Abstract
Flock house virus (FHV) is a small icosahedral insect virus with a bipartite, messenger-sense RNA genome. Its T=3 icosahedral capsid is initially assembled from 180 subunits of a single type of coat protein, capsid precursor protein alpha (407 amino acids). Following assembly, the precursor particles undergo a maturation step in which the alpha subunits autocatalytically cleave between Asn363 and Ala364. This cleavage generates mature coat proteins beta (363 residues) and gamma (44 residues) and is required for acquisition of virion infectivity. The X-ray structure of mature FHV shows that gamma peptides located at the fivefold axes of the virion form a pentameric helical bundle, and it has been suggested that this bundle plays a role in release of viral RNA during FHV uncoating. To provide experimental support for this hypothesis, we generated mutant coat proteins that carried deletions in the gamma region of precursor protein alpha. Surprisingly, we found that these mutations interfered with specific recognition and packaging of viral RNA during assembly. The resulting particles contained large amounts of cellular RNAs and varying amounts of the viral RNAs. Single-site amino acid substitution mutants showed that three phenylalanines located at positions 402, 405, and 407 of coat precursor protein alpha were critically important for specific recognition of the FHV genome. Thus, in addition to its hypothesized role in uncoating and RNA delivery, the C-terminal region of coat protein alpha plays a significant role in recognition of FHV RNA during assembly. A possible link between these two functions is discussed.
Flock house virus (FHV) is a nonenveloped icosahedral insect virus of the family Nodaviridae (for a review, see reference 20). Its genome consists of two positive-sense RNA molecules, RNA1 (3.1 kb) and RNA2 (1.4 kb), which are packaged into a single virion. RNA1 encodes replication functions (4, 11), whereas RNA2 encodes protein alpha (43 kDa), the precursor of the coat protein (8). Following infection of Drosophila cells, the first detectable assembly products are provirions whose T=3 icosahedral shells are composed of 180 alpha protein subunits which encapsidate the two genomic RNAs (9). Assembly catalyzes a maturation event, in which the alpha protein subunits (407 amino acids) cleave between residues Asn363 and Ala364 to yield mature coat proteins beta (363 amino acids) and gamma (44 amino acids) (13). The cleavage usually does not go to completion, and residual protein alpha is commonly observed in any given virus preparation. Maturation of provirions results in increased particle stability and is required for acquisition of virion infectivity (9, 21). It is not known which step in the virus life cycle specifically depends on the cleavage event. However, based on the observation that provirions attach almost as efficiently to susceptible cells as mature virions, it has been suggested that maturation cleavage is required for some step between viral attachment and release of the genome into the cytosol (21).
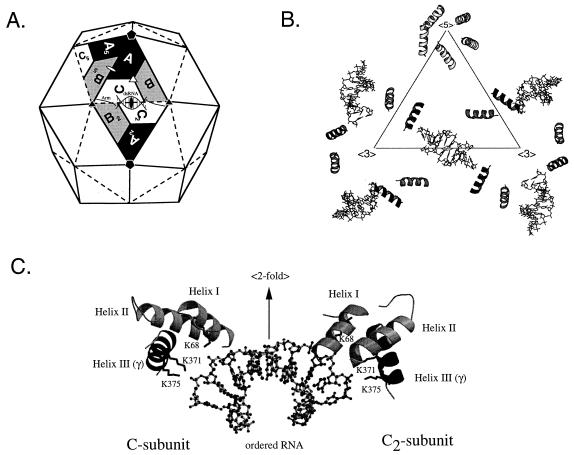
The structure of mature FHV particles was solved to near atomic resolution by X-ray crystallography (6), which shows that the alpha protein cleavage site is located inside the virion near the RNA core. Of the 44 amino acids that comprise the gamma peptide, only the first 18 residues (residues 364 to 381 of the alpha precursor) are visible while the remaining residues lack icosahedral symmetry. The visible portion of gamma forms an amphipathic alpha helix which interacts with its environment in different ways depending on its location in the virus particle (Fig. 1A and B). Gamma helices that are associated with the C subunits at the twofold symmetry axes of the virus particle (Fig. 1A) contact the sugar-phosphate backbone of the encapsidated RNA via two lysine residues at positions 371 and 375 (Fig. 1C). Gamma helices that are associated with the A subunits at the fivefold axes of the particle interact with each other and form a pentameric helical bundle that has a hydrophobic exterior and a hydrophilic core. Cheng et al. (2) have proposed that during viral uncoating this helical bundle is released from the particle to form a channel in a cellular membrane through which the encapsidated RNA could escape into the cytosol. The release of the gamma peptides, as proposed in this model, would explain the requirement for the maturation cleavage. To provide experimental support for this model, we generated gamma peptide deletion mutants to test the effect of the deletion on viral infectivity and more specifically on the release of RNA from the particles. Unexpectedly, we found that C-terminal deletions in the region of protein alpha that represents gamma in mature particles destroyed the ability of the coat precursor to recognize FHV RNA for encapsidation. Instead, the coat protein appeared to package viral and cellular RNAs at random. Using single-site amino acid substitution mutants, we found that the ability of the coat protein to recognize viral RNA was dependent on the presence of three phenylalanine residues located at positions 402, 405, and 407. Thus, in addition to its postulated role in viral uncoating, the gamma peptide, while still an integral component of precursor protein alpha, plays a critical role in specific recognition of the viral RNAs during assembly.
FIG. 1.
Organization of gamma peptides in the FHV particle and their interaction with genomic RNA. (A) Schematic representation of the FHV capsid as a rhombic triacontahedron. Each trapezoid represents a protein subunit which initially consists of 407 amino acids. The labels A, B, and C represent the three subunits in each of the 60 icosahedral asymmetric units in the T=3 particle. Although A, B, and C represent identical gene products, they are not related by strict symmetry and they are structurally slightly different. In mature virions, most subunits are present as proteins beta and gamma, which are generated by autocatalytic cleavage of the precursor protein between residues Asn363 and Ala364. Note that ordered genomic RNA is visible at the icosahedral twofold axes (shown as a solid oval) of the virion. (B) Organization of the internally located gamma peptides and the ordered genomic RNA as seen in the X-ray structure. The triangle represents the central asymmetric unit shown in panel A. The first 18 residues of gamma form an amphipathic alpha helix, while the remaining 26 residues are not visible. Gamma peptides associated with the A subunits form a pentameric helical bundle at the fivefold axes of the virion. This helical bundle has been hypothesized to play a role in FHV uncoating. Gamma peptides associated with the C subunits contact the genomic RNA (shown as stick diagrams), which forms double-helical segments at the icosahedral twofold axes of the virion. (C) Close-up view of the interaction of the gamma peptides with genomic RNA (shown as a ball and stick model). The view is perpendicular to the icosahedral twofold axis, showing specific interactions between the phosphodiester backbone of FHV RNA and the side chains of lysine residues 68, 371, and 375. Lys68 is located on helix I, which consists of residues 61 to 73 of protein beta. Lys371 and Lys375 are located on helix III, which is part of the gamma peptide. Helix II, formed by residues 341 and 353, does not interact with the encapsidated RNA. All interactions are with the C subunit and its twofold related partner (C2 in the diagram shown in panel A). For clarity, only the helical domains of the coat protein subunits are shown.
MATERIALS AND METHODS
Cells.
Drosophila melanogaster cells (Schneider’s line 1) were grown as monolayers in Schneider’s insect medium supplemented with 15% heat-inactivated fetal bovine serum, as described previously (8).
Site-directed mutagenesis of FHV coat protein gene.
Plasmid p2BS(+)-wt (21), containing a cDNA copy of the wild-type (wt) FHV coat protein gene, was used to generate deletion mutants as well as amino acid substitution mutants. Deletions in the open reading frame of coat protein alpha were generated by inverse PCR (14); i.e., oligonucleotide primers were designed in inverted tail-to-tail directions to amplify the entire cloning vector as well as the target sequence except for the area to be deleted. The sequences of all primers used in this study will be provided on request. PCR conditions were as described in reference 5. The expected-size products were purified by agarose gel electrophoresis and the Gene-Clean purification kit (Bio 101), religated with T4 DNA ligase (New England Biolabs), and transformed into DH5α competent cells. Plasmid DNA was isolated from three to five independent clones, and the sequence across the mutated sites was verified by using the Sanger dideoxy sequencing method (19). Amino acid substitution mutants were created by overlap extension PCR (12). Primers were designed such that the final PCR products spanned bases 400 to 1400 of the coat protein gene plus an additional XbaI site following base 1400. The PCR products were purified and digested with XbaI and ClaI, which cuts at position 470 of the RNA2 cDNA. The resulting DNA fragments were used to replace the homologous region in plasmid p2BS(+)-wt. Following ligation and transformation, plasmid DNA was purified and all sequences were verified by the Sanger dideoxy sequencing method.
In vitro transcription of RNA2.
Plasmid p2BS(+)-wt and its mutant versions were linearized with XbaI and used as templates for in vitro transcription of capped RNAs. Specifically, transcripts were synthesized with T3 RNA polymerase by using the mMessage machine kit (Ambion) and protocols provided by the manufacturer. Following RNA synthesis, template DNA was digested with DNase I and RNA transcripts were purified over RNeasy RNA purification columns (Qiagen) by the RNA clean-up protocol. RNAs were eluted from the column with 50 μl of water.
Source of FHV RNA1.
Capped FHV RNA1 was generated by in vitro transcription with T7 RNA polymerase and the mMessage machine kit (Ambion) with the nonlinearized plasmid FHV[1,0] (1), kindly provided by A. Ball, University of Alabama, Birmingham. The transcription reactions yielded highly heterogeneous products which included full-length FHV RNA1. Following purification of the transcripts as described above for RNA2, full-length RNA1 was selectively amplified by transfecting 500 ng of the mixture into 107 Drosophila cells by standard protocols (see below). After 20 to 24 h, total RNA was extracted from the cells and analyzed by agarose gel electrophoresis to determine the approximate amount of RNA1 in a given aliquot. About 0.5 to 1 μg of total RNA, containing roughly 100 ng of RNA1, was used as a source of FHV RNA1 for the generation of FHV particles.
Liposome-mediated RNA transfection of Drosophila cells.
Transfections were carried out in six-well tissue culture plates containing 107 cells per well. To remove serum components, either cells were plated in serum-deficient medium or monolayers were washed twice with 1 ml of serum-free medium and then covered with 1 ml of serum-free medium. RNA-liposome complexes for 107 cells were prepared as follows: a 15-μl volume of Lipofectin (Gibco BRL) at a concentration of 1 mg/ml was diluted to 30 μl with water in a polystyrene tube. A mixture of 0.5 to 1 μg of total Drosophila cell RNA containing approximately 100 ng of FHV RNA1 and 100 to 200 ng of in vitro-synthesized capped FHV RNA2 in 30 μl of water was added. The complexes were allowed to form at room temperature for 15 min and were then applied directly to the cells. After a 2-h incubation period at 27°C, the medium was removed from the cells and replaced with 2 ml of complete growth medium. Incubation at 27°C was continued for 20 to 24 h.
Purification of virus particles from transfected cells.
At 20 to 24 h posttransfection, cells were lysed by the addition of Nonidet P-40 to a final concentration of 0.5% (vol/vol) and incubation on ice for 5 min. Cell debris was then pelleted in a Beckman JA17 rotor at 10,000 rpm (13,800 × g) for 10 min at 4°C, and the clarified supernatant was transferred to a fresh tube. RNase A was added to a final concentration of 10 μg per ml, and the supernatant was incubated at 27°C for 30 min. Virus particles in the supernatant were then pelleted through a 1-ml volume of 30% (wt/wt) sucrose in 50 mM HEPES (pH 7)–5 mM CaCl2–0.1% bovine serum albumin–0.1% 2-mercaptoethanol at 40,000 rpm (274,000 × g) in an SW41 rotor for 2.5 h at 11°C. The pellet was resuspended in 50 mM HEPES (pH 7)–5 mM CaCl2–0.1% 2-mercaptoethanol and layered on a 10-ml 10 to 40% (wt/wt) sucrose gradient in the same buffer. Virus particles were sedimented at 40,000 rpm (274,000 × g) in an SW41 rotor for 1.5 h at 11°C. The gradient was fractionated on an ISCO gradient fractionator at 0.75 ml/min and 0.5 min per fraction.
Plaque assay.
Infectivity titers were determined on monolayers of Drosophila cells as described elsewhere (21).
Electrophoretic analysis of proteins.
Electrophoresis was performed on discontinuous sodium dodecyl sulfate (SDS)-polyacrylamide gels as described in reference 5.
Isolation of RNA from purified particles.
SDS and NaCl were added to gradient-purified FHV particles at final concentrations of 1% (wt/vol) and 0.2 M, respectively. RNA was extracted with an equal volume of acidic phenol-chloroform and precipitated with 3 volumes of ethanol in the presence of 0.3 M sodium acetate and 20 μg of glycogen. After several hours at −20°C, the RNA was pelleted, washed with 70% ethanol, dried, and dissolved in nuclease-free water.
Agarose gel electrophoresis and Northern blot analysis.
Electrophoresis of RNA in agarose-formaldehyde gels and Northern blot analysis were performed as described previously (22). Probes used for hybridization were digoxigenin-UTP-labeled antisense RNAs complementary to various regions of FHV RNA2 and RNA1 (see Results for details). To generate the probes, cDNAs representing these regions were first generated by PCR with plasmid p2BS(+)-wt and amplification conditions as described in reference 5. The resulting products were purified and cloned into pBluescript KSII(+) (Stratagene) prepared with 3′ overhangs (16). After transformation of DH5α competent cells, plasmids containing inserts were selected and the orientation of the insert relative to the T3 and T7 promoters was determined. Following linearization with XbaI or XhoI, in vitro transcription of digoxigenin-UTP-labeled antisense RNA was performed according to the manufacturer’s protocols (Boehringer Mannheim).
RESULTS
Expression of C-terminal deletion mutants Δγ363 and Δγ381.
FHV capsid precursor protein alpha contains 407 amino acids and is cleaved between Asn363 and Ala364 following assembly of provirions. This cleavage results in formation of proteins beta (residues 1 to 363) and gamma (residues 364 to 407), both of which remain associated with the mature virus particle. We initially constructed two C-terminal deletion mutants of protein alpha: in Δγ363 the entire region representing the gamma peptide was eliminated from the precursor protein; in Δγ381 only the portion which is not visible in the X-ray structure of FHV was deleted (note that in the nomenclature of the deletion mutants, the number refers to the location of the C-terminal residue of the mutated coat protein). Deletions were engineered into the cDNA of FHV RNA2, and capped RNAs were generated by in vitro transcription. A mixture of wt RNA1, encoding the replication proteins, and mutant RNA2 was then introduced into Drosophila cells by liposome-mediated transfection. Because the viral RNAs are messenger-sense RNAs, this method effectively imposes one cycle of replication on the cells and thereby allows generation and analysis of even nonviable mutants as long as a sufficient number of cells is transfected.
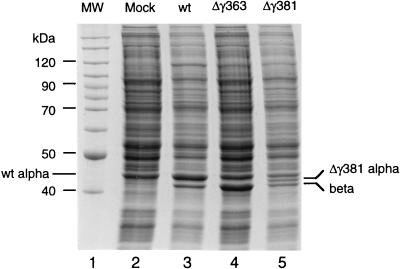
After 24 h, lysates of the transfected cells were analyzed by gel electrophoresis. As shown in Fig. 2, cells transfected with the Δγ363 construct contained a protein that comigrated with beta protein of control cells which had been transfected with wt FHV RNAs. The level of coat protein in the Δγ363-transfected cells appeared to be similar to that in the controls, taking into account that at least 50% of the wt protein was still present in its precursor form alpha. (Note that wt protein alpha comigrates with a cellular protein, probably actin). In lysates transfected with the Δγ381 construct, two viral bands were visible: one that migrated slightly faster than wt alpha protein and presumably represented the mutant alpha protein and one that migrated with wt beta protein and therefore probably represented the cleavage product. The small gamma protein (4.4 kDa in the case of the wt, 1.9 kDa in the case of Δγ381) ran off the gel under the conditions used and was not visible. The fact that the Δγ381 protein appeared to have cleaved into beta and gamma indicated that this protein had assembled into virus particles, since the cleavage reaction is not observed in the monomeric protein subunit (9). For Δγ363, this conclusion could not be drawn at this point because the portion that is cleaved from the precursor protein had been deleted.
FIG. 2.
Gel electrophoresis of lysates prepared from Drosophila cells transfected with wt FHV RNA1 and wt or mutant FHV transcript RNA2. Cell monolayers containing 107 Drosophila cells were transfected with a mixture of approximately 100 ng of RNA1 and 100 ng of RNA2, as described in Materials and Methods. At 24 h after transfection, cells were dislodged into the growth medium, collected by centrifugation, and washed twice with 0.5 ml of phosphate-buffered saline (PBS). The final cell pellet was resuspended in 100 μl of PBS, mixed with an equal volume of 2× electrophoresis buffer, and heated to 95°C for 10 min. Aliquots of 10 μl (approximately 5 × 105 cells) were electrophoresed through an SDS–12% polyacrylamide gel, followed by staining with Coomassie brilliant blue. Gamma peptide migrated off the gel under the conditions used. Lane 1, molecular weight markers; lane 2, lysate from mock-transfected cells; lane 3, lysate from cells transfected with wt RNA1 and wt RNA2 (note that wt protein alpha comigrates with a cellular protein, probably actin); lane 4, lysate from cells transfected with wt RNA1 and Δγ363 RNA2; lane 5, lysate from cells transfected with wt RNA1 and Δγ381 RNA2.
Assembly phenotypes of Δγ363 and Δγ381 coat proteins.
Lysates of cells transfected with the Δγ363 and Δγ381 constructs were further processed to test for the presence of virus particles. To this end, the putative particles were pelleted through a 30% (wt/wt) sucrose cushion and then sedimented through a 10 to 40% (wt/wt) sucrose gradient. The gradients were subsequently fractionated with continuous absorbance at 254 nm. The profile for Δγ363 did not contain any peaks (data not shown), suggesting that this protein was not able to form virus particles. However, upon careful reexamination of the material that had pelleted through the sucrose cushion in the preceding step, we noticed that a small amount of the mutant coat protein was indeed present (data not shown). This suggested that the Δγ363 protein had formed larger assembly aggregates, although with very low efficiency. We were not able to further characterize these assembly products because of their very small amounts.
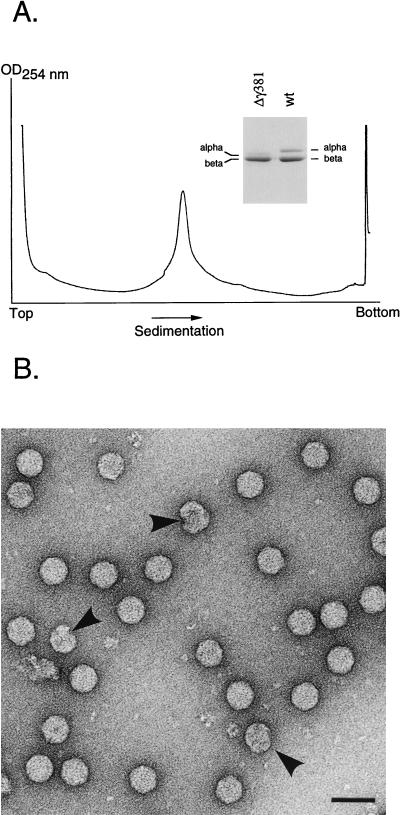
The Δγ381 construct, on the other hand, gave rise to a major peak near the center of the gradient (Fig. 3A). Gel electrophoretic analysis of the peak fraction confirmed that it contained the Δγ381 coat protein. Most of it had been efficiently cleaved because only traces of precursor alpha were visible on the gel (Fig. 3A, inset). The peak was moderately broad at the base, suggesting some heterogeneity in the associated particles. Electron microscopy of negatively stained samples, however, showed that the majority of the particles had the shape and dimensions of native virions (30 nm in diameter), and only a few aberrant structures were seen (Fig. 3B). The ratio of absorbances at 260 and 280 nm was 1.68, which was similar to that of native virions (1.60). To further confirm that the Δγ381 virions did not have other assembly defects, we prepared [35S]methionine-labeled particles. When particles from the gradient fraction containing the highest absorbance were mixed with 3H-labeled native virus, both cosedimented on a 10 to 40% (wt/wt) sucrose gradient (data not shown). Taken together, these results suggested that the Δγ381 particles were structurally very similar to wt virions and that they had maintained a comparable protein-to-RNA ratio. However, the yield of the mutant particles was only 30% of that of wt particles prepared in the same way. This indicated that the Δγ381 coat protein was not able to form particles as efficiently as the full-length coat protein.
FIG. 3.
Sucrose gradient sedimentation profile and electron microscopy of Δγ381 particles. (A) Monolayers of Drosophila cells (approximately 1.2 × 108 cells) were transfected with wt RNA1 and Δγ381 RNA2, as described in Materials and Methods. After 24 h, cells were lysed and virus particles were pelleted through a 30% (wt/wt) sucrose cushion. The resuspended pellet was layered on a linear 10 to 40% (wt/wt) sucrose gradient and centrifuged at 274,000 × g for 1.5 h at 4°C. The gradient was fractionated with continuous absorbance at 254 nm. OD, optical density. (Inset) Electrophoretic analysis of proteins in the peak fraction and comparison with wt FHV. Gamma peptide migrated off the gel under the conditions used. Proteins were stained with Coomassie brilliant blue. (B) Electron micrograph of negatively stained, gradient-purified Δγ381 particles. Arrows indicate aberrant structures. Bar, 100 nm.
Infectivity and RNA contents of Δγ381 particles.
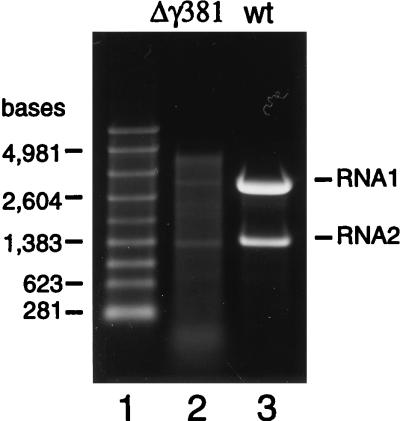
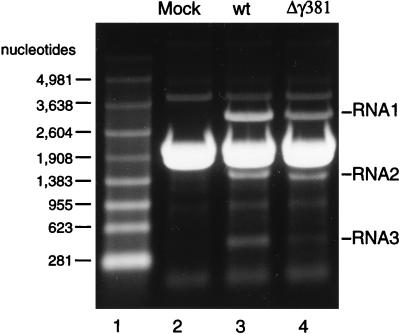
The deletion mutants Δγ363 and Δγ381 had been constructed with the goal of investigating the role of the gamma peptide in viral uncoating. Thus, we first set out to determine the infectivity of the Δγ381 particles by plaque assay. Surprisingly, the particles showed no infectivity at all. This result prompted us to examine the nature of the encapsidated RNAs to confirm that the particles had packaged the normal complement of FHV RNA1 and RNA2. To this end, RNA was extracted from gradient-purified virions with phenol and chloroform and electrophoresed through a denaturing agarose gel. Unexpectedly, instead of containing RNA1 and RNA2, the Δγ381 particles contained a heterogeneous mixture of RNAs that varied in size from about 100 bases to approximately 4,500 bases (Fig. 4). Small amounts of viral RNA1 and RNA2, however, appeared to be present in this mixture. Since the nucleotide sequence removed from RNA2 to generate the Δγ381 deletion mutant did not contain sequences required for encapsidation of this RNA (23), we concluded that removal of the C-terminal end of the coat protein had rendered it unable to specifically recognize FHV RNAs for encapsidation. Instead, a broad mixture of apparently cellular RNAs as well as small amounts of the viral RNAs was packaged into the particles.
FIG. 4.
Gel electrophoretic analysis of RNAs extracted from Δγ381 and wt FHV particles. RNA was extracted with phenol-chloroform from gradient-purified virions and electrophoresed through a denaturing 1% agarose-formaldehyde gel. Nucleic acids were visualized by staining with ethidium bromide. Lane 1, RNA size markers; lane 2, RNA extracted from Δγ381 particles; lane 3, RNA extracted from wt particles.
Northern blot analysis of Δγ381 RNA.
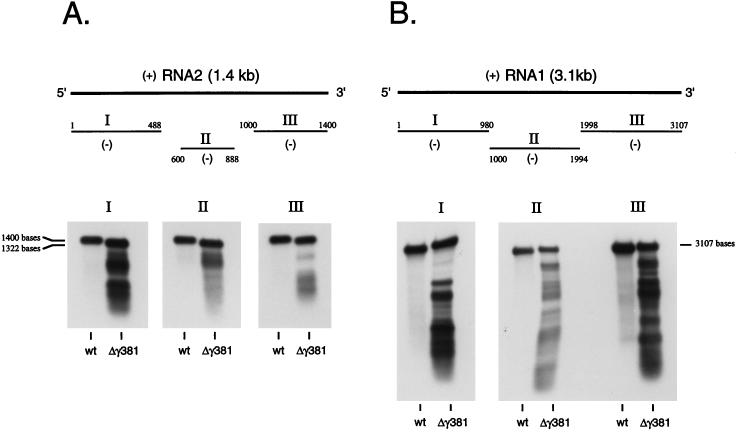
To verify that the RNAs present in Δγ381 particles included FHV RNA1 and mutant RNA2, we performed Northern blot analysis initially with full-length negative-sense RNA1 and RNA2 probes. The results (not shown) indicated that RNA1 and mutant RNA2 were indeed present, but also present were smaller breakdown products of both strands. To determine whether these breakdown products could be mapped to specific regions on the two RNAs, Northern blot analysis was repeated with probes that were complementary to the 5′ end, the 3′ end, and interior regions of RNA1 and RNA2. However, as shown in Fig. 5, the hybridization pattern did not reveal any obvious mechanism by which the smaller products might have been generated. Nonetheless, they appeared to be fairly defined and did not represent a continuous smear of degraded RNAs.
FIG. 5.
Northern blot analysis of RNA extracted from Δγ381 particles and wt FHV particles. RNA (100 ng of Δγ381 RNA per lane; 5 ng of wt RNA per lane) was size fractionated on a 1% agarose-formaldehyde gel and transferred to a nylon membrane. Following UV cross-linking, the immobilized RNAs were hybridized to digoxigenin-UTP-labeled negative-sense RNA probes complementary to three different regions of RNA2 or RNA1. Hybridization complexes were visualized by chemiluminescence. (A) Hybridization with probes complementary to RNA2. Probe I was complementary to nt 1 to 488, probe II was complementary to nt 600 to 888, and probe III was complementary to nt 1000 to 1400. Note that full-length RNA in the Δγ381 sample migrates slightly faster than full-length wt RNA2 due to a deletion of 78 nt. (B) Hybridization with probes complementary to RNA1. Probe I was complementary to nt 1 to 980, probe II was complementary to nt 1000 to 1994, and probe III was complementary to nt 1998 to 3107. Note that samples hybridized with probe I migrated at a slight angle during gel electrophoresis, explaining the apparently larger molecular size of RNA1 in the Δγ381 sample than in the wt sample.
While full-length RNA1 and mutant RNA2 were clearly present in the collection of encapsidated nucleic acids, RNAs larger than the size of RNA1 were not detected, even though such RNAs were visible on the ethidium bromide-stained agarose gel (Fig. 4). This strongly suggested that these larger RNAs represented cellular RNAs.
Biological activity of RNA encapsidated by Δγ381 coat protein.
Despite the fact that Northern blot analysis confirmed that the Δγ381 particles contained small amounts of full-length FHV RNAs, plaque assay analysis repeatedly showed that they were not infectious. To rule out the possibility that the full-length RNAs present inside the particles were defective with regard to transcription or replication, Drosophila cells were transfected with the entire mixture of RNAs extracted from the Δγ381 particles, and total RNA was purified from the cells 24 h later. At this time, the cells showed severe cytopathic effect (not shown) indicative of active replication of the input RNA1 and RNA2. Indeed, analysis of the isolated RNAs on a denaturing agarose gel showed that RNA1, RNA2, and the subgenomic RNA3, which is derived from RNA1, were present (Fig. 6). These RNAs were unlikely to represent the input RNAs because they would not have been stable over the course of 24 h. In addition, the amounts of RNA1 and RNA2 recovered from the transfected cells were substantially higher than the amount originally introduced. Thus, the fact that Δγ381 particles were not infectious was not due to the fact that they lacked biologically active viral RNAs.
FIG. 6.
Electrophoretic analysis of total cellular RNA isolated from Drosophila cells transfected either with wt FHV RNAs or RNAs purified from Δγ381 particles. Drosophila cell monolayers (107 cells) were transfected either with 100 ng of wt RNA1 and 100 ng of capped in vitro-synthesized wt RNA2 or with approximately 1.5 μg of total RNA purified from Δγ381 particles. After 24 h, total cellular RNA was purified from the transfected cells and an aliquot of 3 μg was size fractionated on a 1% agarose-formaldehyde gel. RNAs were visualized with ethidium bromide. Lane 1, RNA molecular size markers; lane 2, total RNA from mock-transfected cells; lane 3, total RNA from cells transfected with wt FHV RNAs; lane 4, total RNA from cells transfected with RNA purified from Δγ381 particles. Note that RNA2 in this lane migrated slightly faster than wt RNA2 in lane 3 due to a deletion of 78 nt. The major band between RNA1 and RNA2 represents the two rRNAs which did not resolve under the electrophoresis conditions used here.
Expression of additional C-terminal deletion mutants.
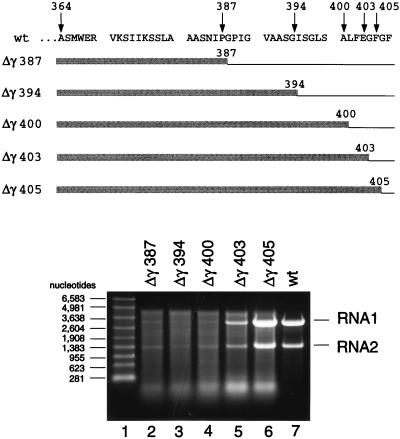
The results so far suggested that the C-terminal region of the coat protein was necessary for specific encapsidation of FHV RNA1 and RNA2. In order to define this region more precisely, we created a series of additional mutants in which the size of the deletion was gradually reduced. As shown in Fig. 7, mutants Δγ387, Δγ394, and Δγ400 had the same RNA encapsidation phenotype as Δγ381. In addition, these proteins assembled as inefficiently as the Δγ381 protein, and the resulting particles were not infectious (Table 1). A change, however, was detected for mutants Δγ403 and Δγ405. Particles assembled from Δγ403 protein contained substantially higher proportions of the viral RNAs than the previous mutants, and this proportion increased even further for the Δγ405 mutant. Remarkably, however, not even the Δγ405 protein packaged FHV RNAs as accurately as the wt protein even though it lacked only two amino acid residues at the C terminus. Both Δγ403 and Δγ405 proteins assembled with the same efficiency as the wt protein, and the resulting particles showed substantial infectivity. The specific infectivities, however, were only 11 and 44% of that of the wt, respectively (Table 1).
FIG. 7.
Electrophoretic analysis of RNA packaged by five different coat protein deletion mutants. (Top) Amino acid sequence of the wt gamma peptide (residues 364 to 407) and schematic representation of the mutant coat proteins. Thin lines indicate deleted areas, whereas shaded boxes represent maintained residues. Numbers refer to terminal residues in the mutant coat proteins. (Bottom) Denaturing agarose gel showing RNAs extracted from gradient-purified mutant particles. Particles were synthesized and purified as described in Materials and Methods, and RNA was extracted with phenol-chloroform. Aliquots were size fractionated on a denaturing 1% agarose-formaldehyde gel, and nucleic acids were visualized by staining with ethidium bromide. Lane 1, RNA molecular size markers; lane 2, RNA extracted from Δγ387 particles; lane 3, RNA extracted from Δγ394 particles; lane 4, RNA extracted from Δγ400 particles; lane 5, RNA extracted from Δγ403 particles; lane 6, RNA extracted from Δγ405 particles; lane 7, RNA extracted from wt particles.
TABLE 1.
Properties of soluble and assembled mutant and wt coat precursor proteins
| Mutant or wt coat protein alpha | % Efficiency of assemblya | Cleavage of alpha protein in assembled particles | Specific encapsidation of FHV RNA1 and RNA2 | % Infectivity of assembled virionsb |
|---|---|---|---|---|
| Δγ363 | NAc | NA | NA | NA |
| Δγ381 | 30 | Normal | + | <(1.5 × 10−6)d |
| Δγ387 | 30 | Normal | + | <(1.5 × 10−6)d |
| Δγ394 | 30 | Normal | + | <(1.5 × 10−6)d |
| Δγ400 | 30 | Normal | + | <(1.5 × 10−6)d |
| Δγ403 | 100 | Normal | ++ | 11 |
| Δγ405 | 100 | Normal | +++ | 44 |
| F402A | 100 | Normal | +++ | 8 |
| E403A | 100 | Normal | ++++ | 55 |
| F405A | 100 | Normal | +++ | 35 |
| F407A | 100 | Normal | +++ | 27 |
| wt | 100 | Normal | ++++ | 100 |
Relative to wt protein.
Relative to specific infectivity of a reference preparation of gradient-purified wt FHV (10.7 × 1010 PFU/mg).
NA, not applicable.
No plaques detected.
Analysis of single amino acid substitution mutants.
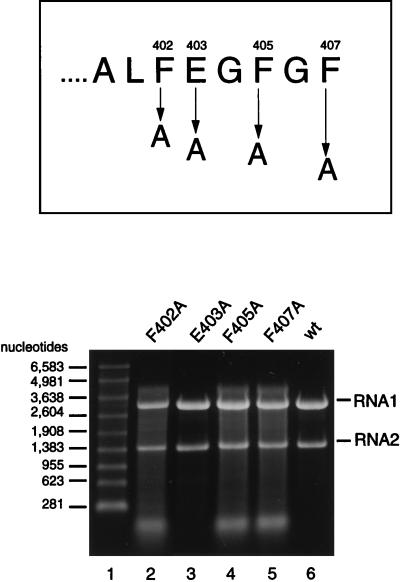
The results from the previous experiments indicated that residues 400 to 407 played a crucial role in specific recognition of RNA1 and RNA2 for assembly. Closer examination of the sequences of these residues revealed four potentially critical amino acids that we suspected might be involved in the interaction with the viral RNA: phenylalanines at positions 402, 405, and 407 and glutamic acid at position 403. To test the roles of these four residues in specific encapsidation of FHV RNAs, they were individually mutated to alanine residues. As shown in Fig. 8, replacement of any of the phenylalanine residues caused the same phenotype as observed for deletion mutants Δγ403 and Δγ405. Replacement of glutamic acid at position 403, on the other hand, had no effect. All of the single-site mutant proteins assembled with high efficiency, and the particles showed specific infectivities that ranged from 8 to 55% of that of wt particles (Table 1).
FIG. 8.
Electrophoretic analysis of RNA packaged by four different single amino acid substitution mutants. (Top) Sequence of the last eight amino acids of the gamma peptide and positions that were changed to alanine. (Bottom) Denaturing agarose gel showing RNAs extracted from gradient-purified particles. Particles were synthesized and purified as described in Materials and Methods, and RNA was extracted with phenol-chloroform. Aliquots were size fractionated on a denaturing 1% agarose-formaldehyde gel, and nucleic acids were visualized by staining with ethidium bromide. Lane 1, RNA molecular size markers; lane 2, RNA extracted from F402A particles; lane 3, RNA extracted from E403A particles; lane 4, RNA extracted from F405A particles; lane 5, RNA extracted from F407A particles; lane 6, RNA extracted from wt particles.
Effect of actinomycin D on RNA encapsidation.
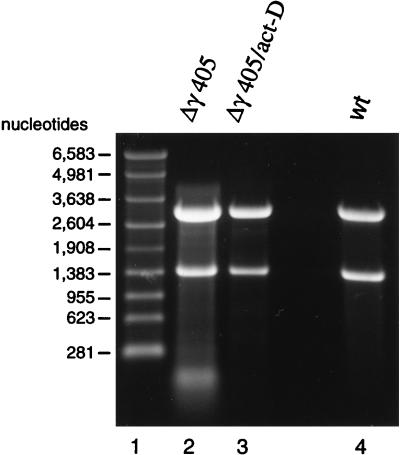
The nonviral RNAs present in the mutant FHV particles had to be of cellular origin. To prove this, we transfected Drosophila cells with FHV RNA1 and Δγ405 RNA2 and incubated the cells in the presence of actinomycin D for 20 h. Actinomycin D intercalates into cellular DNA and thereby inhibits transcription of cellular RNA. Viral RNA synthesis, on the other, is not affected (7). We reasoned that in the absence of any appreciable amount of cellular RNA, only the viral RNAs should be encapsidated into particles. Indeed, as shown in Fig. 9, Δγ405 virions generated in the presence of actinomycin D contained only viral RNAs. However, a secondary effect of the presence of actinomycin D for essentially the entire incubation period was that the yield of particles dropped to approximately 25% of that obtained in the absence of actinomycin D. Therefore, when we attempted to express the Δγ381 mutant under the same conditions, the yield of particles dropped to such low levels that we were unable to obtain sufficient RNA for visualization on ethidium bromide-stained agarose gels.
FIG. 9.
Effect of actinomycin D on RNA encapsidation by Δγ405 coat protein. Monolayers of Drosophila cells (approximately 1.2 × 108 cells) were transfected with wt RNA1 and in vitro-synthesized, capped Δγ405 RNA, as described in Materials and Methods. At 4 h after transfection, actinomycin D was added to the culture medium at a final concentration of 5 μg/ml, and incubation was continued for 20 h. Particles were then gradient purified, and RNA was extracted with phenol and chloroform. An aliquot was analyzed on a denaturing 1% agarose-formaldehyde gel followed by staining with ethidium bromide. Lane 1, RNA molecular size markers; lane 2, RNA extracted from Δγ405 particles grown in the absence of actinomycin D; lane 3, RNA extracted from Δγ405 particles grown in the presence of actinomycin D; lane 4, RNA extracted from wt FHV particles.
DISCUSSION
The initial objective of this study was to examine the role of cleavage peptide gamma in FHV uncoating and RNA delivery. To this end, we decided to use a genetic approach that relied on the generation of mutant viruses which carried alterations in the gamma chain. In the first mutant, Δγ363, the entire region representing the gamma peptide was deleted from the coat precursor protein in order to confirm that the peptide was indeed critical for viral infectivity. Unexpectedly, the Δγ363 protein did not form detectable amounts of virus particles even though it appeared to be synthesized with the same efficiency as wt protein in transfected Drosophila cells. A possible explanation for the inability of the Δγ363 protein to form particles was that the deletion caused incorrect folding of the polypeptide chain, which in turn inhibited assembly. We do not think this was likely, however, because 26 of the 44 amino acids comprising the gamma peptide are naturally flexible and only the N-terminal 18 residues form a stable secondary structure (6). This secondary structure is an amphipathic alpha helix which is located below the central β-barrel motif formed by the major coat protein beta. The helix does not appear to play a role in stabilizing the β-barrel and its interconnecting loops and furthermore does not contribute significantly to the interactions between the coat protein subunits in the assembled virion. It was therefore surprising to find that deletion of the gamma peptide inhibited formation of particles. However, the high-resolution structure of FHV shows that gamma peptides which are associated with the C subunits at the icosahedral twofold axes of the virus particle contact the sugar-phosphate backbone of the encapsidated RNA via side chains of lysine residues 371 and 375 (Fig. 1C). This interaction, which confers a certain, maybe specific, organization on the RNA within the virion, may also be critical for formation of the protein subunit interactions that are established during assembly of the virion.
Indeed, the Δγ381 protein, which contained only the alpha-helical portion of gamma, did assemble into particles, although with lower efficiency than wt protein. The majority of the particles appeared to be structurally indistinguishable from native virions based on sucrose gradient sedimentation analysis and electron micrographs of negatively stained specimens. The Δγ381 particles also matured with the same efficiency as wt virions, indicating that the quaternary interactions of the coat protein subunits in the mutant particles were very similar, if not identical, to those observed in wt virus. Despite this structural resemblance, the Δγ381 particles did not contain the normal complement of FHV RNA1 and RNA2 and were not infectious. The inability of the mutant protein to select FHV RNAs for packaging could not have been due to inadvertent deletion of a packaging signal in RNA2 because the sequences required for specific encapsidation of this RNA are located near the 5′ end and were present in the Δγ381 RNA as well as in all the other mutant RNAs used in this study (23). Furthermore, RNA2-derived defective interfering RNAs that carry substantially larger deletions in the C-terminal region of the open reading frame of protein alpha are efficiently encapsidated when wt coat protein is provided in trans (24). Thus, the logical conclusion was that the Δγ381 coat protein itself lacked elements required for specific recognition of the FHV RNAs. These elements had to be located within the seven C-terminal amino acids of protein alpha because mutants that lacked only these residues had the same phenotype as the Δγ381 protein with regard to assembly, RNA encapsidation, and infectivity.
Closer inspection of the seven C-terminal residues of the coat precursor protein revealed an “aromatic island” consisting of three phenylalanines located at positions 402, 405, and 407. We suspected that these phenylalanines might be involved in specific recognition of the viral RNAs because it is known from the high-resolution structures of other RNA-protein complexes that amino acids bearing aromatic side chains can interact with RNA via base stacking (15, 17, 18). Indeed, site-directed mutagenesis confirmed that the presence of each phenylalanine residue was critically important for specific packaging of viral RNAs into FHV particles.
Detailed insights into how viral coat protein specifically contacts the genomic RNA will have to await high-resolution structural analysis of the recognition complex. The results presented here combined with information from the atomic model of the FHV particle suggest that during assembly two types of interactions are established between the C-terminal region of coat protein alpha and FHV RNA. The first interaction is the specific recognition event and involves the flexible region of the C terminus, which is not visible in the atomic structure of the virion. Most likely the seven C-terminal residues, in particular the three phenylalanines, contact a specific secondary structure on the viral RNA to form a high-affinity RNA-protein complex. Whether such a complex is formed separately with RNA1 and RNA2 or whether recognition of one RNA is sufficient for encapsidation of the other is currently not known.
The second interaction involves the amphipathic alpha helix formed by residues 364 to 381. The side chains of lysine residues 371 and 375 nonspecifically contact the phosphodiester backbone of the viral RNA, and this interaction may be required to position the RNA correctly between the subunits such that optimal protein-protein contacts can be formed. These contacts, however, are observed only for protein subunits located at position C on the icosahedral surface lattice and not for the A and B subunits where RNA is not visible.
The specific recognition complex occurs only once or twice within the virion, and its location is not yet known. We point out, however, that in the C subunits it could be stabilized by the electrostatic interactions between the amphipathic helix and the RNA backbone. It is therefore perhaps more likely that the recognition complex would be associated with one of the twofold axes of the virion.
In addition to the specific interaction of the C terminus with the viral RNA, it is likely that the C-terminal residues of many, if not all, subunits of the virion interact with the encapsidated nucleic acid, but in a nucleotide sequence-independent manner. This hypothesis is based on the fact that the C-terminal end of the alpha protein is flexible and that RNAs can present similar recognition surfaces from distinct primary sequences (3). X-ray analysis combined with cryoelectron microscopy has already shown that all gamma peptides within the virion are in contact with bulk genomic RNA (2). This interaction undoubtedly contributes to the stability of the virus particle, but it probably also increases the efficiency of viral assembly. If so, it would explain why some of the deletion mutants did not form particles as readily as wt protein.
The data accumulated so far suggest that the gamma peptide may have at least two functions in the life cycle of FHV. As an integral component of capsid precursor protein alpha, it is required for specific recognition of the viral RNAs and probably for promoting subunit-subunit interactions during FHV assembly. This function may involve a coat protein oligomer rather than the monomeric subunit. After maturation cleavage, the gamma peptide may provide a means for release of RNA from the virion, although this has yet to be proven experimentally. The domain of gamma that is implicated in RNA release is the amphipathic alpha helix which, at the fivefold axes of the viral particle, forms a pentameric channel-like bundle with its fivefold related partners. This bundle has a hydrophobic surface and a hydrophilic core, and it has been hypothesized that it provides a conduit for the viral RNA such that it can cross a cellular membrane during the process of uncoating (2). Amino acid alignments of gamma peptides from four insect nodaviruses (Fig. 10) show that the region forming the amphipathic helix is indeed highly conserved, as one would expect for a functionally important structural module. In addition, the crystal structures of FHV, black beetle virus (BBV), and nodamuravirus (NoV) show that the viruses do contain helical bundles at equivalent locations in the virions.
FIG. 10.
Multiple alignment of amino acid sequences of gamma peptides from four insect nodaviruses: BBV, FHV, Boolarra virus (BoV), and NoV. Arrow indicates site of maturation cleavage. Shaded residues are identical in all viruses, whereas hatched residues are conserved. In BBV, FHV, and NoV, whose structures are known at high resolution, gamma peptides located at position C on the icosahedral surface lattice contact the phosphodiester backbone of encapsidated RNA via positively charged side chains at positions 371 and 375. Boxed phenylalanine residues at positions 402, 405, and 407 of FHV are required for specific encapsidation of the viral RNAs by coat precursor protein alpha.
In contrast, the amino acid sequence of the region implicated in specific recognition of the viral RNAs diverges considerably between the viruses except for FHV and BBV, which are very closely related. This sequence divergence is consistent with the fact that each coat protein has to recognize different genomic RNAs.
With the exception of the single amino acid substitution mutant E403A, all mutant coat proteins analyzed in this study displayed a reduced ability to recognize the viral nucleic acids for encapsidation. These coat proteins nonetheless packaged RNA such that the final products had the same sedimentation rate on sucrose gradients as wt virions. That the mutant particles maintained a protein-to-RNA ratio similar to that of wt virus was also suggested by the ratios of absorbance at 260 and 280 nm, which remained close to that of native virus. These results supported previous observations indicating that formation of particles requires a “headful” of RNA (10). In fact, analysis of the RNA contents of the mutant particles showed that there was a defined upper size limit close to 4,500 nucleotides (nt), which corresponds to the sum of nucleotides in FHV RNA1 (3,107 nt) and RNA2 (1,400 nt). We did not attempt to determine the identity of the nonviral RNAs, as it was not obvious what insights might be gained from that information. It seemed clear that they represented cellular RNAs, particularly as their encapsidation could be suppressed in the presence of actinomycin D. It was puzzling, however, that particles assembled from Δγ381 protein were not infectious even though they clearly contained full-length viral RNAs, albeit in small relative amounts. The same was true for other mutant particles assembled from coat proteins lacking at least seven amino acids at the C terminus. There are several possible explanations for this phenomenon. The most obvious is that the deletion in the gamma peptide rendered it incapable of releasing RNA from the virion during the uncoating step. Alternatively, RNA1 and RNA2 may have to be packaged into the same particle for productive infection of susceptible cells, and this may not have been the case for the mutant viruses. A third explanation rests on the hypothesis that for RNA to be released from the virion, it has to be packaged properly at the time of assembly. In other words, the RNA has to be precisely organized within the particle, and this organization is normally guaranteed by the fact that the initial interactions between coat protein and RNA during assembly involve a specific nucleotide sequence. In a situation in which viral RNA is randomly “stuffed” into the particle, the resulting virions may not be infectious. Determination of which of these hypotheses, if any, is correct will require further experiments. Studies are now under way in our laboratory to determine the exact nature of the coat protein-RNA recognition complex.
ACKNOWLEDGMENTS
We thank A. Ball for generously providing plasmid FHV[1,0] and M. Canady for performing electron microscopy. We also thank J. E. Johnson for critical reading of the manuscript and V. Reddy and D. Dunsmore for assistance in generating Fig. 1.
This work was supported by NIH grant GM 53491.
Footnotes
Article no. 11623-MB from The Scripps Research Institute.
REFERENCES
- 1.Ball L A. Requirements for the self-directed replication of flock house virus RNA 1. J Virol. 1995;69:720–727. doi: 10.1128/jvi.69.2.720-727.1995. . (Erratum, 69:2722.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng R H, Reddy V S, Olson N H, Fisher A J, Baker T S, Johnson J E. Functional implications of quasi-equivalence in a T=3 icosahedral animal virus established by cryo-electron microscopy and X-ray crystallography. Structure. 1994;2:271–282. doi: 10.1016/s0969-2126(00)00029-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Convery M A, Rowsell S, Stonehouse N J, Ellington A D, Murray H I J B, Peabody D S, Phillips S E V, Stockley P G. Crystal structure of an aptamer-protein complex at 2.8 A resolution. Nat Struct Biol. 1998;5:133–139. doi: 10.1038/nsb0298-133. [DOI] [PubMed] [Google Scholar]
- 4.Dasmahapatra B, Dasgupta R, Ghosh A, Kaesberg P. Structure of the black beetle virus genome and its functional implications. J Mol Biol. 1985;182:183–189. doi: 10.1016/0022-2836(85)90337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong X F, Natarajan P, Tihova M, Johnson J E, Schneemann A. Particle polymorphism caused by deletion of a peptide molecular switch in a quasi-equivalent virus. J Virol. 1998;72:6024–6033. doi: 10.1128/jvi.72.7.6024-6033.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fisher A J, Johnson J E. Ordered duplex RNA controls capsid architecture in an icosahedral animal virus. Nature. 1993;361:176–179. doi: 10.1038/361176a0. [DOI] [PubMed] [Google Scholar]
- 7.Friesen P D, Rueckert R R. Black beetle virus: messenger for protein B is a subgenomic viral RNA. J Virol. 1982;42:986–995. doi: 10.1128/jvi.42.3.986-995.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friesen P D, Rueckert R R. Synthesis of black beetle virus proteins in cultured Drosophila cells: differential expression of RNAs 1 and 2. J Virol. 1981;37:876–886. doi: 10.1128/jvi.37.3.876-886.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallagher T, Rueckert R R. Assembly-dependent maturation cleavage in provirions of a small icosahedral insect ribovirus. J Virol. 1988;62:3399–3406. doi: 10.1128/jvi.62.9.3399-3406.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallagher T M. Ph.D. thesis. University of Wisconsin, Madison; 1987. [Google Scholar]
- 11.Gallagher T M, Friesen P D, Rueckert R R. Autonomous replication and expression of RNA 1 from black beetle virus. J Virol. 1983;46:481–489. doi: 10.1128/jvi.46.2.481-489.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horton R M, Pease L R. Recombination and mutagenesis of DNA sequences using PCR. In: McPherson M J, editor. Directed mutagenesis. A practical approach. Oxford, England: IRL Press; 1991. pp. 217–247. [Google Scholar]
- 13.Hosur M V, Schmidt T, Tucker R C, Johnson J E, Gallagher T M, Selling B H, Rueckert R R. Structure of an insect virus at 3.0 Å resolution. Proteins. 1987;2:167–176. doi: 10.1002/prot.340020302. [DOI] [PubMed] [Google Scholar]
- 14.Imai Y, Matsushima Y, Sugimura T, Terada M. A simple and rapid method for generating a deletion by PCR. Nucleic Acids Res. 1991;19:2785. doi: 10.1093/nar/19.10.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johansson H E, Liljas L, Uhlenbeck O C. RNA recognition by the MS2 phage coat protein. Semin Virol. 1997;8:176–185. [Google Scholar]
- 16.Marchuk D, Drumm M, Saulino A, Collins F S. Construction of T-vector, a rapid and general system for direct cloning of unmodified PCR products. Nucleic Acids Res. 1990;19:1154. doi: 10.1093/nar/19.5.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moras D, Poterszman A. Diverse modes of recognition. The structure of a complex between the RNA-binding domain of the small nuclear ribonucleoprotein U1A and an RNA hairpin stresses the diversity of solutions to the problem of sequence-specific RNA recognition. Curr Biol. 1995;5:249–251. doi: 10.1016/s0960-9822(95)00051-0. [DOI] [PubMed] [Google Scholar]
- 18.Nagai K. RNA-protein complexes. Curr Opin Struct Biol. 1996;6:53–61. doi: 10.1016/s0959-440x(96)80095-9. [DOI] [PubMed] [Google Scholar]
- 19.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneemann A, Reddy V, Johnson J E. The structure and function of nodavirus particles: a paradigm for understanding chemical biology. Adv Virus Res. 1998;50:381–446. doi: 10.1016/s0065-3527(08)60812-x. [DOI] [PubMed] [Google Scholar]
- 21.Schneemann A, Zhong W, Gallagher T M, Rueckert R R. Maturation cleavage required for infectivity of a nodavirus. J Virol. 1992;66:6728–6734. doi: 10.1128/jvi.66.11.6728-6734.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider P A, Schneemann A, Lipkin W I. RNA splicing in Borna disease virus, a nonsegmented, negative-strand RNA virus. J Virol. 1994;68:5007–5012. doi: 10.1128/jvi.68.8.5007-5012.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong W, Dasgupta R, Rueckert R. Evidence that the packaging signal for nodaviral RNA2 is a bulged stem-loop. Proc Natl Acad Sci USA. 1992;89:11146–11150. doi: 10.1073/pnas.89.23.11146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhong W, Rueckert R R. Flock house virus: down-regulation of subgenomic RNA3 synthesis does not involve coat protein and is targeted to synthesis of its positive strand. J Virol. 1993;67:2716–2722. doi: 10.1128/jvi.67.5.2716-2722.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]