Abstract
Central nervous system infiltration by circulating leukemic cells and enhanced in vitro transendothelial migration of promyelocytic leukemia HL-60-derived macrophages through a blood–brain barrier model was recently demonstrated. The intrinsic molecular and signaling mechanisms involved are, however, poorly documented. Drug targeting of such translocation event performed by circulating microbes and immune cells may prevent secondary cerebral infections and development of brain pathologies. In this study, we specifically investigated the in vitro targeting efficacy of the chemopreventive and dietary-derived epigallocatechin-3-gallate (EGCG) molecule on the NF-κB-mediated transcriptional regulation of a panel of 89 biomarkers associated with promyelocytic HL-60 differentiation into macrophages. NF-κB-mediated signaling during HL-60 macrophage differentiation was reversed by EGCG, in part through reduced IκB phosphorylation and led to the inhibition of moderately to highly expressed NF-κB gene targets among which the matrix metalloproteinase (MMP)-9 and the cyclooxygenase (COX)-2. In contrast, EGCG exhibited low efficacy in reversing NF-κB-regulated genes and showed selective antagonism toward COX-2 expression while that of MMP-9 remained high in terminally differentiated macrophages. Decreased expression of the 67-kDa non-integrin Laminin Receptor in terminally differentiated macrophages may explain such differential EGCG efficacy. Our results suggest that terminally differentiated macrophage transendothelial migration associated with neuroinflammation may not be pharmacologically affected by such a specific class of flavonoid. The differentiation status of a given in vitro cell model must therefore be carefully considered for optimized assessment of therapeutic drugs.
Keywords: EGCG, Leukemia, NF-κB, Macrophage differentiation, Blood–brain barrier
Introduction
The identification of transcription factors, such as NF-κB, STAT3, HIF-1α, and their gene products, such as cytokines, chemokines, and chemokine receptors, have laid molecular foundation for the decisive role of inflammation in carcinogenesis [1]. More specifically, NF-κB-mediated signaling can intervene in oncogenesis through its capacity to further regulate the expression of a large number of downstream gene products involved in apoptosis, cell proliferation and differentiation [2]. Impaired NF-κB activity has now been demonstrated not only in solid cancers but also in various types of hematologic malignancies including acute myeloid leukemia (AML), chronic myelogenous leukemia and in a subset of myelodysplastic syndromes [3, 4].
Given that NF-κB-mediated inflammation contributes to survival and proliferation of malignant cells, tumor angiogenesis, metastasis and reduced response to chemotherapy, new therapeutic strategies combining different NF-κB or proteasome inhibitors has, therefore, been proposed in adjuvant therapy for cancer [5]. Unfortunately, the chemotherapeutic treatment outcome of various hematologic disorders, including most adult acute promyelocitic leukemia (APL) and AML, remains unacceptable [6, 7]. Hence, novel avenues for the treatment of leukemia are required.
Considerable attention has recently been focused on identifying naturally occurring chemopreventive substances capable of inhibiting, retarding, or reversing multi-stage carcinogenesis [8]. In fact, it has been demonstrated that some edible phytochemicals alter gene expression, directly or indirectly, thereby regulating carcinogenic processes. (−)-Epigallocatechin-3-gallate (EGCG), a principal antioxidant derived from green tea, has been ascribed proteasome inhibition properties and is one of the most extensively investigated chemopreventive phytochemicals considered in clinical trials [9–11]. EGCG has been shown to block each stage of carcinogenesis by modulating the signal transduction pathways involved in cell proliferation, transformation, inflammation, apoptosis, metastasis, and invasion [12]. Moreover, its antiangiogenic properties make it a good candidate for targeting tumor-associated neovascularization [13]. Since the inclusion of antiangiogenic drugs into treatment protocols for leukemia and for hematologic malignancies is becoming an important task for future clinical studies [14, 15], we sought to investigate the in vitro anti-NF-κB molecular effects of EGCG on the monocytic/macrophagic differentiation processes using a myeloid leukemia cell model.
Among the several leukemic cell lines that have been established over the years, the human promyelocytic HL-60 leukemia cells have proven useful in understanding the process whereby immature cells differentiate into cells of distinct mature myelomonocytic lineages [16]. In particular, HL-60 cells can be induced to differentiate into mature functional monocytic/macrophagic-like cells by the tumor-promoting and protein kinase C activator phorbol 12-myristate 13-acetate (PMA, also known as TPA) [17, 18]. Macrophage differentiation has also been shown to involve secretion and activation of collagenase MMP-9 [19], a crucial matrix metalloproteinase involved in extracellular matrix (ECM) degradation during tumor metastasis and in inflammatory disorders [20].
Since an important aspect in inflammation and tumor progression is the involvement of the inflammatory response mediated by tumor-associated macrophages (TAM) [21], and since polyphenols have been suggested to regulate the anti-tumorigenic properties of TAM [22], we compared the transcriptional chemopreventive efficacy of EGCG on NF-κB targets involved in PMA-mediated signaling and in late stage terminally differentiated HL-60 macrophages.
Materials and methods
Materials
Sodium dodecylsulfate (SDS) and bovine serum albumin (BSA) were purchased from Sigma (Oakville, ON). Cell culture media was obtained from Life Technologies (Burlington, ON). Electrophoresis reagents were purchased from Bio-Rad (Mississauga, ON). The enhanced chemiluminescence (ECL) reagents were from Perkin Elmer (Waltham, MA). Micro bicinchoninic acid protein assay reagents were from Pierce (Rockford, IL). The polyclonal antibodies against IκB and phospho-IκB were purchased from Cell Signaling (Danvers, MA). The polyclonal antibody against COX-2 was from Cayman Chemical (Ann Arbor, MI). The polyclonal antibody against the 67-kDa Laminin Receptor was from Santa Cruz Biotechnology (Santa Cruz, CA). The monoclonal antibody against GAPDH was from Advanced Immunochemical Inc. (Long Beach, CA). Horseradish peroxidase-conjugated donkey anti-rabbit and anti-mouse IgG secondary antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA). EGCG and other reagents were from Sigma-Aldrich Canada.
Cell culture
The HL-60 promyelocytic cell line was purchased from American Type Culture Collection (Manassas, VA) and maintained in Iscove`s modified Dulbecco’s medium (Gibco Invitrogen Cell Culture Systems, Burlington, ON) containing 20 % (v/v) fetal bovine serum (FBS) (HyClone Laboratories, Logan, UT), 2 mM glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin and were cultured at 37 °C under a humidified atmosphere containing 5 % CO2. Slides of PMA-treated HL-60 cells were mounted for light microscopy and air-dried, stained with Diff-Quick (Baxter Healthcare Corp., Miami, FL) and examined for morulae. Given that numerous protocols can be found in the literature to differentiate resting HL-60 cells into “macrophage-like cells” with PMA (between 2 and 8 days with various PMA concentrations, alone or in combination with other molecules), we wish to emphasis that we termed our cell models as follows throughout the text: The “HL-60 macrophage differentiation” condition represents the adherent subpopulation of HL-60 cells immediately harvested upon PMA treatment. We termed “terminally differentiated macrophages” those same adherent cells, which were subsequently maintained in culture for 24–48 h more.
Gelatin zymography
Gelatin zymography was used to assess the extent of proMMP-9 gelatinolytic activity as previously described for proMMP-2 [23]. Briefly, an aliquot (20 μl) of the culture medium was subjected to SDS-PAGE in a gel containing 0.1 mg/ml gelatin, a substrate that is efficiently hydrolyzed by both proMMP-2 and proMMP-9. The gels were then incubated in 2.5 % Triton X-100 and rinsed in nanopure distilled H2O. Gels were further incubated at 37 °C for 20 h in 20 mM NaCl, 5 mM CaCl2, 0.02 % Brij-35, 50 mM Tris–HCl buffer, pH 7.6, then stained with 0.1 % Coomassie Brilliant blue R-250 and destained in 10 % acetic acid, 30 % methanol in H2O. Gelatinolytic activity was detected as unstained bands on a blue background.
Immunoblotting procedures
Proteins from control and treated cells were separated by SDS-PAGE. After electrophoresis, proteins were electrotransferred to polyvinylidene difluoride membranes which were then blocked for 1 h at room temperature with 5 % non-fat dry milk in Tris-buffered saline (150 mM NaCl, 20 mM Tris–HCl, pH 7.5) containing 0.3 % Tween-20 (TBST). Membranes were further washed in TBST and incubated with the primary antibodies (1/1,000 dilution) in TBST containing 3 % bovine serum albumin and 0.1 % sodium azide, followed by a 1 h incubation with horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG (1/2,500 dilution) in TBST containing 5 % non-fat dry milk. Immunoreactive material was visualized by enhanced chemiluminescence (Amersham Biosciences, Baie d’Urfée, QC).
Total RNA isolation, cDNA synthesis, and real-time quantitative RT-PCR
Total RNA was extracted from cell suspensions or monolayers using TriZol reagent (Life Technologies, Gaithersburg, MD). For cDNA synthesis, 2 μg of total RNA was reverse-transcribed using a high-capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA). cDNA was stored at −80 °C prior to PCR. Gene expression was quantified by real-time quantitative PCR using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA). DNA amplification was carried out using an Icycler iQ5 (Bio-Rad, Hercules, CA) and product detection was performed by measuring binding of the fluorescent dye SYBR Green I to double-stranded DNA. The QuantiTect primer sets were provided by Qiagen (Valencia, CA): MMP-9 (QT00040040), COX-2 (QT00040586), β-Actin (QT01136772). GAPDH primer sets were synthesized by Biocorp (Dollard-des-Ormeaux, QC) with the following sequences: forward CCATCACCATCTTCCAGGAG and reverse CCTGCTTCACCACCTTCTTG. The relative quantities of target gene mRNA compared against two internal controls, GAPDH and β-Actin mRNA, were measured by following a ΔCT method employing an amplification plot (fluorescence signal vs. cycle number). The difference (ΔCT) between the mean values in the triplicate samples of target gene and those of GAPDH and β-actin mRNAs were calculated by iQ5 Optical System Software version 2.0 (Bio-Rad, Hercules, CA) and the relative quantified value (RQV) was expressed as 2−ΔCT.
Human NF-κB signaling targets PCR array
The Human NF-κB Signaling Targets RT² Profiler PCR Arrays (PAHS-225, SA Biosciences, Frederick, MD) were used according to the manufacturer’s protocol. The detailed list of these key genes responsive to NF-κB signal transduction can be found on the manufacturer’s website (http://www.sabiosciences.com/rt_pcr_product/HTML/PAHS-225A.html). Using real-time quantitative PCR, we reliably analyzed expression of a focused panel of genes related to NF-κB downstream gene targets. Relative gene expressions were calculated using the 2−ΔΔCT method, in which CT indicates the fractional cycle number where the fluorescent signal reaches detection threshold. The “delta–delta” method uses the normalized ΔCT value of each sample, calculated using a total of five endogenous control genes (B2 M, HPRT1, RPL13A, GAPDH, and ACTB). Fold change values are then presented as average fold change = 2(average ΔΔCT) for genes in adherent PMA-differentiated macrophages relative to control HL-60 cells in suspension. Detectable PCR products were obtained and defined as requiring <35 cycles. The resulting raw data were then analyzed using the PCR Array Data Analysis Template (http://www.sabiosciences.com/pcrarraydataanalysis.php). This integrated web-based software package automatically performs all ΔΔCT based fold change calculations from our uploaded raw threshold cycle data.
Statistical data analysis
Unless otherwise stated, data are representative of three or more independent experiments. Statistical significance was assessed using Student’s unpaired t test. Probability values of less than 0.05 were considered significant and an asterisk identifies such significance in the figures. Error bars in all figures represent standard error means (SEM) values.
Results
EGCG reverses PMA-mediated IκB degradation in adherent HL-60 cells
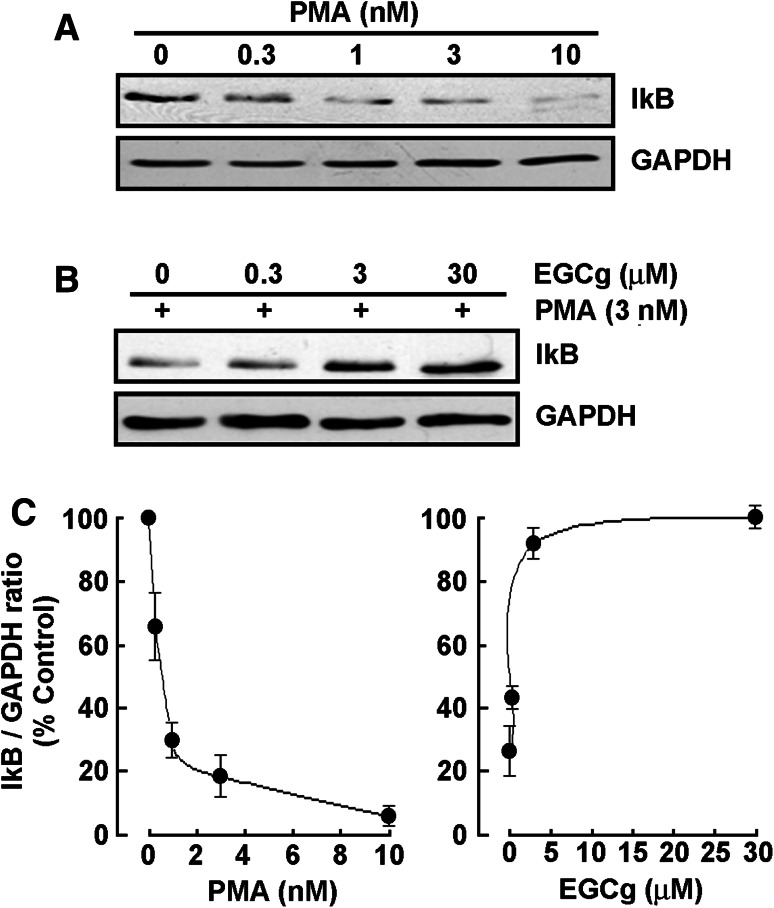
Among the agents well documented to induce differentiation of HL-60 cells, the tumor-promoting agent phorbol-12-myristate-13-acetate (PMA) triggers a terminal differentiated monocytic/macrophage phenotype [17]. We have previously validated that PMA treatment of serum-starved HL-60 cells induced an adhesive phenotype accompanied by macrophage differentiation in these cells which originally remain in suspension [24]. In order to first assess whether any NF-κB signaling was involved upon PMA stimulation, we performed immunoblotting on lysates isolated from adherent cells and found that IκB expression (Fig. 1a) decreased dose-dependently, as a consequence of its prior phosphorylation, and to be almost completely degraded at 10 nM PMA (Fig. 1c, left panel). When increasing EGCG concentrations were added simultaneously to PMA, IκB degradation by PMA was prevented (Fig. 1b) and almost completely reversed at 3 μM EGCG (Fig. 1c, right panel). This evidence suggests that NF-κB signaling is involved in PMA-induced differentiation process and that NF-κB gene targets are potentially modulated.
Fig. 1.
EGCG reverses PMA-mediated IκB degradation in adherent HL-60 cells. a Serum-starved HL-60 cells were treated with increasing PMA concentrations, or b with combined 3 nM PMA and increasing concentrations of EGCG for 18 h. Cells that remained in suspension were discarded and lysates from the adherent macrophage-differentiated cells isolated, electrophoresed via SDS-PAGE and immunodetection of IκB and GAPDH proteins performed as described in the “Materials and methods” section. c Quantification was performed by scanning densitometry of the autoradiogram. Data are representative of four independent experiments and were represented as the percent ( %) expression of untreated HL-60 cells in suspension (left panel) and as the percent ( %) of 3 nM PMA/30 μM EGCG-treated adherent cells (right panel)
EGCG reverses PMA-induced IκB phosphorylation in adherent HL-60 cells
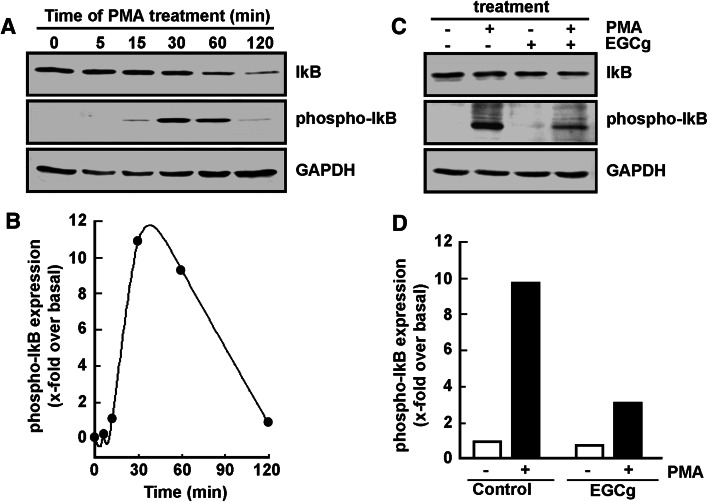
Iκb phosphorylation status was next investigated and the effect of EGCG assessed. PMA is shown to trigger maximal IκB phosphorylation upon 30 min treatment with PMA (Fig. 2a), which phosphorylation is subsequently followed by significant IκB degradation within the next hour (Fig. 2b). When such PMA treatment is performed in the presence of EGCG (Fig. 2c), IκB phosphorylation by PMA is significantly reduced (Fig. 2d, black bars). One may now conclude that EGCG’s significant reduction in PMA-induced IκB phosphorylation may prevent p65/p50 NF-κB translocation to the nucleus that would regulate transcription of NF-κB gene targets during HL60 differentiation.
Fig. 2.
EGCG reverses PMA-induced IκB phosphorylation in adherent HL-60 cells. a Serum-starved HL-60 cells were treated with 3 nM PMA for up to 120 min. Cells that remained in suspension were discarded and lysates from the adherent macrophage-differentiated cells isolated, electrophoresed via SDS-PAGE and immunodetection of IκB, phospho-IκB, and GAPDH proteins performed as described in the “Materials and methods” section. b Quantification was performed by scanning densitometry of the autoradiogram. Data are representative of two independent experiments and were represented as the x-fold expression over untreated HL-60 cells in suspension (time 0). The respective phospho-IkB/GAPDH ratios were blotted. c Similarly as in a, cells were treated for 30 min in the presence of 3 nM PMA, 30 μM EGCG, or a combination of both, and adherent cells harvested. d Quantification was performed as in b
Gene array analysis reveals NF-κB gene targets are associated with macrophage differentiation status and with differential inhibitory potential of EGCG
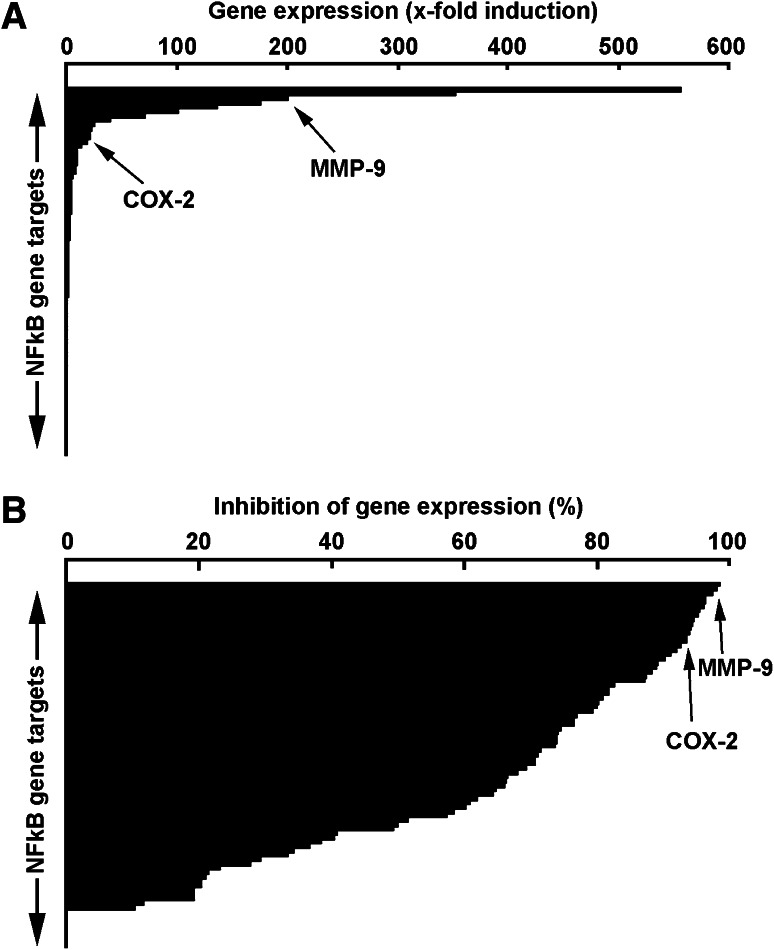
In light of the evidence that PMA-induced macrophage differentiation involves NF-κB signalling, we used a gene array approach to explore some NF-κB downstream gene targets. Transcriptional profiling was performed on genes involved in inflammation, apoptosis, development and cell differentiation, stress response, and immune response. Among the 89 NF-κB downstream gene targets assessed, 25 % were highly (>tenfold, Table 1) induced in differentiated HL-60 macrophage-like cells when compared to vehicle-treated HL-60 cells (Fig. 3a). Among these, COX-2 and MMP-9 gene expression is specifically shown (Fig. 3a, arrows) as these two biomarkers are thought to contribute to transendothelial migration by immune cells. Anti-PMA inhibitory potential of EGCG was subsequently assessed on the gene expression of the similar NF-κB gene targets using gene array strategy. We found that ECGG inhibited, by more than 70 % approximately half of the PMA-inducible NF-κB targets assessed (Fig. 3b) including COX-2 and MMP-9. Collectively, we identified several NF-κB downstream target genes that are significantly induced during PMA-induced HL-60 differentiation into macrophages and demonstrated that EGCG can very efficiently inhibit the expression of those highly induced genes as well as that of moderately induced genes (Table 2).
Table 1.
PMA triggers transcriptional increase in NF-κB gene targets in adherent HL-60 cells
| Gene name | Gene ID | Induction by PMA (x-fold) | Functional grouping |
|---|---|---|---|
| IL1RN | 3557 | 556 | Cytokines/chemokines |
| CCL5 | 6352 | 352 | Differentiation, cytokines/chemokines |
| IL1B | 3553 | 200 | Inflammation, cytokines/chemokines |
| ICAM1 | 3383 | 176 | Immune response |
| MMP9 | 4318 | 137 | Differentiation, apoptosis |
| IL1A | 3552 | 102 | Inflammation, cytokines/chemokines |
| IL2RA | 3559 | 71 | Inflammation |
| INS | 3630 | 40 | Inflammation, apoptosis |
| CD40 | 958 | 27 | Immune response |
| IL8 | 3576 | 24 | Cytokines/chemokines |
| F3 | 2152 | 23 | Anti-apoptosis |
| PDGFB | 5155 | 22 | Stress response |
| TNSF10 | 8743 | 20 | Cytokines/chemokines |
| CD83 | 9308 | 15 | Immune response |
| CCL2 | 6347 | 14 | Immune response, cytokines/chemokines |
| MITF | 4286 | 12 | Differentiation, apoptosis |
| BIRC3 | 330 | 11 | Apoptosis |
| COX2 | 5743 | 10 | Inflammation, apoptosis |
| TNFRSF1B | 7133 | 9.7 | Apoptosis |
Serum-starved HL-60 cells were treated with 3 nM PMA for 18 h. Total RNA was isolated from vehicle-treated cells that remained in suspension and from PMA-treated cells that adhered to the flasks (macrophage-differentiated cells). The identity of only those genes that were induced by tenfold in adherent versus suspension cells is shown and is extracted from Fig. 2a. Data are representative from two independent arrays. Italicized data were further confirmed at the protein and/or activity level
Fig. 3.
Gene array analysis reveals NF-κB gene targets associated with macrophage differentiation and high inhibitory potential of EGCG. a Serum-starved HL-60 cells were treated with 3 nM PMA for 18 h. Total RNA was isolated from vehicle-treated cells that remained in suspension and from PMA-treated cells that adhered to the flasks (macrophage-differentiated cells). a Histogram representation of the 89 NF-κB gene targets array expression levels. b Serum-starved HL-60 cells were treated with either 30 μM EGCG or a combination of 3 nM PMA and 30 μM EGCG for 18 h. Total RNA was isolated from EGCG-treated cells that remained in suspension and from PMA-treated cells that adhered to the flasks (macrophage-differentiated cells). Histogram representation of the gene expression array levels represents the percent of gene inhibition
Table 2.
EGCG reverses PMA-mediated IκB degradation in adherent HL-60 cells
| Gene name | Gene ID | EGCG inhibition (%) | Functional grouping |
|---|---|---|---|
| MMP9 | 4318 | 99 | Differentiation, apoptosis |
| CCR5 | 1234 | 98 | Inflammation |
| IL1RN | 3557 | 97 | Cytokines/chemokines |
| CD40 | 958 | 96 | Immune response |
| VCAM1 | 7412 | 96 | Differentiation |
| CXCL1 | 2919 | 96 | Cytokines/chemokines |
| CCL5 | 6352 | 95 | Cytokines/chemokines, development |
| FASLG | 356 | 95 | Apoptosis |
| IL1B | 3553 | 95 | Inflammation, cytokines/chemokines, |
| COX2 | 5743 | 95 | Inflammation, apoptosis |
| CXCL2 | 2920 | 94 | Cytokines/chemokines |
| IL1R2 | 7850 | 94 | Immune response |
| TNF | 7124 | 94 | Inflammation, development, stress response |
| CXCL10 | 3627 | 94 | Inflammation |
| MITF | 4286 | 93 | Differentiation, apoptosis |
| PDGFB | 5155 | 92 | Stress response |
| IL2RA | 3559 | 91 | Inflammation |
| CCL2 | 6347 | 90 | Immune response, cytokines/chemokines |
| CXCL9 | 4283 | 90 | Inflammation |
Serum-starved HL-60 cells were treated with either 30 μM EGCG or a combination of 3 nM PMA and 30 μM EGCG for 18 h. Total RNA was isolated from EGCG-treated cells that remained in suspension and from PMA-treated cells that adhered to the flasks (macrophage-differentiated cells). The identity of only those genes which expression was inhibited by 90 % and more is shown and is extracted from Fig. 2b. Data are representative from two independent arrays. Italized data were further confirmed at the protein and/or activity level
EGCG antagonizes COX-2 and MMP-9 biomarkers expression induced upon PMA-mediated HL-60 cell differentiation into macrophages
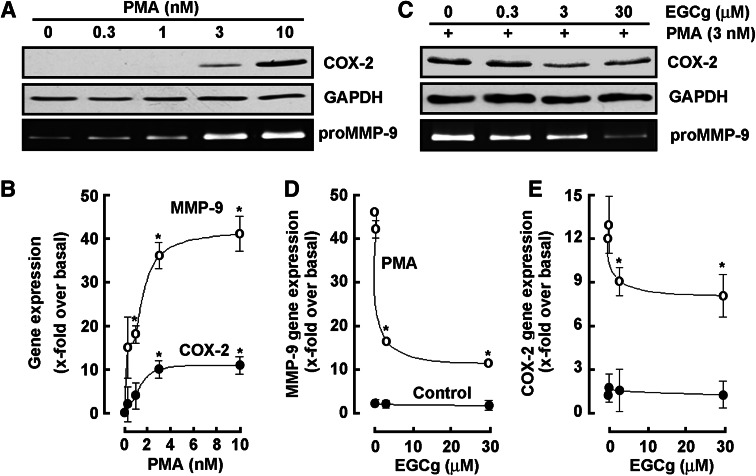
Given that MMP-9 is the major MMP that contributes to BBB disruption [25] and that COX-2 inhibition was demonstrated to limit BBB disruption [26], we next aimed at validating those transcriptional profiling data obtained through gene arrays (Tables 1,2) at the molecular level, and further wished to investigate the chemopreventive efficacy of EGCG on these two biomarkers’ expression regulation. Serum-starved HL-60 cells were therefore treated with increasing PMA concentrations, or with combined 3 nM PMA and increasing concentrations of EGCG for 18 h. Cell lysates from the adherent PMA-differentiated cells increasingly expressed COX-2 (Fig. 4a, upper panel), while MMP-9 secreted into the conditioned media also dose-dependently increased as assessed by gelatin zymography (Fig. 4a, lower panel). Total RNA was isolated from the above-mentioned conditions and qRT-PCR performed to confirm that both MMP-9 and COX-2 transcriptional regulation (Fig. 4b) paralleled that of their respective protein expression. When increasing concentrations of EGCG were added during PMA-induced HL-60 differentiation, we found in accordance with the gene array data that both MMP-9 and COX-2 PMA-mediated induction was reversed (Fig. 4c). Interestingly, EGCG had no effect on neither MMP-9 (Fig. 4d) nor COX-2 (Fig. 4e) basal levels. Given the paralleled effects on MMP-9 gene expression and MMP-9 gelatinolytic activity, one can safely rule out the possible contribution of TIMP-1 on MMP-9. Further, quantification of MMP-9 protein levels, through ELISA measures, may be required to strengthen and complement our enzymatic and gene expression assessment.
Fig. 4.
EGCG antagonizes COX-2 and MMP-9 biomarkers expression induced upon PMA-mediated HL-60 cell differentiation into macrophages. a, b Serum-starved HL-60 cells were treated with increasing PMA concentrations, or c, d, e with combined 3 nM PMA and increasing concentrations of EGCG for 18 h. Cells that remained in suspension were discarded and lysates from the adherent macrophage-differentiated cells isolated, electrophoresed via SDS-PAGE and immunodetection of COX-2 (a, c, upper panels) and GAPDH (a, c, middle panels) proteins performed as described in the “Materials and methods” section. Conditioned media were collected and gelatin zymography (a, c, lower panels) performed as described in the “Materials and methods” section. Total RNA was isolated from similar conditions than in a and b, and qRT-PCR performed as described in the “Materials and methods” section to assess MMP-9 and COX-2 gene expression (b, d, e)
Differential efficacy of EGCG to inhibit NF-κB gene targets between PMA-mediated HL-60 cell differentiation into macrophages and differentiated macrophages
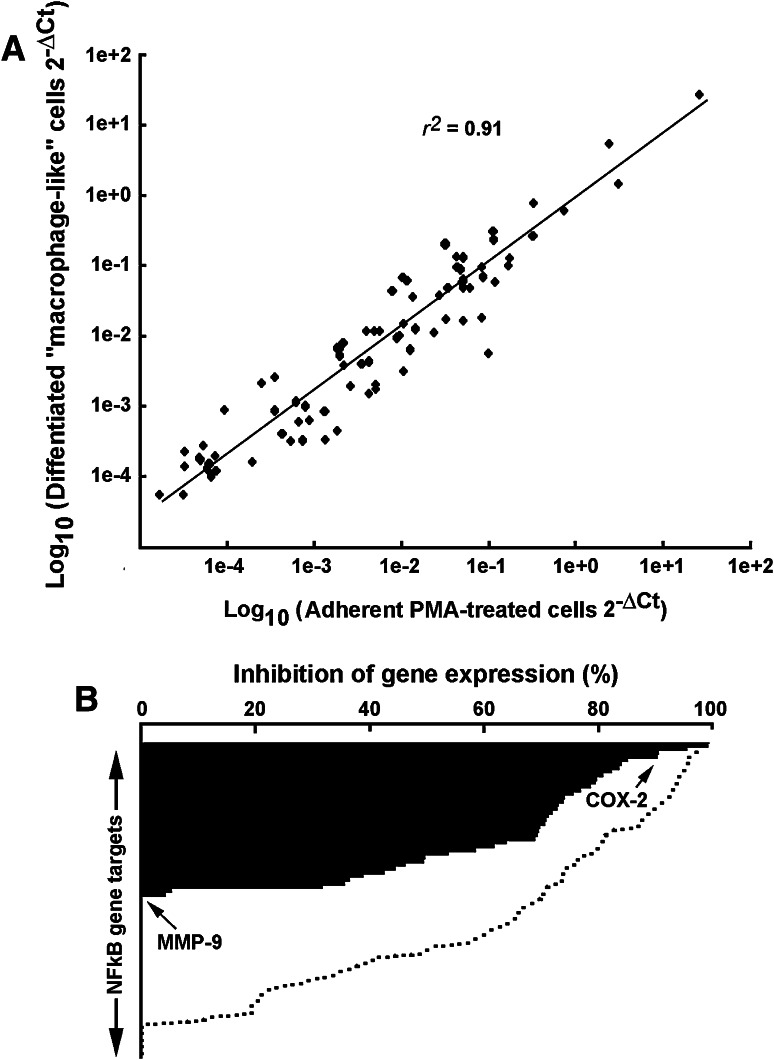
We next sought at investigating the effect of EGCG on terminally differentiated macrophage-like cells. We treated HL-60 cells with PMA for 18 h. Media containing PMA was removed and adherent terminally differentiated cells further cultured for 24 h in serum-free media containing or not 30 μM EGCG. Gene expression levels of key NF-κB targets were compared between adherent terminally differentiated cells and adherent PMA-induced differentiation cells using gene arrays. We found that similar transcriptional profiling characterized both cell populations with a correlation of more than 0.9 (Fig. 5a). When adherent terminally differentiated cells were then treated with EGCG, we found that only 23 % of the NF-κB gene targets were inhibited by more than 70 % (Fig. 5b, black bars) as compared to PMA-induced differentiation cells (Fig. 5b, dotted line). Interestingly, while COX-2 gene expression was still inhibitable by EGCG, that of MMP-9 was found insensitive to EGCG (Fig. 5b, arrows).
Fig. 5.
Differential efficacy of EGCG to inhibit NF-κB gene targets between PMA-mediated HL-60 cell differentiation into macrophages and differentiated macrophages. a Total RNA was isolated from adherent cells obtained upon 3 nM PMA treatment of serum-starved HL-60 cells, and gene expression profiles compared with total RNA isolated from PMA-differentiated cells cultured for 2 days. b EGCG inhibitory effect was evaluated from total RNA isolated from adherent cells obtained upon 3 nM PMA treatment of serum-starved HL-60 cells (dotted lines, data from Fig. 3b), and gene expression profiles compared with total RNA isolated from PMA-differentiated cells cultured for 2 days and then serum-starved in the presence of 30 μM EGCG
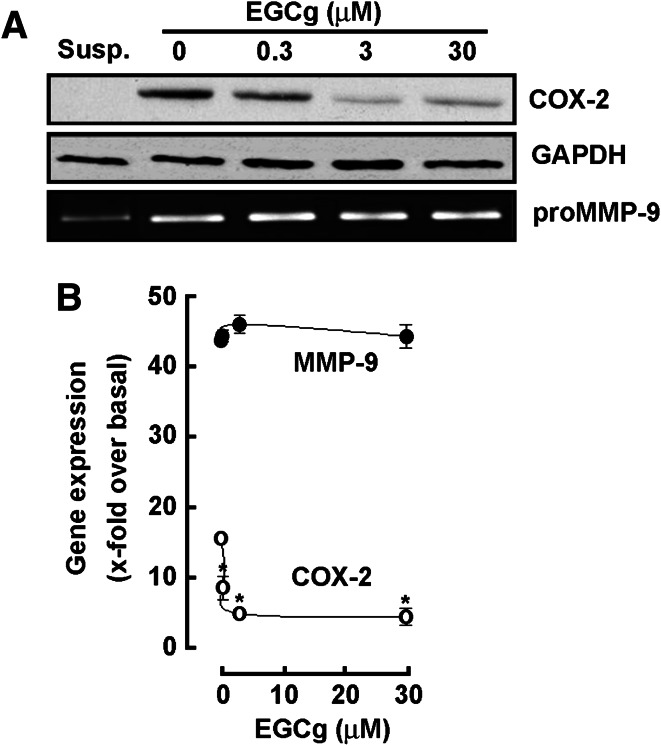
EGCG antagonizes COX-2 but not MMP-9 expression induced upon PMA-mediated HL-60 cell differentiation into macrophages
In order to further validate the gene array data obtained previously, immunoblotting and qPCR were performed to assess the impact of EGCG on COX-2 and MMP-9 protein (Fig. 6a) and gene (Fig. 6b) expression in terminally differentiated HL-60 macrophages. While EGCG efficiently inhibited COX-2 protein (Fig. 6a, upper panel) and gene expression (Fig. 6b, open circles), lack of MMP-9 inhibition by EGCG was observed in terminally differentiated cells both at the protein (Fig. 6a, lower panel) and transcriptional level (Fig. 6b, closed circle).
Fig. 6.
EGCG antagonizes COX-2 but not MMP-9 expression induced upon PMA-mediated HL-60 cell differentiation into macrophages. Serum-starved HL-60 cells were treated for 18 h with 3 nM PMA to induce HL-60 cell differentiation and adhesion. Adherent cells were then further cultured in serum-free conditions and in the presence of increasing concentrations of EGCG for 18 h. a Cell lysates were electrophoresed via SDS-PAGE and immunodetection of COX-2 (upper panels) and GAPDH (middle panels) proteins performed as described in the “Materials and methods” section. Conditioned media were collected and gelatin zymography (lower panels) performed as described in the “Materials and methods” section. b Total RNA was isolated from similar conditions as described earlier, and qRT-PCR performed as described in the “Materials and methods” section to assess MMP-9 and COX-2 gene expression. Susp, untreated and non-differentiated HL-60 cells growing in suspension
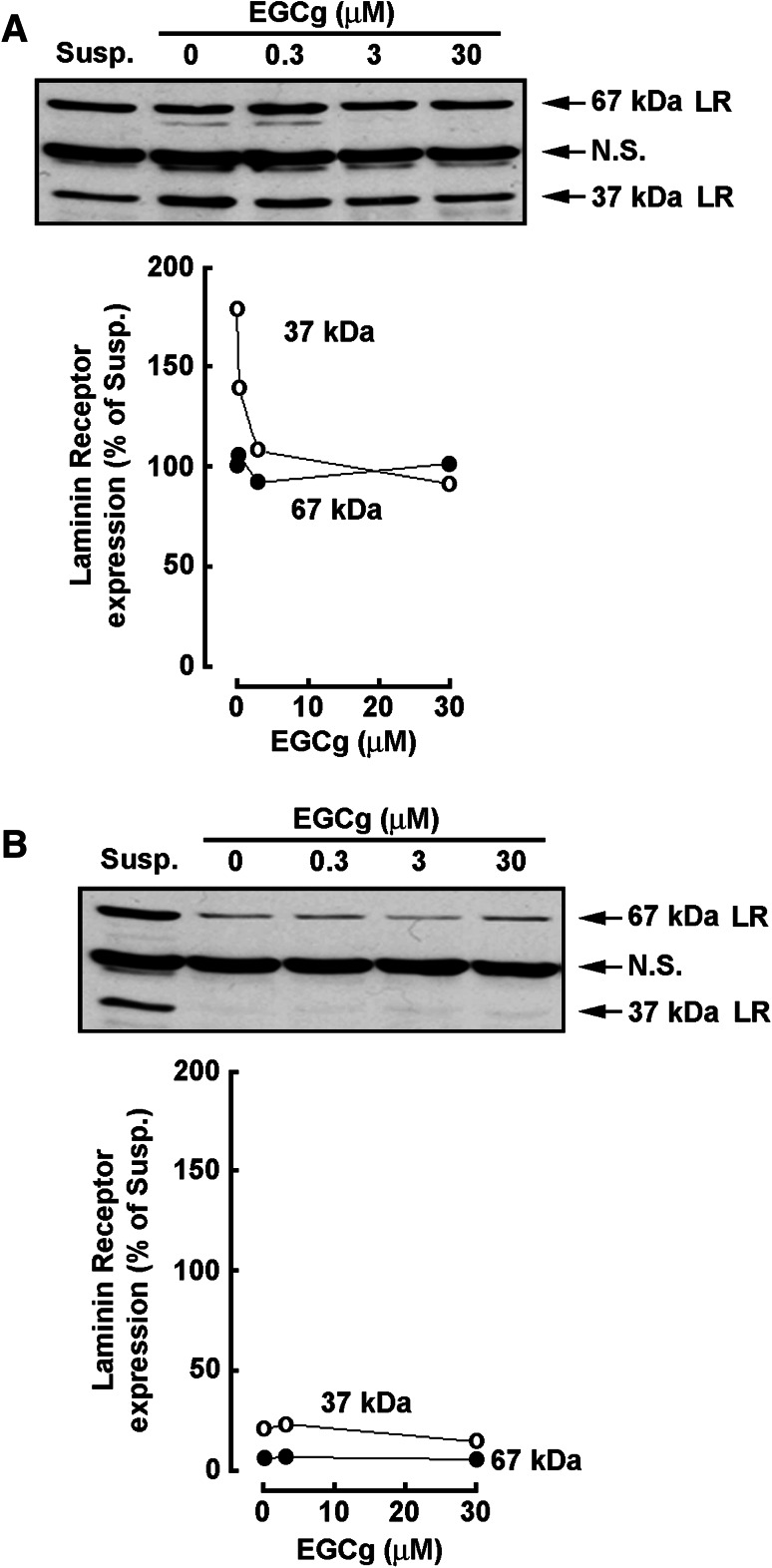
The 67-kDa Laminin Receptor expression is decreased in terminally differentiated HL-60 macrophages
The 67-kDa non-integrin Laminin Receptor (LR) has recently been identified as a direct cell surface receptor for EGCG [27]. We therefore tested the expression of both LR subunits during PMA-induced differentiation as well as in terminally differentiated HL-60 macrophages. We found that the 67-kDa LR expression remained unaffected, as compared to expression in non-differentiated HL-60 cells (Susp.) during PMA treatment regardless of EGCG (Fig. 7a, closed circles). In contrast, EGCG efficiently inhibited the induction of the 37-kDa LR precursor (Fig. 7a, open circle). When PMA alone was first used to terminally differentiate HL-60 cells into macrophages, the expression of both the 37- and 67-kDa LR subunits was significantly reduced (Fig. 7b).
Fig. 7.
The 67-kDa Laminin Receptor expression is decreased in terminally differentiated HL-60 macrophages. a Serum-starved HL-60 cells were treated with 3 nM PMA in the presence of increasing concentrations of EGCG to induce HL-60 cell adhesion for 18 h. b PMA alone was first used to terminally differentiate HL-60 cells into macrophages. Adherent cells were afterward further cultured in serum-free conditions and in the presence of increasing concentrations of EGCG for 18 h. Cell lysates from a and b were electrophoresed via SDS-PAGE and immunodetection of the 37-kDa (open circles)/67-kDa (closed circles) Laminin Receptor (LR) proteins performed as described in the “Materials and methods” section. Data are representative of 2 independent experiments. NS nonspecific immunoreactivity, Susp untreated and non-differentiated HL-60 cells growing in suspension
Discussion
Molecular evidences that demonstrate the various functions of NF-κB during different tumor stages and that supports the rationale to target NF-κB in cancer prevention and therapy have recently been provided [28]. Accordingly, pharmacological targeting of NF-κB-regulated downstream gene products may reasonably be envisioned in the inhibition of inflammatory [1, 29–31] and carcinogenic processes [5, 32, 33]. Recent findings in the anti-inflammatory activity of plant and diet-derived compounds, demonstrate that most of them belong to the chemical group of alkaloids, coumarins, flavonoids, polyphenols, and terpenoids [32]. Although flavonoids have been used in inflammatory pathways targeting [34, 35] and that evidence from cancer genetics and cancer genome studies supports the involvement of NF-κB in human cancer, particularly in multiple myeloma, the therapeutic potential and benefit of targeting NF-κB still remain debatable [36], partly because of the heterogeneous cellular composition and status in cell differentiation within the tumor microenvironment. As such, the recruitment and infiltration of macrophages in the tumor microenvironment activates them to support the malignant progression of cancer cells, and administration of either NF-κB-targeting drugs or COX-2 inhibitors was shown to block both inflammatory angiogenesis and tumor angiogenesis [37].
In this study, we analyzed the transcriptional regulation of 89 NF-κB-regulated genes by assessing their expression in PMA-mediated signaling and in terminally differentiated HL-60 macrophages. The respective gene expression profiles were, not surprisingly, found induced within the cell differentiation, immune response, and inflammation functions (Table 1) and were relatively similar between PMA-treated and terminally differentiated cell populations (r 2 = 0.91; Fig. 4a). In contrast, high EGCG inhibitory activity was found in PMA-mediated pro-carcinogenic stimulation of HL-60 cells (Fig. 3b), while significantly lower inhibitory capacity was attributed to such flavonoid in terminally differentiated macrophages (Fig. 5b). Altogether, this supports the chemopreventive, rather than therapeutic, efficacy of EGCG and provides rational for its crucial role as a signal transduction inhibitor. Moreover, we show that the macrophagic/monocytic differentiation status significantly influences EGCG’s ability to inhibit PMA-induced downstream NF-κB gene targets. The latter may, in part, be explained by the downregulation of the 67-kDa LR expression, which is considered as the EGCG receptor [27]. Although significantly less efficient in terminally differentiated macrophages, the fact that EGCG is still able to abrogate the expression of some NF-κB gene targets is suggestive of pharmacological effects independent of the 67-kDa LR expression as reported elsewhere [38].
The differential targeting efficacy of EGCG is further demonstrated in our study in relation to the NF-κB downstream gene targets COX-2 and MMP-9, both considered as inflammation biomarkers [35, 39]. While COX-2 protein and gene expression is inhibited both during PMA treatment and in terminally differentiated macrophages, we show that MMP-9 in contrast can no longer be inhibited in those cells that acquired macrophagic phenotype. Given the lack of MMP-9 inhibition by EGCG in terminally differentiated macrophages (Fig. 6), our data therefore imply that EGCG may not be efficient in inhibiting the ability of macrophages to disrupt the BBB and to infiltrate the brain. While human brain microvascular endothelial cells (HBMEC) play an essential role as structural and functional components of the BBB, its disruption by the macrophage-secreted MMP-9 may therefore favor secondary cerebral infections and development of brain pathologies [40, 41]. Moreover, although some studies report pharmacological effects of EGCG at submicromolar levels, most experiments require concentrations of above 10–20 μM to demonstrate that effect. Given that in humans, tea polyphenols undergo glucuronidation, sulfation, methylation, and ring fission, the peak plasma concentration of EGCG is believed to approximate 1 μM [42]. Accordingly, IC50 effects in our current study were reached at ~ 3 μM or less in accordance with those plasma concentrations reported, while optimal effects were achieved at 30 μM in accordance with in vitro data reported above (i.e., 10–20 μM). Finally, unmetabolized EGCG was directly tested within an acellular system and on the activity of recombinant MMPs [43]. The authors reported an IC50 value for EGCG of 0.8 μM, again nicely approximating that EGCG concentration we report in this current study. Given the very close IC50 values calculated between cellular and acellular in vitro models, one may therefore safely consider that the impact of EGCG we evaluated may well originate from the parental non-metabolized molecule.
Only few reports documented an association between COX-2 expression and ECM degradation consequent to pro-carcinogenic stimulation. Among the intracellular events that could link PMA-induced signaling to COX-2 induction, NF-κB can contribute to regulate the expression of COX-2 through endoplasmic reticulum stress and, in part, through induction of the endoplasmic reticulum chaperone GRP78/BiP, which is expressed at high levels in a variety of tumors and which confers drug resistance to both proliferating and dormant cancer cells [44]. As for the transcriptional control of MMP-9 expression upon PMA stimulation, there is increasing evidence that its expression can also be regulated at the levels of mRNA stability, translation, and protein secretion. The ability to modulate MMP-9 expression at multiple steps through distinct signaling pathways may be particularly important during malignant conversion, when tumor cells need to induce or maintain MMP-9 levels in response to changing environmental cues. Among the nuclear factors shown to stabilize the mRNA and augment the expression of MMP-9, the RNA-binding protein HuR has been ascribed a pivotal role in the development of tumors [20, 45], and increased nucleocytoplasmic shuttling was also reported to promote COX-2 mRNA stabilization [46]. More importantly, HuR has been found to be a key mediator in PMA-treated HL-60 cells where inhibition of HuR expression by EGCG was reported [24].
In recent years, a large number of mechanisms of action have been attributed to flavonoids commonly found in fruits, vegetables, wine, or tea as they can act as potent antioxidants and free radical scavengers [47, 48]. Flavonoids targeting of NF-κB has also been shown to inhibit in vitro brain endothelial cell tubulogenesis [49]. Accordingly, among strategies developed to jointly inhibit ECM degradation and inflammation processes in carcinogenesis, the design, synthesis and evaluation of flavonoid derivatives has not surprisingly emerged as a potent strategy to also target neurodegenerative disorders including different forms of dementia, as well as Alzheimer’s disease [50, 51]. Our data support the chemopreventive potential of such a class of molecules, but prompt for caution when interpreting the data particularly regarding the differentiation status of a given in vitro experimental cell model. Finally, our data support a crucial role for MMP-9 and provide a molecular rational explaining how the disruption of the BBB [52, 53], and subsequent transendothelial migration of terminally differentiated HL-60 macrophages [54], may lead to cerebral infections and development of brain pathologies.
Acknowledgments
BA holds a Canada Research Chair in Molecular Oncology from the Canadian Institutes of Health Research (CIHR). This study was funded by a grant from the Natural Sciences and Engineering Research Council of Canada (NSERC).
Conflict of interest
The authors declare no conflict or competing financial interest.
Abbreviations
- BBB
Blood–brain barrier
- ECM
Extracellular matrix
- EGCG
Epigallocatechin-3-gallate
- LR
Laminin receptor
- MMP-9
Matrix metalloproteinase-9
- NF-κB
Nuclear factor-kappa B
- PMA
Phorbol 12-myristate 13-acetate
References
- 1.Zhu Z, Zhong S, Shen Z. Targeting the inflammatory pathways to enhance chemotherapy of cancer. Cancer Biol Ther. 2011;12(2):95–105. doi: 10.4161/cbt.12.2.15952. [DOI] [PubMed] [Google Scholar]
- 2.Cilloni D, Martinelli G, Messa F, et al. Nuclear factor kB as a target for new drug development in myeloid malignancies. Haematologica. 2007;92(9):1224–1229. doi: 10.3324/haematol.11199. [DOI] [PubMed] [Google Scholar]
- 3.Breccia M, Alimena G. NF-κB as a potential therapeutic target in myelodysplastic syndromes and acute myeloid leukemia. Expert Opin Ther Targets. 2010;14(11):1157–1176. doi: 10.1517/14728222.2010.522570. [DOI] [PubMed] [Google Scholar]
- 4.Pepper C, Hewamana S, Brennan P, et al. NF-kappaB as a prognostic marker and therapeutic target in chronic lymphocytic leukemia. Future Oncol. 2009;5(7):1027–1037. doi: 10.2217/fon.09.72. [DOI] [PubMed] [Google Scholar]
- 5.Fuchs O. Transcription factor NF-κB inhibitors as single therapeutic agents or in combination with classical chemotherapeutic agents for the treatment of hematologic malignancies. Curr Mol Pharmacol. 2010;3(3):98–122. doi: 10.2174/1874467211003030098. [DOI] [PubMed] [Google Scholar]
- 6.Larson RA, Daley GQ, Schiffer CA, et al. Treatment by design in leukemia, a meeting report, Philadelphia, Pennsylvania, December 2002. Leukemia. 2003;17(12):2358–2382. doi: 10.1038/sj.leu.2403156. [DOI] [PubMed] [Google Scholar]
- 7.Park JH, Tallman MS. Treatment of acute promyelocytic leukemia without cytotoxic chemotherapy. Oncology (Williston Park) 2011;25(8):733–741. [PubMed] [Google Scholar]
- 8.Surh Y. Molecular mechanisms of chemopreventive effects of selected dietary and medicinal phenolic substances. Mutat Res. 1999;428(1–2):305–327. doi: 10.1016/s1383-5742(99)00057-5. [DOI] [PubMed] [Google Scholar]
- 9.Yang H, Landis-Piwowar KH, Chan T, et al. Green tea polyphenols as proteasome inhibitors: implication in chemoprevention. Curr Cancer Drug Targets. 2011;11(3):296–306. doi: 10.2174/156800911794519743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan N, Afaq F, Saleem M, et al. Targeting multiple signaling pathways by green tea polyphenol (-)-epigallocatechin-3-gallate. Cancer Res. 2006;66(5):2500–2505. doi: 10.1158/0008-5472.CAN-05-3636. [DOI] [PubMed] [Google Scholar]
- 11.Mereles D, Hunstein W. Epigallocatechin-3-gallate (EGCG) for clinical trials: more pitfalls than promises? Int J Mol Sci. 2011;12(9):5592–5603. doi: 10.3390/ijms12095592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Na HK, Surh YJ. Intracellular signaling network as a prime chemopreventive target of (-)-epigallocatechin gallate. Mol Nutr Food Res. 2006;50(2):152–159. doi: 10.1002/mnfr.200500154. [DOI] [PubMed] [Google Scholar]
- 13.Demeule M, Michaud-Levesque J, Annabi B, et al. Green tea catechins as novel antitumor and antiangiogenic compounds. Curr Med Chem Anticancer Agents. 2002;2(4):441–463. doi: 10.2174/1568011023353930. [DOI] [PubMed] [Google Scholar]
- 14.Thomas DA, Giles FJ, Cortes J, et al. Antiangiogenic therapy in leukemia. Acta Haematol. 2001;106(4):190–207. doi: 10.1159/000046616. [DOI] [PubMed] [Google Scholar]
- 15.Moehler TM, Hillengass J, Goldschmidt H, et al. Antiangiogenic therapy in hematologic malignancies. Curr Pharm Des. 2004;10(11):1221–1234. doi: 10.2174/1381612043452587. [DOI] [PubMed] [Google Scholar]
- 16.Collins SJ, Ruscetti FW, Gallagher RE, et al. Terminal differentiation of human promyelocytic leukemia cells induced by dimethyl sulfoxide and other polar compounds. Proc Natl Acad Sci USA. 1978;75(5):2458–2462. doi: 10.1073/pnas.75.5.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huberman E, Callaham MF. Induction of terminal differentiation in human promyelocytic leukemia cells by tumor-promoting agents. Proc Natl Acad Sci USA. 1979;76(3):1293–1297. doi: 10.1073/pnas.76.3.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tonetti DA, Henning-Chubb C, Yamanishi DT, et al. Protein kinase C-beta is required for macrophage differentiation of human HL-60 leukemia cells. J Biol Chem. 1994;269(37):23230–23235. [PubMed] [Google Scholar]
- 19.Xie B, Laouar A, Huberman E. Autocrine regulation of macrophage differentiation and 92-kDa gelatinase production by tumor necrosis factor-alpha via alpha5 beta1 integrin in HL-60 cells. J Biol Chem. 1998;273(19):11583–11588. doi: 10.1074/jbc.273.19.11583. [DOI] [PubMed] [Google Scholar]
- 20.McMillan JI, Weeks R, West JW, et al. Pharmacological inhibition of gelatinase B induction and tumor cell invasion. Int J Cancer. 1996;67(4):523–531. doi: 10.1002/(SICI)1097-0215(19960807)67:4<523::AID-IJC11>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 21.Bingle L, Brown NJ, Lewis CE. The role of tumour-associated macrophages in tumour progression: implications for new anticancer therapies. J Pathol. 2002;196(3):254–265. doi: 10.1002/path.1027. [DOI] [PubMed] [Google Scholar]
- 22.D’Alessandro T, Prasain J, Benton MR, et al. Polyphenols, inflammatory response, and cancer prevention: chlorination of isoflavones by human neutrophils. J Nutr. 2003;133(11 Suppl 1):3773S–3777S. doi: 10.1093/jn/133.11.3773S. [DOI] [PubMed] [Google Scholar]
- 23.Belkaid A, Fortier S, Cao J, et al. Necrosis induction in glioblastoma cells reveals a new “bioswitch” function for the MT1-MMP/G6PT signaling axis in proMMP-2 activation versus cell death decision. Neoplasia. 2007;9(4):332–340. doi: 10.1593/neo.07142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Annabi B, Currie JC, Moghrabi A, et al. Inhibition of HuR and MMP-9 expression in macrophage-differentiated HL-60 myeloid leukemia cells by green tea polyphenol EGCg. Leuk Res. 2007;31(9):1277–1284. doi: 10.1016/j.leukres.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 25.Jin R, Yang G, Li G. Molecular insights and therapeutic targets for blood-brain barrier disruption in ischemic stroke: critical role of matrix metalloproteinases and tissue-type plasminogen activator. Neurobiol Dis. 2010;38(3):376–385. doi: 10.1016/j.nbd.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Candelario-Jalil E, Taheri S, Yang Y, et al. Cyclooxygenase inhibition limits blood-brain barrier disruption following intracerebral injection of tumor necrosis factor-alpha in the rat. J Pharmacol Exp Ther. 2007;323(2):488–498. doi: 10.1124/jpet.107.127035. [DOI] [PubMed] [Google Scholar]
- 27.Umeda D, Yano S, Yamada K, et al. Green tea polyphenol epigallocatechin-3-gallate signaling pathway through 67-kDa laminin receptor. J Biol Chem. 2008;283(6):3050–3058. doi: 10.1074/jbc.M707892200. [DOI] [PubMed] [Google Scholar]
- 28.Arkan MC, Greten FR. IKK- and NF-κB-mediated functions in carcinogenesis. Curr Top Microbiol Immunol. 2011;349:159–169. doi: 10.1007/82_2010_97. [DOI] [PubMed] [Google Scholar]
- 29.Cabrini G, Bezzerri V, Mancini I, et al. Targeting transcription factor activity as a strategy to inhibit pro-inflammatory genes involved in cystic fibrosis: decoy oligonucleotides and low-molecular weight compounds. Curr Med Chem. 2010;17(35):4392–4404. doi: 10.2174/092986710793361243. [DOI] [PubMed] [Google Scholar]
- 30.Bamborough P, Morse MA, Ray KP. Targeting IKKβ for the treatment of rheumatoid arthritis. Drug News Perspect. 2010;23(8):483–490. doi: 10.1358/dnp.2010.23.8.1447844. [DOI] [PubMed] [Google Scholar]
- 31.Chen S. Natural products triggering biological targets—a review of the anti-inflammatory phytochemicals targeting the arachidonic acid pathway in allergy asthma and rheumatoid arthritis. Curr Drug Targets. 2011;12(3):288–301. doi: 10.2174/138945011794815347. [DOI] [PubMed] [Google Scholar]
- 32.Chen W, Li Z, Bai L, et al. NF-kappaB in lung cancer, a carcinogenesis mediator and a prevention and therapy target. Front Biosci. 2011;16:1172–1185. doi: 10.2741/3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adams H, Obermann EC, Dirnhofer S, et al. Targetable molecular pathways in classical Hodgkin’s lymphoma. Expert Opin Investig Drugs. 2011;20(2):141–151. doi: 10.1517/13543784.2011.546562. [DOI] [PubMed] [Google Scholar]
- 34.Prasad S, Phromnoi K, Yadav VR, et al. Targeting inflammatory pathways by flavonoids for prevention and treatment of cancer. Planta Med. 2010;76(11):1044–1063. doi: 10.1055/s-0030-1250111. [DOI] [PubMed] [Google Scholar]
- 35.Luqman S, Pezzuto JM. NFkappaB: a promising target for natural products in cancer chemoprevention. Phytother Res. 2010;24(7):949–963. doi: 10.1002/ptr.3171. [DOI] [PubMed] [Google Scholar]
- 36.Baud V, Karin M. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat Rev Drug Discov. 2009;8(1):33–40. doi: 10.1038/nrd2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ono M. Molecular links between tumor angiogenesis and inflammation: inflammatory stimuli of macrophages and cancer cells as targets for therapeutic strategy. Cancer Sci. 2008;99(8):1501–1506. doi: 10.1111/j.1349-7006.2008.00853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hazawa M, Takahashi K, Sugata S, et al. (-)-Epigallocatechin-3-O-gallate induces nonapoptotic cell death in leukemia cells independent of the 67 kDa laminin receptor. J Nat Prod. 2011;74(4):695–700. doi: 10.1021/np1007729. [DOI] [PubMed] [Google Scholar]
- 39.Tergaonkar V. NFkappaB pathway: a good signaling paradigm and therapeutic target. Int J Biochem Cell Biol. 2006;38(10):1647–1653. doi: 10.1016/j.biocel.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 40.Lakka SS, Gondi CS, Rao JS. Proteases and glioma angiogenesis. Brain Pathol. 2005;15(4):327–341. doi: 10.1111/j.1750-3639.2005.tb00118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bonoiu A, Mahajan SD, Ye L, et al. MMP-9 gene silencing by a quantum dot-siRNA nanoplex delivery to maintain the integrity of the blood brain barrier. Brain Res. 2009;1282:142–155. doi: 10.1016/j.brainres.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lambert JD, Yang CS. Mechanisms of cancer prevention by tea constituents. J Nutr. 2003;133(10):3262S–3267S. doi: 10.1093/jn/133.10.3262S. [DOI] [PubMed] [Google Scholar]
- 43.Demeule M, Brossard M, Pagé M, et al. Matrix metalloproteinase inhibition by green tea catechins. Biochim Biophys Acta. 2000;1478(1):51–60. doi: 10.1016/S0167-4838(00)00009-1. [DOI] [PubMed] [Google Scholar]
- 44.Craggs L, Kalaria RN. Revisiting dietary antioxidants, neurodegeneration and dementia. NeuroReport. 2011;22(1):1–3. doi: 10.1097/WNR.0b013e328342741c. [DOI] [PubMed] [Google Scholar]
- 45.Akool el S, Kleinert H, Hamada FM, et al. Nitric oxide increases the decay of matrix metalloproteinase 9 mRNA by inhibiting the expression of mRNA-stabilizing factor HuR. Mol Cell Biol. 2003;23(14):4901–4916. doi: 10.1128/MCB.23.14.4901-4916.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johann AM, Weigert A, Eberhardt W, et al. Apoptotic cell-derived sphingosine-1-phosphate promotes HuR-dependent cyclooxygenase-2 mRNA stabilization and protein expression. J Immunol. 2008;180(2):1239–1248. doi: 10.4049/jimmunol.180.2.1239. [DOI] [PubMed] [Google Scholar]
- 47.Rusak G, Gutzeit HO, Ludwig-Műller J. Structurally related flavonoids with antioxidative properties differentially affect cell cycle progression and apoptosis of human acute leukemia cells. Nutr Res. 2005;25(2):143–155. doi: 10.1016/j.nutres.2004.12.003. [DOI] [Google Scholar]
- 48.Rice-Evans C, Packer L, editors. Flavonoids in health and disease. 2. New York: Marcel Dekker Inc.; 2003. [Google Scholar]
- 49.Tahanian E, Sanchez LA, Shiao TC, et al. Flavonoids targeting of IκB phosphorylation abrogates carcinogen-induced MMP-9 and COX-2 expression in human brain endothelial cells. Drug Des Devel Ther. 2011;5:299–309. doi: 10.2147/DDDT.S19931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams P, Sorribas A, Howes MJ. Natural products as a source of Alzheimer’s drug leads. Nat Prod Rep. 2011;28(1):48–77. doi: 10.1039/c0np00027b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Howes MJ, Perry E. The role of phytochemicals in the treatment and prevention of dementia. Drugs Aging. 2011;28(6):439–468. doi: 10.2165/11591310-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 52.Feng SR, Chen ZX, Cen JN, et al. Disruption of blood brain-barrier by leukemic cells in central nervous system leukemia. Zhonghua Xue Ye Xue Za Zhi. 2011;32(5):289–293. [PubMed] [Google Scholar]
- 53.Feng S, Cen J, Huang Y, et al. Matrix metalloproteinase-2 and -9 secreted by leukemic cells increase the permeability of blood-brain barrier by disrupting tight junction proteins. PLoS ONE. 2011;6(8):e20599. doi: 10.1371/journal.pone.0020599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seidel G, Böcker K, Schulte J, et al. Pertussis toxin permeabilization enhances the traversal of Escherichia coli K1, macrophages, and monocytes in a cerebral endothelial barrier model in vitro. Int J Med Microbiol. 2011;301(3):204–212. doi: 10.1016/j.ijmm.2010.08.018. [DOI] [PubMed] [Google Scholar]