 expression at the mRNA level was found, indicating that miR-375 represses JAK2 at the posttranscriptional level (Fig. 3d). In summary, we conclude that JAK2 is a bona fide target of miR-375.
Fig. 3.
miR-375 directly targets JAK2. a Predicted binding sites for miR-375 in the 3′-UTR of human JAK2 mRNA (wild type and mutant). b HEK-293T cells were co-transfected with recombinant pMIR-Report-JAK2 3′UTR (wild type or mutant), miR-375 mimics or NC, and pMIR-Report β-gal control plasmid for 48 h, then luciferase activity was measured and normalized to β-galactosidase activity. c Western blotting analysis of JAK2 protein expression in GES-1 and BGC-823 cells after transfection of the indicated oligonucleotides at 100 nM for 48 h; β-actin expression was used as an internal control. The results were normalized to the blots transfected with NC of miR-375. d qRT-PCR results of JAK2 mRNA in GES-1 and BGC-823 cells after transfection of the indicated oligonucleotides at 100 nM for 24 h. The results were normalized to the mRNA level transfected with NC of miR-375. The data are presented as the mean ± SD of triplicate samples, *P < 0.05 versus the control group
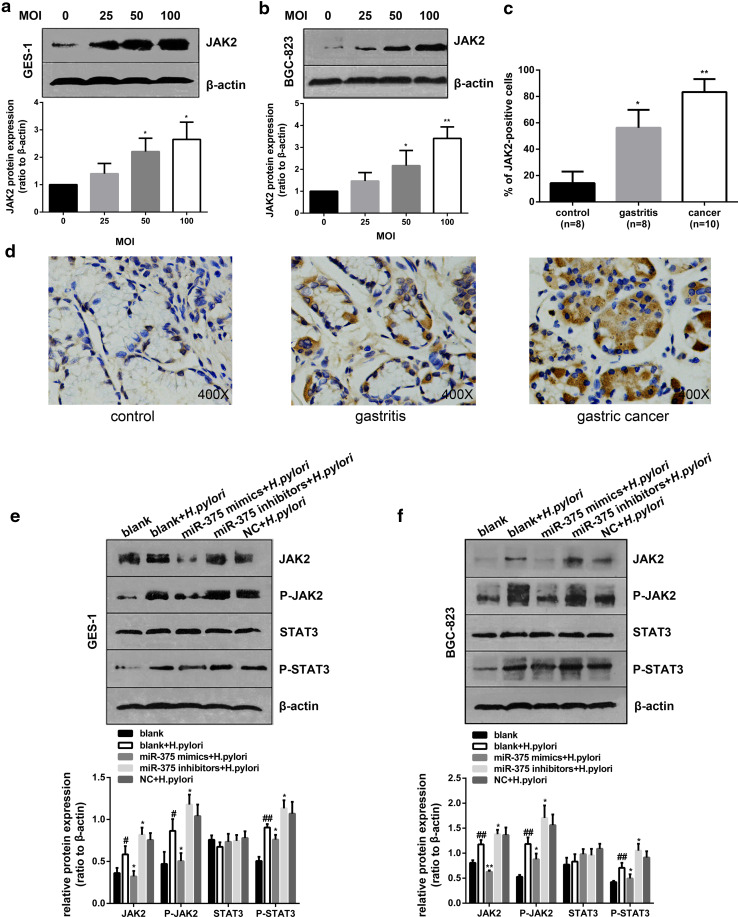
The regulation of JAK2–STAT3 signaling by miR-375 is involved in H. pylori-induced cell proliferation and migration
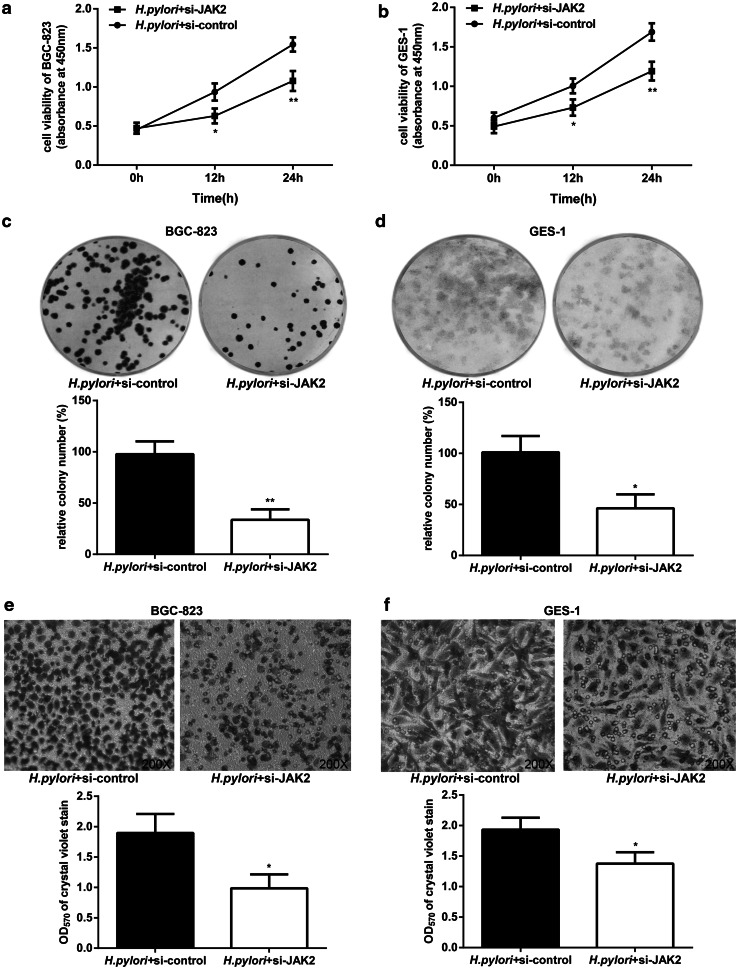
As described above, H. pylori significantly lowered the level of miR-375 in GES-1 and BGC-823 cells. We also found a strong increase in JAK2 protein expression in these two cell lines in response to H. pylori infection (Fig. 4a, b). Meanwhile, we investigated the expression of JAK2 in gastric specimens by IHC assays and observed a significant up-regulation in gastritis (56.3 % positive cells) and cancer (83.7 % positive cells) as compared with matched normal tissues (14.3 % positive cells) (Fig. 4c, d). Because we confirmed that JAK2 is a direct target of miR-375, we further explored whether JAK2 is involved in the anti-cancer effects of miR-375. As shown in Fig. 4e, f, the overexpression of miR-375 could inhibit the H. pylori-induced up-regulation of JAK2 and phospho-JAK2 (P-JAK2) in GES-1 and BGC-823 cells, which implies that H. pylori infection may activate JAK2 signaling by attenuating miR-375 expression. To further confirm the role of JAK2 in the anti-neoplastic effects of miR-375, we silenced JAK2 by RNAi in both GES-1 and BGC-823 cells. The effect of JAK2 siRNA on JAK2 mRNA was determined by qRT-PCR (Supplementary Fig. 4a). As shown in Fig. 5, specific JAK2 siRNAs markedly suppressed cell proliferation and migration. Therefore, we can conclude that JAK2 serves as a direct mediator of the antitumor effects of miR-375 involved in H. pylori-induced gastric carcinogenesis.
Fig. 4.
H. pylori infection elevates JAK2 expression and activates the JAK2–STAT3 signaling pathway mediated by miR-375 regulation. a, b Western blotting analysis and quantification of JAK2 expression over the indicated MOI after H. pylori stimulation of GES-1 (a) and BGC-823 (b) cells for 48 h; β-actin expression was used as an internal control. The results were normalized to the blots at a MOI of 0. c Quantitative immunohistochemistry results of percentage of JAK2-positive cells in control, gastritis, and cancerous gastric tissues. d Immunohistochemistry staining of JAK2 expression in gastric tissues. The data are presented as the mean ± SD of triplicate samples, *P < 0.05, **P < 0.01 versus the control group. e, f The effects of miR-375 regulation on H. pylori-activated JAK2–STAT3 signaling in GES-1 (e) and BGC-823 (f) cells were determined by western blotting and quantitative analysis; β-actin expression was used as an internal control. The data are presented as the mean ± SD of triplicate samples, # P < 0.05, ## P < 0.01 versus the control group of the blank and *P < 0.05, **P < 0.01 versus the control group of the blank + H. pylori
Fig. 5.
The effects of specific JAK2 siRNAs on H. pylori-induced cell proliferation and migration in GES-1 and BGC-823 cells. a, b CCK-8 results of BGC-823 (a) and GES-1 (b) cells after JAK2 silencing and H. pylori infection for the indicated times. c, d The colony numbers of BGC-823 (c) and GES-1 (d) cells after JAK2 silencing and H. pylori infection for 12 h. e, f The migratory ability of BGC-823 (e) and GES-1 (f) cells was evaluated by transwell assays after JAK2 silencing and H. pylori infection for 12 h. Representative image fields of the migratory cells on the membrane stained with crystal violet (×200), and OD570 of crystal violet stain was determined to quantify the transwell assays. The data are represented as the mean ± SD of triplicate samples, *P < 0.05, **P < 0.01 versus the control group
It is widely accepted that the activation of JAK2–STAT3 cascade is involved in a variety of pathologies, including cancer transformation. We therefore determined the expression pattern of phospho-STAT3 (P-STAT3), which was largely consistent with that of P-JAK2, as shown in Fig. 4e, f. As a significant transcription factor, STAT3 can contribute to carcinogenesis by up-regulating target oncogenes, such as BCL-2 and TWIST1 [20, 22]. As expected, both qRT-PCR (Fig. 6a, b) and western blotting analysis (Fig. 6c, d) revealed that a pretreatment with specific JAK2 siRNAs could inhibit the elevation of BCL-2 and TWIST1 caused by H. pylori. Thus, we further knocked down BCL-2 and TWIST1 by RNAi to clarify the molecular mechanism of H. pylori-induced cell proliferation and migration regulated by the JAK2–STAT3 cascade (Supplementary Fig. 4b and 4c). As shown in Fig. 6e–h, a pretreatment with specific BCL-2 siRNAs inhibited cell proliferation, while the knockdown of TWIST1 significantly improved cell migration induced by H. pylori. All together, these results suggested that the regulation of JAK2–STAT3 signaling by miR-375 participates in H. pylori-induced cell proliferation and migration by activating the downstream target genes BCL-2 and TWIST1.
Fig. 6.
The regulation of JAK2–STAT3 signaling pathway is involved in H. pylori-induced cell proliferation and migration by activating the downstream target genes BCL-2 and TWIST1. a, b qRT-PCR results of BCL-2 and TWIST1 mRNA in BGC-823 (a) and GES-1 (b) cells after JAK2 silencing and H. pylori infection for 12 h; GAPDH was used as an internal control. The results were normalized to the mRNA level transfected with negative control of siRNAs. c, d Western blotting and quantitative analysis of BCL-2 and TWIST1 expression in BGC-823 (c) and GES-1 (d) cells after JAK2 silencing and H. pylori infection for 24 h; β-actin expression was used as an internal control. e, f CCK-8 results of BGC-823 (e) and GES-1 (f) cells after BCL-2 silencing and H. pylori infection for the indicated times. g, h The migratory ability of BGC-823 (g) and GES-1 (h) cells was evaluated by transwell assays after TWIST1 silencing and H. pylori infection for 12 h. Representative image fields of the migratory cells on the membrane stained with crystal violet (×200), and OD570 of crystal violet stain was determined to quantify the transwell assays. The data are represented as the mean ± SD of triplicate samples, *P < 0.05, **P < 0.01 versus the control group
Discussion
Helicobacter pylori infection has long been recognized as a critical risk factor for gastritis and gastric cancer. The complex immune responses caused by H. pylori may account for such neoplastic transformation. However, the immune mediators that underlie the response to H. pylori infection have not been precisely defined, and the mechanisms that associate inflammatory responses with gastric carcinogenesis have not been clearly elucidated. Over the past few years, it has become generally accepted that changes in miRNA expression are involved in the onset and progression of most human cancers, where they act as either oncogenes or tumor suppressors [25]. Very recently, miRNAs emerged as immune mediators, cross talking with numerous cytokines [26]. Consistent with previous studies [13], the expression of miR-375 was found to be low in gastric cancer. Notably, our research indicates that miR-375 expression is significantly decreased in gastric cancer cell lines and in response to H. pylori infection. Among the possible target genes, we found that miR-375 intimately associates with inflammation-provoked malignancy by negatively regulating the oncogene JAK2. Though it has been previously reported that JAK2 may be a target gene of miR-375 [13], we confirmed these results in new gastric epithelial cell lines and to our best knowledge, this is the first report indicating the role of the miR-375–JAK2–STAT3 pathway in H. pylori-induced promotion of gastric carcinogenesis in vitro and in vivo.
As with other cancers in the gastrointestinal tract, such as colon cancer and esophageal adenocarcinoma, gastric cancer is related to chronic inflammation. Indeed, inflammatory responses contribute to almost all types of cancer; nevertheless, unique mechanisms that are associated with the specific factors involved in each class of cancer likely provide different cancer signatures [27]. In this sense, during the process of H. pylori-mediated inflammation-provoked malignancy, it is of great importance to seek out molecular alterations that are specifically induced by H. pylori. As indispensable molecules involved in the innate and acquired immune responses [28], miRNAs understandably came into focus in our study. Recently, Teng et al. [29] found that let-7b is involved in the inflammatory and immune responses associated with H. pylori infection by targeting TLR4, supporting the notion that miRNAs may serve as a unique bridge between H. pylori infection and gastric cancer development. Moreover, miR-370 participated in the progress of H. pylori-induced gastritis toward gastric cancer by targeting FoxM1 and thus affecting the expression of P27Kip1 [30]. Another study revealed that the overexpression of miR-222 in H. pylori-associated gastric cancer correlated with tumor progression by promoting cancer cell proliferation and inhibiting RECK [31]. Altogether, these reports suggest that the up-regulation of onco-miRNAs or the down-regulation of tumor-suppressing miRNAs may aggravate inflammation-promoted neoplastic transformation. It should be noted, however, that the newest investigations only examined cell proliferation as the indicator of gastric carcinogenesis. In fact, the initiation of migration, angiogenesis, and the beginning of apoptosis inhibition also serve as markers of carcinogenesis. In this study, we focused on the effects of H. pylori on the initiation of proliferation and migration. The activation of the signaling molecules of BCL-2 and TWIST1 may explain the phenomenon we observed in cell proliferation and migration.
microRNA-375 is known as a multifunctional miRNA involved in pancreatic islet development, glucose homeostasis, cell differentiation, and more importantly, carcinogenesis [32]. The down-regulation of miR-375 has been observed in multiple types of cancer, for instance, gastric cancer, hepatocellular carcinoma, esophageal carcinoma, and head and neck cancer [13, 15, 33, 34]. However, miR-375 has also been shown to be up-regulated in small-cell lung cancer, breast cancer, and prostate carcinoma [14, 17, 35]. In addition, a recent study reported that elevated miR-375 was found in recurring gastric cancer by targeting a known tumor suppressor p53 gene; this result implies that the expression of miR-375 is not always constant, even in the same type of cancer [36]. These controversial results further reinforce the significance of our study. Despite the previous investigation of miR-375 expression in gastric cancer, it remains to be defined whether and how miR-375 plays an irreplaceable role during the neoplastic transformation caused by H. pylori.
In the present study, we mainly resolve the question addressed above. The inflammatory responses induced by H. pylori suppress the expression of miR-375, elevating JAK2 expression in cells and tissue specimens. The targeting relationship between miR-375 and JAK2 was confirmed by luciferase assays and western blotting analysis. Because JAK2 is a member of the JAK–STAT signaling pathway that provides the link between inflammation and cancer, we further investigated whether STAT3, a downstream molecule of JAK2, or the whole signaling pathway participates in H. pylori-induced gastric carcinogenesis regulated by miR-375. Both gain-of-function and loss-of-function experiments show that increased JAK2–STAT3 activation controlled by miR-375 plays an essential role in gastric cancer progression. However, it is also interesting to note that BGC-823 cells were more sensitive to miR-375 mimics than GES-1 cells, most likely because of the relatively low basal level of miR-375 in BGC-823. Once infected with H. pylori, both cell lines displayed high sensitivity to the overexpression of miR-375.
Recently, accumulating evidence has shown that inflammation is a complex process involving different signaling pathways and mediators. During the initiation of malignant transformation and cancer progression, JAK2–STAT3 signaling is responsible for inducing and maintaining a pro-carcinogenic inflammatory microenvironment [19, 37]. Given its central roles in inflammation and cancer, it is not surprising that signaling by JAK2–STAT3 is highly interrelated with other pathways, such as NF-κB [38]. This type of interconnection partially explains our results that BCL-2 and TWIST1 are not completely eliminated by both JAK2 siRNAs and H. pylori. We can infer that the activation of other signaling pathways is also responsible for H. pylori-induced cell proliferation and migration. On the other hand, miR-375 most likely raises BCL-2 and TWIST1 expression by targeting other genes, thus promoting gastric carcinogenesis. Therefore, further studies should be performed to clarify the complex cross talk network between the identified miR-375–JAK2–STAT3 pathway, inflammatory cells, and tumor cells.
Having shown that H. pylori infection contributes to the expression of pro-inflammatory cytokines, the inhibition of miR-375, and the further promotion of carcinogenesis in this investigation, we raise two fundamental questions that remain unanswered: does the expression pattern of pro-inflammatory cytokines have a role in miR-375 suppression? If so, what mediates this biological process? Indeed, Angela et al. [39] had reported that AREB6 (also known as ZEB1) suppressed the transcription of miR-375 using bioinformatics. Here, we predict ZEB1 to be a critical mediator at the cellular level. The expression pattern of ZEB1 is consistent with that of IL-8 and opposite to that of miR-375 (data not shown). Further studies need to be performed to explore these mediators that can facilitate cell proliferation and migration and regulate miR-375.
In summary, our findings demonstrate that H. pylori regulates the expression of miR-375 to activate JAK2–STAT3 signaling, thus offering new insight into the role of miRNAs in the aggravation of H. pylori-induced inflammation-provoked neoplastic transformation. Moreover, the identified miR-375–JAK2–STAT3 signaling suggests a compelling biomarker and therapeutic target for H. pylori-induced gastric cancer.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank the Shanghai Institute of Digestive Disease and Prof. Qinglong Guo for providing the cell lines used in this study and the members of the Tao Xi lab for helpful comments on the manuscript.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Zabaleta J. MicroRNA: a bridge from H. pylori infection to gastritis and gastric cancer development. Front Genet. 2012;3:294. doi: 10.3389/fgene.2012.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Isomoto H, Matsushima K, Inoue N, Hayashi T, Nakayama T, Kunizaki M, Hidaka S, Nakayama M, Hisatsune J, Nakashima M, Nagayasu T, Nakao K, Hirayama T. Interweaving microRNAs and proinflammatory cytokines in gastric mucosa with reference to H. pylori infection. J Clin Immunol. 2012;32:290–299. doi: 10.1007/s10875-011-9626-3. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 6.Belair C, Darfeuille F, Staedel C. Helicobacter pylori and gastric cancer: possible role of microRNAs in this intimate relationship. Clin Microbiol Infect. 2009;15:806–812. doi: 10.1111/j.1469-0691.2009.02960.x. [DOI] [PubMed] [Google Scholar]
- 7.Link A, Kupcinskas J, Wex T, Malfertheiner P. Macro-role of microRNA in gastric cancer. Dig Dis. 2012;30:255–267. doi: 10.1159/000336919. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Z, Li Z, Gao C, Chen P, Chen J, Liu W, Xiao S, Lu H. miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab Invest. 2008;88:1358–1366. doi: 10.1038/labinvest.2008.94. [DOI] [PubMed] [Google Scholar]
- 9.Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini M, Cavazzini L, Volinia S, Alder H, Ruco LP, Baldassarre G, Croce CM, Vecchione A. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 10.Li X, Zhang Y, Zhang H, Liu X, Gong T, Li M, Sun L, Ji G, Shi Y, Han Z, Han S, Nie Y, Chen X, Zhao Q, Ding J, Wu K, Daiming F. miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol Cancer Res. 2011;9:824–833. doi: 10.1158/1541-7786.MCR-10-0529. [DOI] [PubMed] [Google Scholar]
- 11.Xiao B, Liu Z, Li BS, Tang B, Li W, Guo G, Shi Y, Wang F, Wu Y, Tong WD, Guo H, Mao XH, Zou QM. Induction of microRNA-155 during Helicobacter pylori infection and its negative regulatory role in the inflammatory response. J Infect Dis. 2009;200:916–925. doi: 10.1086/605443. [DOI] [PubMed] [Google Scholar]
- 12.Oertli M, Engler DB, Kohler E, Koch M, Meyer TF, Müller A. MicroRNA-155 is essential for the T cell-mediated control of Helicobacter pylori infection and for the induction of chronic gastritis and colitis. J Immunol. 2011;187:3578–3586. doi: 10.4049/jimmunol.1101772. [DOI] [PubMed] [Google Scholar]
- 13.Ding L, Xu Y, Zhang W, Deng Y, Si M, Du Y, Yao H, Liu X, Ke Y, Si J, Zhou T. MiR-375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell Res. 2010;20:784–793. doi: 10.1038/cr.2010.79. [DOI] [PubMed] [Google Scholar]
- 14.Zhao H, Zhu L, Jin Y, Ji H, Yan X, Zhu X. miR-375 is highly expressed and possibly transactivated by achaete-scute complex homolog 1 in small-cell lung cancer cells. Acta Biochim Biophys Sin. 2012;44:177–182. doi: 10.1093/abbs/gmr110. [DOI] [PubMed] [Google Scholar]
- 15.He XX, Chang Y, Meng FY, Wang MY, Xie QH, Tang F, Li PY, Song YH, Lin JS. MicroRNA-375 targets AEG-1 in hepatocellular carcinoma and suppresses liver cancer cell growth in vitro and in vivo. Oncogene. 2012;31:3357–3369. doi: 10.1038/onc.2011.500. [DOI] [PubMed] [Google Scholar]
- 16.Tsukamoto Y, Nakada C, Noguchi T, Tanigawa M, Nguyen LT, Uchida T, Hijiya N, Matsuura K, Fujioka T, Seto M, Moriyama M. MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer Res. 2010;70:2339–2349. doi: 10.1158/0008-5472.CAN-09-2777. [DOI] [PubMed] [Google Scholar]
- 17.de Souza Rocha Simonini P, Breiling A, Gupta N, Malekpour M, Youns M, Omranipour R, Malekpour F, Volinia S, Croce CM, Najmabadi H, Diederichs S, Sahin O, Mayer D, Lyko F, Hoheisel JD, Riazalhosseini Y. Epigenetically deregulated microRNA-375 is involved in a positive feedback loop with estrogen receptor alpha in breast cancer cells. Cancer Res. 2010;70:9175–9184. doi: 10.1158/0008-5472.CAN-10-1318. [DOI] [PubMed] [Google Scholar]
- 18.O’Shea JJ, Holland SM, Staudt LM. JAKs and STATs in immunity, immunodeficiency, and cancer. N Engl J Med. 2013;368:161–170. doi: 10.1056/NEJMra1202117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Butturini E, Carcereri de Prati A, Chiavegato G, Rigo A, Cavalieri E, Darra E, Mariotto S. Mild oxidative stress induces S-glutathionylation of STAT3 and enhances chemosensitivity of tumoural cells to chemotherapeutic drugs. Free Radic Biol Med. 2013;65:1322–1330. doi: 10.1016/j.freeradbiomed.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 21.Kim JK, Kim JY, Kim HJ, Park KG, Harris RA, Cho WJ, Lee JT, Lee IK. Scoparone exerts anti-tumor activity against DU145 prostate cancer cells via inhibition of STAT3 activity. PLoS ONE. 2013;8:e80391. doi: 10.1371/journal.pone.0080391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu KW, Hsieh RH, Huang KH, Fen-Yau Li A, Chi CW, Wang TY, Tseng MJ, Wu KJ, Yeh TS. Activation of the Notch1/STAT3/Twist signaling axis promotes gastric cancer progression. Carcinogenesis. 2012;33:1459–1467. doi: 10.1093/carcin/bgs165. [DOI] [PubMed] [Google Scholar]
- 23.Li CW, Xia W, Huo L, Lim SO, Wu Y, Hsu JL, Chao CH, Yamaguchi H, Yang NK, Ding Q, Wang Y, Lai YJ, LaBaff AM, Wu TJ, Lin BR, Yang MH, Hortobagyi GN, Hung MC. Epithelial-mesenchymal transition induced by TNF-α requires NF-κB-mediated transcriptional upregulation of Twist1. Cancer Res. 2012;72:1290–1300. doi: 10.1158/0008-5472.CAN-11-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen X, Tang J, Hu J, Guo L, Xing Y, Xi T. MiR-424 regulates monocytic differentiation of human leukemia U937 cells by directly targeting CDX2. Biotechnol Lett. 2013;35:1799–1806. doi: 10.1007/s10529-013-1264-9. [DOI] [PubMed] [Google Scholar]
- 25.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schetter AJ, Heegaard NH, Harris CC. Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis. 2010;31:37–49. doi: 10.1093/carcin/bgp272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sepulveda AR. Helicobacter, inflammation, and gastric cancer. Curr Pathobiol Rep. 2013;1:9–18. doi: 10.1007/s40139-013-0009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sonkoly E, Stahle M, Pivarcsi A. MicroRNAs and immunity: novel players in the regulation of normal immune function and inflammation. Semin Cancer Biol. 2008;18:131–140. doi: 10.1016/j.semcancer.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 29.Teng GG, Wang WH, Dai Y, Wang SJ, Chu YX, Li J. Let-7b is involved in the inflammation and immune responses associated with Helicobacter pylori infection by targeting Toll-like receptor 4. PLoS ONE. 2013;8:e56709. doi: 10.1371/journal.pone.0056709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng Y, Wang L, Zeng J, Shen L, Liang X, Yu H, Liu S, Liu Z, Sun Y, Li W, Chen C, Jia J. FoxM1 is overexpressed in Helicobacter pylori-induced gastric carcinogenesis and is negatively regulated by miR-370. Mol Cancer Res. 2013;11:834–844. doi: 10.1158/1541-7786.MCR-13-0007. [DOI] [PubMed] [Google Scholar]
- 31.Li N, Tang B, Zhu ED, Li BS, Zhuang Y, Yu S, Lu DS, Zou QM, Xiao B, Mao XH. Increased miR-222 in H. pylori-associated gastric cancer correlated with tumor progression by promoting cancer cell proliferation and targeting RECK. FEBS Lett. 2012;586:722–728. doi: 10.1016/j.febslet.2012.01.025. [DOI] [PubMed] [Google Scholar]
- 32.Xu Y, Deng Y, Yan X, Zhou T. Targeting miR-375 in gastric cancer. Expert Opin Ther Targets. 2011;15:961–972. doi: 10.1517/14728222.2011.581232. [DOI] [PubMed] [Google Scholar]
- 33.Mathé EA, Nguyen GH, Bowman ED, Zhao Y, Budhu A, Schetter AJ, Braun R, Reimers M, Kumamoto K, Hughes D. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clin Cancer Res. 2009;15:6192–6200. doi: 10.1158/1078-0432.CCR-09-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Avissar M, Christensen BC, Kelsey KT, Marsit CJ. MicroRNA expression ratio is predictive of head and neck squamous cell carcinoma. Clin Cancer Res. 2009;15:2850–2855. doi: 10.1158/1078-0432.CCR-08-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wach S, Nolte E, Szczyrba J, Stöhr R, Hartmann A, Ørntoft T, Dyrskjøt L, Eltze E, Wieland W, Keck B, Ekici AB, Grässer F, Wullich B. MicroRNA profiles of prostate carcinoma detected by multiplatform microRNA screening. Int J Cancer. 2012;130:611–621. doi: 10.1002/ijc.26064. [DOI] [PubMed] [Google Scholar]
- 36.Liu Y, Xing R, Zhang X, Dong W, Zhang J, Yan Z, Li W, Cui J, Lu Y. miR-375 targets the p53 gene to regulate cellular response to ionizing radiation and etoposide in gastric cancer cells. DNA Repair (Amst) 2013;12:741–750. doi: 10.1016/j.dnarep.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 37.Tekpli X, Landvik NE, Anmarkud KH, Skaug V, Haugen A, Zienolddiny S. DNA methylation at promoter regions of interleukin 1B, interleukin 6, and interleukin 8 in non-small cell lung cancer. Cancer Immunol Immunother. 2013;62:337–345. doi: 10.1007/s00262-012-1340-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee H, Herrmann A, Deng JH, Kujawski M, Niu G, Li Z, Forman S, Jove R, Pardoll DM, Yu H. Persistently activated Stat3 maintains constitutive NF-κB activity in tumors. Cancer Cell. 2009;15:283–293. doi: 10.1016/j.ccr.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Re A, Corá D, Taverna D, Caselle M. Genome-wide survey of microRNA-transcription factor feed-forward regulatory circuits in human. Mol BioSyst. 2009;5:854–867. doi: 10.1039/b900177h. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.