Abstract
CD40 is a member of the tumor necrosis factor receptor family. We reveal here a correlation between CD40 expression and colon cancer differentiation. Upon CD40 ligand (CD40L) binding, CD40/CD40L signaling inhibited colon cancer proliferation, induced apoptosis, stalled cells at G0/G1, and influenced cell adhesion and metastasis. Clustering analysis identified the elevation of aryl hydrocarbon receptor repressor (AHRR) expression along with activation of CD40/CD40L signaling. Examination of clinical specimens revealed that both AHR and AHRR levels correlated with colon cancer histological grade. In addition, high expression of AHRR was associated with high expression of CD40 in tumor cells, with CD40L expression being particularly high in the tumor interstitium. Real-time PCR and western blotting analysis showed that AHRR expression in colon cancer cells was up-regulated by CD40L binding. The likely mediating signaling pathways for the effects of CD40 are described herein.
Keywords: Colon cancer, CD40 ligand, CD40, Aryl hydrocarbon receptor repressor, Aryl hydrocarbon receptor
Introduction
CD40, a 48- kDa type I transmembrane glycoprotein, belongs to the tumor necrosis factor receptor superfamily. CD40 was first identified and functionally characterized in an array of immune system-associated cells, such as B cells [1]. After binding the CD40 ligand (CD40L), CD40–CD40L crosslinking activates B cells, providing a second stimulus to promote their proliferation [2]. Additionally, CD40L binding to antigen-presenting cells promotes their maturation, increases the expression of major histocompatibility complex molecules, and activates the proliferation of T cells [3]. Ligation of CD40L to APCs can also increase the cytolytic function of natural killer (NK) cells and cytokine induction via natural killer T cells [4].
Subsequent studies have found that CD40 is expressed in many epithelial cell carcinomas and all B cell malignancies [5–9]. The interaction of CD40L with CD40 in myeloma cells has been shown to induce tumor cell proliferation, migration, and clone formation through activation of the PI3K/AKT/NF-κB signaling pathway [10]. Tai et al. [11] reported that CD40 induces vascular endothelial growth factor secretion and myeloma cell migration, which indicated that CD40 activation plays a functional role in myeloma angiogenesis and homing. These studies suggest that CD40 activation may promote the progression of malignant tumors originating from B cells. However, in contrast to the proliferative effects of CD40 signaling in myeloma, CD40 stimulation in solid tumor cells results in growth inhibition both in vivo and in vitro. Gomes et al. [12] demonstrated CD40 expression in human breast cancer and that CD40L binding could inhibit cell growth. Li et al. [13] reported that activating the CD40 pathway in gastric cancer cells with CD40L inhibited cell proliferation and induced apoptosis. Similarly, CD40 was highly expressed in ovarian cancer, and concomitant use of anti-CD40 antibody and CD40L inhibited the proliferation and induced apoptosis of ovarian cancer cells [7]. Furthermore, Sabel et al. [14] suggested that CD40 expression might be a prognostic marker in lung cancer, where its expression level correlates with metastasis and poor prognosis.
In this study, we investigated the effects of CD40 expression and CD40/CD40L signaling in human colon cancer tissue and cell lines. Using a gene expression microarray, we identified significantly differentially expressed proteins in carcinoma cells treated with CD40L. Furthermore, we characterized the expression of one of the proteins identified via clustering analysis in relation to CD40 expression and signaling.
Materials and methods
Cell lines and colon cancer tissue
The established human colon cancer cell lines HCT116, HT29, SW480, SW620, and Lovo were obtained from the American Type Cell Collection (ATCC, Manassas, VA, USA). All of the colon cancer cell lines were cultured in DMEM (Hyclone, Utah, USA) supplemented with 10 % fetal calf serum (SAFC, St. Louis, MO) at 37 °C under a 5 % CO2 atmosphere. Tissue and blood were collected from the First Affiliated Hospital of Chengdu Medical College. All patients and donors provided signed consent before the study. The study was approved by the institutional ethics committee of Chengdu Medical College (No. CYFYLL2014010).
Reagents
The recombinant soluble human CD40 ligand (rshCD40L) was purchased from Peprotech (Rocky Hill, NJ, USA). The CD40 antibody was obtained from GeneTex (Irvine, CA, USA). The CD40L antibody was obtained from Cell signal (Boston, Massachusetts, USA). The AHRR and AHR antibodies were obtained from Abcam (Cambridge, England, UK). HRP-conjugated secondary antibodies for western blotting were purchased from Zhongshan Biological (Beijing, China). FITC-conjugated AffiniPure goat anti-rabbit IgG (H + L), FITC-conjugated AffiniPure goat anti-mouse IgG (H + L), 3H-indocyanine (Cy3)-conjugated AffiniPure goat anti-rabbit IgG (H + L), Cy3-conjugated AffiniPure goat anti-mouse IgG (H + L), and E-cadherin antibody were all obtained from Proteintech (Chicago, Illinois, USA). Allophycocyanin (APC)-conjugated anti-human CD40L antibody was obtained from eBioscience (San Diego, California, USA).
Immunohistochemistry
Briefly, four consecutive paraffin slices were incubated with the individual primary antibodies (CD40-antibody, CD40L-antibody, AHRR-antibody, and AHR-antibody) overnight at 4 °C. Negative controls were performed with isotype control antibody. Sections were incubated with polymer helper solution (Zhongshan Biological, Beijing, China) for 1 h at 37 °C, followed by washing twice with PBS. The sections were incubated with polyperoxidase-anti-mouse/rabbit IgG (Zhongshan Biological, Beijing, China) at room temperature for 20 min. 3,3-Diaminobenzidine substrate solution was used to determine peroxidase activity, and the slides were counterstained with hematoxylin. The intensity and saturation of immunostained cells were evaluated under a microscope (Leica, Wetzlar, Germany) over random fields. For data analysis, the staining level of the target proteins (CD40, CD40L, AHRR, and AHR) was calculated by multiplying the intensity of staining by the ratio of positively stained cells. The observation and evaluation were performed by two independent investigators.
Immunofluorescence
The frozen sections were fixed using acetone solution and then blocked in goat serum for 30 min. For detection of CD40 expression in colon tumor cells, the sections were incubated with mouse anti-human E-cadherin antibody (1:200) and rabbit anti-human CD40 antibody (1:200) overnight at 4 °C. Next, the Cy3-labeled goat anti-rabbit IgG (1:200) and FITC-labeled goat anti-mouse IgG (1:200) were added and the sections were further incubated for 1 h at 37 °C. For detection of the co-association among CD40L, CD40, and AHRR, one of two consecutive slices was incubated with rabbit anti-human AHRR (1:200) and mouse anti-human CD40 antibody (1:200) overnight at 4 °C. Subsequently, FITC-labeled goat anti-rabbit IgG (1:200) and Cy3-labeled goat anti-mouse IgG (1:500) were added, and the section was further incubated for 1 h at 37 °C. The other section was incubated with APC-labeled mouse anti-human CD40L (1:200) for 1 h at 37 °C. The sections were mounted with DAPI and observed under a fluorescence microscope (Leica, Wetzlar, Germany).
Flow cytometric detection
The expression of CD40 or CD40L on the cell surface was assessed by flow cytometry. Briefly, 1 × 106 colon cancer cells were washed and resuspended with PBS buffer. The cells were then incubated with mouse anti-human CD40 antibody or isotype control antibody at room temperature for 30 min, followed by washing with PBS. The suspensions were incubated with FITC-conjugated sheep anti-mouse IgG at room temperature for another 30 min. For detection of the expression of CD40L in peripheral blood mononuclear cells extracted by density gradient centrifugation, the APC-conjugated mouse anti-human CD40L-antibody was used. A C6 flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) was used to analyze the expression of CD40 or CD40L. CD40L can be expressed in T lymphocytes, granulocytes, and NK cells. Therefore, in the data analysis, excluding cell debris, lymphocytes, monocytes, and granulocytes are all included in the scope of analysis. The four-quadrant gate was set up according to the control group (isotype control antibody). And then the quadrant gate was coped into the experimental group for quantification the expression of CD40L.
Western blotting
The extracted total proteins were separated by 10 % SDS-PAGE and then transferred to a polyvinylidene fluoride membrane. The polyvinylidene fluoride membrane was incubated with primary antibodies (anti-CD40 antibody, 1:1000 and anti-AHRR-antibody, 1:2000). An HRP-conjugated antibody was used for detection. Bands were visualized using the enhanced chemiluminescence system (GE Healthcare, Uppsala, Sweden).
Enzyme-linked immunosorbent assay for measurement of CD40L
Peripheral blood was extracted from colon cancer patients and healthy donors. The colon cancer patients included 19 men (55.9 %) and 15 women (44.1 %) with a median age of 66 years (range 40–87 years). The healthy donors (controls) included 18 men (52.9 %) and 16 women (47.1 %), with a median age of 62 years (range 35–80 years). The blood samples were stored at 4 °C overnight and subsequently were centrifuged at 1200×g for 10 min. Serum was collected and subjected to enzyme-linked immunosorbent assay to determine the serum CD40L concentration. An enzyme-linked immunosorbent assay kit for CD40L (RayBiotech, Atlanta, Georgia, USA) was used, and the assay was performed according to the manufacturer’s protocol. Colorimetric changes were measured using a microplate reader (powerWave XS2, BioTek, Vermont, USA) at a wavelength of 450 nm.
MTT assay
Colon cancer cell growth was measured by MTT assay. Briefly, colon cancer cells (2–5 × 103 cells/well) were plated in 96-well plates. After 16 h, the cells were co-cultured with rshCD40L or vehicle control for 48 h. Subsequently, 20 μL of MTT (10 mg/mL) was added to each well, and the plates were incubated for 4 h in incubator. The supernatant was removed and the formed formazan crystals were dissolved using 200 μL of dimethyl sulfoxide. The absorbance was measured at 450 nm using a BD microplate reader. The inhibition of colon cancer growth was determined from the differences in optical densities between experimental and control wells as a percentage of the control.
Cell cycle and apoptosis analysis by flow cytometry
Flow cytometry was used to analyze cell cycle and apoptosis. Cells were categorized as control, rshCD40L (0.1 μg/mL), or rshCD40L (1 μg/mL) groups. For the apoptosis analysis, after incubation for 48 h, the cells were stained with Annexin V-FITC and propidium iodide buffer in the dark for 20 min. For cell cycle analysis, cells were fixed in 2 mL of cold 70 % ethanol overnight at 4 °C. Subsequently, cell pellets were incubated with RNase and propidium iodide mixture for 30 min at 37 °C. Apoptotic cells and cell cycle distribution were analyzed using a flow cytometer.
Cell migration and adhesion assay
For the adhesion assay, 1 × 105 cells/well were pre-incubated with rshCD40L and plated in a 96-well plate, which was pre-coated with Matrigel. The unattached cells were removed by washing with PBS after incubation for 4 h. Next, the attached cells were fixed with 4 % paraformaldehyde for 30 min. 0.02 % crystal violet dye was used for staining of cells. Four random fields viewed under a Leica DM4000B microscope were photographed. The crystal violet was dissolved with 70 % ethanol and measured using a microplate reader at 570 nm to quantify the number of attached cells. Cell migration assays were performed in transwell plates. Cells were plated into the upper chamber, and cells attached to the lower side were fixed in 4 % paraformaldehyde and stained using 0.5 % crystal violet. The migrated cells were counted in five microscopic fields.
Microarray analysis
The colon cancer cells (HT29) were cultured for 6 h with rshCD40L (1 μg/mL) or without rshCD40L, and the total RNA was extracted from the two groups. RNA quality and quantity were measured using a Fast prep-1 nanophotometer (Implen, Munich, Germany). Agarose gel electrophoresis was used to assess RNA integrity. Total RNA was amplified and labeled using the One-Color Low Input Quick Amp labeling kit (Agilent Technologies, Santa Clara, CA, US), following the manufacturer’s instructions. Labeled cRNA was purified by RNeasy mini kit (Qiagen, GmBH, Germany). Each slide was hybridized with 1.65 μg of Cy3-labeled cRNA using a gene expression hybridization kit (Agilent Technologies, Santa Clara, CA, US) in a hybridization oven. After hybridization for 17 h, slides were washed in staining dishes using a gene expression wash buffer kit (Agilent Technologies, Santa Clara, CA, US). Slides were scanned using an Agilent microarray scanner (Agilent Technologies, Santa Clara, CA, US). Data were extracted with Feature Extraction software 10.7 (Agilent Technologies, Santa Clara, CA, US). Raw data were normalized using a quantile algorithm in Gene Spring Software 11.0 (Agilent Technologies, Santa Clara, CA, US).
Real-time PCR
Real-time PCR was conducted using a 10-μL reaction mixture comprising 5 μL of SYBR® Select Master mix (Invitrogen, Carlsbad, USA), 1 μL of each primer, and 3 μL of cDNA template. All samples were amplified in a 96-tube plate in an CFX96 real-time system (Bio-Rad, Hercules, California, USA).
Statistical analysis
Data were expressed as the mean ± SD from at least three independent experiments. The statistical differences between groups were analyzed using one- or two-way analysis of variance by SPSS version 16.0 (Chicago, IL, USA). p < 0.05 was considered as statistically significant.
Results
CD40 expression in colon cancer
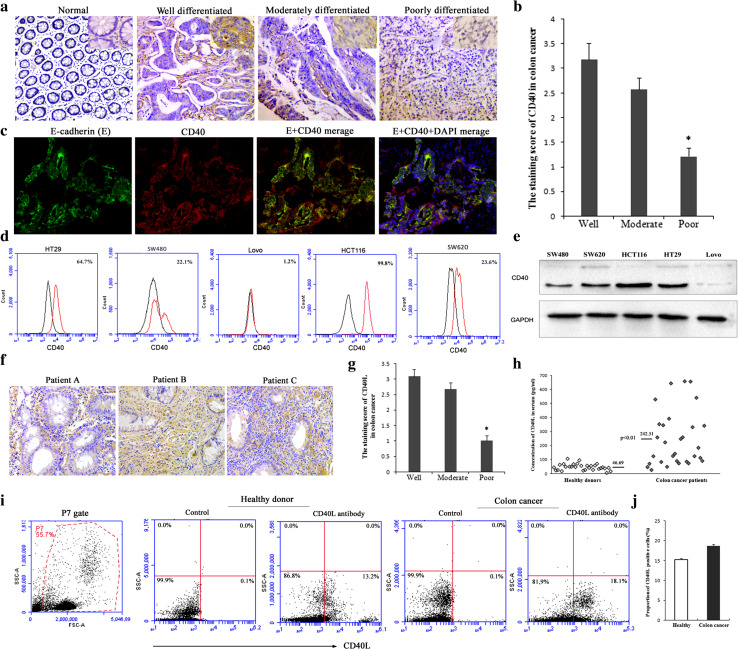
Expression of CD40 in 110 colon cancer tissue samples was investigated by immunohistochemistry and was detected in 87 (79 %) cases, but not in any of the 20 normal colon tissues analyzed (p < 0.001). Figure 1a shows four representative examples of the spectrum of normal colon and colon cancer indifferent stages of differentiation. The staining score for CD40 was 3.18 ± 2.54 in the well-differentiated carcinoma tissues, but only 1.22 ± 2.07 in the poorly differentiated group, implicating that CD40 expression was significantly correlated with cell differentiation (Fig. 1a, b; Table 1, *p < 0.01). However, the expression of CD40 was not correlated with TNM stage or clinical stage (Table 1). CD40, which is widely expressed, is a molecule associated with immunity. In colon cancer tissues, CD40 could be expressed by colon cancer cells or B cells and dendritic cells. To demonstrate that CD40 is expressed in tumor cells, we detected the co-expression of CD40 and E-cadherin by immunofluorescence assay. The result indicated that CD40 was mainly expressed in cancer cells (Fig. 1c). Flow cytometry and western blotting were used to detect the expression of CD40 in colon cancer cell lines. The four colon cancer cell lines (HCT116, HT29, SW620, and SW480) displayed variable but positive CD40 expression levels. In contrast, Lovo cells stained negative (1.2 %) for CD40 (Fig. 1d). Western blotting results confirmed a lack of CD40 expression in the Lovo cell line and positive expression in the other four cell lines (Fig. 1e).
Fig. 1.
Expression of CD40 and CD40L in colon cancer. a CD40 was expressed in colon cancer tissues. Four representative immunohistochemically stained tissues are shown (×100): well-, moderately, and poorly differentiated colon cancer and normal colon tissue. The small map in each upper right corner of the image indicates higher magnification (×200). b Semiquantitative analysis of the expression of CD40 in 110 cases of colon cancer patients at various stages of differentiation. *p < 0.01 versus well- or moderately differentiated colon cancer. c Immunofluorescence analysis of E-cadherin (green) and CD40 (red) co-expression in colon cancer tissue. Nuclei are counterstained with DAPI (blue) (×200). d Surface expression of CD40 in human colon cancer cells was analyzed by flow cytometry. Background staining (black) or CD40 antibody staining (red) is indicated. The proportion of cells expressing CD40 is indicated in each histogram. e CD40 expression in colon cancer cells was checked via western blotting. f Expression of CD40L was detectable in colon cancer tissues. Staining of three representative tissues is shown. g Semiquantitative analysis of the expression of CD40L in 110 cases of colon cancer patients at various stages of differentiation. *p < 0.01 versus well- or moderately differentiated colon cancer. h Expression of CD40L in serum of colon cancer patients was checked via enzyme-linked immunosorbent assay. p < 0.01 versus healthy donors. i Surface expression of CD40L in human peripheral blood mononuclear cell was analyzed by flow cytometry. All peripheral blood mononuclear cells were included in the scope of analysis after exclusion of cell debris. j Quantitative analysis of CD40L expression in peripheral blood cells of healthy volunteers and patients with colon cancer
Table 1.
Statistical analysis of the relationship of CD40, AHRR, or AHR expression with clinical pathological characteristics in colon cancer patients
| Factors | Case (n) | AHR | AHRR | CD40 | |||
|---|---|---|---|---|---|---|---|
| Staining score | p value | Staining score | p value | Staining score | p value | ||
| Age | <0.05 | >0.05 | >0.05 | ||||
| <70 years | 77 | 2.42 ± 2.09 | 1.73 ± 1.84 | 2.37 ± 2.79 | |||
| ≥70 years | 33 | 1.91 ± 1.83 | 1.16 ± 1.67 | 1.93 ± 2.14 | |||
| Differentiation | <0.01 | <0.05 | <0.01 | ||||
| Well | 34 | 3.29 ± 2.54 | 2.28 ± 2.01 | 3.18 ± 2.54 | |||
| Moderate | 40 | 3.18 ± 2.23 | 1.07 ± 1.12 | 2.58 ± 2.23 | |||
| Poor | 22 | 1.22 ± 1.17 | 0.92 ± 1.01 | 1.22 ± 2.07 | |||
| T stage | >0.05 | >0.05 | >0.05 | ||||
| T2 | 6 | 1.26 ± 1.57 | 1.15 ± 1.06 | 1.17 ± 1.47 | |||
| T3 | 50 | 1.86 ± 2.12 | 1.46 ± 1.54 | 1.88 ± 2.53 | |||
| T4 | 54 | 2.13 ± 2.14 | 1.75 ± 1.37 | 2.17 ± 2.41 | |||
| N (lymph node metastasis) | >0.05 | >0.05 | >0.05 | ||||
| N0 | 73 | 1.79 ± 2.31 | 1.51 ± 1.24 | 1.89 ± 2.41 | |||
| N1 | 25 | 2.14 ± 2.21 | 1.53 ± 1.27 | 2.08 ± 2.48 | |||
| N2 | 12 | 2.67 ± 2.25 | 1.95 ± 1.68 | 2.33 ± 2.53 | |||
| Clinical stage | >0.05 | >0.05 | >0.05 | ||||
| I–II | 71 | 2.09 ± 2.21 | 1.08 ± 1.23 | 2.10 ± 2.41 | |||
| III–IV | 39 | 1.98 ± 2.36 | 1.95 ± 2.14 | 1.92 ± 2.44 | |||
CD40L expression in colon cancer tissue and serum of colon cancer patients
The expression of CD40L was investigated by immunohistochemistry in 110 colon cancer tissue samples, and it was detected in 62 (56 %) cases. Figure 1f shows three CD40L staining examples, indicating that CD40L was detectable in colon cancer tissue. Furthermore, staining results revealed that CD40L was mainly expressed in the tumor interstitium, which comprises a large number of infiltrating immune cells, rather than by tumor cells (Fig. 1f). Semiquantitative analysis indicated that expression of CD40L was higher in well- and moderately differentiated colon cancer, but lower expression was observed in poorly differentiated colon cancer (Fig. 1g, *p < 0.01). ELISA and flow cytometry were used to detect the expression of CD40L in peripheral blood of colon cancer patients. ELISA indicated that the serum samples from colon cancer patients had variable CD40L expression levels, but the levels were higher than those from control cases (Fig. 1h, p < 0.01). In addition, flow cytometry analysis was used to detect the expression of CD40L in peripheral blood mononuclear cells, which mainly consisted of lymphocytes, monocytes, and granulocytes. CD40L was expressed by peripheral blood mononuclear cells from both healthy control groups and colon cancer patients with no significant difference (Fig. 1i, j). All of these results indicate that the expression of CD40L was detectable in colon cancer tissue and peripheral blood specimens from colon cancer patients.
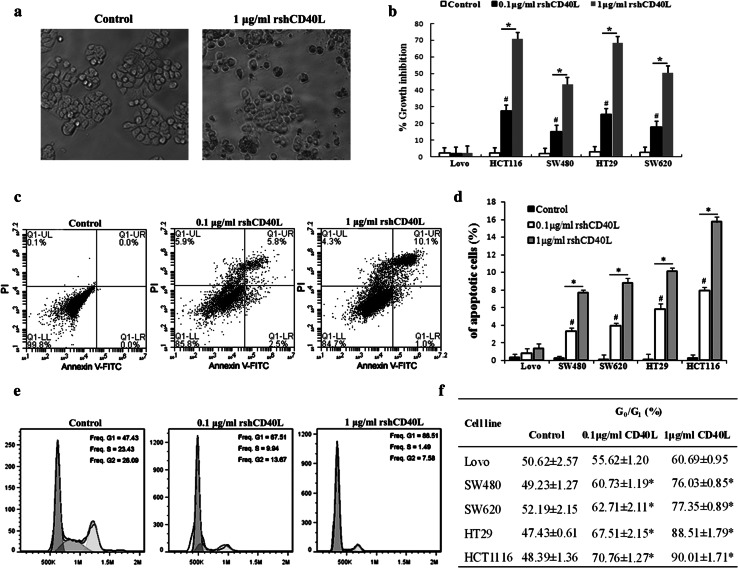
Effect of rshCD40L on cell proliferation, apoptosis, and cell cycle distribution
The expression of CD40 in colon cancer prompted us to examine the effects of the regulation of CD40 signaling on cell growth. MTT assays revealed that treatment of CD40-positive tumor cells with rshCD40L significantly inhibited proliferation (Fig. 2a, b). A significant reduction in the growth of CD40-positive cells was observed after treatment with rshCD40L relative to that in the control group (Fig. 2b, *p < 0.05). As the concentration of rshCD40L increased, the inhibitory effect on proliferation also increased significantly (# p < 0.05). The percentage of apoptotic cells increased after 48 h of treatment with rshCD40L relative to that in the untreated control (Fig. 2c, d, *p < 0.05). As the rshCD40L level increased, the percentage of apoptotic cells also increased significantly (Fig. 2d, # p < 0.05). Treatment with rshCD40L also resulted in a significant and concentration-dependent accumulation of cells in the G0/G1 phases (Fig. 2e, f, *p < 0.05). For HT29 cells, the percentage of cells blocked in the G1/G0 phase was 89 % for the 0.1 μg/mL rshCD40L group and 68 % for the 1 μg/mL rshCD40L group; in contrast, the percentage was 47 % in the control group (Fig. 2e). These results demonstrated that rshCD40L significantly and concentration-dependently inhibited the growth of the CD40-positive colon cancer cells.
Fig. 2.
Growth inhibitory effect of CD40L on colon cancer cells. a Morphological changes of colon cancer HT29 cells in different groups were observed (×100) under an inverted microscope. b The proliferation of colon cancer cells was assessed by MTT assay. The results are expressed as percent growth inhibition. # p < 0.05 versus 1 μg/mL rshCD40L. *p < 0.05 versus control. c For the apoptosis analysis, after incubation for 48 h, the apoptotic cells were analyzed using flow cytometry. An apoptosis flow chart of HT29 cells is shown. d Data are expressed as the percentage of cells that were apoptotic. Data are shown as the mean ± SD of three independent experiments. # p < 0.05 versus 1 μg/mL rshCD40L. *p < 0.05 versus control. e For cell cycle analysis, cells were stained with PI, and flow cytometry was used to analyze the cell cycle distribution. A representative analysis chart for HT29 colon cancer cells is shown. f The percentage of G0/G1 cells in different groups of five colon cancer lines is presented. Data are shown as the mean ± SD of three independent experiments. *p < 0.05 versus control
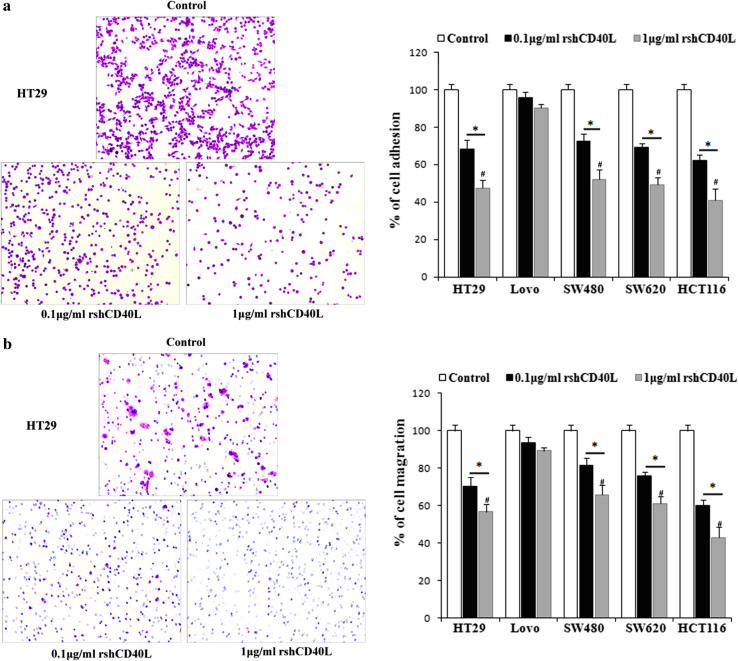
rshCD40L reduces the adhesion and migration of colon cancer cells
The effect of rshCD40L on cell adhesion in cultured colon cancer cells was studied in a 96-well plate pre-coated with Matrigel. The adhesion of cells was significantly inhibited by rshCD40L in a concentration-dependent manner compared with the untreated control (Fig. 3a, *p < 0.05, # p < 0.05). Activation of CD40 by rshCD40L also significantly decreased cell migration in a concentration-dependent manner (Fig. 3b, # p < 0.05). Compared with the control, the migration of HT29 cells was 75 % in the 0.1 μg/mL rshCD40L group and 51 % in the 1 μg/mL rshCD40L group (Fig. 3b, *p < 0.05). These results indicate that rshCD40L exerted anti-migration effects on colon cancer.
Fig. 3.
Treatment with rshCD40L reduces the adhesion and migration of colon cancer cells. a The crystal violet-stained attached cells are shown (×100), and attached cells were quantified by determining the OD at 570 nm. Data are shown as the mean ± SD of three independent experiments. # p < 0.05 versus 0.1 μg/mL rshCD40L. *p < 0.05 versus control. b Migration assays were conducted using modified 24-well transwell plates. Randomly chosen viewing fields were photographed (×100), and the number of cells that migrated was assessed as a percentage of the total cell count. Data are shown as the mean ± SD of three independent experiments. # p < 0.05 versus 0.1 μg/mL rshCD40L *p < 0.05 versus control
Analysis of microarray expression profile
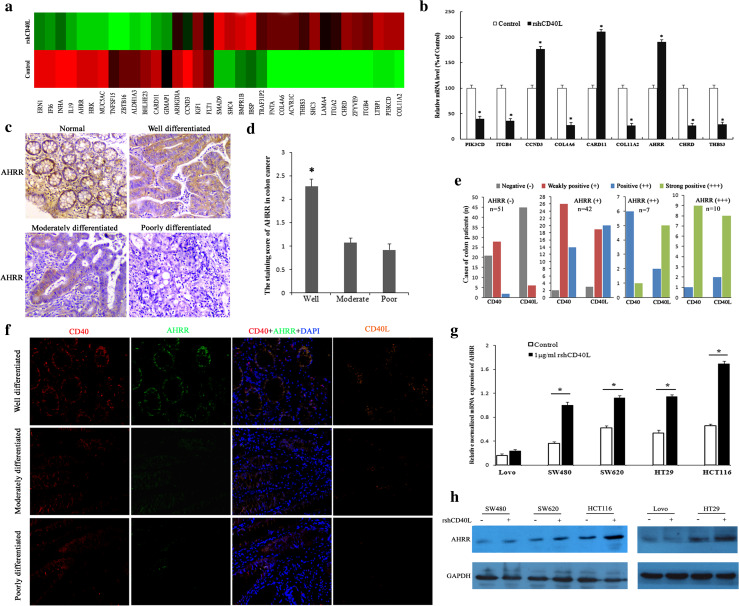
Data from four independent samples (HT29 colon cancer cells treated with or without rshCD40L, two samples from each group) showed that 251 genes were down-regulated and 190 genes were up-regulated by regulation of CD40 signaling compared to controls. A cluster analysis revealed several differentially expressed genes (Fig. 4a). Gene ontology functional class analysis indicated that the most prevalent terms were: cell cytoskeletal, cell cycle, cell recognition, apoptosis, drug binding, cell adhesion, and transforming growth factor-β (TGF-β) signal transduction (Table 2). To verify the results of gene chip screening, the differentially expressed genes associated with the cell cycle, apoptosis, and adhesion were selected for detection by in-depth analysis of the database. The results of the microarray screening were confirmed by PCR. The significantly up-regulated genes were CARD11, AHRR, and CCND3, and the down-regulated genes were PIK3CD, ITGB4, COL4A6, COL11A2, THBS3, and CHRD (Fig. 4b, *p < 0.05).
Fig. 4.
Up-regulation of AHRR by CD40 activation in colon cancer cells. a HT29 colon cancer cells were cultured with or without rshCD40L, and the total RNA was extracted for expression microarray analysis. Heat map for differentially expressed genes in the two groups is shown. b Real-time PCR analysis was used for determining the expression of HRK, CARD11, AHRR, PIK3CD, ITGA2, ITGB4, COL4A6, COL11A2, THBS3, ZFYVE9, and CHRD. *p < 0.01 versus control. c Expression of AHRR was detected by immunohistochemistry in different colon cancer tissues. Staining of four representative tissues is shown (×100): normal colon tissue and well-, moderately, or poorly differentiated colon cancer. d Semiquantitative analysis of the expression of AHRR in 110 cases of colon cancer patients at various stages of differentiation. *p < 0.05 versus moderately or poorly differentiated colon cancer. e Semiquantitative analysis of expression correlation among CD40, CD40L, and AHRR in 110 cases of colon cancer patients. From left to right: first negative staining of AHRR, second weakly positive staining of AHRR, third positive staining of AHRR, fourth, strong positive staining of AHRR. The colon cancer tissues negative for AHRR staining showed negative or weak staining for CD40 and CD40L. The sections showing strong AHRR staining also stained strongly for CD40 and CD40L. f Immunofluorescence analysis of AHRR (green), CD40 (red), and CD40L (orange) expression in colon cancer tissue. Nuclei are counterstained with DAPI (blue) (×200). Top staining of well-differentiated colon cancer tissues is shown, middle staining of moderately differentiated colon cancer tissues is shown, bottom staining of poorly differentiated colon cancer tissues is shown. High AHRR expression was associated with high CD40 expression in tumor cells, and CD40L expression was particularly high in the tumor interstitium in well- and moderately differentiated colon cancer. AHRR expression was weak when expression of CD40 and CD40L was reduced or negative in poorly differentiated colon cancer. Colon cancer cells were treated with rshCD40L or vehicle control for 12 h and harvested for g real-time PCR analysis, or treated for 48 h and harvested for h western blotting analysis of AHRR
Table 2.
Consistently differential expressed genes, along with their functions, in cells treated with rshCD40L
| Brief function | GenBank ID | Gene name | Expression |
|---|---|---|---|
| Cytoskeletal protein | NM_001039141 | TRIOBP | Downregulation |
| NM_032866 | CGNL1 | Downregulation | |
| NM_003279 | TNNC2 | Downregulation | |
| NM_006633 | IQGAP2 | Downregulation | |
| NM_001613 | ACTA2 | Downregulation | |
| NM_004389 | CTNNA2 | Downregulation | |
| Regulation of cell cycle | NM_002027 | FNTA | Upregulation |
| NM_000499 | CYP1A1 | Upregulation | |
| NM_001760 | CCND3 | Upregulation | |
| NM_002341 | LTB | Upregulation | |
| Cell recognition | NM_007155 | ZP3 | Downregulation |
| NM_017573 | PCSK4 | Downregulation | |
| NM_020703 | AMIGO1 | Downregulation | |
| NM_004385 | VCAN | Downregulation | |
| NM_004787 | SLIT2 | Downregulation | |
| Drug binding | NM_000524 | HTR1A | Upregulation |
| NM_001006630 | CHRM2 | Upregulation | |
| NM_033181 | CNR1 | Upregulation | |
| Cell adhesion | NM_005026 | PIK3CD | Downregulation |
| NM_000213 | ITGB4 | Downregulation | |
| NM_033641 | COL4A6 | Downregulation | |
| NM_002019 | FLT1 | Downregulation | |
| NM_016848 | SHC3 | Downregulation | |
| Cell death | NM_003806 | HRK | Upregulation |
| NM_032415 | CARD11 | Upregulation | |
| NM_020731 | AHRR | Upregulation | |
| NM_001433 | ERN1 | Upregulation | |
| NM_005118 | TNFSF15 | Upregulation | |
| NM_080680 | COL11A2 | Upregulation | |
| TGF-β signaling pathway | NM_206943 | LTBP1 | Downregulation |
| NM_005905 | SMAD9 | Downregulation | |
| NM_007323 | ZFYVE9 | Downregulation | |
| NM_015424 | CHRD | Downregulation | |
| NM_007112 | THBS3 | Downregulation |
Expression of AHRR/AHR in colon cancer tissues
According to the microarray and RT-PCR results, the expression of AHRR was significantly increased. AHRR represses the transcription activity of the AHR signaling cascade, which mediates tumor promotion. Furthermore, it was recently reported that AHRR is associated with tumors and may be a new putative tumor suppressor gene [15]. Therefore, the expression of AHRR and AHR in 110 paraffin-embedded colon cancer tissues was investigated by immunohistochemistry. Expression of AHRR was detected in 59 of 110 (54 %) of colon cancer cases. However, expression of AHR was detected in 103 of 110 (94 %) of colon cancer cases. The association of AHR/AHRR expression with clinicopathological parameters is shown in Table 2. Figure 4c shows AHRR expression in the normal colon and in colon cancer at various stages of differentiation. The expression of AHRR was higher in well-differentiated colon cancer, but lower or negative expression was observed in moderately and poorly differentiated colon cancer (Fig. 4c, d; Table 1, *p < 0.05). AHRR/AHR expression was significantly correlated with histological grade (Table 1, p < 0.05), but not with clinical stage and TNM stage. Interestingly, AHR expression was significantly correlated with patient age. The expression of AHR was higher in >70-year-old patients compared than in patients under 70 years of age. Semiquantitative analysis of immunohistochemistry detection for CD40, CD40L, and AHRR (three consecutive paraffin slices) indicated that AHRR histological staining was consistent with the staining of CD40 and CD40L in colon cancer. The sections showing strong AHRR staining also stained strongly for CD40 and CD40L. The AHRR-negative staining colon cancer tissues displayed negative or weak staining for CD40 and CD40L (Fig. 4e). To further confirm the correlation among CD40, CD40L, and AHRR, an immunofluorescence assay was used to detect the co-expression of CD40, CD40L, and AHRR. CD40 and AHRR were expressed in tumor cells. Further high AHRR expression was associated with high CD40 expression, and CD40L expression was particularly high in the tumor interstitium in well and moderately differentiated colon cancer. However, AHRR expression was weak when CD40 and CD40L expression was reduced or negative in poorly differentiated colon cancer. (Fig. 4f). These results suggest that AHRR tumor suppression activity may be correlated with CD40 signal regulation.
Up-regulation of AHRR by CD40 activation in colon cancer cells
To further confirm the effect of the CD40 signaling pathway on AHRR, AHRR mRNA and protein levels were determined in five colon cancer cell lines. Colon cancer cells were treated with rshCD40L or vehicle control for 12 h and harvested for AHRR and CD40 real-time PCR analysis, or treatment was performed for 48 h and cells were then harvested for western blotting analysis. AHRR was up-regulated at both the gene and protein levels following activation of the CD40 signaling pathway (Fig. 4g, h).
Discussion
In this study, a significant correlation was found between the expression of CD40 and colon cancer; cultured colon cancer cell lines also expressed this receptor, albeit at variable levels. In addition, the expression of CD40 was correlated with the histological grade of the cells; therefore, future studies should evaluate the effects of CD40 on cell growth and the progression of colon cancers. Zhou et al. [7] reported that treatment with rshCD40L or anti-CD40 agonist antibody significantly inhibited ovarian carcinoma cell growth and induced apoptosis. Similarly, Wu et al. [16] reported that combination treatment with rshCD40L and Interferon-γ significantly inhibited the proliferation of CD40-positive colon cancer cell lines and induced apoptosis. However, there have been no reports on the growth inhibition mechanism and effectiveness of the regulation of CD40 signaling on the migration and adhesion of colon cancer.
In this study, the functions of CD40/CD40L in colon cancer cell lines were demonstrated by ligation of the receptor with rshCD40L. We found that rshCD40L treatment inhibited cell growth and induced apoptosis, resulting in a significant accumulation of cells arrested in the G0/G1 phases. Furthermore, treatment with rshCD40L reduced cell migration in vitro and inhibited the adhesion of cells onto the Matrigel. These results showed that the growth and malignant progression of colon cancer were inhibited by treatment with rshCD40L in a concentration-dependent manner.
Using microarray and real-time PCR analyses, we found that several cell growth- and malignant progression-associated genes were significantly differentially expressed following regulation of CD40 signaling in colon cancer cells. Among these genes, AHRR has been reported to be a potential tumor suppressor gene in multiple types of human cancers [15]. AHRR inhibits the expression of AHR, which is involved in a signaling pathway that mediates the toxic effects of dioxin, including immunosuppression, teratogenesis, and tumor promotion, as well as the regulation of cell differentiation and growth [17]. As AHR has been discovered to promote cell cycle progression in MCF-7 cells, AHRR over-expression in MCF-7 cells would suppress cell cycle progression [18]. Recently, reduced expression of AHRR was observed in gastric adenocarcinoma, lung cancer, breast cancer, and various other human cancerous tissues, implying that AHRR is associated with multiple human cancers [19–21]. However, to the best of our knowledge, no other report has focused on the expression status and pathological value of AHRR in colon cancer.
In the present study, expression of AHRR was detected in 54 % of colon cancer cases. Additionally, AHRR expression was discovered to be significantly correlated with the histological grade of colon cancer, but not with the clinical or TNM stages. Interestingly, we found that CD40 and AHRR were expressed by tumor cells in colon cancer tissue, and high AHRR expression was associated with high CD40 expression in tumor cells. Simultaneously, the expression of CD40L was particularly high in the tumor interstitium. In addition, when colon cancer cells were treated with rshCD40L, AHRR was up-regulated at both gene and protein levels; CD40 expression was also up-regulated. This finding indicates that activation of the CD40 signaling pathway could inhibit the growth and malignant progression of colon cancer by regulating AHRR expression.
In conclusion, we have demonstrated that colon cancer cells express CD40 at high levels, and that the expression is correlated with the degree of cellular differentiation. In addition, rshCD40L treatment significantly inhibited cell growth, induced apoptosis, stalled cells in the G0/G1 phase, and influenced cell adhesion and metastasis. These data suggest that CD40L binding could inhibit the growth and malignant progression of colon cancer. Moreover, the activation of CD40 signaling elevated the expression of AHRR and enhanced tumor suppression activity. Because the expression of AHRR was significantly correlated with histological grade, we propose that AHRR might have pathological value in colon cancer. Finally, a more thorough investigation of the proposed signaling pathways that may mediate CD40-induced growth and metastasis inhibition could aid in the development of novel treatments for colon cancer.
Acknowledgments
The present study was supported by the National Natural Science Foundation of China (Grant No. 81302170), the National Clinical Medicine Research Foundation of China (Grant No. L2012055), and the Natural Science Foundation of Chengdu Medical College (Grant No. CYZ12–005).
Abbreviations
- AHR
Aryl hydrocarbon receptor
- AHRR
Aryl hydrocarbon receptor repressor
- APC
Allophycocyanin
- CD40L
CD40 ligand
- Cy3
3H-indocyanines
- rshCD40L
Recombinant soluble human CD40 ligand
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Elgueta R, Benson MJ, de Vries VC, Wasiuk A, Guo Y, Noelle RJ. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol Rev. 2009;229:152–172. doi: 10.1111/j.1600-065X.2009.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Satpathy S, Shenoy GN, Kaw S, Vaidya T, Bal V, Rath S, et al. Inhibition of terminal differentiation of B cells mediated by CD27 and CD40 involves signaling through JNK. J Immunol. 2010;185:6499–6507. doi: 10.4049/jimmunol.0903229. [DOI] [PubMed] [Google Scholar]
- 3.Ma DY, Clark EA. The role of CD40 and CD154/CD40L in dendritic cells. Semin Immunol. 2009;21:265–272. doi: 10.1016/j.smim.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bush TJV. Bishop GA, TLR7 and CD40 cooperate in IL-6 production via enhanced JNK and AP-1 activation. Eur J Immunol. 2008;38:400–409. doi: 10.1002/eji.200737602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hill SC, Youde SJ, Man S, Teale GR, Baxendale AJ, Hislop A, et al. Activation of CD40 in cervical carcinoma cells facilitates CTL responses and augments chemotherapy-induced apoptosis. J Immunol. 2005;174:41–50. doi: 10.4049/jimmunol.174.1.41. [DOI] [PubMed] [Google Scholar]
- 6.Kim SW, Lee HS, Yoon SK, Chung WC, Cho YS, Jeong JJ, et al. Expression of CD40 in gastric cancer and its effect on the apoptosis of gastric cancer cells. Korean J Gastroenterol. 2003;42:274–282. [PubMed] [Google Scholar]
- 7.Zhou Y, He J, Gou LT, Mu B, Liao WC, Ma C, et al. Expression of CD40 and growth-inhibitory activity of CD40 agonist in ovarian carcinoma cells. Cancer Immunol Immunother. 2012;61:1735–1743. doi: 10.1007/s00262-011-1194-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolen Y, Yilmaz G, Esendagli G, Guler NE, Guc D. CD40–1C> T single nucleotide polymorphism and CD40 expression on breast tumors. Cytokine. 2010;50:243–244. doi: 10.1016/j.cyto.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 9.Fanale M, Assouline S, Kuruvilla J, Solal-Celigny P, Heo DS, Verhoef G, et al. Phase IA/II, multicentre, open-label study of the CD40 antagonistic monoclonal antibody lucatumumab in adult patients with advanced non-Hodgkin or Hodgkin lymphoma. Br J Haematol. 2014;164:258–265. doi: 10.1111/bjh.12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tai YT, Podar K, Mitsiades N, Lin B, Mitsiades C, Gupta D, et al. CD40 induces human multiple myeloma cell migration via phosphatidylinositol 3-kinase/AKT/NF-kappa B signaling. Blood. 2003;101:2762–2769. doi: 10.1182/blood-2002-09-2813. [DOI] [PubMed] [Google Scholar]
- 11.Tai YT, Podar K, Gupta D, Lin B, Young G, Akiyama M, et al. CD40 activation induces p53-dependent vascular endothelial growth factor secretion in human multiple myeloma cells. Blood. 2002;99:1419–1427. doi: 10.1182/blood.V99.4.1419. [DOI] [PubMed] [Google Scholar]
- 12.Gomes EM, Rodrigues MS, Phadke AP, Butcher LD, Starling C, Chen S, et al. Antitumor activity of an oncolytic adenoviral-CD40 ligand (CD154) transgene construct in human breast cancer cells. Clin Cancer Res. 2009;15:1317–1325. doi: 10.1158/1078-0432.CCR-08-1360. [DOI] [PubMed] [Google Scholar]
- 13.Li R, Chen WC, Pang XQ, Tian WY, Zhang XG. Influence of sCD40L on gastric cancer cell lines. Mol Biol Rep. 2011;38:5459–5464. doi: 10.1007/s11033-011-0702-9. [DOI] [PubMed] [Google Scholar]
- 14.Sabel MS, Yamada M, Kawaguchi Y, Chen FA, Takita H, Bankert RB. CD40 expression on human lung cancer correlates with metastatic spread. Cancer Immunol Immunother. 2000;49:101–108. doi: 10.1007/s002620050608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zudaire E, Cuesta N, Murty V, Woodson K, Adams L, Gonzalez N, et al. The aryl hydrocarbon receptor repressor is a putative tumor suppressor gene in multiple human cancers. J Clin Investig. 2008;118:640–650. doi: 10.1172/JCI30024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Y, Wang L, He X, Xu H, Zhou L, Zhao F, et al. Expression of CD40 and growth-inhibitory activity of CD40 ligand in colon cancer ex vivo. Cell Immunol. 2008;253:102–109. doi: 10.1016/j.cellimm.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 17.Powell JB, Ghotbaddini M. Cancer-promoting and inhibiting effects of dietary compounds: role of the aryl hydrocarbon receptor (AhR) Biochem Pharmacol (Los Angel) 2014 doi: 10.4172/2167-0501.1000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Englert NA, Turesky RJ, Han W, Bessette EE, Spivack SD, Caggana M, et al. Genetic and epigenetic regulation of AHR gene expression in MCF-7 breast cancer cells: role of the proximal promoter GC-rich region. Biochem Pharmacol. 2012;84:722–735. doi: 10.1016/j.bcp.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cauchi S, Stucker I, Cenee S, Kremers P, Beaune P, Massaad-Massade L. Structure and polymorphisms of human aryl hydrocarbon receptor repressor (AhRR) gene in a French population: relationship with CYP1A1 inducibility and lung cancer. Pharmacogenetics. 2003;13:339–347. doi: 10.1097/00008571-200306000-00005. [DOI] [PubMed] [Google Scholar]
- 20.Kanno Y, Takane Y, Izawa T, Nakahama T, Inouye Y. The inhibitory effect of aryl hydrocarbon receptor repressor (AhRR) on the growth of human breast cancer MCF-7 cells. Biol Pharm Bull. 2006;29:1254–1257. doi: 10.1248/bpb.29.1254. [DOI] [PubMed] [Google Scholar]
- 21.Li YF, Wang DD, Zhao BW, Wang W, Yuan SQ, Huang CY, et al. Poor prognosis of gastric adenocarcinoma with decreased expression of AHRR. PLoS One. 2012;7:e43555. doi: 10.1371/journal.pone.0043555. [DOI] [PMC free article] [PubMed] [Google Scholar]