Dear Editors,
A recently published paper found that a polymorphism, rs10815225, in the PD-L1 promoter region was associated with PD-L1 overexpression in gastric cancer cells [1]. The polymorphism offered a binding-site for a transcriptional factor (SP1), resulting in elevated expression of PD-L1 mRNA and protein in gastric cancer. However, our data showed otherwise.
Our team has been investigating the immunology of gastric cancer, including the role of PD-L1, which is one key immune inhibitory factor during cancer-immune interaction. The expression of PD-L1 in tumor microenvironment can aid in predicting patients’ prognosis and outcome of PD-1 blockade. The intrinsically elevated PD-L1 expression resulting from single nucleotide polymorphism (SNP) mentioned in the study [1] raised our interest. We conducted genotyping for PD-L1 gene in 152 gastric cancer patients who were enrolled in our previous study. The method of amplification refractory mutation system (ARMS) based PCR combined with quantitative PCR was used for genotyping, which has advantages of high specificity and sensitivity. The sequence of 5′-AAGTCCAACGCCCGGCAAACTG-3′ was used as probe (TaqMan-FAM). Three primers, 5′-CGCCGATTTCACCGAAGGTCAGG-3′ (forward), 5′-AGCGTTGCGCCAGGCGC-3′ (reverse for G allele) and 5′-AGCGTTGCGCCAGGCGG-3′ (reverse for C allele) were used to amplify corresponding variants. The interpretation of different genotypes was done according to the Ct values of quantitative PCR. Sanger sequencing was also conducted for validating. The sequencing primers were 5′-GTGCGTTCAGATGTTGGCTTGTTGT-3′ (forward) and 5′-GTAGAGACCCTCCGTCCTAAAGTGC-3′ (reverse). The expression of PD-L1 was measured by immunohistochemistry (the primary antibody: the rabbit anti-PD-L1 monoclonal antibody [28-8], ab205921, UK; the specific antigen buffer for FFPE tissues: universal HIER antigen retrieval reagent (10×), ab208572 http://www.abcam.cn) and then classified strictly by Immune Reactive Score (IRS) system, which multiplied the staining intensity by the percentage of PD-L1 positive cells [2]. Different percentages of positive expression (1, 5, 10, and 20%) were also valued. SPSS 19.0 software was used for statistical analysis.
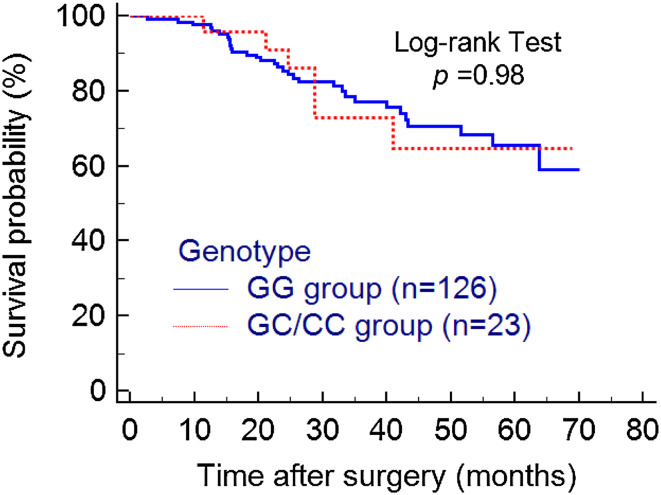
There were 149 samples enrolled in the final analysis, among which 126 were GG carriers, 20 were GC carriers and only 3 were CC carriers. A goodness-of-fit Chi-square test showed that the genotype distribution in our cohort met Hardy–Weinberg equilibrium. The minor allele frequency (MAF) was 8.72%. Even though almost all of the PD-L1 positive tissues are GG homozygotes (38/41), statistical analysis did not find any significant difference of IRS between GG group and GC group (OR = 0.68, 95% CI 0.21–2.27; p = 0.53) or GG group vs C allele carriers (GC and CC groups) (OR = 0.57, 95% CI 0.18–1.86; p = 0.36). No significant association of different genotypes with 1, 5, 10 or 15% PD-L1 positivity was found either (Table 1). Other clinical features (e.g. drinking history, onset position, differentiation, TNM stage, CEA, CA19-9) did not significantly correlate with the polymorphism rs10815225 either as is shown in Table 1, except for the differentiation (GG vs GC, OR = 0.28, 95% CI 0.09–0.84, p = 0.02). Next, we also analyzed if rs10815225 had any effect on patients’ prognosis just as the authors questioned at the end of their article. The result showed that GC/CC group showed a similar overall survival as the GG group (HR = 0.99, 95% CI 0.42–2.46; p = 0.98) (Fig. 1).
Table 1.
The association of rs10815225 with clinical features and PD-L1 expression of gastric cancer (adjusted by age and gender)
| Characteristics | G/G | G/C | C/C | C carrier | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | n | OR (95% CI) | p | n | OR (95% CI) | p | n | OR (95% CI) | p | |
| Drinking History | ||||||||||
| Yes | 33 | 6 | 0.90 (0.27–3.00) | 0.8 | 2 | – | – | 8 | 1.20 (0.40–3.55) | 0.74 |
| No | 93 | 14 | 6 | 1 | 15 | |||||
| Onset position | ||||||||||
| Cardia/fundus | 9 | 4 | 2.75 (0.71–10.69) | 0.14 | 1 | – | – | 5 | 3.24 (0.92–11.41) | 0.07 |
| Gastric body | 30 | 3 | 0.55 (0.15–2.08) | 0.38 | 1 | 4 | 0.69 (0.21–2.27) | 0.55 | ||
| Pylorus | 85 | 15 | 1.47 (0.48–4.44) | 0.50 | 2 | 17 | 1.34 (0.48–3.75) | 0.58 | ||
| Differentiation | ||||||||||
| Poor differentiation | 60 | 15 | 0.28 (0.09–0.84) | 0.02 | 2 | – | – | 17 | 0.30 (0.11–0.83) | 0.02 |
| Other (moderate/well) | 60 | 5 | 1 | 6 | ||||||
| TNM stage | ||||||||||
| II | 29 | 6 | 0.61 (0.20–1.82) | 0.3 | 0 | – | – | 6 | 0.78 (0.27–2.25) | 0.64 |
| III | 97 | 14 | 8 | 3 | 17 | |||||
| CEA | ||||||||||
| Normal | 76 | 13 | 1.27 (0.12–13.11) | 0.84 | 2 | – | – | 15 | 1.00 (0.10–9.68) | 1.00 |
| Abnormal | 6 | 1 | 0 | 1 | ||||||
| CA19-9 | ||||||||||
| Normal | 71 | 12 | – | 1.0 | 2 | – | – | 14 | – | 1.00 |
| Abnormal | 5 | 0 | 0 | 0 | 0 | |||||
| Mean overall survival (months) | 55.87 | 56.94 | – | – | 40.68 | – | – | 54.40 | HR: 0.99 (0.42–2.46) | 0.98 |
| IRS score | ||||||||||
| Low | 93 | 16 | 0.68 (0.21–2.27) | 0.5 | 3 | – | – | 19 | 0.57 (0.18–1.86) | 0.36 |
| High | 33 | 4 | 3 | 0 | 4 | |||||
| ≥1% PD-L1 positivity | ||||||||||
| Yes | 38 | 3 | 0.44 (0.12–1.61) | 0.2 | 0 | – | – | 3 | 0.37 (0.10–1.36) | 0.13 |
| No | 88 | 17 | 1 | 3 | 20 | |||||
| ≥5% PD-L1 positivity | ||||||||||
| Yes | 24 | 1 | 0.23 (0.03–1.84) | 0.1 | 0 | – | – | 1 | 0.20 (0.03–1.59) | 0.13 |
| No | 102 | 19 | 7 | 3 | 22 | |||||
| ≥10% PD-L1 positivity | ||||||||||
| Yes | 18 | 1 | 0.36 (0.04–2.90) | 0.3 | 0 | 1 | 0.31 (0.04–2.47) | 0.27 | ||
| No | 108 | 19 | 3 | 3 | 22 | |||||
| ≥20% PD-L1 positivity | ||||||||||
| Yes | 14 | 0 | – | 1.0 | 0 | – | 0 | 1.00 | ||
| No | 112 | 20 | 0 | 3 | 23 | |||||
Fig. 1.
Kaplan–Meier analysis of overall survival in different genotypes
The most possible explanation for this inconsistency is that rs10815225 may alter PD-L1 mRNA expression but not PD-L1 protein expression. The evidences the authors provided to prove G-allelic PD-L1 had an elevated expression of PD-L1 are insufficient and questionable. Overall, in the context of this new data, we may need to re-consider the previous statement that the polymorphism in the promoter region of PD-L1 was associated with PD-L1 overexpression. Considering the reported inconsonant expression between PD-L1 mRNA and protein, unknown posttranscriptional-regulation mechanisms may be involved [3]. Further studies are needed to unveil this discrepancy.
Weili Wang
Ping Liao
Yijing He
Acknowledgement
This work was supported by the National Natural Science Foundation of China (Grant Nos. 81403022 and 81673517) and National key research and development program (No. 2016YFC0905000).
Compliance with ethical standards
Conflict of interest
All authors declare that they have no conflict of interest.
References
- 1.Tao LH, Zhou XR, Li FC, et al. A polymorphism in the promoter region of PD-L1 serves as a binding-site for SP1 and is associated with PD-L1 overexpression and increased occurrence of gastric cancer. Cancer Immunol Immunother. 2017;66(3):309–318. doi: 10.1007/s00262-016-1936-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Specht E, Kaemmerer D, Sanger J, Wirtz RM, Schulz S, Lupp A. Comparison of immunoreactive score, HER2/neu score and H score for the immunohistochemical evaluation of somatostatin receptors in bronchopulmonary neuroendocrine neoplasms. Histopathology. 2015;67(3):368–377. doi: 10.1111/his.12662. [DOI] [PubMed] [Google Scholar]
- 3.Wang W, Sun J, Li F, et al. A frequent somatic mutation in CD274 3′-UTR leads to protein over-expression in gastric cancer by disrupting miR-570 binding. Hum Mutat. 2012;33(3):480–484. doi: 10.1002/humu.22014. [DOI] [PubMed] [Google Scholar]