Abstract
Evaluation of biological effects of adjuvants on immune cells has been assessed in a limited number of studies. Moreover, no data are available on samples derived from cancer patients who may have a severe immune impairment. The effects of a novel RNA-based adjuvant (RNAdjuvant® developed by CureVac) were assessed in an ex vivo setting on PBMCs obtained from 8 healthy volunteers and 17 HCC patients, using a multiparametric approach to analyze network dynamics of early immune responses. Evaluation of CD80, CD86 and HLA-DR expression, cytokine production as well as gene expression was performed. Moreover, the downstream effect on CD4+ T cell phenotyping was evaluated. Treatment with RNAdjuvant® showed comparable effects on PBMCs of both HCC and healthy subjects. In particular, CD80, CD86 and HLA-DR expression was found up-regulated in circulating dendritic cells, which promoted a CD4+ T cell differentiation toward an effector phenotype. A mixed Th1/Th2 cytokine pattern was induced, although a more predominant production of TNFα and IFNγ was observed in HCC patients versus healthy controls. The cytokine profile was further confirmed by gene transcriptional analysis, which showed up-regulation of several genes involved in innate and adaptive immune-related pathways. The present study is the first demonstration that HCC patients and healthy subjects are equally responsive to an adjuvant. This may suggest that the same vaccine formulation including the RNAdjuvant® might have similar potency in healthy subjects and cancer patients.
Electronic supplementary material
The online version of this article (doi:10.1007/s00262-016-1923-5) contains supplementary material, which is available to authorized users.
Keywords: RNAdjuvant, Hepatocellular carcinoma, Immune response, Cancer vaccine
Introduction
Since the discovery of human tumor antigens, the cancer immunotherapy field has rapidly progressed and several active (e.g., therapeutic cancer vaccine) as well as passive strategies (e.g., adoptive T cell transfer) are being investigated [1]. The anti-tumor efficacy of such strategies, however, is extremely variable and several lines of research are pursued in order to improve clinical outcome in cancer patients [2–4]. Several reasons may be taken into account for such low efficacy. Among others, the low immunogenicity of antigens delivered as peptides. Indeed, as for preventive anti-microbial vaccines, employment of the minimal antigenic determinant (e.g., epitope), devoid of all potentially risky adjuvanting “contaminants,” results in a dramatically lower immunogenicity [4, 5]. This has required and continuously requires the need for developing adjuvants to improve the immune response elicited by vaccines. However, to date only a few number of adjuvants have been approved for use in humans [6, 7].
The increasing detailed knowledge about the central role of the innate immunity in igniting and directing the adaptive immune response has been crucial to the adjuvant field. In particular, the discovery of pattern recognition receptors (PRRs) in cells of the innate immune system, including Toll-like receptors (TLRs), Nod-like Receptors (NLRs), Retinoic acid-Inducible Gene 1-Like Receptor (RIG-I) or Stimulator of interferon genes (STING), has promoted an explosion in the discovery of novel adjuvants (reviewed in [8]). Indeed, they act as sensors for bacterial and viral components (Pathogen-Associated Molecular Patterns; PAMPs) and induce the innate immune response by activation of a signaling cascade leading to up-regulation of various immune genes, including inflammatory cytokines, chemokines, and type I IFNs. This will eventually end up in triggering a robust adaptive immune response [9]. Therefore, PRRs represent the ideal target for promoting an effective immune response.
In particular, several TLR ligands have been extensively evaluated as vaccine adjuvants in preclinical as well as human clinical setting (reviewed in [10]). Nevertheless, only the TLR4-ligand monophosphoryl lipid A (MPL) is licenced for human use as adjuvant of the preventive vaccines for hepatitis B virus (HBV) and human papilloma virus (HPV) [11, 12].
Moreover, most if not all of the licenced adjuvants induce only weak T cell immune response and their application in cancer immunotherapy would be very limited. Therefore, there is a continuous quest for new adjuvants and in particular for molecules able to elicit a balanced humoral and cellular immune response with potential application in both preventive and therapeutic vaccines.
For such a reason, a novel RNA-based adjuvant (RNAdjuvant®) has been recently developed by CureVac AG (Tübingen, Germany). The RNA component consists of a single-stranded, noncoding, non-capped RNA sequence containing several polyU-repeats. The RNA molecule is complexed with a polymeric carrier formed by a disulfide-crosslinked cationic peptide in order to increase its stability against RNase degradation. It was recently shown that RNAdjuvant® exhibited a favorable preclinical safety profile and induced Th1/Th2 balanced, long-lasting immune responses to HPV-16 E7 protein-derived long peptides, resulting in a strong activity against syngeneic TC-1 tumor model [13]. In this study, an extensive multiparametric analysis was conducted to clarify the immunological and molecular mechanisms underlying the biological effect observed in the animal model. Antigen-specific CD8+ effector and memory T cells were induced only in the presence of the adjuvant. A gene expression analysis at the site of injection showed induction of Type I IFN and Type I IFN-stimulated genes (ISGs) as well as the up-regulation of inflammasome-associated genes. Moreover, distinct chemokines and immune activation markers were found up-regulated [13]. Overall, such data suggest that RNAdjuvant® is a safe and potent immunostimulator for cancer vaccines.
Vaccines have been shown to elicit similar antibody titers in healthy subjects as well as in cancer patients not undergoing chemotherapy although it is still uncertain whether such immune response is fully protective (reviewed in [14]).
Adjuvants, which are an essential component of vaccine formulations, are assumed to have comparable adjuvanting efficacy in healthy subjects and cancer patients. However, in order to prove that the same effective vaccine formulation in healthy subjects may be implemented in cancer patients as well, an experimental validation is needed. For such a reason, we took advantage of a multiparametric platform, developed and fully validated by our group, to assess ex vivo the innate and early adaptive response in human PBMCs [15–21].
The immune stimulating effects of the RNAdjuvant® were evaluated in PBMCs from healthy subjects and patients affected by hepatocellular carcinoma (HCC). Indeed, RNAdjuvant® is going to be employed in the therapeutic cancer vaccine formulation developed by the EU-funded HEPAVAC Consortium (www.hepavac.eu) specifically targeting liver cancer [22]. The overall results showed that HCC patients and healthy subjects are equally responsive to RNAdjuvant®.
Materials and methods
Enrolled subjects
Peripheral blood was obtained by venipuncture from 8 healthy volunteers and 17 HCV chronically infected HCC patients. All human specimens were obtained and processed at the National Cancer Institute in Naples under informed consent, as approved by the Institutional Review Board.
PBMC isolation
Fresh human PBMCs were isolated by Ficoll-Hypaque density gradient centrifugation and plated in six-well plates at a concentration of approximately 1 × 107 cells/well in a maximum volume of 3 ml/well. Isolated PBMCs were incubated for 24 h (short-term culture) or 7 days (medium-term culture) in RPMI 1640 medium.
Cell culture medium
PBMCs culture medium consisted of RPMI 1640 medium (Life Technologies, Carlsbad, CA) supplemented with 2 mM l-glutamine (Sigma), 10% fetal calf serum (Life Technologies) and 2% penicillin/streptomycin (5000 I.U./5 mg per ml, MP Biomedicals). Recombinant interleukin-2 (rIL-2; R&D Systems, Minneapolis, Minn.) was added at a concentration of 75 U/ml for medium-term culture (every 3 days).
Cell treatment
PBMCs were incubated for 24 h (short-term culture) with 20 μg/ml of RNAdjuvant® provided by CureVac AG (Tübingen, Germany). In parallel, cells were pulsed with 4 μg/ml of lipopolysaccharide (LPS), as positive control (from Salmonella enterica serotype Minnesota, purity >99.0%). Alternatively, PBMCs were incubated for 7 days (medium-term culture) with the same concentration of RNAdjuvant® or LPS, plus IL-2 added at day 0, 3 and 6. PBS was used as negative control. At the end of the incubation, PBMCs were harvested, washed with PBS 1X (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.2) without calcium and magnesium and analyzed by flow cytometry. All cell supernatants were collected for quantification of cytokine production by enzyme-linked immunosorbent assay (ELISA).
Flow cytometry
Short-term culture PBMCs were incubated for 30 min at 4 °C with human monoclonal antibodies specific for CD80, CD83, CD86, HLA-DR, CD123, CD11c and CD14 (BD Pharmingen, San Diego, CA), washed and then analyzed with a FACScalibur flow cytometer (BD Pharmingen). Data analysis was carried out with WinMDI2.8 Software. A Paired t test was performed, all p values were considered significant if less than 0.05.
T-maturation analysis
Medium-term culture PBMCs were analyzed for markers of CD4+ T cell phenotyping by flow cytometry. A T cell phenotyping was obtained after gating on the CD3+/CD4+ cells. Naïve CD4+ T cells were identified as RA+/62L+/CD27+; Effector cells as RA+/62L−/CD27−; Central Memory cells as RO+/62L+/CD27+; Effector Memory cells as RO+/62L−/CD27−.
Cytokine analysis
At the time the cells were harvested, the supernatants were also collected and stored frozen until analyzed. IL-6, TNF, IFNg and IL-10 production was assessed using “Human ELISA MAX™ Deluxe (Biolegend)” for quantitative detection of each human cytokine, according to the manufacturer’s instructions. Data acquisition was performed using the Sirio-S ELISA reader. A Paired t test was performed, and all p-values were considered significant if less than 0.05.
RNA extraction and PCR array
Total RNA was extracted using Qiagen RNEasy Mini Kit (Qiagen, Germany) according to the manufacturer’s protocol and checked for quantity and quality using a NanoDrop 2000c. 250 ng of RNA of each sample was converted into cDNA using the RT2 First Strand Kit (SABiosciences, Frederick, USA). cDNA was added to the RT2 SYBR Green qPCR Master Mix (SABiosciences, Frederick, USA) and then aliquoted on the “Human Innate e Adaptive immune responses RT2 Profiler PCR Array” (Quiagen) plat. All steps were done according to the manufacturer’s protocol for the CFX96 Real time System (Bio-rad laboratories, Inc.)
Statistical analysis
Statistical analysis was performed using Graphpad Prism 6 software (Graphpad Software, La Jolla, CA, USA), and the results were considered statistically significant at a level of p < 0.05. The t Test, nonparametric Wilcoxon matched-pairs signed-rank test, was used to analyze intergroup comparisons. PCR array data were analyzed by the RT2 profiler PCR array Data Analysis software which calculated two normalized average Ct values, a paired t test P value and a fold change. Genes were identified to be differentially expressed on a significance level of p < 0.05.
Results
Clinical parameters of subjects included in the analysis
Twenty-five subjects were enrolled in the present study. Seventeen subjects were HCV chronically infected HCC patients, of whom eight were males and nine were females. Eight healthy donors were enrolled as controls matched for age and life style. Clinical parameters of enrolled subjects are described in Table 1.
Table 1.
Clinical parameters of enrolled subjects
| Sex | HCV | HCC | |
|---|---|---|---|
| Healthy 1 | M | Neg | No |
| Healthy 2 | M | Neg | No |
| Healthy 3 | F | Neg | No |
| Healthy 4 | F | Neg | No |
| Healthy 5 | F | Neg | No |
| Healthy 6 | F | Neg | No |
| Healthy 7 | F | Neg | No |
| Healthy 8 | F | Neg | No |
| HCC 1 | M | Pos | Yes |
| HCC 2 | M | Pos | Yes |
| HCC 3 | M | Pos | Yes |
| HCC 4 | M | Pos | Yes |
| HCC 5 | M | Pos | Yes |
| HCC 6 | M | Pos | Yes |
| HCC 7 | M | Pos | Yes |
| HCC 8 | M | Pos | Yes |
| HCC 9 | F | Pos | Yes |
| HCC 10 | F | Pos | Yes |
| HCC 11 | F | Pos | Yes |
| HCC 12 | F | Pos | Yes |
| HCC 13 | F | Pos | Yes |
| HCC 14 | F | Pos | Yes |
| HCC 15 | F | Pos | Yes |
| HCC 16 | F | Pos | Yes |
| HCC 17 | F | Pos | Yes |
Maturation markers induced in circulating APCs by the RNAdjuvant®
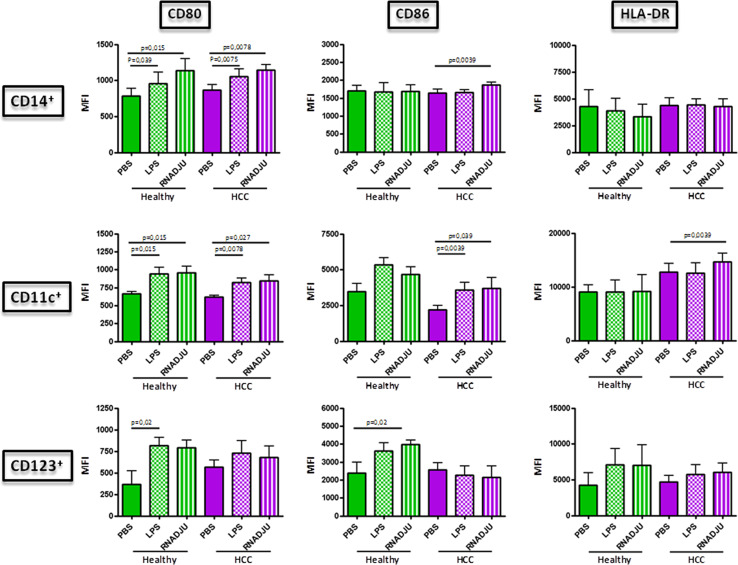
In order to evaluate the maturation markers induced by RNAdjuvant®, the expression of CD80, CD86 and HLA-DR molecules was examined in circulating antigen-presenting cells (APCs) by flow cytometry. In particular, HLA-DR+CD14+ monocytes, HLA-DR+CD14−CD11c+ myeloid dendritic cells (mDCs) and HLA-DR+CD14−CD123+ plasmacytoid dendritic cells (pDCs) were evaluated in parallel.
The basal expression of the evaluated markers was largely comparable between healthy and HCC subjects in all the three cell types (Suppl. Fig. 1). On the contrary, treatment with RNAadjuvant® in samples from the HCC subjects induced a statistically significant up-regulation of CD80 and CD86 in CD14+ cells and all three markers in CD11c+ cells. The up-regulation in the CD123+ cell subset showed a trend which did not reach the statistical significance. On the contrary, the RNAdjuvant® treatment induced an up-regulation of the three markers in all cell subset from healthy subjects. Such effect was comparable to the one induced by control LPS (Fig. 1).
Fig. 1.
Expression of surface circulating APC markers in PBMCs. Mean Fluorescence Index (MFI) for each of the indicated markers was evaluated on cells after 24 h treatment with RNAdjuvant and LPS by flow cytometry. Healthy group green bars; HCC group violet bars
T cell maturation induced by the RNAdjuvant
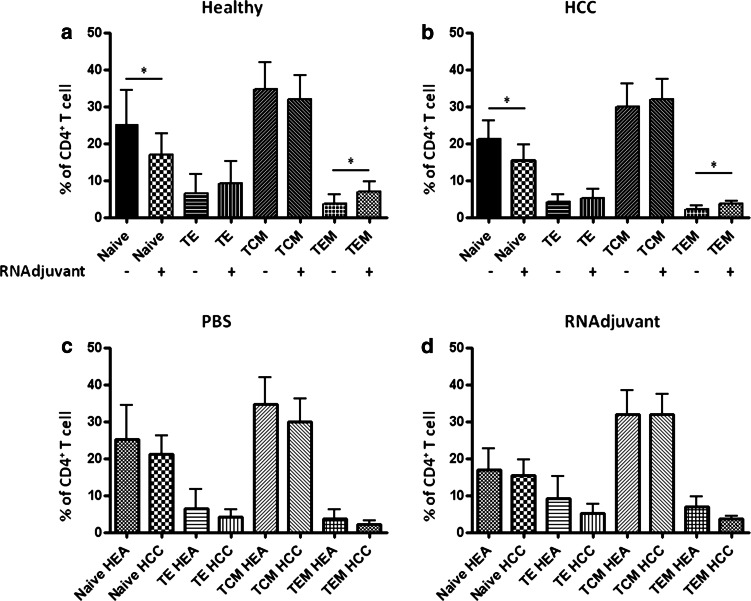
The effect on CD4+ T cell phenotyping induced by APCs treated with RNAdjuvant® was evaluated in PBMCs, after 6 days post-treatment. APCs treated with RNAdjuvant® induced a reduction of naïve CD4+ T cells and a parallel increase in both effector (TE) and effector memory (TEM) cells. Such effects were comparable in samples from both healthy subjects and HCC patients. In both groups, the effect reached the statistical significance only for the naïve and TEM cells. On the contrary, the central effector memory (TCM) cells showed a trend to increase only in HCC patients (Fig. 2a, b).
Fig. 2.
Effect of RNAdj on ex vivo differentiation of naïve CD4+ T cells from healthy subjects and HCC patients. Isolated PBMCs from healthy subjects (a) and HCC patients (b) were incubated with 20 μg/ml of RNAdjuvant. IL-2 was added every 3 days. After 6 days phenotype of CD4+ T cells was assessed by flow cytometry. Naïve CD4+ T cells were identified as RA+/62L+/CD27+; Effector as RA+/62L−/CD27−; Central Memory as RO+/62L+/CD27+; Effector Memory as RO+/62L-/CD27-. Side-by-side comparison between PBS and RNAdj in healthy subjects and HCC patients is shown in panels (c, d)
The inter-group comparative analysis, both at baseline and after RNAdjuvant® treatment, showed a comparable percentage of all the CD4+ T cell sub-populations in HCC patients versus healthy subjects. The lower levels observed in HCC patients, indeed, did not reach the statistical significance (Fig. 2c, d). The only exception was the TCM subpopulation, which was perfectly comparable after RNAdjuvant® treatment in both groups of samples.
Cytokine pattern induced in circulating APCs by the RNAdjuvant
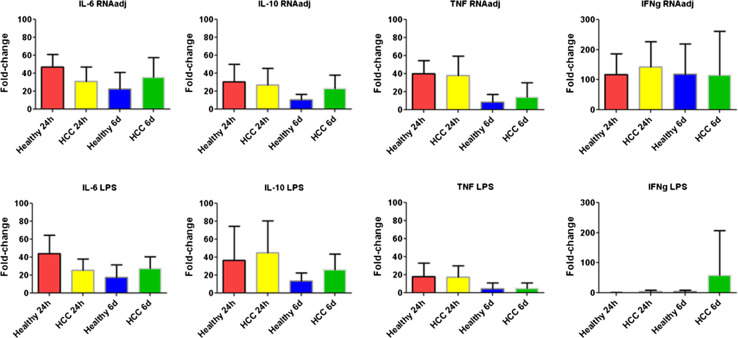
The level of Th1 (IFNγ and TNFα), Th2 (IL-10) and pro-inflammatory (IL-6) cytokines was assessed by ELISA in supernatants of PBMCs after treatment with RNAdjuvant® (Fig. 3). The evaluation was performed after incubation for 24 h and 6 days to assess the persistence of the cytokine release.
Fig. 3.
Analysis of cytokine production in supernatants of PBMCs from HCC and healthy subjects. Isolated PBMCs were incubated for 24 h or 6 days with 20 μg/ml of RNAdjuvant or with 4 μg/ml of LPS), as positive control. IL-6, TNF, IFNg and IL-10 production was assessed in the cell culture supernatant using “Human ELISA MAX™ Deluxe (Biolegend)”
The first observation was the comparable baseline production of all four cytokines by PBMCs from healthy subjects and HCC patients (Suppl. Fig. 2).
Treatment with RNAdjuvant® induced a balanced production of both Th1 and Th2 cytokines in both HCC and healthy subjects (p value <0.01). In particular, the highest fold change over the basal production was observed for the Th1 cytokine IFNγ (average of 117.8 in healthy and 142.6 in HCC) (Fig. 3). The increased production of the cytokines showed persistent fold changes for 6 days, especially in HCC samples. The only exception was the Th1 TNFα cytokine whose fold change dramatically dropped at 6 days in both groups of samples (Fig. 3). Interestingly, the levels of Th2 IL-10 and pro-inflammatory IL-6 cytokines were comparable to those induced by the LPS, used as positive control. On the contrary, RNAdjuvant® induced a statistically significant higher level of the Th1 cytokines TNFα (at 24 h) and IFNγ (at both 24 h and 6 days) compared to LPS.
Genes transcriptionally activated by RNadjuvant in PBMCs
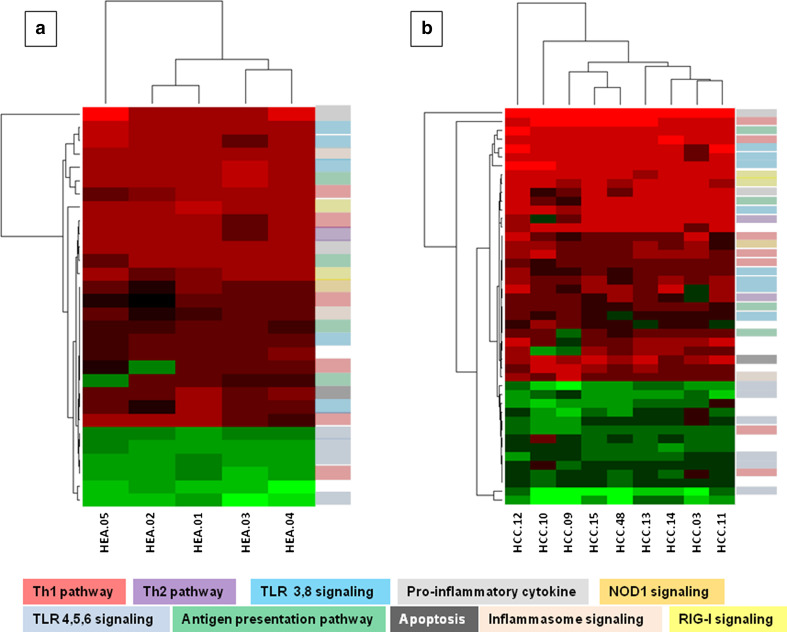
To further characterize the immunological effects induced by the RNAdjuvant® in stimulated PBMCs, the gene expression profile was analyzed. Normalized array expression data were evaluated to assess the effect of the RNAdjuvant® treatment over the control (e.g., PBS). Samples from HCC subjects and healthy controls were collected after 24 h of treatment.
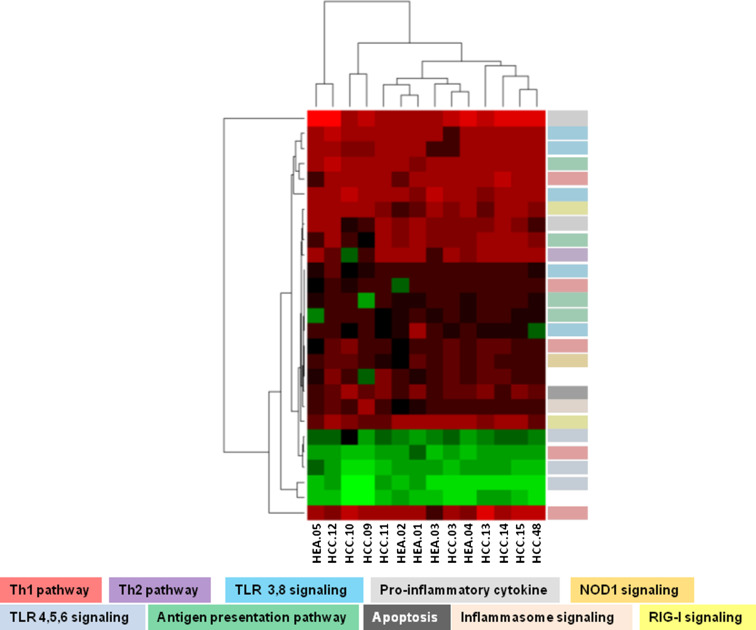
The analysis identified 30 differentially expressed genes in the healthy subjects (24 up-regulated and 6 down-regulated) and 45 differentially expressed genes in the HCC patients (31 up-regulated and 14 down-regulated) (Fig. 4a, b; Suppl. Table 1). The same pathways/signaling were modulated in the two groups (Fig. 4a, b). The comparative analysis showed that 27 differentially expressed genes were common to both groups, 22 up-regulated and 5 down-regulated (Fig. 5). In particular, the heatmap and the clustering generated with the expression values of shared genes showed that HCC subjects and healthy controls do not segregate into two different groups (Fig. 5). This suggests that they respond similarly to the adjuvant.
Fig. 4.
Gene expression profile after RNAdjuvant treatment. a Heatmap and the clustering generated with the expression values of 30 differentially expressed genes in Healthy group; b heatmap and the clustering generated with the expression values of 45 differentially expressed genes in HCC group
Fig. 5.
Gene expression profile after RNAdjuvant treatment. Heatmap and the clustering generated with the expression values of 27 shared genes in healthy and HCC groups
Overall, shared genes involved in several immune pathways were found up-regulated (Suppl. Table 1). In particular, the IL-6 and IFNγ genes were the most strongly up-regulated ones. The intra-cellular TLRs 3, 7 and 8 and their downstream mediators (e.g., MyD88, TICAM1, NFkB, Type I IFNs), the RIG-I (e.g., DDX58 and IRF7) as well as the inflammasome (e.g., Caspase 1) were found up-regulated. These findings suggest that the RNAdjuvant® treatment is able to activate the innate immunity via more than one Pathogen-Related Receptor (PRR) signaling. Moreover, in support of the cytokine ELISA data, both Th1 (e.g., IFNγ) and Th2 (e.g., IL-10) cytokines were activated along with pro-inflammatory cytokines (e.g., IL-6).
Expression of co-stimulatory CD40 and CD80 genes was up-regulated, along with HLA-A and E molecules, indicating a potentiated antigen-presentation activity to effector T cells.
Concerning the down-regulated genes, it was very interesting to find a set of genes correlated with the TLR4 and 2 signaling (e.g., CD14, LY96 and MAPK1). Of note, the down-regulation of TLR 5 and 6 was specifically identified only in samples from HCC patients (Fig. 4b; Suppl. Table 1).
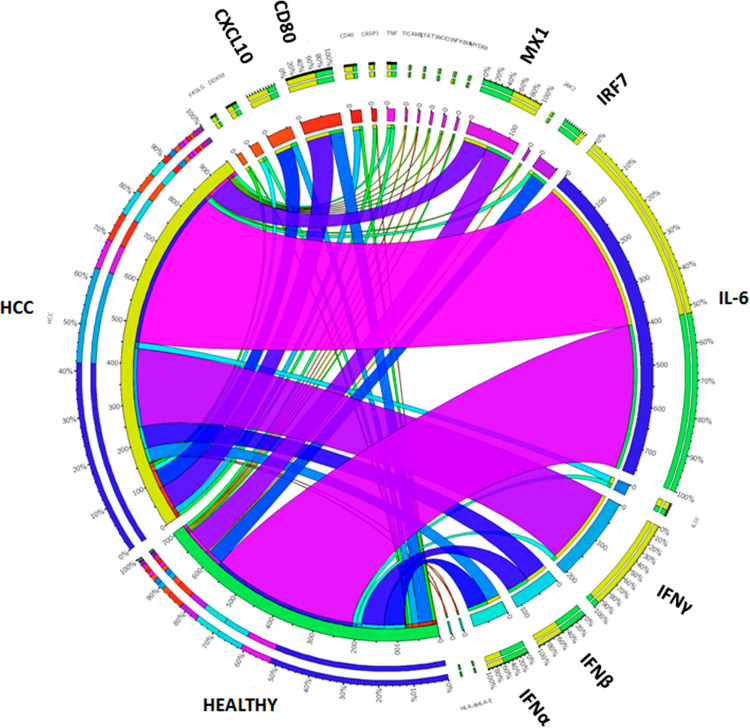
The expression levels of the described genes in the two groups of samples are further described in a Circos image (Fig. 6). The IL-6 and IFNγ are confirmed to be the most expressed genes upon RNAdjuvant® treatment of PBMCs. However, their differential expression pattern is striking. Indeed, IL-6 is equally expressed in PBMCs from healthy and HCC subjects, while IFNγ is mostly expressed in PBMCs from HCC subjects. Considering the other highly expressed genes, type I IFNs and MX1 are equally expressed in both groups; CXCL10 and CD80 are mostly expressed in HCC samples; IRF7 is mostly expressed in healthy samples.
Fig. 6.
Circos image. Circos plot showing log fold change of shared up-regulated genes in HCC and Healthy group. Log fold changes for the up-regulated genes are indicated by the width of ribbons and the presence of each gene is indicated by connection between genes and groups
Discussion
Several tumor-associated antigens (TAAs) have been characterized to date for development of anti-cancer vaccines. However, due to their low intrinsic immunogenicity peptides are not efficient in eliciting immune responses and require formulations with adjuvants to both potentiate immunogenicity and induce a Th1/Th2 balanced immunity.
Most of the adjuvants are developed for preventive vaccines, mainly aiming at eliciting a Th2-type response. On the contrary, the purpose of therapeutic cancer vaccines is to elicit a Th1-type response and very few adjuvants are available for this purpose. Given the availability of very limited information, in the present study we aimed at verifying the responsiveness to an adjuvant in cancer patients versus healthy subjects. This was performed employing a multiparametric analysis in an ex vivo setting, exposing PBMCs from liver cancer patients and healthy controls to a novel RNA-based adjuvant (RNAdjuvant®, CureVac AG).
Results in samples from the HCC subjects showed that the CD80 and CD86 were statistically up-regulated in CD14+ monocyte as well as in CD11c+ myeloid DC (mDC), together with HLA-DR. The up-regulation in the CD123+ cell subset showed a trend which did not reach the statistical significance. These results are largely comparable between HCC and control groups.
Such observations confirm that RNAdjuvant® is a strong immune potentiator activating maturation of APCs and expression of co-stimulatory molecules to promote an efficient adaptive response. Moreover, they suggest that the HCC status does not impair the responsiveness of circulating APC populations to the immunogenic stimulus of RNAdjuvant®.
Cells from HCC patients and healthy subjects showed a comparable basal production of Th1/Th2 cytokines. The RNAdjuvant® treatment induced a balanced production of both Th1 and Th2 cytokines in both HCC and healthy subjects which persisted for 6 days. This would strongly suggest an in vivo long-lasting driving milieu for development of a balanced immune response [23]. In particular, when compared to LPS, RNAdjuvant® induced a much higher level of the Th1 cytokines, confirming that it has the required properties to be an eligible adjuvant for a therapeutic cancer vaccine formulation. The induction of the anti-inflammatory IL-10, as previously reported by our group, suggests the activation of a positive homeostatic mechanism to neutralize an excessive pro-inflammatory status which would be detrimental to an effective immune response [18].
The ability of APCs treated with RNAdjuvant® to induce activation of naïve CD4+ T cells into effector cells was assessed ex vivo. The results showed that, in samples from both healthy subjects and HCC patients, reduction of naïve CD4+ T cells was associated with a parallel increase in both effector (TE) and effector memory (TEM) cells. The central effector memory (TCM) cells showed a trend to increase only in HCC patients. All the CD4+ T cell sub-populations showed a comparable percentage in HCC patients versus healthy subjects, both at baseline and after RNAdjuvant® treatment. Such observation strongly suggests that RNAdjuvant® is able to induce a shift in the phenotype of CD4+ T cells from naïve to effector, which are the main actors in helping and boosting the T and B cell immune response. In particular, the observed increase in both effector memory (TEM) and central memory (TCM) is of high relevance, suggesting that vaccines formulated in RNAdjuvant® is able to generate subsets capable of circulating in lymphoid (TCM) as well as non-limphoid compartments (TEM) [24, 25]. Upon contact with the appropriate antigen, effector memory cells can instantly execute effector functions, whereas central or lymphoid memory cells can rapidly proliferate, expanding and acquiring effector functions. This is of high relevance for vaccine development.
The gene expression analysis performed on PBMCs treated with RNAdjuvant® showed in both healthy subjects and HCC patients the modulation of genes involved in several innate and adaptive immune-related pathways. Overall, the results showed a similar gene expression profile in the two groups, with overlapping pathways. The RNAdjuvant® was able to induce the antigen-presentation activity of APCs to effector T cells and to activate the innate immunity probably via more than one Pathogen-Related Receptor (PRR) signaling (e.g., TLRs, RIG-I and inflammasome). In particular the signaling of intra-cellular TLRs (e.g., TLR 3, 7, 8) was activated, according to the RNA nature of the RNAdjuvant® [13, 26]. Interestingly, the signaling of the cell-surface TLRs (e.g., TLR 2, 4, 5, 6) was down-regulated. This observation suggests that the RNAdjuvant® might turn on a signaling and, at the same time, shut down a possible “interfering” signaling. Further experimental evidences on a larger sample size representing different cancers are required to validate such observation.
The balanced activation of Th1/Th2 pathways assessed by ELISA was confirmed also by the gene expression analysis although, on average, the Th1 pathway was much more activated in cells from HCC patients than healthy subjects. Moreover, activation of type I IFNs is of high relevance due to their key role in activating effector and memory Th1 cells and counteracting Th2 and Th17 differentiation [27].
Collectively, the data show that RNAdjuvant® is a very promising candidate as a potent immunostimulator in therapeutic cancer vaccination formulations. Nonetheless, given the induction of a balanced Th1/Th2 response, its inclusion in preventive vaccine formulations is foreseeable, as currently evaluated in a safety Phase I clinical trial conducted in healthy subjects in combination with rabies virus vaccine (ClinicalTrials.gov Identifier: NCT02238756).
Moreover, our results showed that the effects of RNAdjuvant® are comparable in HCC and healthy subjects, although specific minor differences were observed. In this respect, the present study is the first demonstration that cancer patients and healthy subjects are equally responsive to an adjuvant, suggesting that the same vaccine formulation may have similar potency in healthy subjects and cancer patients.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The study was funded by the European Union´s 7th Framework Programme for Research, Project “Cancer Vaccine development for Hepatocellular Carcinoma—HEPAVAC” (Grant Nr. 602893) and the Italian Ministry of Health through Institutional “Ricerca Corrente.” Luisa Circelli, Maria Tagliamonte and Annacarmen Petrizzo are HEPAVAC fellows. We are indebted to Dr. Maria Napolitano for the technical support provided at flow cytometer analysis in the first set of experiments.
Authors’ contributions
LC, MT and AP carried out all experimental procedures. RH, MLT and FMB participated in the design of the study and analysis of data. LB conceived the study, participated in its design and coordination. LC and LB drafted the manuscript. All authors read and approved the final manuscript.
Abbreviations
- APC
Antigen-presenting cell
- HBV
Hepatitis B virus
- HCC
Hepatocellular carcinoma
- HCV
Hepatitis C virus
- HPV
Human papilloma virus
- IFN-γ
Interferon gamma
- IL-6
Interleukin 6
- IL-10
Interleukin 10
- ISGs
IFN-stimulated genes
- LPS
Lipopolysaccharide
- mDC
Myeloid dendritic cell
- MPL
Monophosphoryl lipid A
- NLRs
Nod-like Receptors
- PAMPs
Pathogen-associated molecular patterns
- PBMCs
Peripheral blood mononuclear cells
- PBS
Phosphate-buffered saline
- pDC
Plasmcytoid dendritic cell
- PRRs
Pattern recognition receptors
- RIG-I
Retinoic acid-inducible gene 1-like receptor
- STING
Stimulator of interferon genes
- TAA
Tumor-associated antigen
- TE
Effector T cell
- TCM
Central memory T cell
- TEM
Effector memory T cell
- Th1/2
T helper 1/2
- TLRs
Toll-like receptors
- TNFα
Tumor necrosis factor alpha
Compliance with ethical standards
Conflict of interest
Regina Heidenreich is employee of CureVac AG. All other authors declare that they have no competing interests.
References
- 1.Topalian SL, Weiner GJ, Pardoll DM. Cancer immunotherapy comes of age. J Clin Oncol. 2011;29:4828–4836. doi: 10.1200/JCO.2011.38.0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bloy N, Pol J, Aranda F, Eggermont A, Cremer I, Fridman WH, Fucikova J, Galon J, Tartour E, Spisek R, Dhodapkar MV, Zitvogel L, Kroemer G, Galluzzi L. Trial watch: dendritic cell-based anticancer therapy. Oncoimmunology. 2014;3:e963424. doi: 10.4161/21624011.2014.963424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aranda F, Buque A, Bloy N, Castoldi F, Eggermont A, Cremer I, Fridman WH, Fucikova J, Galon J, Spisek R, Tartour E, Zitvogel L, Kroemer G, Galluzzi L. Trial watch: adoptive cell transfer for oncological indications. Oncoimmunology. 2015;4:e1046673. doi: 10.1080/2162402X.2015.1046673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pol J, Bloy N, Buque A, Eggermont A, Cremer I, Sautes-Fridman C, Galon J, Tartour E, Zitvogel L, Kroemer G, Galluzzi L. Trial watch: peptide-based anticancer vaccines. Oncoimmunology. 2015;4:e974411. doi: 10.4161/2162402X.2014.974411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delany I, Rappuoli R, De GE. Vaccines for the 21st century. EMBO Mol Med. 2014;6:708–720. doi: 10.1002/emmm.201403876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pasquale AD, Preiss S, Silva FT, Garcon N. Vaccine adjuvants: from 1920 to 2015 and Beyond. Vaccines (Basel) 2015;3:320–343. doi: 10.3390/vaccines3020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moyer TJ, Zmolek AC, Irvine DJ. Beyond antigens and adjuvants: formulating future vaccines. J Clin Invest. 2016;126:799–808. doi: 10.1172/JCI81083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olive C. Pattern recognition receptors: sentinels in innate immunity and targets of new vaccine adjuvants. Expert Rev Vaccines. 2012;11:237–256. doi: 10.1586/erv.11.189. [DOI] [PubMed] [Google Scholar]
- 9.Gurtler C, Bowie AG. Innate immune detection of microbial nucleic acids. Trends Microbiol. 2013;21:413–420. doi: 10.1016/j.tim.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toussi DN, Massari P. Immune adjuvant effect of molecularly-defined toll-like receptor ligands. Vaccines (Basel) 2014;2:323–353. doi: 10.3390/vaccines2020323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levie K, Gjorup I, Skinhoj P, Stoffel M. A 2-dose regimen of a recombinant hepatitis B vaccine with the immune stimulant AS04 compared with the standard 3-dose regimen of Engerix-B in healthy young adults. Scand J Infect Dis. 2002;34:610–614. doi: 10.1080/00365540110080881. [DOI] [PubMed] [Google Scholar]
- 12.Angelo MG, Zima J, Da Tavares SF, Baril L, Arellano F. Post-licensure safety surveillance for human papillomavirus-16/18-AS04-adjuvanted vaccine: more than 4 years of experience. Pharmacoepidemiol Drug Saf. 2014;23:456–465. doi: 10.1002/pds.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heidenreich R, Jasny E, Kowalczyk A, Lutz J, Probst J, Baumhof P, Scheel B, Voss S, Kallen KJ, Fotin-Mleczek M. A novel RNA-based adjuvant combines strong immunostimulatory capacities with a favorable safety profile. Int J Cancer. 2015;137:372–384. doi: 10.1002/ijc.29402. [DOI] [PubMed] [Google Scholar]
- 14.Shehata MA, Karim NA. Influenza vaccination in cancer patients undergoing systemic therapy. Clin Med Insights Oncol. 2014;8:57–64. doi: 10.4137/CMO.S13774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buonaguro L, Tornesello ML, Tagliamonte M, Gallo RC, Wang LX, Kamin-Lewis R, Abdelwahab S, Lewis GK, Buonaguro FM. Baculovirus-derived human immunodeficiency virus type 1 virus-like particles activate dendritic cells and induce ex vivo T-cell responses. J Virol. 2006;80:9134–9143. doi: 10.1128/JVI.00050-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buonaguro L, Tornesello ML, Gallo RC, Marincola FM, Lewis GK, Buonaguro FM. Th2 polarization in peripheral blood mononuclear cells from human immunodeficiency virus (HIV)-infected subjects, as activated by HIV virus-like particles. J Virol. 2009;83:304–313. doi: 10.1128/JVI.01606-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buonaguro L, Tornesello ML, Jewis GK, Buonaguro FM. Short communication: limited induction of IL-10 in PBMCs from HIV-infected subjects treated with HIV-VLPs. AIDS Res Hum Retroviruses. 2009;25:819–822. doi: 10.1089/aid.2008.0304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buonaguro L, Petrizzo A, Tornesello M, Napolitano M, Martorelli D, Castello G, Beneduce G, De RA, Perrella O, Romagnoli L, Sousa V, De RV, Dolcetti V, Buonaguro FM. Immune signatures in human PBMCs of idiotypic vaccine for HCV-related lymphoproliferative disorders. J Transl Med. 2010;8:18. doi: 10.1186/1479-5876-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Circelli L, Petrizzo A, Tagliamonte M, Tornesello ML, Buonaguro FM, Buonaguro L. Systems biology approach for cancer vaccine development and evaluation. Vaccines (Basel) 2015;3:544–555. doi: 10.3390/vaccines3030544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petrizzo A, Tornesello ML, Napolitano M, D’Alessio G, Salomone MA, Dolcetti R, De RV, Wang E, Marincola FM, Buonaguro FM, Buonaguro L. Multiparametric analyses of human PBMCs loaded ex vivo with a candidate idiotype vaccine for HCV-related lymphoproliferative disorders. PLoS ONE. 2012;7:e44870. doi: 10.1371/journal.pone.0044870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petrizzo A, Tagliamonte M, Tornesello ML, Buonaguro FM, Buonaguro L. Prediction of individual immune responsiveness to a candidate vaccine by a systems vaccinology approach. J Transl Med. 2014;12:11. doi: 10.1186/1479-5876-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buonaguro L, HEPAVAC Consortium Developments in cancer vaccines for hepatocellular carcinoma. Cancer Immunol Immunother. 2016;65:93–99. doi: 10.1007/s00262-015-1728-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belardelli F, Ferrantini M. Cytokines as a link between innate and adaptive antitumor immunity. Trends Immunol. 2002;23:201–208. doi: 10.1016/S1471-4906(02)02195-6. [DOI] [PubMed] [Google Scholar]
- 24.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 25.Farber DL, Yudanin NA, Restifo NP. Human memory T cells: generation, compartmentalization and homeostasis. Nat Rev Immunol. 2014;14:24–35. doi: 10.1038/nri3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 27.Huber JP, Farrar JD. Regulation of effector and memory T-cell functions by type I interferon. Immunology. 2011;132:466–474. doi: 10.1111/j.1365-2567.2011.03412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.