Abstract
The forkhead transcription factor Foxp3 is the only definitive marker of CD4+CD25+ regulatory T cells (Tregs) and has been identified as a key regulator in the development and function of Tregs. Foxp3 expression has been reported in a variety of solid tumors, including melanoma. In this study, we validated Foxp3 expression in both tumor-infiltrating Tregs and melanoma cells by performing immunohistochemical analysis of human melanoma tissue sections. Further, we assessed Foxp3 expression in melanoma cell lines by performing flow cytometry, confocal microscopic analysis, reverse transcription-polymerase chain reaction (RT–PCR), and Western blotting. Inhibition of Foxp3 expression in melanoma cells using small interfering RNA (siRNA) resulted in downregulation of B7-H1 and transforming growth factor (TGF)-β expression; in contrast, Foxp3 overexpression resulted in the upregulation of the expression of these proteins. Coculture of Foxp3-expressing melanoma cells with naive CD4+CD25− T cells resulted in strong inhibition of T-cell proliferation. This antiproliferative effect was partially abrogated by specific inhibition of Foxp3 expression and was effectively enhanced by overexpression of Foxp3. We observed an attenuated antiproliferative effect even when melanoma cells and T cells in the coculture were separated using Transwell inserts. These findings indicated that melanoma cells could have Foxp3-dependent Treg-like suppressive effects on T cells and suggested that the mimicking of Treg function by melanoma cells may represent a possible mechanism of tumor resistance to immune destruction in the melanoma tumor microenvironment.
Keywords: Melanoma, Foxp3, Treg-like, Tumor resistance
Introduction
The immune system is subjected to many levels of control, and regulatory T cells (Tregs) play a major role in the regulation of the immune system [1]. The importance of Tregs lies in their pivotal role in maintaining immunologic tolerance and preventing autoimmune diseases [2, 3]. When activated, Tregs suppress proliferation of and cytokine production in normal CD4+CD25− T cells, CD8+ T cells, and established Th1 and Th2 cells [4, 5]. Despite playing an essential role in the prevention of autoimmunity, Tregs may have a negative effect on the health of a patient by downregulating the beneficial immune responses against tumors [6]. Numerous animal studies have shown that removal or inhibition of Tregs dramatically improves tumor clearance and survival [7, 8]. Further, depletion of Tregs enhances the efficiency of cell-based tumor vaccination [9]. Elevated Treg levels in human melanoma cells are considered as a marker for metastasis and are negatively correlated with survival [10, 11]. The forkhead transcription factor Foxp3 has been identified as a key regulator in the development and function of Tregs and is, at present, the only definitive marker of Tregs [12, 13]. Mutations in the Foxp3 gene lead to fatal autoimmune or inflammatory diseases (the scurfy disease in mice [14] and the immunodysregulation, polyendocrinopathy, enteropathy, X-linked (IPEX) syndrome in humans [15]), which are caused by deficiency or malfunction of Tregs. Furthermore, ectopic expression of Foxp3 in conventional human T cells results in partial acquisition of the phenotypic characteristics and functions of a Treg, although some studies have shown that the suppressive activity of induced Foxp3 + T cells is inferior to that of bona fide Tregs [16, 17].
Melanoma cells are resistant to a wide range of chemotherapeutic and radiotherapeutic treatments [18]. Numerous immune-based therapies (involving cytokines, antibodies, cancer vaccines, adoptive immunotherapy, and combinations of these therapeutic agents and modalities) are the focus of studies on alternative therapeutic approaches [19]. Nevertheless, promising preclinical data from immune modulation trials do not always translate into clinically meaningful results in phase 2 or randomized phase 3 clinical trials [20]. Although cancer vaccines and adoptive T-cell transfer have been shown to increase the levels of the circulating tumor antigen–specific T cells, these approaches produce clinical responses in only a few patients [21]. These observations indicate that tumor resistance to immune destruction may be the dominant process in the melanoma tumor microenvironment [21]. Recent studies have suggested that the presence of Foxp3 + Tregs in the tumor microenvironment [11], the expression of inhibitory ligands (such as B7-H1 and Fas ligand) on melanoma cells [22], the secretion of immunosuppressive factors (such as transforming growth factor (TGF)-β and IL-10) by melanoma cells [23], and the activity of nutrient-catabolizing enzymes may contribute to the resistance of the tumor to immune destruction. To improve the current concepts of immune-based therapies, a broader understanding of the local and systemic tumor-resistant mechanisms in cancer patients is important. Until recently, Foxp3 expression has been thought to be restricted to the T-cell lineage. However, many reports have shown that Foxp3 is expressed by a variety of tumor cells [24–26]. In this report, we have shown another variation of this theme: Foxp3 in the melanoma cells modulated the expression of immune-associated molecules, and Foxp3 expression correlated with Treg-like suppressive activity on T-cell proliferation; thus, the melanoma cells could suppress T cells in vitro via direct inhibition of their proliferation rather than indirect inhibition through Tregs.
Materials and methods
Cells and culture
A375, A2058, 1205Lu, 451Lu, SK-MEL-1, SK-MEL-5, and Hs 294T (ATCC) human melanoma cells were cultured in complete RPMI 1640 medium supplemented with 10% fetal calf serum (FCS), 2 mmol/L of glutamine, and 10 mmol/L of 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES; Invitrogen) at 37°C in the presence of 5% CO2. Normal epidermal melanocytes were used in the primary cell culture; they were derived from human foreskin and were cultured in Keratinocyte-SFM (Gibco) supplemented with 10% FCS, 10 ng/mL of 12-O-tetradecanoylphorbol-13-acetate (TPA), and 10 ng/mL of basic fibroblast growth factor (bFGF; Invitrogen).
Tissue sample preparation and immunohistochemical analysis
The melanoma and melanocytic nevus tissues, which were collected after surgical resection, were fixed in formalin and embedded in paraffin. Collection of tissues for this study was approved by the Committee of Ethics at Fourth Military Medical University, Xi’an, China, and informed consent was obtained from all patients. The criterion for sample selection was almost complete absence or paucity of melanin, which was necessary for good immunohistochemical evaluation. All diagnoses were made on the basis of the histopathologic features observed after staining with hematoxylin and eosin (HE) and were confirmed by performing immunohistochemical analysis with anti-S100 and anti-HMB45 antibodies. Immunohistochemical staining of deparaffinized tissue sections was done according to the standard protocols using anti-Foxp3 antibodies (PCH101; eBioscience). The immunostaining intensities were graded as strong, moderate, and weak by a pathologist specializing in melanoma.
Laser scanning confocal microscopic analysis
For double-label immunofluorescence analysis, cultured cell slides were fixed in acetone for 10 min at 4°C. The slides were treated in blocking solution and then incubated with fluorescein isothiocyanate (FITC)-conjugated rat anti-human Foxp3 monoclonal antibodies (PCH101; eBioscience) for 1 h at room temperature. After rinsing with phosphate-buffered saline (PBS), the slides were treated with diamidino-phenyl-indole (DAPI, Sigma) for 30 min, mounted using an anti-fade mounting medium, and examined under a Leica TCS-SP laser scanning confocal microscope (Leica).
Flow cytometric analysis
Fluorochrome-conjugated antibodies to the human Foxp3 protein and isotype control antibodies (eBioscience) were used in this analysis. Before antibody staining, the cells were washed in PBS containing 1% bovine serum albumin (BSA) and 0.1% NaN3, followed by fixation with 1% paraformaldehyde. Foxp3 staining was performed using the Foxp3 buffer set from eBioscience. A total of 105 events were assessed using FACSCalibur (BD Biosciences), and the data were analyzed using the Cell Quest program (BD Biosciences).
RNA extraction and reverse transcription-polymerase chain reaction (RT–PCR)
Total RNA extraction from the cells was performed using Trizol reagent (Invitrogen). Reverse transcription was performed using Superscript II reverse transcriptase and random primers (Invitrogen). PCR products were analyzed using 1% agarose gel electrophoresis and documented using Alpha Imager. Relative mRNA levels were determined using semiquantitative PCR with the standard SYBR Green RT–PCR kit (Takara) and analyzed using the GraphPad Prism software (GraphPad Software). Semiquantitative PCR was performed under standard conditions. The relative levels of the mRNA transcripts were normalized to the levels of the internal control β-actin. The specific primer pairs were as follows: Foxp3 forward, 5′-CACAACATGCGACCCCCTTTCACC-3′; Foxp3 reverse, 5′-AGGTTGTGGCGGATGGCGTTCTTC-3′; β-actin forward, 5′-GGCACCACACCTTCTACA-3′; β-actin reverse, 5′-AGGAAGGCTGGAAGAGTG-3′; TGF-β1 forward, 5′-ATGAACTCATTCAGTCACCATAGC-3′; TGF-β1 reverse, 5′-CTATCCCCCACTAAAGCAGG-3′; TGF-β2 forward, 5′-AATGATGATGATAATGATGATGACG-3′; TGF-β2 reverse, 5′-GTACAGCAACTCCACTTAATGGG-3′; B7-H1 forward, 5′-CTGCAGGGCATTCCAGAAA-3′; and B7-H1 reverse, 5′-GGTTGAGAATCCCTGCTTGA-3′.
Western blot analysis
Protein extracts were prepared in sodium dodecyl sulfate (SDS) lysis buffer containing protease inhibitors, separated with 12% SDS–polyacrylamide gel electrophoresis (PAGE), and transferred onto polyvinylidene fluoride membranes. The membranes were blocked with Tris-buffered saline and Tween 20 (TBST) containing 5% nonfat dry milk for 2 h at room temperature and then incubated with anti-Foxp3 (eBioscience), anti-B7-H1 (R&D), or anti-β-actin (R&D) primary antibodies overnight at 4°C, followed by incubation with the corresponding horseradish peroxidase–conjugated secondary antibodies for 1 h at room temperature. Immunoreactive bands were detected using the enhanced chemiluminescence detection kit (Amersham Biosciences).
Cytokine production assay
The levels of TGF-β1 and TGF-β2 in the culture supernatants of the melanoma cells were quantified with the TGF-β1 and TGF-β2 Quantikine ELISA kits (R&D). Melanoma cells were seeded in 6-well plates, and the culture supernatants were harvested 72 h later. Cytokine concentrations were normalized to the cell numbers.
Foxp3 silencing in melanoma cells was performed using small interfering RNA (siRNA)
Three siRNAs specific for the Foxp3 gene were designed and synthesized by Takara. Transfection was performed using the pSilencer3.1-H1 neo expression plasmid. The A2058 melanoma cells were resuspended in serum-free medium, seeded in 6-well plates, and transfected with Foxp3 siRNA or control siRNA using Lipofectamine2000 (Invitrogen). Normal medium was used 6 h later. Cells were harvested 48 h after transfection for performing an mRNA assay or after 72 h for performing a protein assay. Three different Foxp3-specific siRNAs were screened, and the more efficient and less cytotoxic one (5′-CAGCACATTCCCAGAGTTCCT-3′) was chosen.
Overexpression of Foxp3 in melanoma cells
The Foxp3 gene was subcloned into the pRc/CMV6 expression plasmid and identified by DNA sequencing. The recombinant vector was then used to transfect melanoma cells for overexpression of Foxp3, and the empty vector was used as the negative control.
Isolation of T cells and coculture experiments
Peripheral blood mononuclear cells (PBMCs) were isolated from the peripheral blood of healthy donors using Ficoll-Hypaque gradient centrifugation. To acquire CD4+CD25− T cells, PBMCs were isolated via positive selection using the CD4+ T-cell isolation kit (Miltenyi Biotec); then, negative selection was performed by adding anti-CD25-conjugated magnetic beads and performing magnetic separation on MACS columns to remove the CD25+ cells. A2058 cells, which showed silencing or overexpression of Foxp3 (3 days after transfection), were seeded into 6-well plates at 2 × 105 cells/well and cocultured with 2 × 106 CD4+CD25− carboxyfluorescein succinimidyl ester (CFSE)-labeled T cells (stimulated with anti-CD3/anti-CD28) for 3 days in X-Vivo 15 medium. Proliferation of the T cells was measured on the basis of CFSE dilution using flow cytometry and calculated as the ratio of the number of proliferating T cells to the total number of T cells.
Statistical analysis
Statistical analysis of the data was performed using GraphPad Prism software. The difference between the 2 groups was analyzed by the two-paired t-test. P values less than 0.05 were considered statistically significant.
Results
Foxp3 was expressed by tumor cells in melanoma tissues and by melanoma cell lines
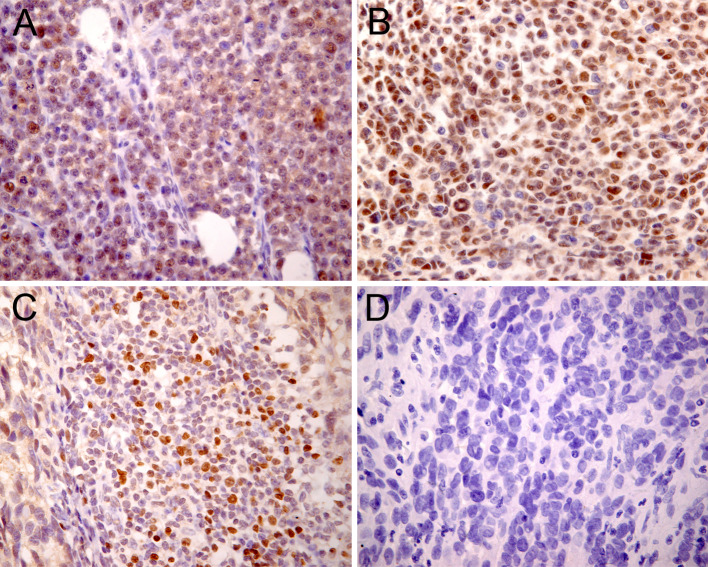
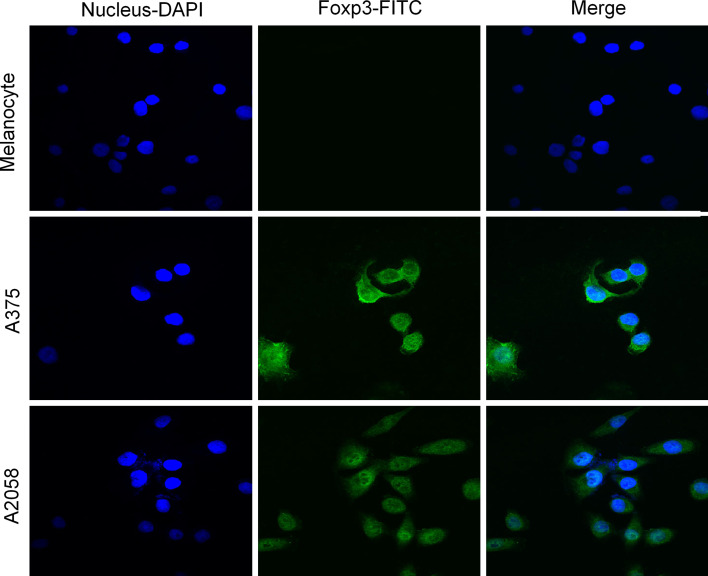
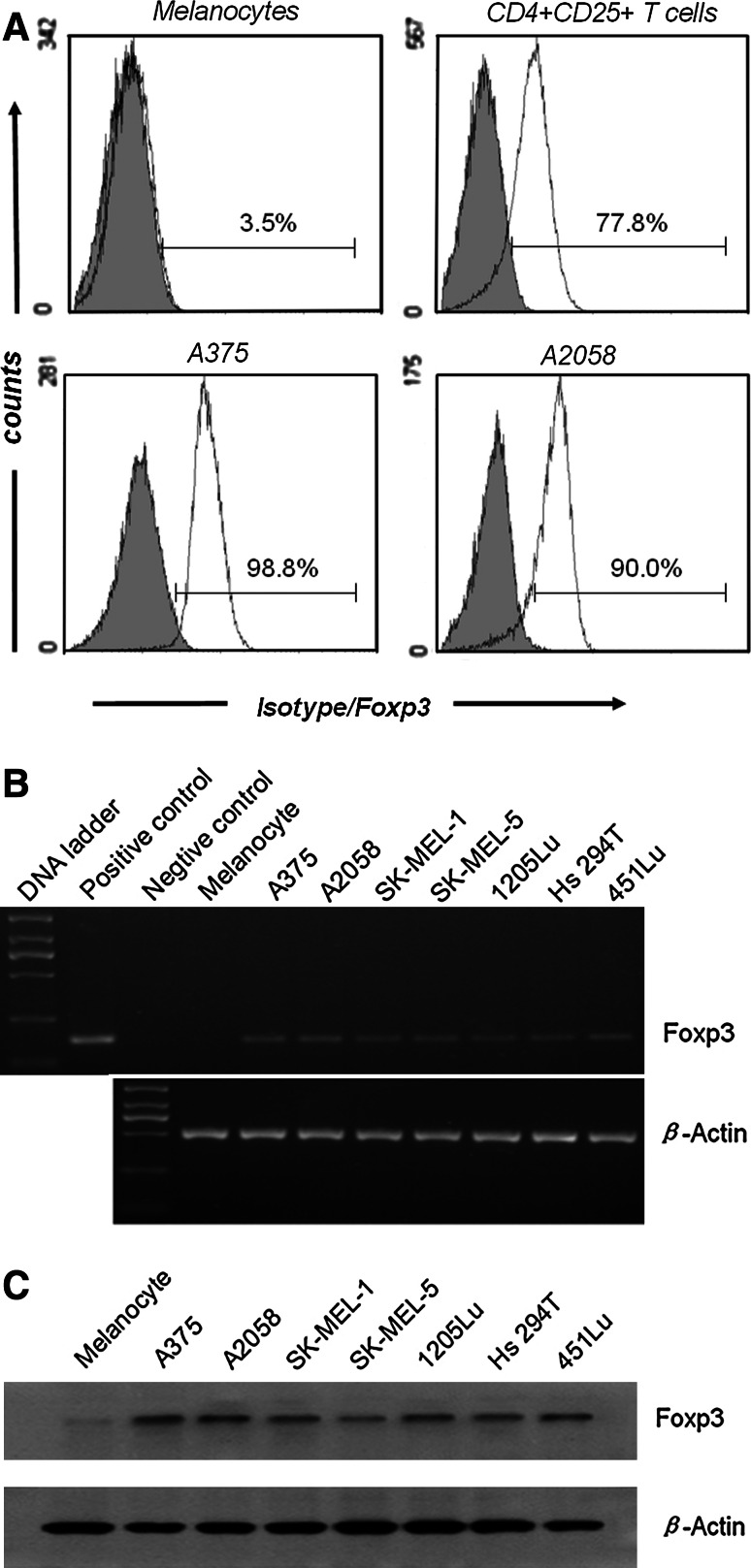
We detected Foxp3 expression in the tumor cells of 14/18 primary melanoma and 12/16 metastatic melanoma specimens but not in the tumor cells of the melanocytic nevus specimens (Table 1). In addition to the immunostaining in cells with lymphocyte morphology (small, round, dense nucleus, and little cytoplasm), immunostaining was also surprisingly apparent in cells with several morphologic features of tumor cells, such as a large, irregular nucleus, and abundant cytoplasm. Foxp3 subcellular staining was usually observed in the nucleus and cytoplasm of some specimens (Fig. 1a); in the other specimens, however, it seemed to be found predominantly in the nucleus (Fig. 1b). In all melanoma specimens, we detected Foxp3-positive tumor-infiltrating lymphocytes (Fig. 1c), which presumably represented Tregs. Nonmalignant melanocytic cells did not show Foxp3 expression (Fig. 1d). The subcellular localization of Foxp3 in melanoma cell lines was confirmed by double-label immunofluorescence confocal microscopic analysis. In this experiment, the nuclei were stained with DAPI to facilitate analysis. The A375 and A2058 cells exhibited intense Foxp3 expression in the nucleus and less intense expression in the cytoplasm; the melanocytes did not exhibit Foxp3 expression (Fig. 2). In accordance with the immunohistochemical data indicating Foxp3 expression in the tumor cells of melanoma tissues, Foxp3 expression was clearly detected in at least 2 melanoma cell lines. The results of Foxp3 staining in melanoma cell lines, which were analyzed by flow cytometry, were uniformly positive, and the results of Foxp3 staining in normal melanocytes were negative (Fig. 3a). Staining with anti-Foxp3 resulted in a shift in the fluorescence of the entire cell population, with reference to the fluorescence in the isotype control. The melanoma cell lines depicted in Fig. 3b showed positive results for Foxp3 transcription. The vector containing the Foxp3 gene was used as the positive control. Moreover, as shown in Fig. 3c, Foxp3 protein could be detected by Western blotting. In contrast, cultured normal epidermal melanocytes did not express Foxp3. These results showed that Foxp3 was expressed both ex vivo and in vitro by the melanoma cells. However, the intensity of Foxp3 expression was variable at both mRNA and protein levels.
Table 1.
Foxp3 expression in melanocytic tissues
| Number of samples | |
|---|---|
| Primary melanoma | |
| Strong expression | 2/18 |
| Moderate expression | 6/18 |
| Weak expression | 6/18 |
| No expression | 4/18 |
| Foxp3-positive lymphocytes | 18/18 |
| Metastatic melanoma | |
| Strong expression | 4/16 |
| Moderate expression | 5/16 |
| Weak expression | 3/16 |
| No expression | 4/16 |
| Foxp3-positive lymphocytes | 16/16 |
| Melanocytic nevus | |
| No expression | 10/10 |
| Foxp3-positive lymphocytes | 0/10 |
Fig. 1.
Immunohistochemical staining of paraffin-embedded melanoma tissues revealed Foxp3 expression in the melanoma cells. Nuclear/cytoplasmic (a) and nuclear (b) Foxp3 staining patterns were observed. Foxp3-positive lymphocytes (c) were probably tumor-infiltrating T regulatory cells (Tregs). In nonmalignant melanocytic nevus tissues, no Foxp3-positive cells were detected (d)
Fig. 2.
Double-label immunofluorescence analysis of Foxp3 expression in melanoma cell lines. The nuclei of melanocytes, A375 cells, and A2058 cells were identified using blue fluorescence. A375 and A2058 cells, but not melanocytes, expressed Foxp3 in both nuclei and cytoplasm (green fluorescence). Superimposed images confirmed colocalization
Fig. 3.
Foxp3 was widely expressed in the melanoma cell lines. a Cells were stained using anti-Foxp3 monoclonal antibodies (mAb) (white curve) or isotype-matched negative control (gray curve) and analyzed using flow cytometry. Two representative melanoma cell lines are shown together with peripheral blood CD4+CD25+ T cells and melanocytes for comparison. b, c Foxp3 expression was analyzed using RT–PCR (b) and Western blot (c)
Foxp3 in melanoma cells modulated the expression of B7-H1, TGF-β1, and TGF-β2
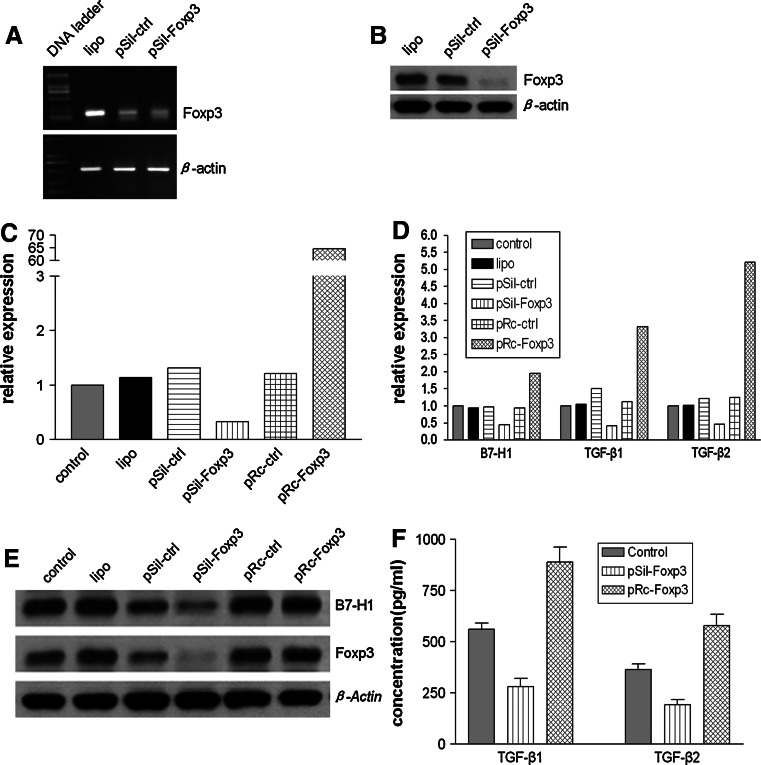
A key function of Foxp3 is the control of inflammatory cytokine production in Tregs. To analyze whether Foxp3 modulated the production of immune-associated molecules in melanoma cells, Foxp3 expression was downregulated via siRNA transfection and upregulated via expression vector transfection. For this experiment, the A2058 cell line was chosen over other cell lines because it showed higher Foxp3 expression and transfection efficiency with Lipofectamin2000. Treatment of A2058 cells with Foxp3 siRNA resulted in a strong downregulation of both Foxp3 mRNA (Fig. 4a) and protein (Fig. 4b). Specific analyses of Foxp3, B7-H1, TGF-β1, and TGF-β2 mRNA transcription by semiquantitative RT–PCR indicated that the expression of the 4 molecules was downregulated following Foxp3 silencing and upregulated following Foxp3 overexpression. The data analyzed using the 2-ΔΔCT method indicated that in comparison with the Foxp3 mRNA transcription levels in the control cells, the transcription level was 0.33-fold lower after silencing and 64.7-fold higher after overexpression (Fig. 4c). Similarly, the transcription of B7-H1, TGF-β1, and TGF-β2 was downregulated 0.44-, 0.41-, and 0.47-fold, respectively, in Foxp3-silenced cells, in comparison with the transcription levels in the control cells; inversely, overexpression of Foxp3 resulted in 1.95-, 3.32-, and 5.22-fold higher levels of B7-H1, TGF-β1, and TGF-β2 transcription (Fig. 4d). Analysis of Foxp3 and B7-H1 expression by Western blotting (Fig. 4e) and that of TGF-β1 and TGF-β2 levels by ELISA (Fig. 4f) indicated that the 4 molecules were regulated in accordance with the changes in the mRNA levels.
Fig. 4.
Foxp3 expression was positively correlated with the expression of immunity-associated molecules. a, b A2058 cells were transfected using only Lipofectamin2000 (lipo), control siRNA (pSil-ctrl), or Foxp3 siRNA (pSil-Foxp3). Cells were recovered 48 h before the analysis of Foxp3 expression by RT–PCR (a) and 72 h before Western blotting (b). c–f relative expression of Foxp3, B7-H1, TGF-β1, and TGF-β2 after specific silencing or overexpression of Foxp3. Following transfection with lipo, pSil-ctrl, pSil-Foxp3, pRc-ctrl (control expression vector), or pRc-Foxp3 (Foxp3 expression vector), the mRNA transcription levels of Foxp3 (c), B7-H1, TGF-β1, and TGF-β2 (d) were measured using semiquantitative PCR. Foxp3 and B7-H1 proteins (e) were analyzed using Western blotting, and the TGF-β1 and TGF-β2 concentrations in the culture supernatants (f) were determined by ELISA
Inhibition of T-cell proliferation in coculture systems was dependent on Foxp3 expression in A2058 cells
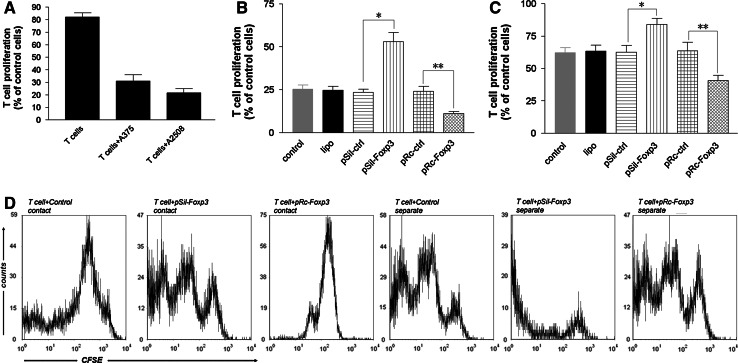
To evaluate the effect of Foxp3-expressing melanoma cells on T-cell proliferation, A375 and A2058 cells were cocultured with naive CD4+CD25− T cells in the preliminary experiments. After 3 days of coculture with anti-CD3/anti-CD28 stimulus, proliferation of T cells was measured using CFSE labeling and flow cytometry analysis. In the cocultures, the A375 and A2058 melanoma cells strongly inhibited the proliferation of the anti-CD3/anti-CD28-activated T cells (Fig. 5a). To verify the presumed Foxp3-induced suppression of T-cell proliferation by melanoma cells, we repeated the coculture experiments with A2058 cells in which Foxp3 expression had been silenced by siRNA transfection or Foxp3 had been overexpressed by expression vector transfection. We observed that the percentage of proliferating T cells in the coculture system increased 1.09-fold after Foxp3 silencing and decreased 0.56-fold after Foxp3 overexpression, in comparison with the corresponding values in the control system (Fig. 5b). To explore whether the proliferation arrest in the coculture system was because of cell–cell contact, a membrane was used to separate the A2058 cells from the T cells in the coculture, and we observed an attenuated antiproliferative effect. The percentage of proliferating T cells in the coculture system increased 0.35-fold after Foxp3 silencing and decreased 0.35-fold after Foxp3 overexpression, in comparison with the percentage of proliferating T cells observed in the control system (Fig. 5c). The antiproliferative effect was observed even when the melanoma cells and T cells were not in contact with each other in the coculture system, suggesting that the melanoma cells inhibited the proliferation of activated T cells by both direct cell–cell contact and indirect mechanisms.
Fig. 5.
a Proliferation of CD4+CD25− T cells after stimulation and coculture with melanoma for 3 days cells was measured using CFSE staining. The number of proliferating T cells was normalized to the number of activated T cells. b A2058 cells were transfected with lipo, pSil-ctrl, pSil-Foxp3, pRc-ctrl, or pRc-Foxp3 and cocultured with activated CD4+CD25− T cells. Columns, mean values of 3 independent experiments; bars, standard deviation. Statistical significance was confirmed using the two-paired t-test. *P < 0.001; **P < 0.05. There was no difference between the levels of the control and those of lipo, pSil-ctrl, or pRc-ctrl, suggesting that the suppressive activity was not because of any nonspecific effect. c Results similar to those in b were found even when the A2058 cells and T cells were separated using Transwell inserts. *P < 0.05; **P < 0.05. There was no difference between the levels of the control and those of lipo, pSil-ctrl, or pRc-ctrl. d Representative flow cytometry histograms of the CFSE-labeled T-cell proliferation cocultured with A2058 cells (control, transfected with pSil-Foxp3 or pRc-Foxp3)
Discussion
Our study used a wide variety of approaches to validate the following findings: Foxp3 was expressed by melanoma cells, Foxp3 modulated the expression of immunity-associated molecules, and Foxp3 expression in melanoma cells positively correlated with Treg-like suppressive activity on T-cell proliferation. Until recently, it was believed that Foxp3 expression was restricted to the T-cell lineage [12, 14]. However, when our study was in progress, some studies reported Foxp3 expression in tumor cells other than those of the T-cell lineage. Immunohistochemical analysis of melanoma and melanocytic nevus tissue sections showed Foxp3 expression in the tumor cells of 26/34 melanoma samples but not in melanocytic nevus samples. Foxp3 expression in melanoma cells was confirmed by analyzing a panel of established melanoma cell lines using flow cytometry, RT–PCR, and Western blotting. Moreover, Foxp3 expression patterns were either nuclear/cytoplasmic or nuclear, as verified by immunohistochemical analysis of the melanoma tissue sections and by double-label immunofluorescence analysis of the melanoma cell lines. These results showed that Foxp3 was expressed both ex vivo and in vitro by the melanoma cells. This finding suggested that Foxp3 expression by melanoma cells was neither a culture artifact nor a transient phenomenon. The activation of Tregs has been reported to change the cytoplasmic/perinuclear expression pattern of Foxp3 to a nuclear expression pattern [27]. The change in the Foxp3 expression pattern may be a result of post-translational modifications. Therefore, heterogeneous subcellular localization of Foxp3 in melanoma cells may reflect the presence of different modified forms of Foxp3. In this small series of samples with different clinicopathologic characteristics, we were unable to find a correlation between the grade of Foxp3 expression or subcellular localization and tumor stage. The functional relevance of this finding requires further investigation.
The specific downregulation of Foxp3 expression in pancreatic carcinoma cells results in the upregulation of IL-6 [24]; this suggests that Foxp3 operates in a way similar to the mode of operation of Tregs. Similar to its role in directing Treg activity, Foxp3 expression in melanoma cells may endow them with Treg-like activity and enable them to resist immune destruction. Even in T cells, Foxp3 expression does not automatically lead to acquisition of full regulatory activity since human T cells can transiently induce Foxp3 expression without acquisition of Treg function [28]. To allow induction of Treg activity, Foxp3 expression needs to be stabilized by promoter demethylation [29]. The observation that aberrant DNA methylation and demethylation patterns are common events in melanoma [30] suggests that the Foxp3 promoter may become demethylated in melanoma cells, leading to stable Foxp3 expression and potential acquisition of regulatory activity. The effects of Foxp3 in tumor cells may be much more subtle because recent evidence suggests that Foxp3 transcriptionally downregulates intracellular phosphodiesterase 3B (PDE3B) in Tregs, and Tregs may show enhanced metabolic fitness, better survival, and reduced apoptosis by reducing the expression levels of this protein [31]. It may be speculated that Foxp3 confers a survival advantage on melanoma cells with normal tumor physiology and under chemotherapeutic stress.
B7-H1 is found abundantly on melanoma cells and has been described to negatively regulate T-cell function by binding with PD-1 [21, 32]. TGF-β has been shown to control the growth of several normal cell populations, including human B and T lymphocytes. Furthermore, the growth of melanoma cells is not inhibited by any of the 3 TGF-β isoforms, and these cells secrete significantly higher levels of TGF-β as immunosuppressive factors [23, 33]. In our study, we discovered that Foxp3 expression positively correlated with the expression of B7-H1 and the secretion of TGF-β. Downregulation of Foxp3 in melanoma cells resulted in lower levels of B7-H1, TGF-β1, and TGF-β2, whereas upregulation of Foxp3 resulted in higher expression levels of these molecules. This indicated that Foxp3 could regulate the expression of inhibitory ligands (such as B7-H1) on melanoma cells and the secretion of immunosuppressive factors (such as TGF-β) by melanoma cells in some way. The functional significance of Foxp3-mediated promotion of B7-H1 and TGF-β in melanoma cells is currently unknown. Given the physiological function of B7-H1 and TGF-β, Foxp3 expression may endow melanoma cells with some degree of suppressive activity by regulating the expression of immune-associated molecules.
The Foxp3-expressing pancreatic carcinoma cell lines have been shown to inhibit proliferation (rather than activation) of T cells, and knockdown of Foxp3 could effectively reduce the suppressive activity of these cells [24]. In the coculture experiments, A375 and A2058 cells strongly inhibited the proliferation of anti-CD3/anti-CD28-activated T cells. Moreover, the Treg-like suppressive activity was strongly associated with the expression of Foxp3. Downregulation or upregulation of Foxp3 effectively reduced or enhanced the suppressive activity. Our results were consistent with the results for the pancreatic carcinoma cells. In addition, an attenuated antiproliferative effect was also observed even when the melanoma cells and T cells were separated by a membrane using Transwell inserts in the cocultures. We inferred that the inhibition of T-cell proliferation by the melanoma cells in the coculture system was by both direct cell–cell contact and indirect mechanisms. The expression of inhibitory ligands and the secretion of immunosuppressive factors by the melanoma cells may partly explain the inhibition of T-cell proliferation by both these approaches. Meanwhile, the results suggested that not only is antitumor immunity influenced by the tumor-infiltrating Tregs but the melanoma cells themselves might modulate T-cell function via expression of Foxp3.
In conclusion, this study showed that Foxp3 was expressed by melanoma cells. Foxp3 expression and function can no longer be considered to be restricted to the T-cell lineage but may play a wider role in tumor cells. Moreover, this study revealed that Foxp3 modulated the expression of immune-associated molecules, and Foxp3 expression positively correlated with the Treg-like suppressive activity on T cells. The expression of Foxp3 by melanoma cells, which enabled the melanoma cells to mimic Treg function, may represent a possible mechanism of tumor resistance to immune destruction in the melanoma tumor microenvironment. More experiments and clinical studies will have to be performed to discover the precise effects of Foxp3 in tumor cells.
Acknowledgments
We would like to thank the colleagues from the Department of Pathology, Xijing Hospital of Fourth Military Medical University for their excellent technical support.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
The authors J. Niu and C. Jiang contributed equally to this work.
References
- 1.Sakaguchi S. Naturally arising CD4 + regulatory t cells for immunologic self-tolerance and negative control of immune responses. Annu Rev Immunol. 2004;22:531–562. doi: 10.1146/annurev.immunol.21.120601.141122. [DOI] [PubMed] [Google Scholar]
- 2.Zheng Y, Rudensky AY. Foxp3 in control of the regulatory T cell lineage. Nat Immunol. 2007;8:457–462. doi: 10.1038/ni1455. [DOI] [PubMed] [Google Scholar]
- 3.Sakaguchi S. Naturally arising Foxp3-expressing CD25 + CD4 + regulatory T cells in immunological tolerance to self and non-self. Nat Immunol. 2005;6:345–352. doi: 10.1038/ni1178. [DOI] [PubMed] [Google Scholar]
- 4.Xu D, Liu H, Komai-Koma M, Campbell C, McSharry C, Alexander J, Liew FY. CD4 + CD25 + regulatory T cells suppress differentiation and functions of Th1 and Th2 cells. Leishmania major infection, and colitis in mice. J Immunol. 2003;170:394–399. doi: 10.4049/jimmunol.170.1.394. [DOI] [PubMed] [Google Scholar]
- 5.Suvas S, Kumaraguru U, Pack CD, Lee S, Rouse BT. CD4 + CD25 + T cells regulate virus-specific primary and memory CD8 + T cell responses. J Exp Med. 2003;198:889–901. doi: 10.1084/jem.20030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curiel TJ, Coukos G, Zou L, Alvarez X, Cheng P, Mottram P, Evdemon-Hogan M, Conejo-Garcia JR, Zhang L, Burow M, Zhu Y, Wei S, Kryczek I, Daniel B, Gordon A, Myers L, Lackner A, Disis ML, Knutson KL, Chen L, Zou W. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med. 2004;10:942–949. doi: 10.1038/nm1093. [DOI] [PubMed] [Google Scholar]
- 7.Zou W. Regulatory T cells, tumour immunity and immunotherapy. Nat Rev Immunol. 2006;6:295–307. doi: 10.1038/nri1806. [DOI] [PubMed] [Google Scholar]
- 8.Beyer M, Schultze JL. Regulatory T cells in cancer. Blood. 2006;108:804–811. doi: 10.1182/blood-2006-02-002774. [DOI] [PubMed] [Google Scholar]
- 9.Dannull J, Su Z, Rizzieri D, Yang BK, Coleman D, Yancey D, Zhang A, Dahm P, Chao N, Gilboa E, Vieweg J. Enhancement of vaccine-mediated antitumor immunity in cancer patients after depletion of regulatory T cells. J Clin Invest. 2005;115:3623–3633. doi: 10.1172/JCI25947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mourmouras V, Fimiani M, Rubegni P, Epistolato MC, Malagnino V, Cardone C, Cosci E, Nisi MC, Miracco C. Evaluation of tumour-infiltrating CD4 + CD25 + FOXP3 + regulatory T cells in human cutaneous benign and atypical naevi, melanomas and melanoma metastases. Br J Dermatol. 2007;157:531–539. doi: 10.1111/j.1365-2133.2007.08057.x. [DOI] [PubMed] [Google Scholar]
- 11.Viguier M, Lemaitre F, Verola O, Cho MS, Gorochov G, Dubertret L, Bachelez H, Kourilsky P, Ferradini L. Foxp3 expressing CD4 + CD25(high) regulatory T cells are overrepresented in human metastatic melanoma lymph nodes and inhibit the function of infiltrating T cells. J Immunol. 2004;173:1444–1453. doi: 10.4049/jimmunol.173.2.1444. [DOI] [PubMed] [Google Scholar]
- 12.Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, Rudensky AY. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22:329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 13.Chatila TA. Molecular mechanisms of regulatory T-cell development. Chem Immunol Allergy. 2008;94:16–28. doi: 10.1159/000154853. [DOI] [PubMed] [Google Scholar]
- 14.Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, Yasayko SA, Wilkinson JE, Galas D, Ziegler SF, Ramsdell F. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27:68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- 15.Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, Whitesell L, Kelly TE, Saulsbury FT, Chance PF, Ochs HD. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001;27:20–21. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- 16.Yagi H, Nomura T, Nakamura K, Yamazaki S, Kitawaki T, Hori S, Maeda M, Onodera M, Uchiyama T, Fujii S, Sakaguchi S. Crucial role of FOXP3 in the development and function of human CD25 + CD4 + regulatory T cells. Int Immunol. 2004;16:1643–1656. doi: 10.1093/intimm/dxh165. [DOI] [PubMed] [Google Scholar]
- 17.Allan SE, Passerini L, Bacchetta R, Crellin N, Dai M, Orban PC, Ziegler SF, Roncarolo MG, Levings MK. The role of 2 FOXP3 isoforms in the generation of human CD4 + Tregs. J Clin Invest. 2005;115:3276–3284. doi: 10.1172/JCI24685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.La Porta CA. Mechanism of drug sensitivity and resistance in melanoma. Curr Cancer Drug Targets. 2009;9:391–397. doi: 10.2174/156800909788166574. [DOI] [PubMed] [Google Scholar]
- 19.Alexandrescu DT, Ichim TE, Riordan NH, Marincola FM, Di Nardo A, Kabigting FD, Dasanu CA. Immunotherapy for melanoma: current status and perspectives. J Immunother. 2010;33:570–590. doi: 10.1097/CJI.0b013e3181e032e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korn EL, Liu PY, Lee SJ, Chapman JA, Niedzwiecki D, Suman VJ, Moon J, Sondak VK, Atkins MB, Eisenhauer EA, Parulekar W, Markovic SN, Saxman S, Kirkwood JM. Meta-analysis of phase II cooperative group trials in metastatic stage IV melanoma to determine progression-free and overall survival benchmarks for future phase II trials. J Clin Oncol. 2008;26:527–534. doi: 10.1200/JCO.2007.12.7837. [DOI] [PubMed] [Google Scholar]
- 21.Gajewski TF. Identifying and overcoming immune resistance mechanisms in the melanoma tumor microenvironment. Clin Cancer Res. 2006;12:2326s–2330s. doi: 10.1158/1078-0432.CCR-05-2517. [DOI] [PubMed] [Google Scholar]
- 22.Dong H, Strome SE, Salomao DR, Tamura H, Hirano F, Flies DB, Roche PC, Lu J, Zhu G, Tamada K, Lennon VA, Celis E, Chen L. Tumor-associated B7–H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med. 2002;8:793–800. doi: 10.1038/nm730. [DOI] [PubMed] [Google Scholar]
- 23.Zou W. Immunosuppressive networks in the tumour environment and their therapeutic relevance. Nat Rev Cancer. 2005;5:263–274. doi: 10.1038/nrc1586. [DOI] [PubMed] [Google Scholar]
- 24.Hinz S, Pagerols-Raluy L, Oberg HH, Ammerpohl O, Grussel S, Sipos B, Grutzmann R, Pilarsky C, Ungefroren H, Saeger HD, Kloppel G, Kabelitz D, Kalthoff H. Foxp3 expression in pancreatic carcinoma cells as a novel mechanism of immune evasion in cancer. Cancer Res. 2007;67:8344–8350. doi: 10.1158/0008-5472.CAN-06-3304. [DOI] [PubMed] [Google Scholar]
- 25.Ebert LM, Tan BS, Browning J, Svobodova S, Russell SE, Kirkpatrick N, Gedye C, Moss D, Ng SP, MacGregor D, Davis ID, Cebon J, Chen W. The regulatory T cell-associated transcription factor FoxP3 is expressed by tumor cells. Cancer Res. 2008;68:3001–3009. doi: 10.1158/0008-5472.CAN-07-5664. [DOI] [PubMed] [Google Scholar]
- 26.Karanikas V, Speletas M, Zamanakou M, Kalala F, Loules G, Kerenidi T, Barda AK, Gourgoulianis KI, Germenis AE. Foxp3 expression in human cancer cells. J Transl Med. 2008;6:19. doi: 10.1186/1479-5876-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C, Rowell EA, Thomas RM, Hancock WW, Wells AD. Transcriptional regulation by Foxp3 is associated with direct promoter occupancy and modulation of histone acetylation. J Biol Chem. 2006;281:36828–36834. doi: 10.1074/jbc.M608848200. [DOI] [PubMed] [Google Scholar]
- 28.Allan SE, Crome SQ, Crellin NK, Passerini L, Steiner TS, Bacchetta R, Roncarolo MG, Levings MK. Activation-induced FOXP3 in human T effector cells does not suppress proliferation or cytokine production. Int Immunol. 2007;19:345–354. doi: 10.1093/intimm/dxm014. [DOI] [PubMed] [Google Scholar]
- 29.Baron U, Floess S, Wieczorek G, Baumann K, Grutzkau A, Dong J, Thiel A, Boeld TJ, Hoffmann P, Edinger M, Turbachova I, Hamann A, Olek S, Huehn J. DNA demethylation in the human FOXP3 locus discriminates regulatory T cells from activated FOXP3(+) conventional T cells. Eur J Immunol. 2007;37:2378–2389. doi: 10.1002/eji.200737594. [DOI] [PubMed] [Google Scholar]
- 30.De Smet C, De Backer O, Faraoni I, Lurquin C, Brasseur F, Boon T. The activation of human gene MAGE-1 in tumor cells is correlated with genome-wide demethylation. Proc Natl Acad Sci U S A. 1996;93:7149–7153. doi: 10.1073/pnas.93.14.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gavin MA, Rasmussen JP, Fontenot JD, Vasta V, Manganiello VC, Beavo JA, Rudensky AY. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 2007;445:771–775. doi: 10.1038/nature05543. [DOI] [PubMed] [Google Scholar]
- 32.Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, Fitz LJ, Malenkovich N, Okazaki T, Byrne MC, Horton HF, Fouser L, Carter L, Ling V, Bowman MR, Carreno BM, Collins M, Wood CR, Honjo T. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med. 2000;192:1027–1034. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krasagakis K, Kruger-Krasagakes S, Fimmel S, Eberle J, Tholke D. m. von der Ohe, U. Mansmann, C.E. Orfanos, Desensitization of melanoma cells to autocrine TGF-beta isoforms, J Cell Physiol. 1999;178:179–187. doi: 10.1002/(SICI)1097-4652(199902)178:2<179::AID-JCP7>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]