Abstract
Frequent expression of cancer testis antigens (CTA) has been consistently observed in head and neck squamous cell carcinomas (HNSCC). For instance, in 52 HNSCC patients, MAGE-A3 and -A4 CTA were expressed in over 75% of tumors, regardless of the sites of primary tumors such as oral cavity or hypopharynx. Yet, T-cell responses against these CTA in tumor-bearing patients have not been investigated in detail. In this study, we assessed the naturally acquired T-cell response against MAGE-A3 and -A4 in nonvaccinated HNSCC patients. Autologous antigen-presenting cells pulsed with overlapping peptide pools were used to detect and isolate MAGE-A3 and MAGE-A4 specific CD4+ T cells from healthy donors and seven head and neck cancer patients. CD4+ T-cell clones were characterized by cytokine secretion. We could detect and isolate MAGE-A3 and MAGE-A4 specific CD4+ T cells from 7/7 cancer patients analyzed. Moreover, we identified six previously described and three new epitopes for MAGE-A3. Among them, the MAGE-A3111–125 and MAGE-A3161–175 epitopes were shown to be naturally processed and presented by DC in association with HLA-DP and DR, respectively. All of the detected MAGE-A4 responses were specific for new helper epitopes. These data suggest that naturally acquired CD4+ T-cell responses against CT antigens often occur in vivo in HNSCC cancer patients and provide a rationale for the development of active immunotherapeutic approaches in this type of tumor.
Keywords: HNSCC, MAGE-A3, MAGE-A4, CD4+ T cells
Introduction
Squamous cell carcinoma of the head and neck (HNSCC) represents the fifth most common cancer worldwide and is a significant cause of cancer morbidity and mortality. A majority of patients with HNSCC present with advanced disease at the time of first evaluation. While early stages of HNSCC are generally curable with surgery and chemoradiation, the overall survival for patients with HNSCC is poor and has not appreciably improved in the past three decades. Therefore, novel therapeutic approaches are urgently needed and recent research has focused on the development of immunotherapy to complement conventional treatments.
Frequent expression of tumor-associated antigens (TAA) has been repeatedly observed in head and neck carcinomas. These observations have led to the design of targeted cancer vaccine trials [1]. TAA are derived from a broad spectrum of intracellular proteins. One of the most promising categories of TAAs is transcriptionally reactivated genes, not expressed in normal adult host tissues but only present in germline cells and cancer cells and referred as cancer testis antigens (CTA). Their highly restricted tissue expression patterns make them ideal targets for immunotherapy of numerous cancers. Tumor-specific vaccines are thus designed to generate T-helper and T-cytotoxic lymphocytes responsive to TAAs. Evidence for the presence of naturally acquired T-cell responses specific for these antigens have been demonstrated in patients with various cancers but, to our knowledge, were never studied in HNSCC patients [2–5]. Generally, the frequency of natural T-cell responses against CT antigens seems to be relatively low in peripheral blood of cancer patients. The natural responses are for this reason difficult to detect and often require at least one round of in vitro expansion by stimulation with the antigenic peptide on appropriate antigen-presenting cells to be detected. Both, the use of overlapping peptide pools and the screening of responding T cells using different cytokines tend to increase the level of natural responses detected in cancer patients that were probably until recently underestimated.
Tumor antigens encoded by the family of MAGE-A genes are of particular interest in cancer immunotherapy, because they are expressed in a variety of malignancies. Among others, these include head and neck squamous cell carcinoma, melanoma or non-small cell lung carcinoma [6, 7]. In a survey performed by our group, tumor biopsies of 52 HNSCC patients showed that MAGE-A3 or -A4 CTA were expressed in 75% of the cases, regardless of the sites of primary tumors such as oral cavity or hypopharynx, confirming previous results [8]. Since MAGE-A3 is most frequently expressed in major tumor types, as mentioned above, it has been the focus of numerous studies on specific T cell-mediated immune responses pre- and post-antigen-specific immunotherapy [9]. A small proportion of cancer bearing patients naturally develop antibody responses to MAGE-A3, indicating that this tumor antigen is capable of evoking spontaneous immune responses [10, 11]. Moreover, epitope-specific memory CD8+ and CD4+ T-cell responses were rarely detected in cancer patients at relatively low frequencies [12–14]. Very few reports to date have documented the precise frequency of MAGE-3 specific CD4+ T cells naturally arising in cancer patients [10, 15, 16]. MAGE-A4 is as frequently expressed as MAGE-A3, at least in HNSCC, but has been far less studied. Despite some data indicating an inverse correlation of the expression of MAGE-A4 protein with patient survival [8], no immunotherapeutic protocols have been conducted, mainly because MAGE-A4 specific T-cell epitopes have not been described yet.
Naturally acquired immunity against these CTA epitopes in HNSCC patients before vaccination or treatment have not been investigated in detail; yet, the overall prevalence of these responses would be informative to optimize further peptide vaccination clinical trials. In this regard, based on the MAGE-A3 and -A4 protein tumor expression, a small panel of HNSCC patients recruited before any therapy were screened for spontaneous T-cell responses against MAGE-A3 and MAGE-A4 proteins. Without pre-existing knowledge of epitopes and or restriction elements, total patients’ PBMCs were analyzed for MAGE-A3 and -A4 specific CD4+ T cells. We could detect specific CD4+ T-cell responses against several known as well as undefined MAGE-A3 and MAGE-A4 epitopes in PBMC from seven out of seven patients analyzed after three rounds of in vitro stimulation with overlapping peptides encoding both proteins. Some of the responding T-cell populations were cloned and analyzed for MHC restriction and avidity of antigen recognition. Using specific CD4+ T-cell clones, we could also show that MAGE-A3111–125 and MAGE-A3161–175 epitopes are naturally processed and presented. We identified the DPβ1*04 allele as a new MHC II-restricting element for the MAGE-A3111–125 epitope and we could successfully generate MAGE-A3111–125/DPβ1*04 multimer to screen PBMCs and TILs from an HNSCC patient. Altogether, these findings indicate that MAGE-A3 and -A4 antigens are promising targets for specific immunotherapy of HNSCC.
Materials and methods
Cells and tissue culture
Peripheral blood mononuclear cells (PBMCs) were obtained from HNSCC patients (Lausanne University Hospital) after informed consent and from seven healthy donors who served as controls (Blood Transfusion Center, Berne, Switzerland). Clinical characteristics of the patients included in this study are described in Table 1. None of the patients received immunotherapy before sampling of blood. HNSCC autologous tumor cell lines were generated from surgically removed primary tumor biopsies. Epstein–Barr virus-transformed lymphoblastoid B-cell lines (LCLs) were established in our laboratory. LCLs and tumor cells were retrovirally transduced with a pMFG vector encoding huIi80MAGE-3-Ires-tNGFR [17]. Tumor and LCLs cells were maintained in continuous in vitro culture in RPMI 1640 medium (Invitrogen Life Technologies) supplemented with 10% heat-inactivated FCS (GIBCO, Invitrogen) l-glutamine (2 mM), penicillin (100 U/ml) and streptomycin (50 μg/ml; Biowhittaker, Walkersville, MD, USA).
Table 1.
Patient characteristics and tumor antigen expression
| Patients | Agea | Gender | Tumor localization | Stage | MAGE expressionb | |
|---|---|---|---|---|---|---|
| A3 | A4 | |||||
| LAU 2032 | 72 | M | Oral cavity | IV | + | +++ |
| LAU 2042 | 69 | F | Oral cavity | III | ++ | +++ |
| LAU 2068 | 68 | F | Oral cavity | IV | +++ | +++ |
| LAU 2069 | 64 | F | Oropharynx | III | +++ | ++ |
| LAU 2073 | 54 | M | Oropharynx | IV | +++ | ++ |
| LAU 2086 | 68 | M | Larynx | III | − | +++ |
| LAU 2091 | 68 | M | Oropharynx | II | − | ++ |
−, negative; +, 1–5%; ++, 5–50%; +++, >50% level of expression
aCalculated at the surgery date
bEvaluated by RT–PCR (quantitative assessment was performed as previously described, using 1:10 dilutions of SK-Mel-37 RNA as reference [38])
HLA typing
HLA class II typing was performed at Geneva’s University Hospital by J. M. Tiercy, on genomic DNA extracted from frozen PBLs isolated from patient’s blood samples. HLA-DRβ1 and -DQβ1 low-resolution typing (two-digit) was performed by reverse PCR sequence-specific oligonucleotide hybridization using Luminex technology (OneLambda; Ingen). High-resolution typing (four digit) for HLA-DRβ1, -DPβ1 and -DQβ1 loci was achieved by PCR sequence-specific primers using Genovision reagents (Milan Analytica).
Synthesis of MAGE-A3 and MAGE-A4 peptides and peptide pools
MAGE-A3 and MAGE-A4 15 mers peptides overlapping by 10 amino acids were synthesized by Proimmune (UK). Selected sequences were synthesized by the stepwise solid phase method and synthetic peptides were purified by semi-preparative reverse-phase HPLC. All peptides were >90% pure as indicated by analytic HPLC. The peptides were lyophilized, then reconstituted in DMSO at 10 mg/ml or aliquots of 1 mg/ml and stored at −20°C. To reduce the number of individual peptide stimulation, we generated six peptide pools with ten peptides each for MAGE-A3 or MAGE-A4 overlapping peptides.
In vitro propagation of specific CD4+ T cells
Twelve pools of 10 MAGE-A3 or MAGE-A4 synthetic peptides were used to stimulate the PBMCs from the HNSCC patients and HDs. Twenty million PBMCs, depleted for CD25+ T cells using anti-CD25 microbeads and MiniMACS magnetic separation columns (Miltenyi Biotec), were cultured for 14 days in RPMI 1640 (Gibco, Grand Island, NY, USA), supplemented with heat-inactivated pooled human serum (8%), l-glutamine (2 mM), penicillin (100 U/ml) and streptomycin (50 μg/ml; Biowhittaker, Walkersville, MD, USA), containing MAGE-A3 or MAGE-A4 peptide pools (2 μM of each peptide). On day two, half medium volume from each well was removed and replenished with fresh tissue culture medium (TCM) containing interleukin (IL)-2 (150 UI/ml) (Chiron), without any further antigen stimulation. Cultures were expanded for 14 days and restimulated two more times with the same amount of MAGE-A3 or MAGE-A4 peptide pools pulsed on irradiated (3,000 rad) autologous PBMCs as antigen-presenting cells (APCs).
IFN-γ/TNF-α intracellular cytokine staining
On days 7–10 after the third round of simulation, cultures were tested for the presence of specific CD4+ T cells with the same pool of peptides (2 μM each) in an IFN-γ and TNF-α intracellular cytokine staining assay performed as follows. Cells were stimulated for 18 h at 37°C in the presence or absence of the peptide pools and 10 μg/ml brefeldin A (Sigma–Aldrich) was added after the first hour of incubation. After incubation and washing, cells were stained with anti-CD4-PE-Cy7 and anti-CD3-APC-Cy7 mAb (Beckman Dickinson) for 20 min at 4°C, and then fixed with 1% formaldehyde, 2% BSA and 5 mM azide in PBS. Cells were then stained intracellularly with anti-IFN-γ-PE and anti-TNF-α-APC mAb (Beckton Dickinson) in the presence of 0.1% saponin (Sigma–Aldrich). Samples were analyzed by flow cytometry on a Facs Array and data were analyzed using Flow Jo software (Beckton Dickinson).
Generation of clones and antigen recognition assay
CD4+ T cells producing IFN-γ in the presence of the pool of peptides were cloned using IFN-γ and TNF-α secretion assays (Miltenyi Biotec) after 6 h of peptide stimulation by flow cytometry-based sorting using a FACSVantage SE (Becton–Dickinson, Sunnyvale, CA, USA). Sorted cells were cloned by limiting dilution and growing clones were screened for IFN-γ and TFN-α release in the presence or absence of indicated MAGE-A3 or MAGE-A4 peptides.
For each functional assay, 5,000–20,000 of growing MAGE-A3 or MAGE-A4 specific CD4+ T-cell clones were used. For recognition of indicated target cells, equal numbers of antigen-presenting cells or tumor cell lines were pulsed or not with indicated peptides, extensively washed and then added to the T cells. In peptide titration experiments, the following concentrations of peptide were added: 50–25–13–6–1.5 and 0.7 μM. Where indicated, supernatants of anti-HLA-DR (D1.12), -DP (B7.12.1) or -DQ (BT3.4) were used to specifically block MHC class II recognition. IFN-γ release was assessed in the culture supernatant after 24 h of incubation at 37°C by ELISA (hu IFN-γ Cytoset kit; BioSource Europe) following the manufacturer’s instructions and using flat-bottom 96-well Nunc-Immuno plates (Apogent).
Generation of HLA-DPβ1*04 multimers and multimer labeling
The generation of HLA-DPβ1*04 multimers was performed as already described [18] using the MAGE-A3111–125 peptide. Multimer labeling was performed for 1 h at 37°C before staining with fluorescent mAbs directed against surface molecules (anti-CD4-FITC) (20 min at 4°C). PI labeling was used to exclude dead cells. Cells were analyzed by flow cytometry on a FACScan (BD Biosciences) using Cell Quest software (BD Biosciences).
Results
MAGE-A3 and MAGE-A4 specific CD4+ T-cell responses in head and neck cancer patients
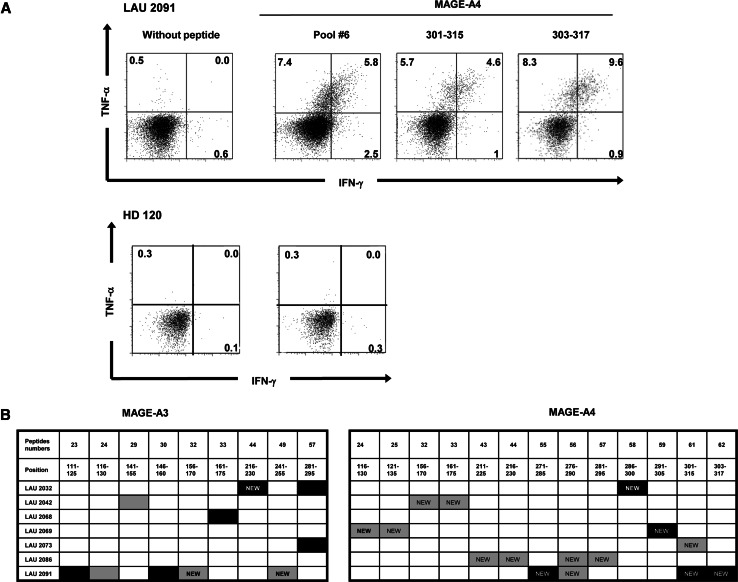
To assess the naturally acquired anti-tumor T-cell responses taking place in head and neck squamous cell carcinomas patients (HNSCC), tumor biopsies were first screened for tumor-associated antigen expression by RT–PCR and immunohistochemistry to identify putative immunogenic antigens. MAGE-A3 and -A4 tumor antigens were specifically expressed in about 70% of the patients (MAGE-A3 in 51%, MAGE-A4 in 60% [8]). We assessed T-cell responses specific for these two well-represented antigens in peripheral blood from seven HNSCC patients, selected upon their MAGE-A3 and/or -A4 high tumor expression (Table 1). Briefly, CD25+ cell-depleted PBMCs from these patients were stimulated with overlapping peptides covering the entire sequence of the two proteins. These peptides were 15 amino acids long, overlapping by 10 amino acids and grouped into six pools containing 10–11 peptides. CD25+ T-cell depletion was performed prior to stimulation since these cells have been reported to limit the in vitro expansion of antigen-specific CD4+ T cells [19]. A side-by-side comparison of CD25+ replete and depleted PBMCs samples was not possible due to the limited amount of blood allowed by the clinical protocol. After three rounds of stimulation, the repertoire of epitopes recognized by polyclonal CD4+ T cells was determined by testing their reactivity to each pool and then each peptide forming the pool in a 6-h IFN-γ/TNF-α intracellular cytokine staining (ICS) assay using a five-color immunofluorescence labeling protocol. The frequency of CD4+ T cells producing IFN-γ or TNF-α in the responding patients ranged from 2 to 64% after the third stimulation. In contrast, undetectable or very low frequency of IFN-γ or TNF-α positive cells was found in healthy controls and in patients’ PBMCs stimulated with irrelevant or no peptides (range 0–2%). An example of the data obtained following this analysis for one patient and one healthy donor are shown in Fig. 1a.
Fig. 1.
Pre-existing CD4+ T-cell responses against MAGE-A3 and MAGE-A4 antigens in HNSCC patients. a PBLs from HNSCC patients were stimulated in vitro with MAGE-A3 and MAGE-A4 overlapping peptide pools. After three rounds of stimulation, cultures were tested by intracellular cytokine staining for IFN-γ and TNF-α secretion in response to the presence of peptide pools. Cytokine secretion of CD4+ CD3+ lymphocyte gated populations from one HNSCC patient and one healthy donor in response to peptide pool and individual peptide are shown as example. b Summary of the responses detected after in vitro stimulation in seven analyzed HNSCC patients. The responses that could be further cloned by cell sorting and limited dilutions are indicated as black boxes. Undescribed epitopes are depicted as “NEW”
Among the seven HNSCC patients tested, all of them showed detectable CD4+ T-cell responses against at least one MAGE-A3 or MAGE-A4 peptide. Four patients, LAU 2032, LAU 2042, LAU 2073 and LAU 2091, showed responses against both MAGE-A3 and MAGE-A4 peptides. Responses against MAGE-A3281–295 peptide and MAGE-A4301–315 peptide appeared simultaneously in two different patients despite their different class II MHC alleles, suggesting recognition of promiscuous epitopes. Most of the detected MAGE-A3 responses were specific to previously found and/or described epitopes, such as 111–125 and 116–130 [17, 20], region 141–155 [20], 146–160 [20, 21], 161–175 [20, 22] and 281–295 [20, 21]. In contrast, three responses were specific to new epitopes spanning the regions 156–170, 216–230 and 241–255. All of the detected MAGE-A4 responses were specific to new epitopes covering a large part of the protein, from region 116 to 317 (Fig. 1b).
To further characterize these epitopes, clones were derived from the primary cultures that were able to respond to short-term stimulation with the selected MAGE-A3 and -A4 peptides using the IFN-γ/TNF-α capture assay and flow cytometry-assisted cell sorting.
CD4+ T-cell responses against MAGE-A3 and -A4 peptides are mainly HLA-DR and -DQ restricted
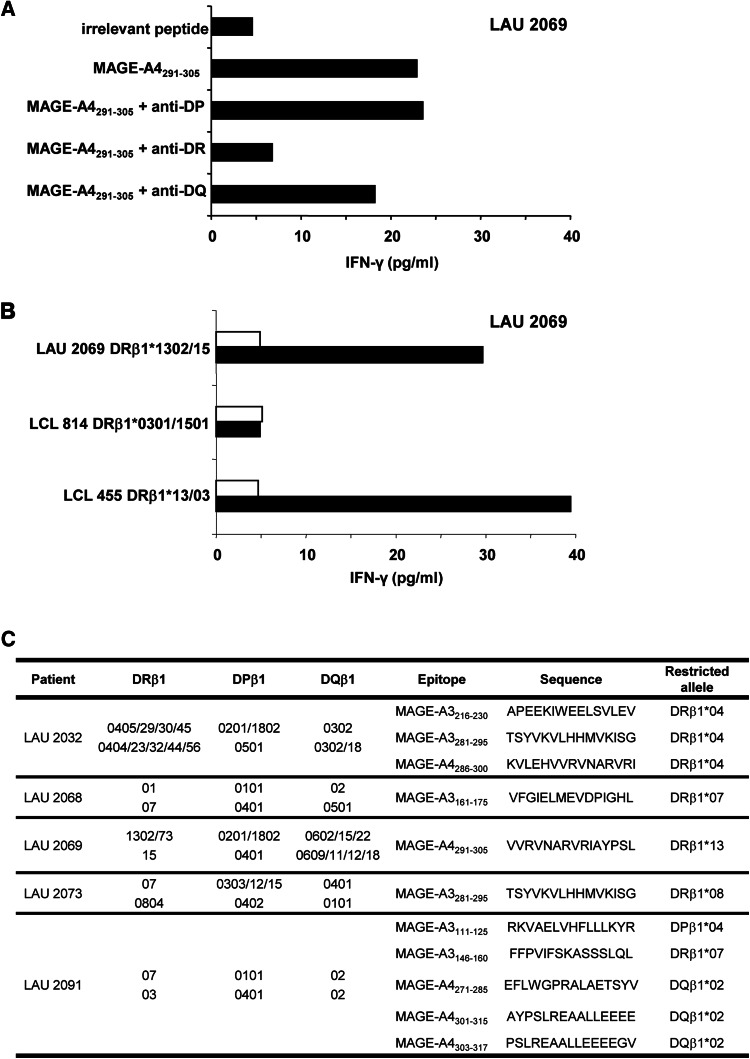
After cloning, ten growing clones that recognized distinct MAGE-A3 peptides (111–125, 146–160, 161–175, 216–230, 281–295) and MAGE-A4 peptides (271–285, 286–300, 291–305, 301–315, 303–317) could be maintained in culture and characterized. To identify the HLA restricting allele of the different peptide epitopes, growing CD4+ T-cell clones were tested in short-term stimulation with the selected MAGE-A3 or -A4 peptides in the presence of either anti-HLA-DR-, -DP- or -DQ-specific Abs known to specifically block antigen recognition. The MAGE-A4291–305 specific CD4+ T-cell clone from LAU 2069 recognized the peptide in an HLA-DR restricted manner (Fig. 2a). In addition, to identify the restricting allele, we challenged CD4+ T-cell clones with a panel of LCLs with identified class II MHC, pulsed with individual peptides. Using different combinations of presenting LCLs, we could determine that the clone from LAU 2069 recognized the MAGE-A4291–305 peptide presented by HLA-DRβ1*13 restricted allele (Fig. 2b).
Fig. 2.
MHC class II restriction of MAGE-A3 and -A4 specific CD4+ T clones. a Example of class II restricted allele assessment on LAU 2069 CD4+ T-cell clone 1 specific for MAGE-A4291–305. Where indicated, anti-DP, DQ, DR purified antibodies were pre-incubated with cells prior to peptide stimulation. IFN-γ release was measured in the supernatant by ELISA. b Example of fine HLA II restriction performed on LAU 2069 clone 1. HLA class II matched LCL cell lines were pulsed (black bars) or not (white bars) with MAGE-A4291–305 peptide and presented to the specific clone. Recognition of the LCL cells with the restricting allele was measured by ELISA IFN-γ. c Summary table of MHC class II restriction of the cloned responses
Clones from LAU 2091 recognized the MAGE-A3146–160 peptide in an HLA-DRβ1*07 restricted manner, while they recognized the MAGE-A4271–285, MAGE-A4301–315 and MAGE-A4305–317 peptides presented by HLA-DQβ1*02. In addition, MAGE-A3111–125 peptide, described as an HLA-DRβ1*13 epitope by Chaux et al. [17], appeared to also be present by the HLA-DPβ1*04 allele. The clone from LAU 2073 recognized the MAGE-A3281–295 peptide presented by HLA-DRβ1*08 and the clone from LAU 2032 recognized the same peptide presented by HLA-DRβ1*04. The MAGE-A3216–230 and MAGE-A4286–300 peptides, recognized by CD4+ T-cell clones from LAU 2032, are presented by HLA-DRβ1* 04. Finally, clones from LAU 2068 and LAU 2069 recognized the MAGE-A3161–175 and MAGE-A4291–305 peptides presented by HLA-DRβ1*07 and HLA-DRβ1*13, respectively (Fig. 2c).
Characterization of MAGE-A3 and MAGE-A4 specific CD4+ T-cell clones
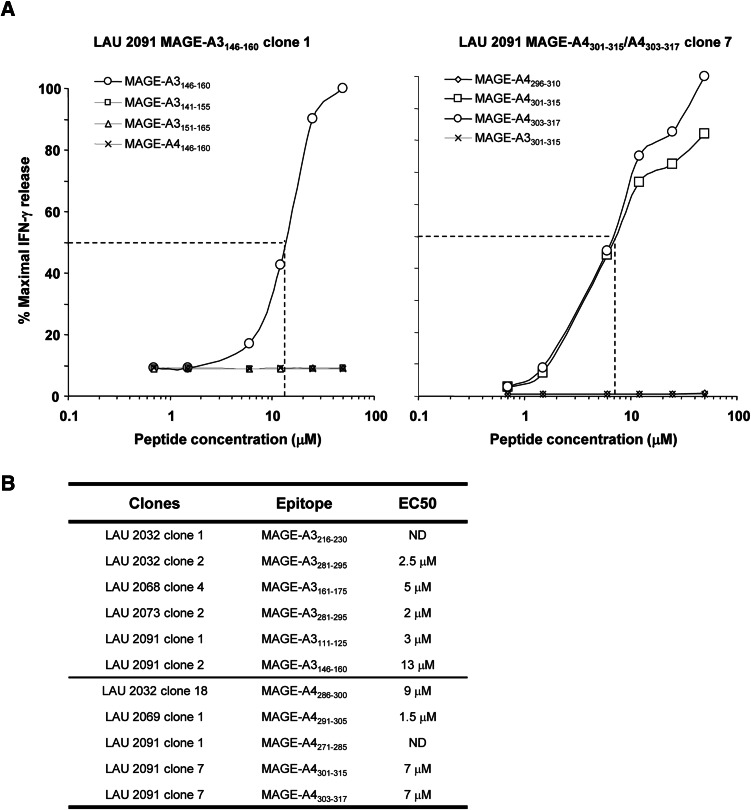
To more precisely define the epitopes recognized by the clones from head and neck cancer patients, we stimulated the MAGE-A3 and MAGE-A4 specific CD4+ T-cell clones with graded amount of peptides followed by assessment of IFN-γ production in the culture supernatants. Peptide titration curves of MAGE-A3146–160 and MAGE-A4301–315 specific CD4+ T-cell clones from LAU 2091 are shown in Fig. 3a. Both clones recognized their corresponding synthetic peptides with relatively low avidity in the micromolar range (EC50 > 10 μM and EC50 > 7 μM, respectively). MAGE-A3146–160 specific CD4+ T-cell clone failed to significantly recognize the neighboring overlapping peptides and the corresponding peptide in the MAGE-A4 protein. The MAGE-A4301–315 specific CD4+ T-cell clone recognized also the 303–317 region and failed to significantly recognize the neighboring overlapping peptides and the corresponding peptide region in the MAGE-A3 protein suggesting that the minimal epitope for this clone was located between the amino acids 303 and 315. The analysis of the other MAGE-A3 and MAGE-A4 specific CD4+ T-cell clones showed that those clones recognized their respective peptides with low avidity in a micromolar range (Fig. 3b).
Fig. 3.
Peptide titration and cross reactivity of MAGE-A3 and -A4 specific CD4+ T-cell clones. a Peptide titration experiments were performed using clone-specific MAGE-A3 sequence (open symbol) and compared to both neighboring overlapping sequences and the MAGE-A4 or MAGE-A3 homologous sequence. Shown are IFN-γ titration curves expressed as percentage maximal IFN-γ release of two representative clones generated from peripheral blood of patient LAU 2091. b Summary of the EC50 of the different clones
Both MAGE-A3111–125 and MAGE-A3161–175 epitopes are naturally processed and presented by the MHC class II pathway
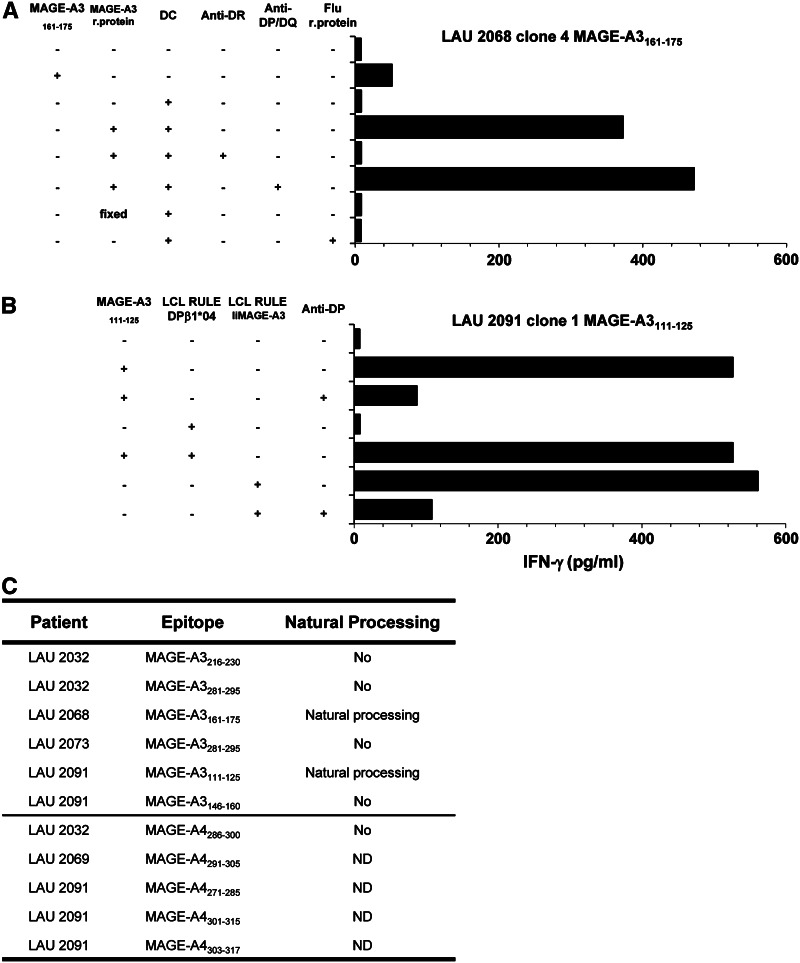
We wanted to determine whether the identified epitopes were naturally processed and presented by MHC class II-positive cells. Autologous monocyte-derived DC, generated after CD14+ cell selection and maturation were used as APC in peptide recognition assays and pre-incubated with the MAGE-A3 recombinant protein or with an irrelevant Flu recombinant protein used as control. As illustrated in Fig. 4a, monocyte-derived DC were able to efficiently present the recombinant MAGE-A3 protein to the CD4+ T-cell clone from LAU 2068 specific for MAGE-A3161–175 peptide on HLA-DR dependent manner, indicating that this epitope was naturally processed and presented. Unfortunately, MAGE-A4 epitopes could not be studied with this method since no MAGE-A4 recombinant protein was available.
Fig. 4.
Natural processing and presentation of identified MAGE-A3 and -A4 peptides. a Mature HLA-matched dendritic cells were activated with LPS and pulsed with 5 μM of recombinant MAGE-A3 protein. After washing, DCs were cocultured with CD4+ T-cell clones at 1:1 ratio. IFN-γ production was measured after 20 h in the supernatant by ELISA. Shown are example of MAGE-A3 161–175 peptide processing by HLA-DRB1*07 mature DC and presentation to LAU 2068 clone 4. b Recognition of the endogenous MAGE-A3 antigen was assessed upon HLA-matched EBV cells transduction with a retroviral construct encoding Ii.MAGE-A3 and coculture with MAGE-A3 peptide-specific clones. Shown is example of LAU 2091 clone 1 specific for epitope 111–125 cocultured with HLA-matched RULE LCL transduced with MAGE-A3. c Summary table of the described epitopes and natural processing
The natural processing of different epitopes could also be assessed using HLA-matched LCLs engineered to express either MAGE-A3 or MAGE-A4 in the endosomal–lysosomal compartment [17]. The MAGE-A3111–125 specific CD4+ T-cell clone from LAU 2091 recognized the peptide in DPβ1*04 LCLs transduced with MAGE-A3, confirming that the MAGE-A3111–125 peptide was processed through the class II presentation pathway (Fig. 4b). In addition, MAGE-A3146–160, MAGE-A3216–230 and MAGE-A3281–295 epitopes were not processed (Fig. 4c). The MAGE-A4286–300 peptide identified on CD4+ T cells from LAU 2032 was not processed by LCLs engineered to express MAGE-A4. Finally, the processing of MAGE-A4271–285, 301–315 and 303–317 peptides identified on CD4+ T cells from LAU 2091 could not be addressed, since no HLA-DQ LCLs engineered to express MAGE-A4 were generated. MAGE-A4291–305 epitope could not be addressed since the specific clone could not be maintained in culture (Fig. 4c).
Memory CD4+ T cells specific for MAGE-A3111–125/DP4 are present after in vitro stimulation of PBMCs of a HNSCC patient
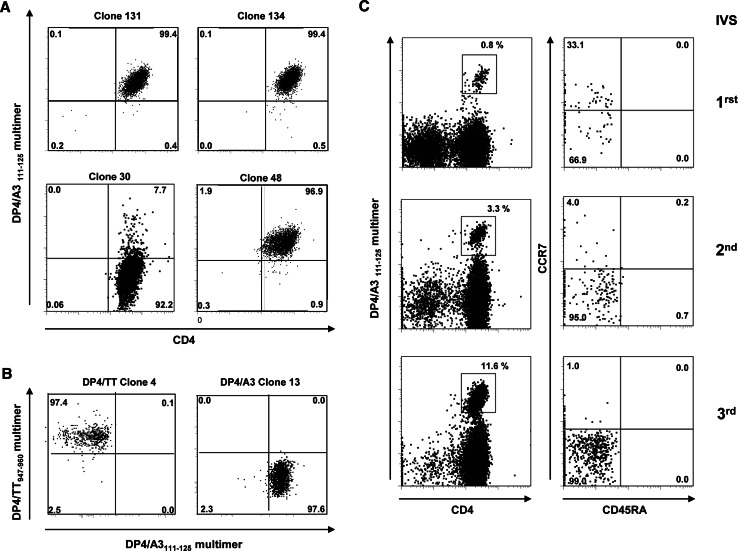
The MAGE-A3111–125 peptide was used to prepare HLA-DPβ1*04 multimer, which was tested on clones generated from LAU 2091 and additional nonspecific clones or negative controls. Several independent CD4+ T-cell clones specific for MAGE-A3111–125 peptide were stained by the multimer and no staining was observed on negative clone 30 (Fig. 5a). In addition, no staining was observed on irrelevant DP4/tetanus toxoid-specific clone 4 (Fig. 5b). We next analyzed ex vivo with the MAGE-A3111–125 multimer, PBMCs from DPβ1*04 LAU 2091 patient in which we detected specific CD4+ T cells after in vitro stimulation (Fig. 5c, left panel). No ex vivo detectable responses against DP4/MAGE-A3111–125 could be detected in PBMCs and TILs of this patient nor in the other head and neck cancer patients (data not shown). Finally, combining the multimer staining with phenotypic marker analysis, we analyzed the three in vitro PBMCs stimulation from LAU 2091 used to generate the specific clones against MAGE-A3111–125 peptide. The specific CD4+ T cells, which were initially detected after three rounds of in vitro stimulation by cytokine secretion assays, could already be detected after the first round of in vitro stimulation by using the HLA-DPβ1*04/A3111–125 multimer. The specific population increased by approximately fourfold after each stimulation with the peptide pools from 0.8 to 11%. We found that all MAGE-A3111–125/DPβ1*04 tetramer-positive cells had a memory phenotype, as they were all CD45RA−. In addition, the CCR7+ expression was downregulated upon antigenic stimulation concomitant with the differentiation of central memory to effector memory cells (Fig. 5c, right panel).
Fig. 5.
Detection of MAGE-A3111–125 specific CD4+ T-cell clones with MAGE-A3 111–125/DPB1*04 multimer. a Specific (clones 131, 134, 48) and nonspecific (clone 30) clones from LAU 2091 generated against MAGE-A3111–125 were labeled with DP4/MAGE-A3111–125 -PE multimer and anti-CD4-FITC antibodies. Cells were gated on live CD3+ cells. b Fine specificity of DP4 tetramer shown by distinct staining of tetanus toxoid947–960 and MAGE-A3111–125 CD4+ T-cell clones (clone 4 and 13, respectively) with a combination of DP4/TT947–960-APC and DP4/MAGE-A3111–125-PE multimers. Cells were gated on live CD4+ T cells. c DP4/MAGE-A3111–125 specific CD4+ T cells detected after 1, 2 and 3 rounds of in vitro stimulation of the LAU 2091 PBLs with MAGE-A3 overlapping peptide pools. Left dot plots PBLs were screened with the tetramer and gated on live CD4+ T cells. Right dot plots phenotype of CD4+ tetramer T-cell populations analyzed by CD45RA and CCR7 staining. All dot plots are gated on live cells
Discussion
The main finding in this study is the demonstration of frequent naturally acquired T-cell immunity in advanced HNSCC patients to the two most frequently expressed shared tumor-specific antigens in HNSCC, namely MAGE-A3 and -A4. Indeed, specific CD4+ T cells could be readily expanded from PBLs from all seven HNSCC chosen based on the expression of the MAGE genes by their tumors. This was not the case when PBLs from a group of healthy donors were subjected to the same in vitro stimulation protocol. Together, our data suggest the existence of memory responses to MAGE-derived antigens. In support of this notion, monitoring of MAGE-specific T cells using fluorescent multimers during the consecutive in vitro stimulations showed the presence of expanded T cells already 10 days after the first round of stimulation with peptides. As is the case for numerous epitopes derived from tumor antigens, the frequency of memory T cells is below the detection limit by multimers-guided flow cytometry, but can be readily revealed by in vitro expansion of specific cells driven by the antigen. Exact determination of the actual frequency of MAGE-specific T cells in these patients would require either limiting dilution analysis coupled to multimer labeling, or enrichment of rare multimer events from a large number of PBLs, as reported in mouse models [23].
While there are several reports identifying multiple epitopes recognized by MHC class I restricted MAGE-A4 specific cytotoxic T lymphocytes [24–26], only one paper identified helper epitopes in a peptide derived from the MAGE-A4 tumor antigen [27]. In the present study, up to 13 distinct class II MAGE-A4 epitopes were recognized by CD4+ T cells in six to seven patients analyzed. With the exception of the 281–295 peptide described by Ohkuri et al., all are undescribed MAGE-A4 derived T-cell epitopes. We obtained clonal CD4+ T-cell populations specific for five epitopes (MAGE-A4: 271–285, 286–300, 291–305, 301–315 and 303–317) presented by HLA class II DQβ1*02, DRβ1*04, DRβ1*13, DQβ1*02 and DQβ1*02, respectively. Some of these peptides may, however, be presented by more than one MHC class II molecule. For instance, specific CD4+ T cells against the MAGE-A4301–315 epitope were observed in two cancer patients. In one patient, LAU 2091, the epitope was presented by HLA-DQβ1*02. However, the second patient, LAU 2073, was HLA-DQβ1*02 negative, thus implying that specific T-cell recognition involves a different MHC class II allele.
Only one HLA-matched LCL engineered to express MAGE-A4 in the endosomal–lysosomal compartment could be tested for natural processing and presentation of MAGE-A4 epitopes. However, the MAGE-A4286–300 specific CD4+ T-cell clone did not detectably recognize this MAGE-A4 expressing LCL. These results might suggest that the peptide is not efficiently processed in the endosomal compartment of transduced B cells. In this regard, the other report on MAGE-4 helper-specific T cells used autologous monocytes-derived dendritic cells pulsed with the recombinant MAGE-A4 protein to determine specific MAGE-A4 CD4+ T cells and used overlapping peptides to identify the minimal MAGE-A4284–293 epitope [27]. This finding indicates that peptides in this region of the protein can be processed for MHC class II loading and presentation to T cells. Thus, an alternative possibility is that the CD4+ T-cell clones obtained from cancer patients used in this study are of too low avidity to pick up processed peptides in MAGE-A4 transduced B cells.
In contrast to MAGE-A4 epitopes, MAGE-A3 epitopes have been extensively studied in terms of specific T-cell responses. Indeed, several class I and class II epitopes have been described both in healthy donor and in cancer patients. In this study, we could detect CD4+ T-cell responses against six previously described MAGE-A3 epitopes (111–125, 116–130, 141–155, 146–160, 161–175 and 281–295) as well as three new epitopes (MAGE-A3: 156–170, 216–230 and 241–255). The MAGE-A3111–125 and MAGE A3161–175 peptides appeared to be immunodominant epitopes. Indeed, these were shown to be naturally processed by different types of APCs and presented by HLA-DPβ1*04 and HLA-DRβ1*07, respectively. The MAGE-A3111–125 epitope has been identified first as an epitope presented by HLA-DR13 [17], then also by HLA-DR1, HLA-DR4, HLA-DR11 and HLA-DR7 molecules [20]. The MAGE-A3111–125 peptide is therefore another example of a promiscuous CD4+ T-cell epitope. Our data show that in addition to presentation by multiple HLA-DR allelic products, this peptide can also be presented by HLA-DP4. The HLA-DPβ1*04 allele is expressed by approximately 60% of the Caucasian population and covers, together with the frequent HLA-DRβ1*01 (20%), HLA-DRβ1*04 (27%), HLA-DRβ1*07 (27%), HLA-DRβ1*11 (19%) and HLA-DRβ1*13 (21%) alleles, a significant fraction of Caucasian patients. Moreover, the MAGE-A3 sequence 111–125 contains also the HLA-A2 (MAGE-A3112–120) binding epitope [28]. We demonstrated here the ability of fluorescent DP4/MAGE-A3111–125 peptide multimers to identify specific CD4+ T cells propagated in vitro, thus opening the way to monitoring of these responses.
Concerning the MAGE-A3161–175 sequence, several predicted MHC class II binding epitopes have been described [22]. It also contains HLA-A1 (MAGE-A3168–176) and HLA-B44 (MAGE-A3167–175) restricted epitopes [29], suggesting an interesting peptide for vaccination. However, natural processing of the class II peptide could not be demonstrated in one study [20]. Another group reported that it was poorly formed and did not find evidence of specific CD4+ T-cell responses in advanced melanoma patients [22]. Our results are in sharp contrast with these previous reports since we could detect specific CD4+ T cells and could also show recognition of processed recombinant MAGE-A3 protein. These discrepancies may be due to the different types of cancer.
In contrast to the former epitopes, the MAGE-A3146–160 specific, MAGE-A3216–230 specific and MAGE-A3281–295 specific CD4+ T cells failed to recognize in vitro the MAGE-A3 protein after processing by autologous dendritic cells. However, reports from other groups showed efficient processing of the MAGE-A3146–160 and MAGE-A3281–295 epitopes [20, 21, 30]. Again, it might be that the CD4 T-cell clones recovered from this series of head and neck cancer patients bear TCRs of relatively low avidity.
Despite recognition of processed peptides in recombinant protein fed, or MAGE-A3 transduced antigen-presenting cells, no tumor recognition could be demonstrated in the few CD4+ T-cell clone/tumor combinations that could be tested (Table 2 and data not shown). This could not be explained by lack of expression of MHC class II molecules, as expression could be detected at the cell surface and their levels could be increased upon IFN-γ treatment. These results suggest that priming of the memory responses detected in cancer patients probably required processing and presentation by professional APCs from internalized apoptotic tumor cells. They also imply indirect antigen recognition by effector CD4+ T cells at the tumor sites. However, it cannot be excluded that high avidity CD4+ T cells against at least some of the MAGE-derived epitopes can directly recognize MHC class II tumor cells. This possibility is reinforced by reports from other groups who have succeeded in showing direct tumor antigen recognition by MAGE antigen-specific CD4+ T cells. Concerning the relationship between MAGE-A3 or -A4 expression and the presence of detectable specific CD4 T-cell responses, all but one responding patients expressed the target gene product (Table 1). The single exception may simply reflect the incomplete covering of antigen expression by the small biopsy that may not be representative of the entire tumor mass.
Table 2.
Tumor cell recognition by specific CD4+ T-cell clones from head and neck cancer patients
| Clones | Epitope | Tumor recognition |
|---|---|---|
| LAU 2032 clone 1 | MAGE-A3216–230 | ND |
| LAU 2032 clone 2 | MAGE-A3281–295 | ND |
| LAU 2068 clone 4 | MAGE-A3161–175 | Negative |
| LAU 2073 clone 2 | MAGE-A3281–295 | Negative |
| LAU 2091 clone 1 | MAGE-A3111–125 | Negative |
| LAU 2091 clone 2 | MAGE-A3146–160 | Negative |
| LAU 2032 clone 18 | MAGE-A4286–300 | ND |
| LAU 2069 clone 1 | MAGE-A4291–305 | ND |
| LAU 2091 clone 1 | MAGE-A4271–285 | ND |
| LAU 2091 clone 7 | MAGE-A4301–315 | ND |
| LAU 2091 clone 7 | MAGE-A4303–317 | ND |
Epitope-specific CD8 T-cell responses were rarely detected from freshly isolated peripheral blood in cancer patients and was found to be quite low before vaccination [12, 14, 31]. However, only after in vitro culture or 3 weeks post-vaccination, CD8+ T cell could be expanded and their functional status estimated [10, 32]. The apparent paucity of MAGE-specific CD8+ T-cell responses in this series of HNSCC patients might suggest the existence of CD8+ T-cell tolerance to these antigens. However, we favor the possibility that this paucity only reflects the inadequacy of peptide pools made of 15-mer peptides to efficiently expand not only CD4+ T cells, but also MHC class I restricted CD8+ T cells. To address this issue, we are planning to revisit MAGE-specific CD8+ T-cell responses in these patients using pools of nine and ten amino acid long synthetic peptides covering the MAGE-A3 and -A4 proteins.
In all, very few reports to date have documented the precise frequency of MAGE-A3 specific CD4+ T cells naturally arising in cancer patients and none for MAGE-A4. By studying a large panel of cytokine produced by peripheral PBLs in responses to tumor-associated antigens, Inokuma et al. [15] were able to detect ex vivo significant CD8 and CD4 T-cell responses against MAGE-A3 in 17/23 breast cancer patients naïve to immunotherapy. Similarly, another study reported the baseline level of CD4+ T-cell responses prior MAGE-A3 protein and adjuvant immunization of melanoma patients and could detect MAGE-A3 specific helper T-cell responses in 3/16 patient before vaccination [16]. In contrast, Atanackovic et al. [10] found only 1 out of 18 non-small cell lung cancer patients vaccinated with MAGE-A3 protein and adjuvant, having a pre-existing antibody response against MAGE-A3 protein as well as CD4 T-cell responses to MAGE-A3 DP4 epitope (p243–258). Globally, spontaneous and detectable anti-MAGE-A3 responses seem to be a rare event in cancer patients; however, some patients may have mounted a response below the detection level of the techniques used at the time of analysis. In our experiments, the requirement of three consecutive rounds of in vitro stimulation to detect MAGE-A3/A4 specific CD4+ T cells reflect a relatively low frequency of these cells as confirmed by the ex vivo absence of detectable DP4/MAGE-A3111–125 peptide multimer+ CD4+ T cells. However, it is also likely that these CD4+ T cells were anergic in vivo and reacquired full functional competence upon repeated in vitro stimulation with specific peptide and IL-2. In agreement with this notion, HNSCC have indeed been shown to be highly immunosuppressive tumors. Among the many immunosuppressive mechanisms, the impairment of T-cell activation and the production of immune inhibitory mediators by the tumors have been frequently observed in HNSCC [33–35] and would fit with the generation of low avidity of CD4+ T cells as detected in this study.
Vaccination with class II MAGE-A3 specific peptide or recombinant MAGE-A3 proteins have shown no significant toxicity and has led to the induction of strong tumor-specific CD4+ T-cell responses [10, 16, 36, 37]. Thus, targeting MAGE-A3 and/or -A4 for immunotherapy is an attractive possibility in patients with HNSSC tumors. Sequences such as MAGE-A3111–125 and MAGE-A3161–175 bear naturally processed epitopes that are recognized by both MHC class II restricted CD4+ T cells and MHC class I restricted CD8+ T cells. Approaches to antigen delivery such as recombinant protein or long synthetic peptides may provide potent immunogens able to induce broad T-cell responses in a large segment of the patient population, thus alleviating the constraints imposed by MHC restriction. These results also provide a useful baseline for future immunomonitoring studies in vaccinated patients.
Acknowledgments
We thank Ms. Eliane Cottin for her precious technical assistance and Dr. Donata Rimoldi for assessment of TAA expression in tumor cells and her valuable suggestions on tumor cell explants and culture.
Abbreviations
- pMHC
Peptide-MHC
- HNSCC
Head and neck squamous cell carcinomas
- HD
Healthy donor
References
- 1.Hoffmann TK, Bier H, Donnenberg AD, Whiteside TL, De Leo AB. p53 as an immunotherapeutic target in head and neck cancer. Adv Otorhinolaryngol. 2005;62:151–160. doi: 10.1159/000082505. [DOI] [PubMed] [Google Scholar]
- 2.Jager E, Jager D, Karbach J, Chen YT, Ritter G, Nagata Y, Gnjatic S, Stockert E, Arand M, Old LJ, Knuth A. Identification of NY-ESO-1 epitopes presented by human histocompatibility antigen (HLA)-DRB4*0101–0103 and recognized by CD4(+) T lymphocytes of patients with NY-ESO-1-expressing melanoma. J Exp Med. 2000;191(4):625–630. doi: 10.1084/jem.191.4.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gnjatic S, Atanackovic D, Jager E, Matsuo M, Selvakumar A, Altorki NK, Maki RG, Dupont B, Ritter G, Chen YT, Knuth A, Old LJ. Survey of naturally occurring CD4+ T cell responses against NY-ESO-1 in cancer patients: correlation with antibody responses. Proc Natl Acad Sci USA. 2003;100(15):8862–8867. doi: 10.1073/pnas.1133324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McNeel DG, Nguyen LD, Disis ML. Identification of T helper epitopes from prostatic acid phosphatase. Cancer Res. 2001;61(13):5161–5167. [PubMed] [Google Scholar]
- 5.Bioley G, Jandus C, Tuyaerts S, Rimoldi D, Kwok WW, Speiser DE, Tiercy JM, Thielemans K, Cerottini JC, Romero P. Melan-a/MART-1-specific CD4 T cells in melanoma patients: identification of new epitopes and ex vivo visualization of specific T cells by MHC class II tetramers. J Immunol. 2006;177(10):6769–6779. doi: 10.4049/jimmunol.177.10.6769. [DOI] [PubMed] [Google Scholar]
- 6.Groeper C, Gambazzi F, Zajac P, Bubendorf L, Adamina M, Rosenthal R, Zerkowski HR, Heberer M, Spagnoli GC. Cancer/testis antigen expression and specific cytotoxic T lymphocyte responses in nonsmall cell lung cancer. Int J Cancer. 2007;120(2):337–343. doi: 10.1002/ijc.22309. [DOI] [PubMed] [Google Scholar]
- 7.Brasseur F, Rimoldi D, Lienard D, Lethe B, Carrel S, Arienti F, Suter L, Vanwijck R, Bourlond A, Humblet Y, et al. Expression of mage genes in primary and metastatic cutaneous melanoma. Int J Cancer. 1995;63(3):375–380. doi: 10.1002/ijc.2910630313. [DOI] [PubMed] [Google Scholar]
- 8.Cuffel C, Rivals JP, Zaugg Y, Salvi S, Seelentag W, Speiser DE, Liénard D, Monnier P, Romero P, Bron L, Rimoldi D (2010) Pattern and clinical significance of cancer-testis gene expression in head and neck squamous cell carcinoma. Int J Cancer [Epub ahead of print] [DOI] [PubMed]
- 9.Speiser DE, Romero P. Molecularly defined vaccines for cancer immunotherapy, and protective T cell immunity. Semin Immunol. 2010;22:144–154. doi: 10.1016/j.smim.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Atanackovic D, Altorki NK, Stockert E, Williamson B, Jungbluth AA, Ritter E, Santiago D, Ferrara CA, Matsuo M, Selvakumar A, Dupont B, Chen YT, Hoffman EW, Ritter G, Old LJ, Gnjatic S. Vaccine-induced CD4+ T cell responses to MAGE-3 protein in lung cancer patients. J Immunol. 2004;172(5):3289–3296. doi: 10.4049/jimmunol.172.5.3289. [DOI] [PubMed] [Google Scholar]
- 11.Stockert E, Jager E, Chen YT, Scanlan MJ, Gout I, Karbach J, Arand M, Knuth A, Old LJ. A survey of the humoral immune response of cancer patients to a panel of human tumor antigens. J Exp Med. 1998;187(8):1349–1354. doi: 10.1084/jem.187.8.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanagiri T, van Baren N, Neyns B, Boon T, Coulie PG. Analysis of a rare melanoma patient with a spontaneous CTL response to a MAGE-A3 peptide presented by HLA-A1. Cancer Immunol Immunother. 2006;55(2):178–184. doi: 10.1007/s00262-005-0063-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scheibenbogen C, Lee KH, Mayer S, Stevanovic S, Moebius U, Herr W, Rammensee HG, Keilholz U. A sensitive elispot assay for detection of CD8+ T lymphocytes specific for HLA class I-binding peptide epitopes derived from influenza proteins in the blood of healthy donors and melanoma patients. Clin Cancer Res. 1997;3(2):221–226. [PubMed] [Google Scholar]
- 14.Dhodapkar MV, Young JW, Chapman PB, Cox WI, Fonteneau JF, Amigorena S, Houghton AN, Steinman RM, Bhardwaj N. Paucity of functional T-cell memory to melanoma antigens in healthy donors and melanoma patients. Clin Cancer Res. 2000;6(12):4831–4838. [PubMed] [Google Scholar]
- 15.Inokuma M, dela Rosa C, Schmitt C, Haaland P, Siebert J, Petry D, Tang M, Suni MA, Ghanekar SA, Gladding D, Dunne JF, Maino VC, Disis ML, Maecker HT. Functional T cell responses to tumor antigens in breast cancer patients have a distinct phenotype and cytokine signature. J Immunol. 2007;179(4):2627–2633. doi: 10.4049/jimmunol.179.4.2627. [DOI] [PubMed] [Google Scholar]
- 16.Vantomme V, Dantinne C, Amrani N, Permanne P, Gheysen D, Bruck C, Stoter G, Britten CM, Keilholz U, Lamers CH, Marchand M, Delire M, Gueguen M. Immunologic analysis of a phase I/II study of vaccination with MAGE-3 protein combined with the AS02B adjuvant in patients with MAGE-3-positive tumors. J Immunother. 2004;27(2):124–135. doi: 10.1097/00002371-200403000-00006. [DOI] [PubMed] [Google Scholar]
- 17.Chaux P, Vantomme V, Stroobant V, Thielemans K, Corthals J, Luiten R, Eggermont AM, Boon T, van der Bruggen P. Identification of MAGE-3 epitopes presented by HLA-DR molecules to CD4(+) T lymphocytes. J Exp Med. 1999;189(5):767–778. doi: 10.1084/jem.189.5.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guillaume P, Dojcinovic D, Luescher IF. Soluble MHC–peptide complexes: tools for the monitoring of T cell responses in clinical trials and basic research. Cancer Immun. 2009;9:7. [PMC free article] [PubMed] [Google Scholar]
- 19.Nishikawa H, Jager E, Ritter G, Old LJ, Gnjatic S. CD4+ CD25+ regulatory T cells control the induction of antigen-specific CD4+ helper T cell responses in cancer patients. Blood. 2005;106(3):1008–1011. doi: 10.1182/blood-2005-02-0607. [DOI] [PubMed] [Google Scholar]
- 20.Consogno G, Manici S, Facchinetti V, Bachi A, Hammer J, Conti-Fine BM, Rugarli C, Traversari C, Protti MP. Identification of immunodominant regions among promiscuous HLA-DR-restricted CD4+ T-cell epitopes on the tumor antigen MAGE-3. Blood. 2003;101(3):1038–1044. doi: 10.1182/blood-2002-03-0933. [DOI] [PubMed] [Google Scholar]
- 21.Manici S, Sturniolo T, Imro MA, Hammer J, Sinigaglia F, Noppen C, Spagnoli G, Mazzi B, Bellone M, Dellabona P, Protti MP. Melanoma cells present a MAGE-3 epitope to CD4(+) cytotoxic T cells in association with histocompatibility leukocyte antigen DR11. J Exp Med. 1999;189(5):871–876. doi: 10.1084/jem.189.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marturano J, Longhi R, Casorati G, Protti MP. MAGE-A3(161–175) contains an HLA-DRBETA4 restricted natural epitope poorly formed through indirect presentation by dendritic cells. Cancer Immunol Immunother. 2008;57(2):207–215. doi: 10.1007/s00262-007-0364-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chu HH, Moon JJ, Takada K, Pepper M, Molitor JA, Schacker TW, Hogquist KA, Jameson SC, Jenkins MK. Positive selection optimizes the number and function of MHCII-restricted CD4+ T cell clones in the naive polyclonal repertoire. Proc Natl Acad Sci USA. 2009;106(27):11241–11245. doi: 10.1073/pnas.0902015106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyahara Y, Naota H, Wang L, Hiasa A, Goto M, Watanabe M, Kitano S, Okumura S, Takemitsu T, Yuta A, Majima Y, Lemonnier FA, Boon T, Shiku H. Determination of cellularly processed HLA-A2402-restricted novel CTL epitopes derived from two cancer germ line genes, MAGE-A4 and sage. Clin Cancer Res. 2005;11(15):5581–5589. doi: 10.1158/1078-0432.CCR-04-2585. [DOI] [PubMed] [Google Scholar]
- 25.Graff-Dubois S, Faure O, Gross DA, Alves P, Scardino A, Chouaib S, Lemonnier FA, Kosmatopoulos K. Generation of CTL recognizing an HLA-A*0201-restricted epitope shared by MAGE-A1, -A2, -A3, -A4, -A6, -A10, and -A12 tumor antigens: implication in a broad-spectrum tumor immunotherapy. J Immunol. 2002;169(1):575–580. doi: 10.4049/jimmunol.169.1.575. [DOI] [PubMed] [Google Scholar]
- 26.Duffour MT, Chaux P, Lurquin C, Cornelis G, Boon T, van der Bruggen P. A MAGE-A4 peptide presented by HLA-A2 is recognized by cytolytic T lymphocytes. Eur J Immunol. 1999;29(10):3329–3337. doi: 10.1002/(SICI)1521-4141(199910)29:10<3329::AID-IMMU3329>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 27.Ohkuri T, Wakita D, Chamoto K, Togashi Y, Kitamura H, Nishimura T. Identification of novel helper epitopes of MAGE-A4 tumour antigen: useful tool for the propagation of Th1 cells. Br J Cancer. 2009;100(7):1135–1143. doi: 10.1038/sj.bjc.6604966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawashima I, Hudson SJ, Tsai V, Southwood S, Takesako K, Appella E, Sette A, Celis E. The multi-epitope approach for immunotherapy for cancer: identification of several CTL epitopes from various tumor-associated antigens expressed on solid epithelial tumors. Hum Immunol. 1998;59(1):1–14. doi: 10.1016/S0198-8859(97)00255-3. [DOI] [PubMed] [Google Scholar]
- 29.Van Der Bruggen P, Zhang Y, Chaux P, Stroobant V, Panichelli C, Schultz ES, Chapiro J, Van Den Eynde BJ, Brasseur F, Boon T. Tumor-specific shared antigenic peptides recognized by human T cells. Immunol Rev. 2002;188:51–64. doi: 10.1034/j.1600-065X.2002.18806.x. [DOI] [PubMed] [Google Scholar]
- 30.Kobayashi H, Song Y, Hoon DS, Appella E, Celis E. Tumor-reactive T helper lymphocytes recognize a promiscuous MAGE-A3 epitope presented by various major histocompatibility complex class II alleles. Cancer Res. 2001;61(12):4773–4778. [PubMed] [Google Scholar]
- 31.Scheibenbogen C, Lee KH, Stevanovic S, Witzens M, Willhauck M, Waldmann V, Naeher H, Rammensee HG, Keilholz U. Analysis of the T cell response to tumor and viral peptide antigens by an IFNgamma-ELISPOT assay. Int J Cancer. 1997;71(6):932–936. doi: 10.1002/(SICI)1097-0215(19970611)71:6<932::AID-IJC3>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 32.Zhang HG, Chen HS, Peng JR, Shang XY, Zhang J, Xing Q, Pang XW, Qin LL, Fei R, Mei MH, Leng XS, Chen WF. Specific CD8(+)T cell responses to HLA-A2 restricted MAGE-A3 p271–279 peptide in hepatocellular carcinoma patients without vaccination. Cancer Immunol Immunother. 2007;56(12):1945–1954. doi: 10.1007/s00262-007-0338-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alhamarneh O, Amarnath SM, Stafford ND, Greenman J. Regulatory T cells: what role do they play in antitumor immunity in patients with head and neck cancer? Head Neck. 2008;30(2):251–261. doi: 10.1002/hed.20739. [DOI] [PubMed] [Google Scholar]
- 34.Lang S, Whiteside TL, Lebeau A, Zeidler R, Mack B, Wollenberg B. Impairment of T-cell activation in head and neck cancer in situ and in vitro: strategies for an immune restoration. Arch Otolaryngol Head Neck Surg. 1999;125(1):82–88. doi: 10.1001/archotol.125.1.82. [DOI] [PubMed] [Google Scholar]
- 35.Young MR. Protective mechanisms of head and neck squamous cell carcinomas from immune assault. Head Neck. 2006;28(5):462–470. doi: 10.1002/hed.20331. [DOI] [PubMed] [Google Scholar]
- 36.Atanackovic D, Altorki NK, Cao Y, Ritter E, Ferrara CA, Ritter G, Hoffman EW, Bokemeyer C, Old LJ, Gnjatic S. Booster vaccination of cancer patients with MAGE-A3 protein reveals long-term immunological memory or tolerance depending on priming. Proc Natl Acad Sci USA. 2008;105(5):1650–1655. doi: 10.1073/pnas.0707140104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marchand M, Punt CJ, Aamdal S, Escudier B, Kruit WH, Keilholz U, Hakansson L, van Baren N, Humblet Y, Mulders P, Avril MF, Eggermont AM, Scheibenbogen C, Uiters J, Wanders J, Delire M, Boon T, Stoter G. Immunisation of metastatic cancer patients with MAGE-3 protein combined with adjuvant SBAS-2: a clinical report. Eur J Cancer. 2003;39(1):70–77. doi: 10.1016/S0959-8049(02)00479-3. [DOI] [PubMed] [Google Scholar]
- 38.Rimoldi D, Rubio-Godoy V, Dutoit V, Lienard D, Salvi S, Guillaume P, Speiser D, Stockert E, Spagnoli G, Servis C, Cerottini JC, Lejeune F, Romero P, Valmori D. Efficient simultaneous presentation of NY-ESO-1/LAGE-1 primary and nonprimary open reading frame-derived CTL epitopes in melanoma. J Immunol. 2000;165(12):7253–7261. doi: 10.4049/jimmunol.165.12.7253. [DOI] [PubMed] [Google Scholar]