Abstract
Background
Human leukocyte antigens (HLAs) play an important role in host defense against viral infection and tumorigenesis. Human cytomegalovirus (HCMV) has been linked to glioma development. This study investigated the relationship between HLA distribution, presence of HCMV, and glioma development in a Han Chinese population.
Methods
The study population included 150 glioma patients and 150 tumor-free brain injury control subjects (control-A) matched according to geography, ethnicity, age, and gender. HLA allele frequency was compared between the two groups using peripheral blood samples by PCR sequence-based typing. These data were also compared with HLA frequencies obtained from a Northern Chinese Han population database (control-B). HCMV DNA was detected in the peripheral blood of glioma patients and control group-A by nested PCR. The expression of HCMV proteins IE1-72 and pp65 in tumor tissues was evaluated by immunohistochemistry.
Results
The frequency of HLA-A*02:01 was decreased in glioma patients as compared to control group-A and -B (P < 0.001 and P = 0.001, respectively). The age/sex-adjusted odds ratio for HLA-A*02:01 positivity vs. negativity was 0.392 (95% confidence interval 0.225–0.683). HCMV was more frequently detected in the peripheral blood and tumor tissue of HLA-A*02:01-negative glioma patients. HLA-A*02:01 and HCMV were not associated with overall survival.
Conclusion
There is a correlation between decreased HLA-A*0201 allele frequency and glioma susceptibility.
Electronic supplementary material
The online version of this article (doi:10.1007/s00262-017-2018-7) contains supplementary material, which is available to authorized users.
Keywords: Glioma, HLA, HCMV, Immunology, Immunotherapy
Introduction
Glioma is the most common primary brain tumor and has very poor prognosis [1]. To improve clinical outcome, the mechanism of gliomagenesis must be clarified so that new treatment approaches can be developed. Although it is controversial [2, 3], our research group and others have reported the presence of human cytomegalovirus (HCMV) components in the tumor tissue and peripheral blood of glioma patients [4–8]. HCMV has also been suggested to play a role in glioma pathogenesis [5, 9–11].
Human leukocyte antigens (HLAs) are widely expressed cell surface molecules responsible for antigen presentation and initiation of immune responses to viral infection [12], autoimmune diseases [13–16], and cancer [17–19]. HLA distribution has been investigated in glioma patients, and several HLA class I and II alleles were found to be positively or negatively associated with glioma occurrence and prognosis [13, 20–23]. However, these studies were mainly based on western Caucasian populations and the findings have been inconsistent; in fact, significant ethnic differences in the distribution of HLA alleles have been reported [24, 25].
To address this issue, the present study investigated HLA allele frequencies in a large cohort of glioma patients and control subjects in Northern China closely matched according to sex, age, ethnicity, and geography. We also examined the relationship between HLA genotype and other genetic factors as well as the correlation between HLA distribution, glioma occurrence, and presence of HCMV.
Materials and methods
Study population
Between January 2012 and December 2014, 209 consecutive patients were diagnosed with glioma at the Neurosurgery Department of the First Hospital of China Medical University. Patients with other diseases, including diabetes mellitus, acquired immunodeficiency syndrome, multiple sclerosis, polyarthritis, ankylosing spondylitis, and other cancers were excluded. Ultimately, 150 newly diagnosed patients were included in the study. According to the World Health Organization 2007 criteria, astrocytoma (Grade II) was diagnosed in 33 patients, anaplastic astrocytoma (Grade III) in 22 patients, and glioblastoma (GBM, Grade IV) in 95 patients by two neuropathologists from the Department of Pathology of China Medical University. Tumor samples were analyzed by Sanger sequencing and pyrosequencing to detect isocitrate dehydrogenase (IDH)1/2 mutation status and O-6 methylguanine DNA methyltransferase (MGMT) promoter methylation. All GBM patients received postoperative radio-/chemotherapy according to the Stupp protocol [1]; their overall survival (OS) was recorded and used for survival analysis.
The control group (control-A) included 150 patients diagnosed with traumatic brain injury matched with glioma patients according to age, gender, ethnicity, and geography, who underwent a computed tomography and/or magnetic resonance imaging brain scan and were found to be free of brain tumors. Exclusion criteria were the same as those applied to glioma patients.
For the second control group (control-B), HLA frequencies for healthy Northern Chinese subjects were obtained from a web database (http://www.allelefrequencies.net). HLA-A, -B, and -DRB1 typing was carried out in 618 randomly selected healthy individuals of Han ethnicity in Northern China [26]. HLA-C frequencies were determined from a dataset of 567 bone marrow donors from a Northern Chinese Han population [27]. HLA-DQB1 and -DPB1 frequencies of 171 Northern Chinese subjects have been previously reported (HLA 1991, 11th International Histocompatibility Conference Yokohoma, Japan, 1997, p. 237).
Peripheral blood was collected from each patient included in the study before any medical and surgical treatment. The study protocol, which was in compliance with the Helsinki Declaration, was approved by the institutional review board of our hospital, and written informed consent for the use of tumor tissue, blood, and clinical data for future research was obtained from each patient.
HLA genotyping
HLA genotypes were determined by PCR sequence-based typing. Briefly, DNA was extracted from whole blood using the QIAamp DNA Blood Mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Class I and II locus typing was performed using the SeCore HLA SBT kit (Invitrogen, Carlsbad, CA, USA) and results were analyzed with Sequencing Analysis software v.5.1 (Applied Biosystems, Foster City, CA, USA) and Invitrogen uTYPETM SBT software (Invitrogen).
Nested PCR analysis
HCMV DNA in peripheral blood was detected as previously described [4, 5] by nested PCR using primers specific for the HCMV glycoprotein B (UL55) gene. The sequences for external (E-1/E-2) and internal (I-1/I-2) primers were as follow: E-l, 5′-TCC AAC ACC CAC AGT ACC CGT-3′; E-2, 5′-CGG AAA CGA TGG TGT AGT TCG-3′; I-1, 5′-TGA CGG TCA AGG ATC AGT GGC-3′; and I-2, 5′-GTA AAC CAC ATC ACC CGT GGA-3′. The size of PCR products was confirmed by 2.0% agarose gel electrophoresis with ethidium bromide staining. The identity of the products was confirmed by direct DNA sequencing [4]. Each sample was tested at least in triplicate.
Immunohistochemistry
Expression of the HCMV proteins IE1-72 and pp65 in tissue samples was detected by immunohistochemistry as previously described [28, 29]. Briefly, after rehydration in a graded series of alcohol, tumor sections were digested with trypsin and antigen retrieval was carried out in citrate buffer at 90 °C for 4 min followed by 45 °C for 2.5 h. Endogenous peroxidase was blocked by incubation with 3% H2O2 for 12 min at room temperature. After Fc receptor blocking, primary antibodies against IE1-72 (1:40, MAB810; Chemicon, Temecula, CA, USA) and pp65 (1:50, clones 2 and 6; Novocastra, Newcastle-Upon-Tyne, UK) were added at 4 °C followed by overnight incubation. The results were independently analyzed by Jian Deng and Zixun Wang, who were blinded to the other results and to the clinical background of the patients. Tissue samples were visualized and photographed under a light microscope (BX-51; Olympus, Tokyo, Japan).
Serology
Anti-HCMV IgG and anti-HCMV IgM in the serum was examined using specific ELISA kits (ARG80536 and ARG80537; Arigobio, Hsinchu, Taiwan), according to the manufacturer’s instruction. Samples were diluted 1:101 with sample diluent. OD value was determined with a microplate reader at 450 nm.
Statistical analysis
HLA class I and II phenotype frequencies were determined by dividing the number of alleles of each HLA type by the total number of chromosomes. Phenotype frequency distributions of HLA alleles in patients and controls were compared with the χ 2 test with Yates continuity correction for small numbers or with Fisher’s exact test. Odds ratio (OR) with a 95% confidence interval (CI) was calculated by multivariable logistic regression to determine the association between HLA phenotype and glioma risk, adjusted for age and sex. Kaplan–Meier survival curves were generated to determine the distribution of OS according to HLA phenotype or the presence of HCMV; these were analyzed with the log-rank test. The χ 2 test, Student’s t test, and analysis of variance were used to evaluate differences between groups. Analyses were carried out using SPSS v.19.0 (SPSS Inc., Chicago, IL, USA), and a two-tailed P value <0.05 was considered statistically significant.
Results
Characteristics of study subjects
The study area was restricted to Northern China, and all subjects were of Han ethnicity. Glioma patients and control subjects were matched with respect to age and sex (Table 1). The age ranges were 22–76 years (mean 47.4 years) and 22–79 years (mean 46.5 years) for cases and controls, respectively. The male-to-female ratio was 56.7–43.3% for both groups.
Table 1.
Characteristics of glioma patients and controls
| Clinical features | Glioma cases | Controls | P b | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All | A*02:01 (+) | A*02:01 (−) | P a | All | A*02:01 (+) | A*02:01 (−) | P a | ||||||||||||||
| No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | ||||||||||
| Total | 150 | 100 | 24 | 16 | 126 | 84 | – | 150 | 100 | 49 | 32.7 | 101 | 67.3 | – | 0.001 | ||||||
| Age | 0.509 | ||||||||||||||||||||
| Mean | 47.4 | 43.8 | 47.0 | 0.208 | 46.5 | 49.6 | 46.4 | 0.139 | |||||||||||||
| SD | 12.3 | 9.7 | 11.6 | 11.4 | 13.0 | 12.0 | |||||||||||||||
| Sex | – | ||||||||||||||||||||
| Male | 85 | 56.7 | 15 | 62.5 | 70 | 55.6 | 0.529 | 85 | 56.7 | 29 | 59.2 | 56 | 55.4 | 0.665 | |||||||
| Female | 65 | 43.3 | 9 | 37.5 | 56 | 44.4 | 65 | 43.3 | 20 | 40.8 | 45 | 44.6 | |||||||||
| Gliomas | |||||||||||||||||||||
| Low-grade | 33 | 22 | 5 | 20.8 | 28 | 22.2 | 0.880 | – | – | – | – | – | – | – | |||||||
| High-grade | 117 | 78 | 19 | 79.2 | 98 | 77.8 | – | – | – | – | – | – | |||||||||
| IDH1/2 | |||||||||||||||||||||
| Mutated | 51 | 34 | 9 | 37.5 | 42 | 33.3 | 0.693 | – | – | – | – | – | – | – | |||||||
| Wild-type | 99 | 66 | 15 | 62.5 | 84 | 66.7 | – | – | – | – | – | – | |||||||||
| MGMT | |||||||||||||||||||||
| Methylated | 59 | 39.3 | 11 | 45.8 | 48 | 38.1 | 0.477 | – | – | – | – | – | – | – | |||||||
| Unmethylated | 91 | 60.7 | 13 | 54.2 | 78 | 61.9 | – | – | – | – | – | – | |||||||||
| IE1-72 | |||||||||||||||||||||
| Positive | 117 | 78 | 6 | 25 | 111 | 88.1 | 0.001 | – | – | – | – | – | – | – | |||||||
| Negative | 33 | 22 | 18 | 75 | 15 | 11.9 | – | – | – | – | – | – | |||||||||
| pp65 | |||||||||||||||||||||
| Positive | 99 | 66 | 6 | 25 | 93 | 73.8 | 0.001 | – | – | – | – | – | – | – | |||||||
| Negative | 51 | 44 | 18 | 75 | 33 | 26.2 | – | – | – | – | – | – | |||||||||
| HCMV DNA | |||||||||||||||||||||
| Positive | 48 | 32 | 2 | 8.3 | 46 | 36.5 | 0.007 | – | – | – | – | – | – | – | |||||||
| Negative | 102 | 68 | 22 | 91.7 | 80 | 63.5 | – | – | – | – | – | – | |||||||||
| HCMV IgG | 0.602 | ||||||||||||||||||||
| Positive | 112 | 74.7 | 15 | 62.5 | 97 | 77.0 | 0.135 | 108 | 72.0 | 32 | 65.3 | 76 | 75.2 | 0.203 | |||||||
| Negative | 38 | 25.3 | 9 | 37.5 | 29 | 23.0 | 42 | 28.0 | 17 | 34.7 | 25 | 24.8 | |||||||||
| HCMV IgM | 0.442 | ||||||||||||||||||||
| Positive | 28 | 18.7 | 1 | 4.2 | 27 | 21.4 | 0.047 | 23 | 15.3 | 3 | 6.1 | 20 | 19.8 | 0.029 | |||||||
| Negative | 122 | 81.3 | 23 | 95.8 | 99 | 78.6 | 127 | 84.7 | 46 | 93.9 | 81 | 80.2 | |||||||||
Statistically significant P values <0.05 are italicized
aA*02:01 (+) vs A*02:01 (−)
bCases vs controls
Association between HLA allele frequencies and glioma incidence
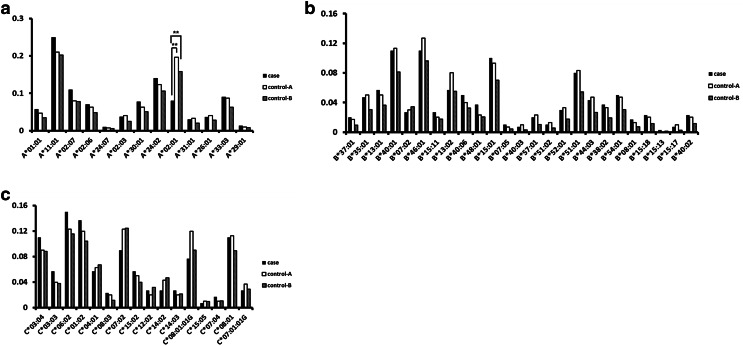
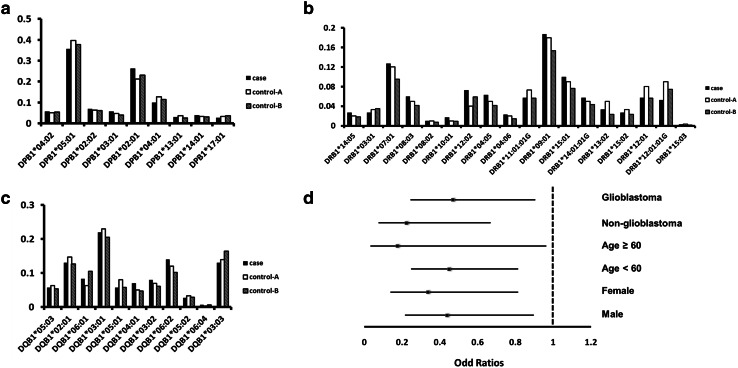
The distribution of genotype frequencies in the study population is summarized in Supplementary Tables 1–6. An analysis of HLA class I (Fig. 1) and class II (Fig. 2a–c) allele frequencies revealed a lower frequency of HLA-A*02:01 in glioma patients (0.080) as compared to control-A (0.197; P < 0.001) and control-B (0.158; P = 0.001) subjects (Fig. 1a). The case–control OR was 0.392 (95% CI 0.225–0.683), even after adjusting for age and sex (Table 2). Subsequent stratified analyses according to histological type, age, and sex showed that the HLA-A*02:01 allele was negatively associated with glioma risk in all subgroups (Fig. 2d). The distribution of HLA-A*02:01 did not vary with age or sex for either cases or controls (Table 1); in the former, HLA-A*02:01 frequency was not associated with tumor grade, IDH1/2 mutation status, or MGMT promoter methylation (Table 1).
Fig. 1.
Distribution of HLA class I alleles. The frequencies of HLA-A (a), HLA-B (b), and HLA-C (c) are shown. Control-A: 150 brain tumor-free patients diagnosed with traumatic brain injury matched with glioma patients according to age, gender, ethnicity, and geography; control-B: HLA frequencies for healthy Northern Chinese subjects obtained from a web database (http://www.allelefrequencies.net). **P < 0.01
Fig. 2.
Distribution of HLA class II alleles. The frequencies of HLA-DPB1 (a), HLA-DRB1 (b) and HLA-DQB1 (c) are shown. d Stratified analyses of case–control ORs for the HLA-A*02:01 allele expressed as categorical data. ORs were adjusted for age and sex, and are shown with 95% CIs
Table 2.
Multivariate case/control odd ratios for A*02:01 allele
| Glioma | Glioblastoma | |||||||
|---|---|---|---|---|---|---|---|---|
| Cases | Controls | ORa (95% CI) | P | Cases | Controls | ORa (95% CI) | P | |
| A*02:01 | ||||||||
| Negative | 126 | 101 | 1 | 0.001 | 76 | 62 | 1 | 0.024 |
| Positive | 24 | 49 | 0.392 (0.225–0.683) | 19 | 33 | 0.470 (0.244–0.906) | ||
Statistically significant P values <0.05 are italicized
aORs (odd ratios) were adjusted for gender, age
The frequencies of HLA-C*08:01:01G (P = 0.075), HLA-DRB1*12:02 (P = 0.077), and HLA-DRB1*12:01:01G (P = 0.082) showed marginally significant differences between cases and control-A, but not between cases and control-B (Supplementary Tables 3, 6).
Association between HLA-A*02:01 and HCMV
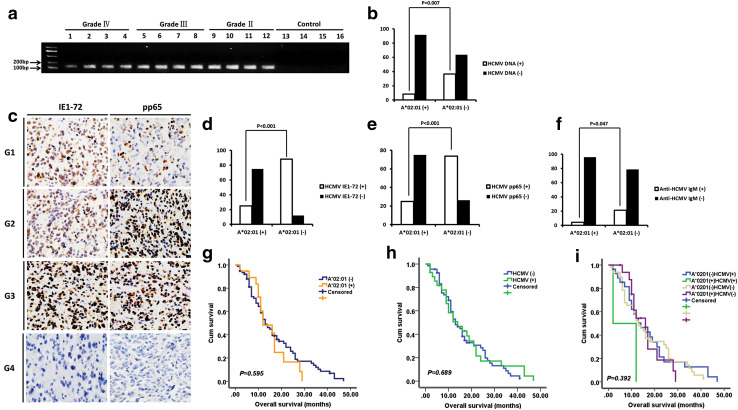
HCMV DNA was detected in the peripheral blood of 48 (32%) of 150 glioma patients (Table 1), including 11/33 (33.3%) astrocytoma, 7/22 (31.8%) anaplastic astrocytoma, and 30/95 (31.6%) GBM patients. IE1-72 immunoreactivity was observed in 117 of 150 (78%) glioma specimens, including 24/33 (72.7%) astrocytoma, 15/22 (68.2%) anaplastic astrocytoma, and 78/95 (82.1%) GBM patients; and pp65 expression was detected in 99 of 150 (66%) glioma patients, including 21/33 (63.6%) astrocytoma, 13/22 (59.1%) anaplastic astrocytoma, and 65/95 (68.4%) GBM patients. Consistent with our previous findings [5], there were no differences between different glioma grades in terms of the presence of HCMV in peripheral blood and tumor tissue, and there was no HCMV DNA detected in the control group (Fig. 3a). Notably, HCMV was more frequently detected in HLA-A*02:01-negative than in HLA-A*02:01-positive glioma patients (Fig. 3b–e; Table 1).
Fig. 3.
Association between HLA-A*02:01 and HCMV. a Detection of HCMV UL55 gene in the peripheral blood of glioma patients and control subjects by nested PCR. Grade II, Grade III, and Grade IV are different grades of glioma. b HCMV DNA was more frequently detected in the peripheral blood of HLA-A*02:01-negative than -positive glioma patients (36.5 vs. 8.3%, P = 0.007). c Representative micrographs from glioma cases showing IE1-72 and pp65 positivity (G1, G2, and G3) and negativity (G4). d, e Expression of HCMV proteins IE1-72 (d, 88.1 vs. 25%, P < 0.001) and pp65 (e, 73.8 vs. 25%, P < 0.001) was more frequently detected in tumor tissue of HLA-A*02:01-negative than -positive glioma patients. f Anti-HCMV IgM was more frequently detected in the serum of HLA-A*0201-negative than -positive glioma patients (21.4 vs. 4.2%, P = 0.047). g–i Prognostic value of HLA-A*02:01 allele and presence of HCMV DNA in 95 GBM patients, as determined by Kaplan–Meier analyses. Neither HLA-A*02:01 (g) nor HCMV (h) was associated with OS. The status of HCMV in conjunction with HLA-A*02:01 was also not correlated with GBM patient survival (i)
Anti-HCMV IgG was detected in the serum of 112 (74.7%) of 150 glioma patients [26/33 (78.8%) astrocytoma, 17/22 (77.3%) anaplastic astrocytoma and 69/95 (72.6%) GBM] and in the serum of 108 (72.0%) of 150 control subjects (P = 0.602). Anti-HCMV IgM was found in the serum of 28 (18.7%) of 150 glioma patients [5/33 (15.2%) astrocytoma, 4/22 (18.2%) anaplastic astrocytoma, 19/95 (20.0%) GBM] and in the serum of 23 (15.3%) of 150 control subjects (P = 0.442). There were no differences between different glioma grades in terms of the positivity of anti-HCMV IgG and IgM in the serum. In consistence with previous reports, neither anti-HCMV IgG nor IgM positivity was significantly associated with glioma incidence [30, 31]. However, anti-HCMV IgM was more frequently detected in HLA-A*02:01-negative than in HLA-A*02:01-positive glioma patients and control subjects (Fig. 3f; Table 1).
Prognostic value of HLA-A*02:01 allele and HCMV
OS information was available for 95 GBM patients, including 55 males (57.9%) and 40 females (42.1%) ranging in age from 24 to 76 years (mean ± SD: 47.2 ± 11.8). The mean OS was 15.2 ± 10.6 months. For GBM, the case–control OR of the HLA-A*02:01 allele was 0.470 (95% CI 0.244–0.906, P = 0.024; Table 2). HCMV DNA was detected in the peripheral blood of 30 GBM patients (31.6%). The Kaplan–Meier survival curve revealed that neither the HLA-A*02:01 allele nor the presence of HCMV DNA was correlated with GBM patient survival (Fig. 3g–h) or associated with the OS of patients, as determined by Cox regression analysis. Similar results were obtained for the HCMV proteins IE1-72 and pp65 and anti-HCMV IgG and IgM (data not shown). In addition, the status of HCMV in conjunction with HLA-A*02:01 was also not correlated with GBM patient survival (Fig. 3i).
Discussion
Multiple factors contribute to gliomagenesis, including immune and inflammatory mechanisms [32, 33]. Given that HLA plays a critical role in host immune responses, it is possible that glioma is associated with specific genotypes of the HLA system. In the present study, we found that HLA-A*02:01 frequency was reduced in glioma patients relative to control subjects. HLA-A*0201 has been detected in all populations, with a predominance in Western Caucasians. In fact, HLA-A*0201 is the most prevalent HLA type in Caucasians (about 40% as compared to 15.5% in Asians) [25]. The ubiquity of the A*0201 allele underscores its importance in protective immunity; indeed, it can specifically present glioma-associated antigenic epitopes derived from interleukin 13 receptor alpha 2 chain [34], cluster of differentiation (CD)133 [35], and 3-beta-hydroxysteroid dehydrogenase type 7 genes [36] to cytotoxic T lymphocytes (CTLs). Since HLA-A*0201 was not associated with IDH1/2 mutation status or MGMT promoter methylation, glioma is predicted to arise via other mechanisms.
A positive association has been reported between HLA-DRB1*14 and the presence of symptomatic cerebral glioma in a Northern Italian population [21]. HLA-A*11 and lower frequencies of HLA-B*07 and HLA-C*04 were associated with high-grade glioma incidence in eastern Sicily [13]. B*13 and the B*07-Cw*07 haplotype were found to be positively associated and Cw*01 was negatively associated with GBM in the San Francisco Bay area; moreover, A*32 and B*55 showed prognostic significance [22]. Another study found that HLA-A*32 was negatively associated with GBM occurrence in Midwestern US [20], and DQB1*06, DQB1*05, and DRB1*13 alleles have been linked to glioma susceptibility in the US [23]. Two studies of Japanese patients reported associations between HLA-A*24 and HLA-B*61 and glioma incidence.
There are several possible explanations for the discrepancies between these studies. (1) The subtypes of HLA-A2 and other HLA alleles were not specified in some of the studies. HLA-A2 subtypes differ by one or a few amino acids, which can have a profound effect on peptide binding characteristics and on the specificity of HLA-A2-restricted CTL clones. HLA-A2 subtype mismatch can elicit strong allogeneic responses in unrelated bone marrow transplantation [25]. Therefore, analysis of HLA allele subtype is important. (2) The glioma cases examined in each study were of different grades. The transition from low- to high-grade glioma is associated with genetic mutations; therefore, distinct pathogenetic mechanisms may underlie different grades of glioma. However, we found no significant differences in HLA-A*0201 distribution among different glioma grades; similar results were obtained when we separately analyzed GBM cases and controls. (3) The selection of control subjects differed among studies; some used an online database with a large sample size, and others recruited closely matched patients. To eliminate this confound, we compared our cases to closely matched control patients (control-A) as well as to data from an online database (control-B), with no significant difference observed between the two control groups. (4) Previous studies have examined glioma cases and controls from different populations and ethnic groups; it is well established that there are ethnic as well as geographic differences in the distribution of HLA alleles, with the latter possibly arising from selection pressures in response to local pathogens [25].
The HLA region is known for its linkage disequilibrium, therefore, another genetic entity linked with HLA-A could be responsible for decreased HLA-A*0201 frequency. For example, TNFA and TNFB is located on chromosome 6 and is in close proximity of HLA-A. Decreased TNF alleles, such as TNFB4, have been associated with glioma risk [37]. Thus, linkage disequilibrium of TNF with HLA-A might influence HLA-A*0201 allele frequency. Other potential glioma-related genes located in the proximity chromosome 6 that might affect HLA-A*0201 frequency include VEGFA [38], PARK2 [39] and CDKN1A [40]. Further research is required to explore the possible internal correlation.
HCMV infection and expression have been linked to glioma initiation and progression [5, 10, 32], although the underlying mechanism is not well understood. We found that HCMV was present in the tumor tissue and peripheral blood of HLA-A*0201-negative glioma patients. The HLA-A*0201 gene product presents peptides from several intracellular pathogens, including influenza, Epstein–Barr, and hepatitis B viruses as well as malaria [12, 25]. HLA-A2 mutations negatively affected the binding of various viral peptides [41]. Whether the absence of the HLA-A*0201 allele facilitates HCMV infection and promotes gliomagenesis remains to be determined. The presence of anti-HCMV IgM in the serum is known to indicate a recent HCMV infection or reactivation. We found that anti-HCMV IgM was more frequently detected in HLA-A*0201-negative individuals. Thus, our results support that HLA-A*0201-negative individuals may be more susceptible to HCMV, possibly due to inadequate HLA-A*0201-restricted T cell immunity. In this study, we cannot ascertain whether the HCMV components were present before the development of glioma or whether they may be a product of glioma progression. We showed that the positivity of anti-HCMV IgM was similar between glioma patients and control subjects, and so was anti-HCMV IgG, in consistency with previous reports [30, 31]. Therefore, it seems unlikely that the presence of HCMV components in the patients was due to the onset of HCMV secondary to the appearance of glioma, in which case we would expect more anti-HCMV IgM positive cases in the glioma patients than in control subjects. The fact that the frequency of the HLA-A*0201 allele and presence of HCMV components did not vary with tumor grade and were not associated with glioma prognosis suggests that these two factors are associated with glioma pathogenesis but not progression. On the other hand, HCMV is known to inhibit HLA-A and -B expression and impair viral antigen presentation to CD8+ T cells [42]. Hence, it is possible that HCMV may be involved in the development of HLA polymorphisms in glioma patients.
Preclinical studies of glioma immunotherapy have attempted to identify HLA-A*02:01-restricted CTL epitopes [34–36]. Our findings indicate that these epitopes are not suitable for immunotherapy of Northern Chinese glioma patients in whom HLA-A*02:01 allele frequency was markedly decreased. Since HCMV is detected at a higher frequency in HLA-A*02:01-negative glioma patients, these individuals may be more responsive to HCMV-specific anti-tumor immunotherapy [43, 44] or antiviral therapy [45] than those who are positive for the HLA-A*02:01 allele. Consistent with our observations, one group described a GBM patient expressing HCMV pp65 in tumor tissue who developed a robust HCMV-specific CD8+ T cell response; however, another HLA-A*02:01 + GBM patient failed to show an increased CMV-specific anti-tumor immune response after vaccination, and HCMV pp65 was not detected in the tumor sample [43].
Conclusion
The results of this study indicate a possible correlation between decreased HLA-A*0201 allele frequency and glioma susceptibility.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Jingpu Shi at the Department of Clinical Epidemiology, The First Affiliated Hospital of China Medical University, for assistance with statistical and survival analyses.
Abbreviations
- CI
Confidence interval
- GBM
Glioblastoma
- HCMV
Human cytomegalovirus
- HLA
Human leukocyte antigen
- IDH
Isocitrate dehydrogenase
- IgG
Immunoglobulin G
- IgM
Immunoglobulin M
- MGMT
O-6 methylguanine DNA methyltransferase
- OR
Odds ratio
- OS
Overall survival
Compliance with ethical standards
Funding
This work was supported by Grants from National Natural Science Foundation of China (Grant Nos. 81472360 and 81402045) and the Science and Technology Department of Liaoning Province (No. 2011225034).
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
References
- 1.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Baumgarten P, Michaelis M, Rothweiler F, Starzetz T, Rabenau HF, et al. Human cytomegalovirus infection in tumor cells of the nervous system is not detectable with standardized pathologico-virological diagnostics. Neuro Oncol. 2014;16:1469–1477. doi: 10.1093/neuonc/nou167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strong MJ, Blanchard E, 4th, Lin Z, Morris CA, Baddoo M, et al. A comprehensive next generation sequencing-based virome assessment in brain tissue suggests no major virus—tumor association. Acta Neuropathol Commun. 2016;4:71. doi: 10.1186/s40478-016-0338-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchell DA, Xie W, Schmittling R, Learn C, Friedman A, et al. Sensitive detection of human cytomegalovirus in tumors and peripheral blood of patients diagnosed with glioblastoma. Neuro Oncol. 2008;10:10–18. doi: 10.1215/15228517-2007-035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding D, Han S, Wang Z, Guo Z, Wu A. Does the existence of HCMV components predict poor prognosis in glioma. J Neurooncol. 2014;116:515–522. doi: 10.1007/s11060-013-1350-9. [DOI] [PubMed] [Google Scholar]
- 6.Dziurzynski K, Chang SM, Heimberger AB, Kalejta RF, McGregor DSR, et al. Consensus on the role of human cytomegalovirus in glioblastoma. Neuro Oncol. 2012;14:246–255. doi: 10.1093/neuonc/nor227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohammad AA, Rahbar A, Lui WO, Davoudi B, Catrina A, et al. Detection of circulating hcmv-miR-UL112-3p in patients with glioblastoma, rheumatoid arthritis, diabetes mellitus and healthy controls. PLoS One. 2014;9:e113740. doi: 10.1371/journal.pone.0113740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.dos Santos CJ, Stangherlin LM, Figueiredo EG, Correa C, Teixeira MJ, da Silva MC. High prevalence of HCMV and viral load in tumor tissues and peripheral blood of glioblastoma multiforme patients. J Med Virol. 2014;86:1953–1961. doi: 10.1002/jmv.23820. [DOI] [PubMed] [Google Scholar]
- 9.Fajfr M, Stepanova V. Cytomegalovirus and its relationship to chronic inflammatory bowel diseases and tumors. Klin Mikrobiol Infekc Lek. 2013;19:103–106. [PubMed] [Google Scholar]
- 10.Cobbs CS. Cytomegalovirus and brain tumor: epidemiology, biology and therapeutic aspects. Curr Opin Oncol. 2013;25:682–688. doi: 10.1097/CCO.0000000000000005. [DOI] [PubMed] [Google Scholar]
- 11.Price RL, Song J, Bingmer K, Kim TH, Yi JY, et al. Cytomegalovirus contributes to glioblastoma in the context of tumor suppressor mutations. Cancer Res. 2013;73:3441–3450. doi: 10.1158/0008-5472.CAN-12-3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang HG, Pang XW, Shang XY, Xing Q, Chen WF. Functional supertype of HLA-A2 in the presentation of Flu matrix p58–66 to induce CD8+ T-cell response in a Northern Chinese population. Tissue Antigens. 2003;62:285–295. doi: 10.1034/j.1399-0039.2003.00102.x. [DOI] [PubMed] [Google Scholar]
- 13.La Torre D, Maugeri R, Angileri FF, Pezzino G, Conti A, et al. Human leukocyte antigen frequency in human high-grade gliomas: a case–control study in Sicily. Neurosurgery. 2009;64:1082–1088. doi: 10.1227/01.NEU.0000345946.35786.92. [DOI] [PubMed] [Google Scholar]
- 14.Hou TY, Chen HC, Chen CH, Chang DM, Liu FC, Lai JH. Usefulness of human leucocyte antigen-B27 subtypes in predicting ankylosing spondylitis: Taiwan experience. Intern Med J. 2007;37:749–752. doi: 10.1111/j.1445-5994.2007.01450.x. [DOI] [PubMed] [Google Scholar]
- 15.Ballerini C, Guerini FR, Rombola G, Rosati E, Massacesi L, et al. HLA-multiple sclerosis association in continental Italy and correlation with disease prevalence in Europe. J Neuroimmunol. 2004;150:178–185. doi: 10.1016/j.jneuroim.2004.01.015. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez-Gay MA, Garcia-Porrua C, Hajeer AH. Influence of human leukocyte antigen-DRB1 on the susceptibility and severity of rheumatoid arthritis. Semin Arthritis Rheum. 2002;31:355–360. doi: 10.1053/sarh.2002.32552. [DOI] [PubMed] [Google Scholar]
- 17.Luongo V, Pirozzi G, Caraco C, Errico S, de Angelis F, et al. HLA allele frequency and clinical outcome in Italian patients with cutaneous melanoma. Tissue Antigens. 2004;64:84–87. doi: 10.1111/j.0001-2815.2004.00250.x. [DOI] [PubMed] [Google Scholar]
- 18.Ohtani M, Azuma T, Yamazaki S, Yamakawa A, Ito Y, et al. Association of the HLA-DRB1 gene locus with gastric adenocarcinoma in Japan. Dig Liver Dis. 2003;35:468–472. doi: 10.1016/S1590-8658(03)00218-4. [DOI] [PubMed] [Google Scholar]
- 19.Younger AR, Amria S, Jeffrey WA, Mahdy AE, Goldstein OG, et al. HLA class II antigen presentation by prostate cancer cells. Prostate Cancer Prostatic Dis. 2008;11:334–341. doi: 10.1038/sj.pcan.4501021. [DOI] [PubMed] [Google Scholar]
- 20.Song W, Ruder AM, Hu L, Li Y, Ni R, et al. Genetic epidemiology of glioblastoma multiforme: confirmatory and new findings from analyses of human leukocyte antigen alleles and motifs. PLoS One. 2009;4:e7157. doi: 10.1371/journal.pone.0007157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guerini FR, Agliardi C, Zanzottera M, Delbue S, Pagani E, et al. Human leukocyte antigen distribution analysis in North Italian brain Glioma patients: an association with HLA-DRB1*14. J Neurooncol. 2006;77:213–217. doi: 10.1007/s11060-005-9032-x. [DOI] [PubMed] [Google Scholar]
- 22.Tang J, Shao W, Dorak MT, Li Y, Miike R, et al. Positive and negative associations of human leukocyte antigen variants with the onset and prognosis of adult glioblastoma multiforme. Cancer Epidemiol Biomark Prev. 2005;14:2040–2044. doi: 10.1158/1055-9965.EPI-05-0136. [DOI] [PubMed] [Google Scholar]
- 23.Bassig BA, Inskip PD, Burdette L, Shapiro WR, Selker RG, et al. Selected human leukocyte antigen class II polymorphisms and risk of adult glioma. J Neuroimmunol. 2011;233:185–191. doi: 10.1016/j.jneuroim.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krausa P, Brywka M, 3rd, Savage D, Hui KM, Bunce M, et al. Genetic polymorphism within HLA-A*02: significant allelic variation revealed in different populations. Tissue Antigens. 1995;45:223–231. doi: 10.1111/j.1399-0039.1995.tb02444.x. [DOI] [PubMed] [Google Scholar]
- 25.Liang B, Zhu L, Liang Z, Weng X, Lu X, et al. A simplified PCR-SSP method for HLA-A2 subtype in a population of Wuhan, China. Cell Mol Immunol. 2006;3:453–458. [PubMed] [Google Scholar]
- 26.Yang G, Deng YJ, Hu SN, Wu DY, Li SB, et al. HLA-A, -B, and -DRB1 polymorphism defined by sequence-based typing of the Han population in Northern China. Tissue Antigens. 2006;67:146–152. doi: 10.1111/j.1399-0039.2006.00529.x. [DOI] [PubMed] [Google Scholar]
- 27.Deng Z, Wang D, Xu Y, Gao S, Zhou H, et al. HLA-C polymorphisms and PCR dropout in exons 2 and 3 of the Cw*0706 allele in sequence-based typing for unrelated Chinese marrow donors. Hum Immunol. 2010;71:577–581. doi: 10.1016/j.humimm.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 28.Scheurer ME, Bondy ML, Aldape KD, Albrecht T, El-Zein R. Detection of human cytomegalovirus in different histological types of gliomas. Acta Neuropathol. 2008;116:79–86. doi: 10.1007/s00401-008-0359-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cobbs CS, Matlaf L, Harkins LE. Methods for the detection of cytomegalovirus in glioblastoma cells and tissues. Methods Mol Biol. 2014;1119:165–196. doi: 10.1007/978-1-62703-788-4_11. [DOI] [PubMed] [Google Scholar]
- 30.Sjostrom S, Hjalmars U, Juto P, Wadell G, Hallmans G, et al. Human immunoglobulin G levels of viruses and associated glioma risk. Cancer Causes Control. 2011;22:1259–1266. doi: 10.1007/s10552-011-9799-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amirian ES, Marquez-Do D, Bondy ML, Scheurer ME. Anti-human-cytomegalovirus immunoglobulin G levels in glioma risk and prognosis. Cancer Med. 2013;2:57–62. doi: 10.1002/cam4.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soroceanu L, Cobbs CS. Is HCMV a tumor promoter. Virus Res. 2011;157:193–203. doi: 10.1016/j.virusres.2010.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hambardzumyan D, Gutmann DH, Kettenmann H. The role of microglia and macrophages in glioma maintenance and progression. Nat Neurosci. 2016;19:20–27. doi: 10.1038/nn.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okano F, Storkus WJ, Chambers WH, Pollack IF, Okada H. Identification of a novel HLA-A*0201-restricted, cytotoxic T lymphocyte epitope in a human glioma-associated antigen, interleukin 13 receptor alpha2 chain. Clin Cancer Res. 2002;8:2851–2855. [PubMed] [Google Scholar]
- 35.Ji J, Judkowski VA, Liu G, Wang H, Bunying A, et al. Identification of novel human leukocyte antigen-A*0201-restricted, cytotoxic T lymphocyte epitopes on CD133 for cancer stem cell immunotherapy. Stem Cells Transl Med. 2014;3:356–364. doi: 10.5966/sctm.2013-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Myers CE, Hanavan P, Antwi K, Mahadevan D, Nadeem AJ, et al. CTL recognition of a novel HLA-A*0201-binding peptide derived from glioblastoma multiforme tumor cells. Cancer Immunol Immunother. 2011;60:1319–1332. doi: 10.1007/s00262-011-1032-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frigerio S, Ciusani E, Pozzi A, Silvani A, Salmaggi A, Boiardi A. Tumor necrosis factor microsatellite polymorphisms in Italian glioblastoma patients. Cancer Genet Cytogenet. 1999;109:172–174. doi: 10.1016/S0165-4608(98)00161-7. [DOI] [PubMed] [Google Scholar]
- 38.Li R, Zhao Y, Fan W, Chen H, Chen Y, et al. Possible association between polymorphisms of human vascular endothelial growth factor A gene and susceptibility to glioma in a Chinese population. Int J Cancer. 2011;128:166–175. doi: 10.1002/ijc.25306. [DOI] [PubMed] [Google Scholar]
- 39.Yin D, Ogawa S, Kawamata N, Tunici P, Finocchiaro G, et al. High-resolution genomic copy number profiling of glioblastoma multiforme by single nucleotide polymorphism DNA microarray. Mol Cancer Res. 2009;7:665–677. doi: 10.1158/1541-7786.MCR-08-0270. [DOI] [PubMed] [Google Scholar]
- 40.Tenan M, Carrara F, DiDonato S, Finocchiaro G. Absence of mutations and identification of two polymorphisms in the SSCP and sequence analysis of p21CKI gene in malignant gliomas. Int J Cancer. 1995;62:115–117. doi: 10.1002/ijc.2910620121. [DOI] [PubMed] [Google Scholar]
- 41.Tussey LG, Matsui M, Rowland-Jones S, Warburton R, Frelinger JA, McMichael A. Analysis of mutant HLA-A2 molecules. Differential effects on peptide binding and CTL recognition. J Immunol. 1994;152:1213–1221. [PubMed] [Google Scholar]
- 42.Zou Y, Bresnahan W, Taylor RT, Stastny P. Effect of human cytomegalovirus on expression of MHC class I-related chains A. J Immunol. 2005;174:3098–3104. doi: 10.4049/jimmunol.174.5.3098. [DOI] [PubMed] [Google Scholar]
- 43.Prins RM, Cloughesy TF, Liau LM. Cytomegalovirus immunity after vaccination with autologous glioblastoma lysate. N Engl J Med. 2008;359:539–541. doi: 10.1056/NEJMc0804818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schuessler A, Smith C, Beagley L, Boyle GM, Rehan S, et al. Autologous T-cell therapy for cytomegalovirus as a consolidative treatment for recurrent glioblastoma. Cancer Res. 2014;74:3466–3476. doi: 10.1158/0008-5472.CAN-14-0296. [DOI] [PubMed] [Google Scholar]
- 45.Stragliotto G, Rahbar A, Solberg NW, Lilja A, Taher C, et al. Effects of valganciclovir as an add-on therapy in patients with cytomegalovirus-positive glioblastoma: a randomized, double-blind, hypothesis-generating study. Int J Cancer. 2013;133:1204–1213. doi: 10.1002/ijc.28111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.