Abstract
The purpose of this study was to determine the clonal relationship between B cells within a breast cancer and the B cells in the tumor-draining lymph node (TDLN). We also determined the binding capacity of antibodies derived from these sources to autologous cancer and autologous noncancer breast tissue. Antibody clonality of B cells derived from tumor and lymph node was determined by analyzing heavy and light chain immunoglobulin sequences. The number of shared clonal groups observed between tumor and lymph node antibodies was significant for both heavy (p = 0.004) and light chain (p = 0.012) populations. Panning with phage-displayed single-chain variable fragment libraries derived from the tumor and lymph node B cells resulted in multiple antibodies that bound autologous tumor. Sequence analysis of enriched antibodies recovered after the third round of panning the tumor and TDLN libraries against autologous tumor lysates had a genetic relationship. These results indicate that B cells infiltrating a patient’s breast cancer and B cells present in the tumor-draining lymph node are clonally and functionally related.
Electronic supplementary material
The online version of this article (doi:10.1007/s00262-014-1612-1) contains supplementary material, which is available to authorized users.
Keywords: Tumor-infiltrating lymphocytes, B lymphocytes, Invasive ductal carcinoma, Axillary lymph node, Phage display, Antibody
Introduction
The role of B cells in the immune response to breast cancer is not fully understood [1, 2]. Multiple studies have reported that B cells are arranged in ectopic germinal centers within patient tumors and are likely associated with an antigen-mediated immune response [3–6]. Genetic analyses of tumor-infiltrating B lymphocytes (TIL-B cells) in these studies have demonstrated clonality. The presence of somatic mutations indicates that these TIL-B cells have undergone antigen-mediated selection. Phage-displayed antibodies derived from TIL-B cells in breast cancer bind to tumor-associated antigens [3, 7–9]. Tumor antigen-binding antibodies have also been successfully isolated from phage libraries constructed from tumor-draining lymph nodes (TDLN) of patients with breast cancer [10–12].
The anatomic distribution of tumor-specific B cells in patients with breast cancer has not been fully characterized. Radiotracer-guided surgery of sentinel nodes was developed by our group and provides an accurate method to identify the lymph nodes receiving drainage from a cancer [13]. Imaging studies of radiolabeled nodes receiving drainage from a breast cancer have demonstrated that a small set of interconnected lymph nodes receive this drainage [14]. The typical drainage pattern shows primary, secondary, and tertiary nodes [14]. Recent studies suggest that the immune response in the sentinel node may be suppressed by the tumor and exhibit abnormal architecture [15, 16]. Secondary and tertiary TDLN are also important sites of lymphocytic activation due to antigen exposure from the draining tissue [17–19].
The clonal and functional relationship of B cells in the TDLN to B cells in the tumor has not been adequately explored and is the focus of this report. In this study, we observed a clonal relationship between TIL-B cells and B cells in the TDLN in a patient with breast cancer. A functional relationship was demonstrated by binding of single-chain variable fragment (scFv) antibodies derived from both of these tissue sources to autologous tumor.
Methods
Tissue procurement and RNA isolation
Under a protocol approved by the University of Vermont Institutional Review Board, tissues were obtained from a patient with invasive breast cancer at the time of surgical treatment and included the primary tumor, sentinel node, a secondary lymph node adjacent to the sentinel node, and blood. The cut surface of the sentinel node was gently scraped with a scalpel to obtain cells. Sentinel node cells and tumor were flash frozen and stored at −80 °C. A section of the secondary node was removed with a scalpel, placed in RNA Later (QIAGEN), and stored at −20 °C. Peripheral blood mononuclear cells (PBMCs) were separated by density centrifugation (Ficoll-Paque plus, GE Healthcare) and stored at −80 °C. RNA was extracted with an RNeasy Kit (QIAGEN) and evaluated with an Agilent Bioanalyzer (UVM Microarray Facility). Insufficient RNA was obtained from the sentinel node and was not analyzed further.
Immunohistochemistry
Formalin-fixed breast tumor tissue was stained with mouse anti-CD20 (Clone L26, Thermo Scientific), rabbit anti-CD3 (Clone SP7, Thermo Scientific), anti-CD4 (Clone 4B12, Vector Labs), and rabbit anti-CD8 (Clone SP16, Thermo Scientific).
scFv library construction
Phage libraries were constructed in the pComb3X vector received from Carlos Barbas with cDNA created from tumor, TDLN, and PBMCs according to previously described methods using VL and VH primers [20]. Heavy and light chain DNA was PCR amplified from cDNA and gel purified (QIAGEN). To select the class-switched B cell population, only IgG heavy chains were amplified from IgG constant regions. Light chains were not restricted to this immunoglobulin isotype. Heavy and light chain genes were randomly combined in a second overlap PCR, gel purified, and ligated into the pComb3X vector. Vector containing the full insert was gel purified and transformed into TG1 electrocompetent cells (Agilent Technologies) by electroporation. The library was amplified overnight with K07 helper phage, polyethylene glycol (PEG) purified, and stored at −80 °C.
IMGT sequence analysis
Phage clones were randomly picked and grown overnight in 2xYT with carbenicillin. Phage DNA was isolated using miniprep and sequenced by Elim Biopharmceuticals with forward primers ompseq, HRML-F, and back primer gback [20]. Sequences of heavy and light chains were submitted to the ImMunoGeneTics information system (IMGT/V-QUEST) [21]. Each heavy and light chain was individually examined for quality (Supplemental Tables 1 and 2). If the gene identity was poor (<80 % identity), the third complementarity determining region (CDR3) was of indeterminate length, stop codons were present, or the amino acid translation was out of frame, and the heavy or light chain was removed from subsequent analyses. Identical heavy or light chain sequences from the same library were considered duplicates and removed from the analyses. To determine gene frequency, a source-specific prevalence of each VDJ recombination was calculated along with exact 95 % confidence intervals using the software on http://statpages.org/confint.html. We used nonparametric statistics to evaluate differences in the gene frequency in each sample. Our null hypothesis was that the prevalence of each individual gene in the sample was identical among tumor, lymph node, and blood samples. The alternative hypothesis assumed heterogeneity among the three sources. Due to the low observed frequencies for some gene segments, exact 3 × 2 contingency table methods were used for the overall test as implemented in StatXact-4 (Cytel Software Corporation, Cambridge MA, 1989). These overall p values were determined for each gene segment. If the null hypothesis was rejected at the 5 % significance level, only then were paired comparisons between the three sources conducted using Fisher’s exact test to minimize inflating the type I error levels. To determine mutation frequency, the total number of nucleotide base pairs and mutations in each framework region and CDR were calculated with IMGT/V-QUEST for each heavy or light chain, and the number of replacement and silent mutations in each domain were each divided by the total number of base pairs per region [21]. Replacement to silent ratio was calculated by dividing the frequency of replacement mutations to silent mutations.
Heavy or light chains were considered clonal between libraries if they either shared identical sequences but originated from different libraries or originated from the same library, shared the same junctional region, and had >2 nucleotide differences in the rest of the gene [22]. The number of clonal groups overlapping between libraries was evaluated with an exact homogeneity goodness-of-fit test implemented in StatXact-4 [23].
Lysate preparation
Lysates were prepared from BT-474 and MCF 10A cells (American Type Culture Collection), autologous tumor tissue or from heterologous normal colon tissue by subjecting samples to homogenization, if necessary, six freeze/thaw cycles, and sonication in phosphate-buffered saline (PBS). Lysate protein was quantitated using a BCA assay (Pierce).
Phage panning
Each library was panned against autologous tumor lysate [3, 20]. During the first round of enrichment, casein-blocked tumor lysate was incubated with 4 × 1012 pfu of phage. On subsequent rounds, incubation was with 106 phage amplified from the previous round. After each round, phage was washed, acid eluted, and amplified overnight in TG1 cells with K07 helper phage. The harvested phage was PEG purified and biologically titered (Supplemental Figure 1).
Binding analyses
After panning, randomly selected scFvs from each library were individually sequenced, and results were examined with aid of the IMGT database to avoid redundant binding assays on duplicate clones (Supplemental Tables 3 and 4) [21]. Supernatants were used for binding assessment.
The patient’s tumor was in limited quantity, so BT-474 cells, which had similar phenotypic features to the patient’s tumor, were chosen as a target for initial binding studies [24]. BT-474 lysates (20 μg/well) were added to a 96-well Maxisorp plate (Nunc), blocked with Blocker Casein in Tris-buffered saline (TBS, Thermo), incubated with phage supernatant or Blocker Casein for 2 h at room temperature. Plates were washed with 0.1 % Tween-TBS, incubated with horseradish peroxidase-conjugated anti-M13 antibody (1:5,000, GE Healthcare), and developed with 3,3′,5,5′-tetramethylbenzidine (TMB, EMD Millipore). The relative concentration of binding phage antibody was determined by the mean slope or velocity of OD650 readings taken every 3 min over a 15-min period (i.e., mOD/min). A linear regression was created for each triplicate samples, and the value of the mean slope (Mean V) was calculated with the KC4 v2.7 software (Biotek), which accounts for assay noise. The average Mean V of triplicate samples was compared to a blank sample containing the same scFv and casein (and no lysates). Thus, values reported are mean slope relative to a sample-specific blank.
BT-474 lysate-binding clones were evaluated for binding to autologous tumor and nontumor breast tissue. The concentration values of the phage supernatants were normalized using a chemiluminescent enzyme-linked immunosorbent assay (ELISA) [25]. Histological sections were mounted on 12-mm round poly-d-lysine-coated coverslips (BD Bioscience) as described earlier [26]. Clones were visually assessed for binding with fluorescence microscopy. A pan-binding scFv antibody discovered by our group, 799, was used as a positive control [26].
Results
Evidence of ectopic germinal centers in the primary cancer
The study patient from whom the libraries were derived had an invasive ductal carcinoma. The tumor was 1.6 cm in largest dimension, estrogen receptor positive, progesterone receptor positive, human epidermal growth factor receptor 2 (HER2) positive, moderately to poorly differentiated, and positive for the presence of lymphovascular invasion. Two sentinel lymph nodes were negative for tumor metastases. The patient received no chemotherapy prior to surgery when tissue samples were obtained. CD20+ B cells and CD3+ T cells were scattered separately throughout the tumor and together in multiple clusters adjacent to tumor cells (Fig. 1). These clusters also contained both CD4+ and CD8+ T cells (data not shown) and were characteristic of ectopic germinal centers [3, 4].
Fig. 1.
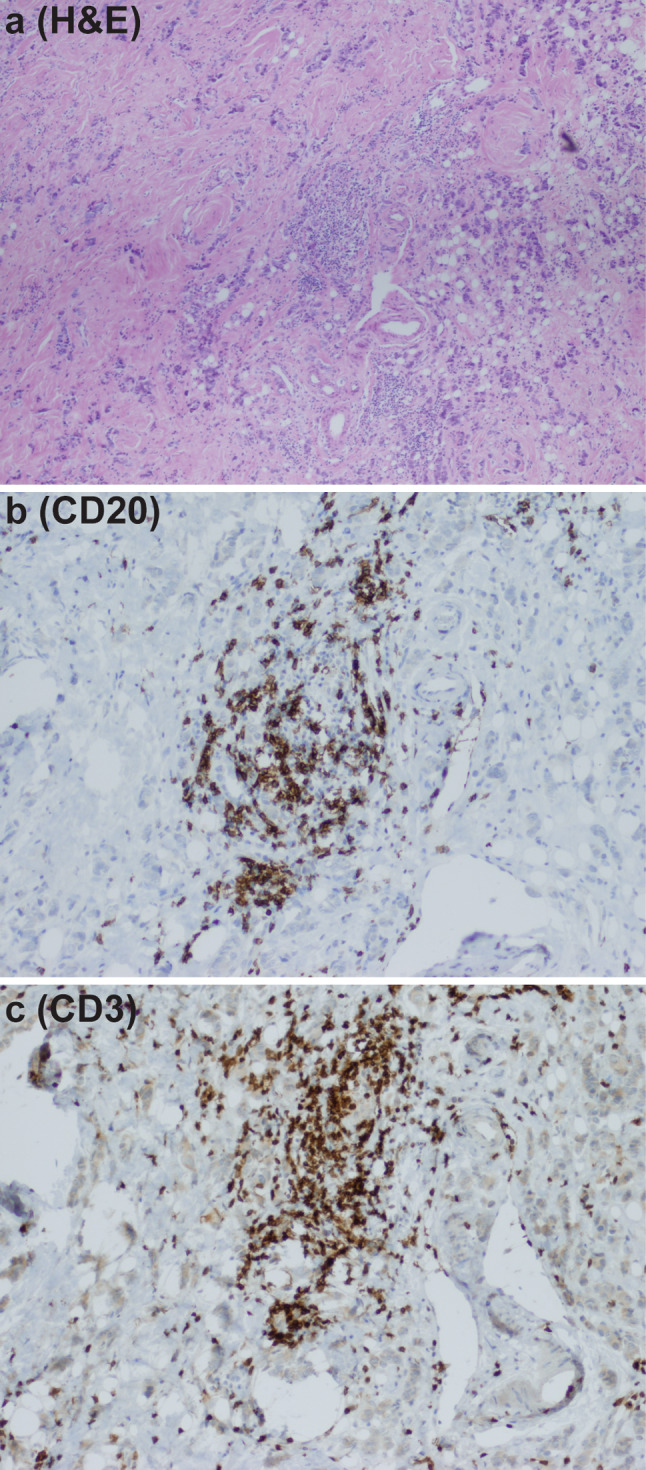
Representative sample of ectopic germinal centers within the breast cancer of the study patient. Hematoxylin and eosin stain demonstrated the presence of lymphocytes within tumor tissue. a When stained by immunohistochemistry, it was observed that lymphocyte clusters contained CD20+ B cells (b) and CD3+ T cells (c)
Tumor and axillary lymph node heavy and light chains were genetically similar
The frequencies of heavy chain VH1 through VH6 genes from randomly picked clones are shown numerically in Supplemental Table 5. A significantly higher frequency of VH5 (p = 0.03) and lower frequency of VH3 (p = 0.01) were observed in the lymph node when compared to blood. No significant difference in proportion of any VH genes was observed between tumor and lymph node or between tumor and blood. The proportion of JH genes from each tissue source is shown in Supplemental Table 6. The proportion of JH4 gene segments in the lymph node was significantly lower than in the blood (p = 0.01). No significant difference in proportion of any JH genes was observed between tumor and lymph node or between tumor and blood. These data indicate that VH and JH gene frequencies of the lymph node were different from the blood but not distinguishable from the tumor.
Kappa and lambda light chain sequences of randomly sampled clones from the three antibody libraries were analyzed. There were no significant differences observed for the kappa or lambda light chain gene frequencies between the clones from each library (Supplemental Tables 7 and 8). Potential differences in light chain gene frequencies among tumor, lymph node, and blood were below the threshold of detection in this sample size, and bias was not detected.
We looked for selection based on CDR3 length among all three libraries and found no significant difference in CDR3 length among the heavy or light chain populations (data not shown). Any differences were below the threshold of detection, and bias was not identified on the basis of CDR3 length among tumor, lymph node, or blood.
Mutations in light chain indicate B cells in the tumor and lymph node are antigen experienced
Mutation analysis of the heavy and light chains of randomly sampled phage clones from each of the three libraries indicated that the B cells from which the libraries were derived were antigen experienced. We considered a heavy or light chain positive for evidence for the presence of antigen-experienced B cells when replacement mutations were enriched over silent mutations in CDR regions compared to framework regions [27]. Heavy chains in each library were based on IgG subtype, and as expected, we observed that replacement mutations were enriched over silent mutations in the CDR regions compared to the framework regions of heavy chains indicative of somatic hypermutation (data not shown) [28, 29]. We observed that replacement mutations were enriched over silent mutations in the CDR regions compared to the framework regions of light chains in the tumor and lymph node libraries suggesting that a portion of B cells from tumor and TDLN had undergone antigen-mediated selection.
Clonal groups of B cells were present within each tissue source
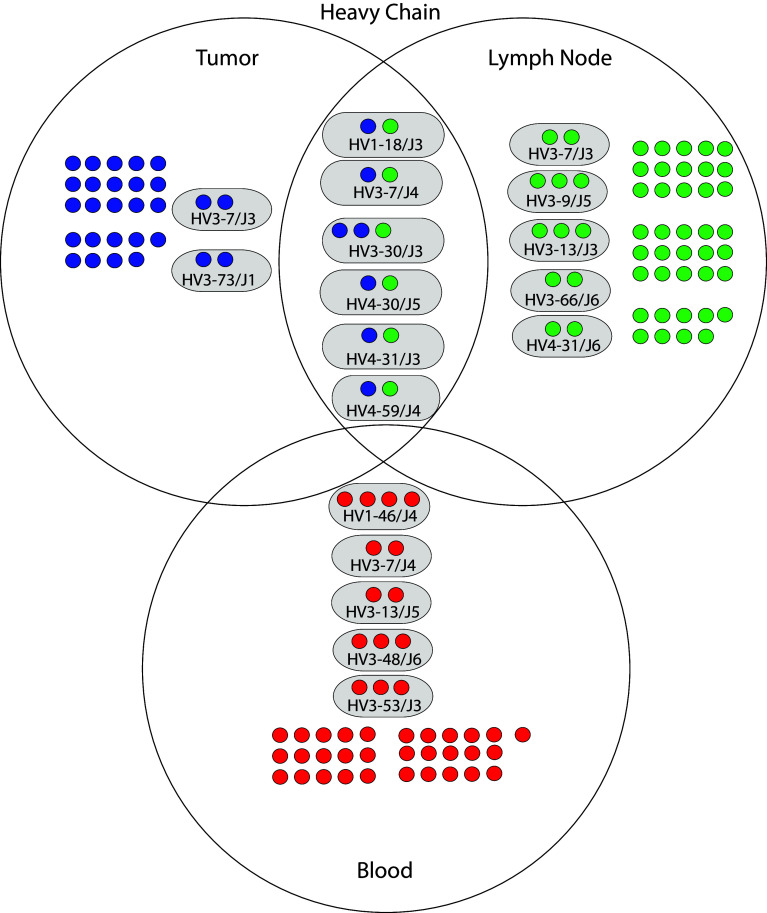
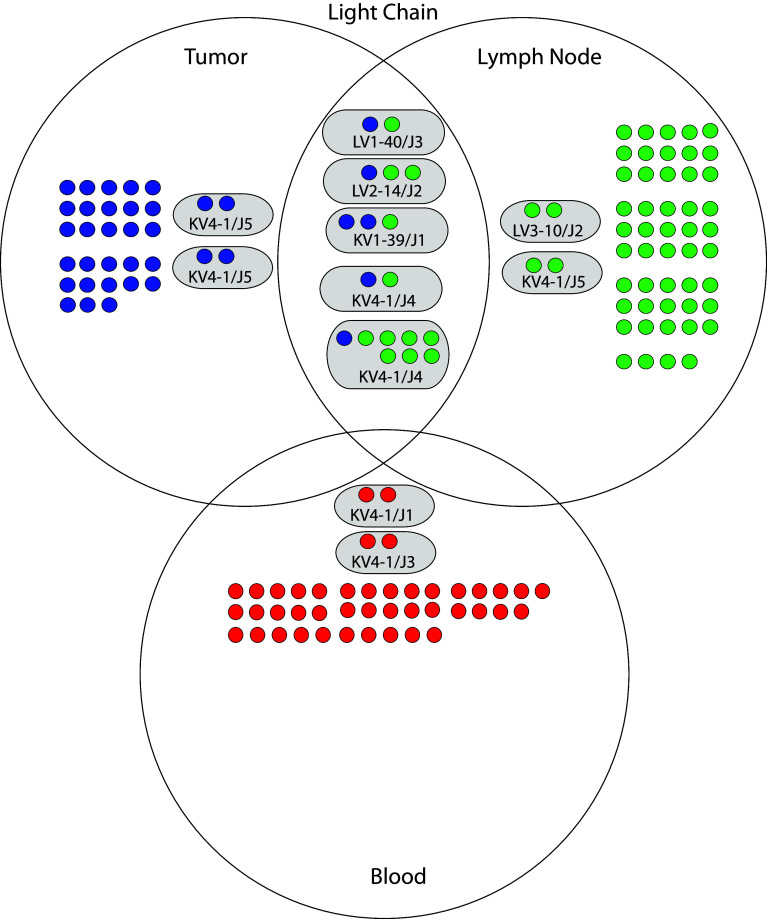
We compared the junctional regions of heavy and light chains of the randomly sampled clones in each of the three libraries to identify clonal groups of B cells within each library. Within each library, heavy or light chain genes were considered clonal if they contained identical junctional regions and exhibited >2 nucleotide differences in the rest of the gene [22]. Approximately one-third of the heavy chains in each library were clonal (Supplemental Table 9). We identified two heavy chain clonal groups exclusive to the tumor library, five heavy chain groups exclusive to the lymph node library, and five heavy chain groups exclusive to the blood library (Fig. 2). Roughly, one-third of the light chains in tumor and lymph node libraries were clonal, whereas less than 10 % of light chains in the blood library were clonal (Supplemental Table 10). We also identified two light chain clonal groups exclusive to the tumor library, two light chain groups exclusive to the lymph node library, and two light chain groups exclusive to the blood library (Fig. 3). These data indicate within the set of randomly sampled clones, and multiple heavy and light chain clonal groups were identified in each library that were not also present in the other two libraries.
Fig. 2.
Genetic relationships of heavy chains derived from tumor, lymph node, and blood of the study patient. The three large circles represent the heavy chains derived from each library. Individual heavy chain sequences are depicted as single smaller solid circles colored blue (tumor), green (lymph node), and red (blood). Heavy chain sequences that were nonclonal and unique to a library are contained in the nonoverlapping area of each library circle. Heavy chain sequences that had a clonal relationship are depicted within smaller oval circles having a gray background. Clonal groups were labeled according to their shared variable and joining gene segments. For example, the designation “HV3-7/J3” indicates that the sequences were identified with the heavy chain variable segment VH3-7 and the heavy chain joining segment JH3. Clonal groups within a library are contained within the nonoverlap region of each library circle. Clonal groups shared between libraries are contained within the overlap region of the library circles
Fig. 3.
Genetic relationships of light chains derived from tumor, lymph node, and blood of the study patient. The three large circles represent the light chains derived from each library. Individual light chain sequences are depicted as single smaller solid circles colored blue (tumor), green (lymph node), and red (blood). Light chain sequences that were nonclonal and unique to a library are contained in the nonoverlapping area of each library circle. Light chain sequences that had a clonal relationship are depicted within smaller oval circles having a gray background. Clonal groups were labeled according to their shared variable and joining gene segments. For example, the designation “LV3-10/J2” indicates that the sequences were identified with the lambda chain variable segment LV3-10 and the light chain joining segment J2. Clonal groups within a library are contained within the nonoverlap region of each library circle. Clonal groups shared between libraries are contained within the overlap region of the library circles
Clonal groups of B cells were present between the tumor and lymph node
We compared the junctional regions of heavy and light chains of the randomly sampled clones among the three libraries to identify clonal groups of B cells between libraries. Heavy or light chain genes were considered clonal between libraries if they shared identical junctional regions. Six heavy chain clonal groups were shared between the tumor and lymph node libraries (Fig. 2). The number of heavy chain clonal groups shared between the tumor and lymph node libraries was significantly greater than expected by chance (exact goodness-of-fit test p value 0.004) (Supplemental Table 11). We also identified five light chain clonal groups shared between the tumor and lymph node libraries (Fig. 3). The number of light chain clonal groups shared between tumor and lymph node libraries was also significantly greater than expected by chance (Exact Goodness-of-Fit p value 0.01) (Supplemental Table 12). We did not identify any heavy or light chain clonal groups shared between the blood library and the other two libraries. We conclude that several heavy and light chain clonal groups were shared between the libraries derived from the tumor and lymph node. These data indicate that in this patient, B cells present in the tumor and lymph node were clonally related to each other.
Binding analysis to cancer cells
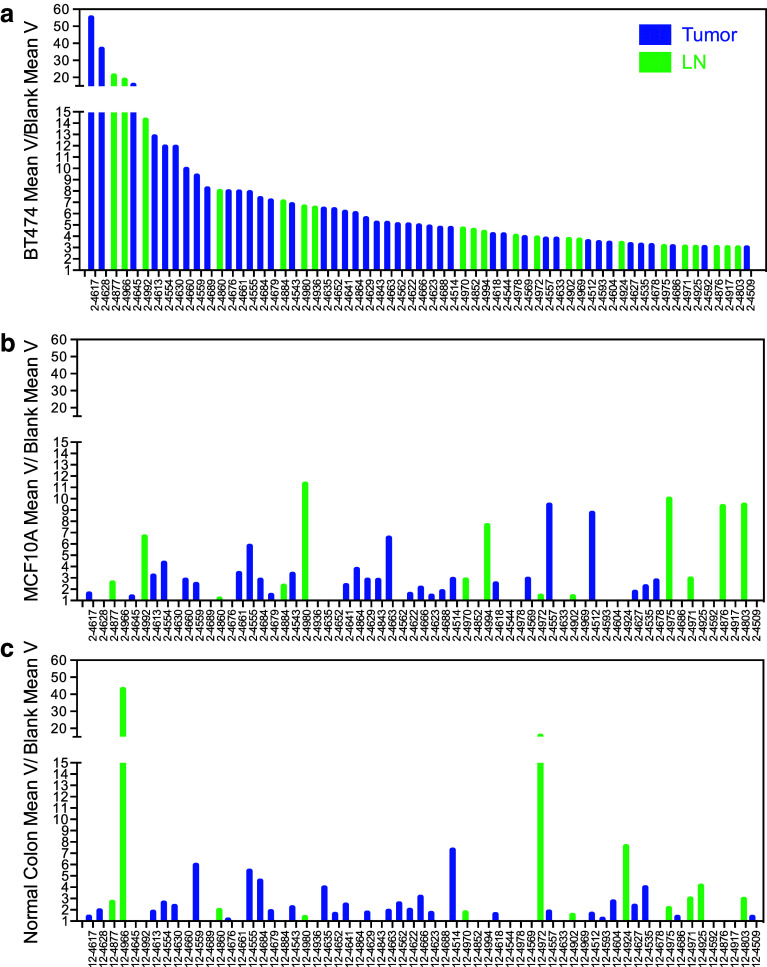
Each phage library was enriched for binding to autologous tumor lysates with three rounds of panning. After the final round of enrichment, randomly selected sets of clones from all three libraries were first tested for binding to BT474 breast cancer cell line lysates. Approximately 54 % of the sampled clones had intermediate binding and 21 % showed no binding over background (data not shown). The top 25 % nonduplicative BT474 binding clones among the three libraries (total 66 clones) had at least threefold higher binding than background (Fig. 4). Of these clones, 42 were from the tumor library, 24 were from the lymph node library, and none were from the blood library. These results demonstrate that tumor and lymph node provided the most tumor-binding scFvs after enrichment for binding to tumor cell lysates.
Fig. 4.
Binding enrichment of individual scFvs from each library. Each sample was tested in triplicate by ELISA against BT474 cell line lysates or negative control (casein). Only high binders are shown (a). Each high binder was tested in triplicate against normal breast cell line (MCF10A) lysates (b), normal colon tissue lysates (c), or negative control (casein). Bars represent the average of the calculated mean variance for three samples divided by the mean variance of the same scFv reading in a blank well to normalize for phage titer and plate variation
Specificity of the top 25 % tumor-binding clones
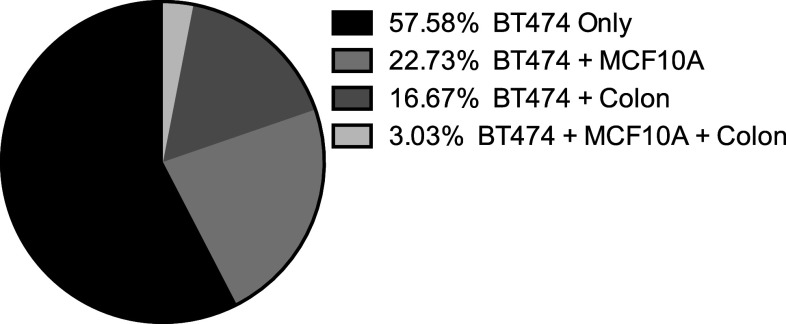
The top 25 % of BT474 binding clones were tested for binding to lysates from a noncancer breast cell line (MCF10A) and normal colon tissue (Fig. 4b, c). The majority (38/66) of these clones showed preferential binding to breast cancer cells, and these clones were evaluated further for binding to autologous tumor tissue (Fig. 5). Twelve clones bound with variable intensity to autologous tumor. Four of these 12 clones that bound autologous tumor had preferential binding to autologous tumor compared to autologous nontumor breast tissue (Fig. 6).
Fig. 5.
Evaluation of high binders after enrichment to various targets. Of the high binders tested, the majority was specific for BT474 cells. The percent of scFv binding to normal breast cell line lysates (MCF10A) and/or normal colon tissue lysates is shown
Fig. 6.
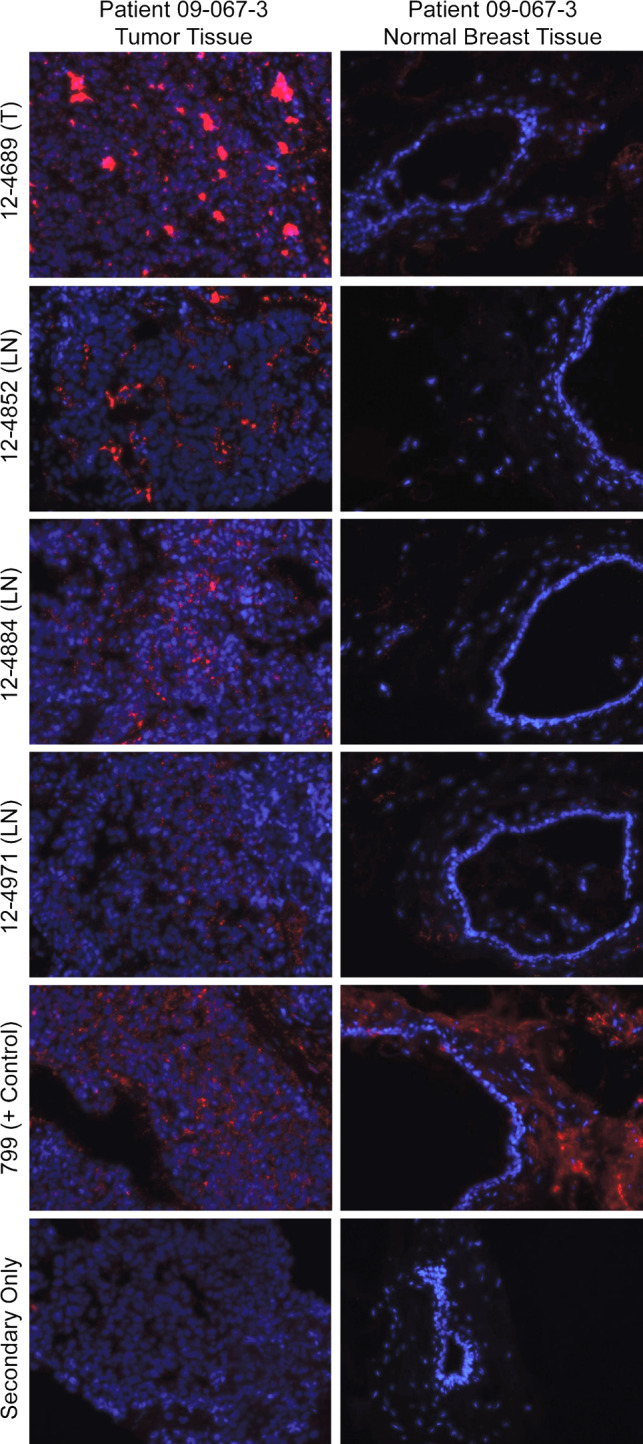
Binding of four different scFv to autologous patient tumor and normal tissues. One tumor (T) and three lymph node (LN)-derived antibodies demonstrated specific binding to tumor tissue and noncancer tissue of the same breast cancer patient. 799, a pan binder identified by our laboratory is shown as positive control, and tissue stained with secondary antibody only is used as a negative control
Clonal analysis of enriched scFv
Randomly selected phage clones from the libraries after the final round of enrichment to tumor cell lysate were sequenced to identify genetic relationships. Within the lymph node library, nine heavy chain and nine light chain clonal groups were identified (Supplemental Table 13). Most of the heavy chain and light chain groups were novel. However, one heavy chain group (HV3-9/J5) was also present in the pre-enrichment lymph node population. Another heavy chain group (HV4-31/J3) found only in the lymph node population after enrichment was identified as a clonal group shared between tumor and lymph node in the pre-enrichment analysis. Similarly, one light chain group (KV1-39/J1) found in the lymph node population after enrichment was identified as a clonal group shared between tumor and lymph node in the pre-enrichment analysis. One light chain group (KV3-20/J3) found only in the lymph node population after enrichment was identified in the tumor population in the pre-enrichment analysis. Within the tumor library, no heavy chain groups were identified after enrichment, whereas one light chain clonal group was identified. Within the blood library, no heavy or light chain clonal groups were identified after enrichment.
Between the tumor and lymph node libraries, five heavy chain and five light chain clonal groups were identified (Supplemental Table 14). Three heavy chain groups shared between tumor and lymph node pre-enrichment populations were also identified in the post-enrichment population (HV1-18/J3, HV1-2/J5, HV3-30/J3). One novel heavy chain clonal group (HV4-B/J4) was identified between tumor and lymph node after enrichment, whereas this heavy chain was only identified in the tumor pre-enrichment population. Another novel heavy chain clonal group (HV3-66/J6) was identified between tumor and lymph node after enrichment, whereas this heavy chain was only identified in the lymph node pre-enrichment population. Two light chain groups shared between tumor and lymph node pre-enrichment populations were also identified in the post-enrichment population (LV3-10/J2 and KV4-1/J4). One novel light chain clonal group (LV6-57/J2) was identified between tumor and lymph node after enrichment, whereas this light chain was only identified in the tumor pre-enrichment population. Another novel light chain clonal group (LV6-57/J2 with a different CDR3 region) was identified between tumor and lymph node after enrichment, whereas this light chain was only identified in the lymph node pre-enrichment population. Between the tumor and blood libraries or between the lymph node and blood libraries, no heavy or light chain clonal groups were identified after enrichment (Supplemental Table 14). These results demonstrate a genetic relationship between tumor and lymph node libraries after selecting for functional binding.
Discussion
In this study, we identified a genetic and functional relationship between B cells infiltrating a patient’s breast tumor and B cells present in the TDLN. B cells were observed in the tumor in scattered formation and in multiple ectopic germinal centers; these observations extend previous reports in which ectopic germinal centers were identified in breast cancers [3–6].
Heavy chain diversity of B cells in the tumor was similar to B cells in the TDLN and blood. The distribution of VH gene segments of B cells in the tumor was consistent with a previous report on invasive ductal carcinoma [3, 6]. A 2-mm core of tumor was used to create the tumor antibody library in this study, and we observed greater heavy chain gene diversity than found in other studies that used much smaller microdissected fragments of tumor [4, 5]. Together these observations indicate that several independent populations of B cells exist within the tumor tissue, particularly when larger pieces of tumor are studied.
We observed that replacement mutations were enriched over silent mutations in the CDR regions compared to the framework regions of light chains in the tumor and lymph node libraries that suggested antigen-mediated selection. A study on plasma cell infiltrated invasive ductal carcinomas observed similar results in a mutation analysis of light chains [6]. Other studies on TIL-B cells in breast cancer using phage-displayed libraries did not examine light chains but did observe evidence of antigen-mediated selection in unrestricted heavy chain populations [3–5]. Together, these results are important because they suggest that at least a portion of TIL-B cells have undergone somatic hypermutation and affinity maturation associated with antigen-mediated selection.
Clonal heavy and light chain groups were shared between the tumor and lymph libraries derived from the study patient. This study is the first to report a shared B cell lineage in two different locations in the same breast cancer patient. Only one other study has looked for overlap in the B cell populations between breast tumor and TDLN and was unable to find any relationship, possibly due to the small number of sequences examined [3]. The clonal relationship between tumor and lymph node in both the heavy and light chain populations was enhanced after enrichment for binding to autologous tumor lysates. This observation is important because it indicates that the relationship shared between tumor and lymph node B cells in this patient may be characterized by selection for tumor-associated antigens.
Our results suggest that tumor and TDLN are both good sources of B cells that express tumor-targeting antibodies. Even though the libraries constructed from the three different tissue sources were comparable in size and diversity, the tumor and the lymph node libraries yielded the greatest number of tumor-binding antibodies. To our knowledge, this is the first study to compare phage-displayed antibodies derived from different tissue sources in the same breast cancer patient. Other libraries derived from patients with breast cancer have provided tumor-binding phage-displayed antibodies. Antibodies to common tumor targets, such as epithelial mucin (MUC1) and HER2, or breast cancer cell lines have been isolated from libraries derived from TIL-B cells, TDLN, or blood [9–12, 30]. Antibodies that bind actin and ganglioside D3 in a tumor have been identified from libraries constructed from TIL-B cells in a medullary carcinoma [8, 31, 32]. Our study confirms previous reports that phage antibody libraries derived from the tissue of patients with breast cancer can be a rich resource of tumor-binding antibodies, a proportion of which showed specificity for autologous tumor over autologous nontumor breast tissue.
Within our top 25 % of BT474 lysate-binding antibodies, multiple clones bound to the patient’s own cancer. Furthermore, multiple phage-displayed antibodies bound specifically to the tumor tissue and not to corresponding normal breast tissue from the same patient. This is the first study to test tumor and lymph node-derived phage-displayed antibodies on matched normal and tumor tissue from the same breast cancer patient. Antibodies that bind selectively to cancer and not corresponding normal breast tissue may be particularly promising for therapy.
This study is the first report describing a shared B cell lineage in two different sources in the same patient with breast cancer. Heavy and light chains selected for tumor binding from the tumor and lymph node libraries were clonally related to each other indicating a physiologic relationship that may be important relative to the tumor-specific immune response. Very little published data are available describing the genetic relationship between B cells localized to tumor and B cells in the TDLN of patients with cancer. Only one other study looked for overlap in clonal groups identified within the tumor and TDLN and did not observe any among 58 tumor and 26 lymph node heavy chains examined [3]. Next-generation sequencing may provide a more thorough analysis of the B cell repertoire shared between tumor and TDLN [33].
Analysis of tissues derived from one patient allowed the observation of relationships between these tissues. Future experiments should expand the study of genetic relationships between tumor and lymph node to more patients.
A limitation of this study is that combinatorial phage-displayed antibodies do not recapitulate the original B cell heavy and light chain combinations.
In conclusion, we found that B cells from the tumor of a patient with breast cancer were clonally related to B cells in her TDLN. We enriched phage-displayed antibody libraries derived from TIL-B cells and B cells from the TDLN of this patient for antibodies that bound autologous tumor lysates. Multiple phage-displayed antibodies from tumor and lymph node libraries bound with specificity to the same patient’s tumor tissue. We also identified clonal genetic relationships between antibodies selected from the tumor and lymph node phage libraries after enrichment suggesting a functional relationship between the patient’s B cells and her breast cancer. Antibodies that bind preferentially to a cancer and not the corresponding normal tissue, such as the ones identified in this report, have potential value for research on mechanisms of immune responses and possibly therapy.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We acknowledge Stephanie Pero, Girja Shukla, Yujing Sun, and Donald Weaver for helpful discussions and Chelsea Carmen for reagent preparation. We also thank Seth Harlow, Ted James, and Patti Lutton for their help in recruiting patients for this study. Finally, we recognize the administrative support of Shelley Bissonnette, Eileen Caffrey, and Sarah Howe in securing financial support and maintaining regulatory obligations. This research was supported by the SD Ireland Cancer Research Fund, the University of Vermont Department of Surgery, and Department of Defense Predoctoral Training Award #W81XWH-08-1-0756.
Conflict of interest
The authors of this paper report no conflict of interest with regard to this paper.
Abbreviations
- CDR
Complementarity determining region
- ELISA
Enzyme-linked immunosorbent assay
- HER2
Human epidermal growth factor receptor 2
- IMGT
ImMunoGeneTics information system
- Mean V
Value of the mean slope
- MUC1
Epithelial mucin
- PBMCs
Peripheral blood mononuclear cells
- PBS
Phosphate-buffered saline
- PEG
Polyethylene glycol
- scFv
Single-chain variable fragment
- TBS
Tris-buffered saline
- TDLN
Tumor-draining lymph nodes
- TIL-B cells
Tumor-infiltrating B lymphocytes
- TMB
3,3′,5,5′-Tetramethylbenzidine
References
- 1.Spaner D, Bahlo A. B lymphocytes in cancer immunology. In: Medin J, Fowler D, editors. Experimental and applied immunotherapy. New York: Humana Press; 2011. pp. 37–57. [Google Scholar]
- 2.Nielsen JS, Nelson BH. Tumor-infiltrating B cells and T cells: working together to promote patient survival. Oncoimmunology. 2012;1:1623–1625. doi: 10.4161/onci.21650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coronella JA, Spier C, Welch M, et al. Antigen-driven oligoclonal expansion of tumor-infiltrating B cells in infiltrating ductal carcinoma of the breast. J Immunol. 2002;169:1829–1836. doi: 10.4049/jimmunol.169.4.1829. [DOI] [PubMed] [Google Scholar]
- 4.Nzula S, Going JJ, Stott DI. Antigen-driven clonal proliferation, somatic hypermutation, and selection of B lymphocytes infiltrating human ductal breast carcinomas. Cancer Res. 2003;63:3275–3280. [PubMed] [Google Scholar]
- 5.Simsa P, Teillaud J-L, Stott DI, et al. Tumor-infiltrating B cell immunoglobulin variable region gene usage in invasive ductal breast carcinoma. Pathol Oncol Res. 2005;11:92–97. doi: 10.1007/BF02893374. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Ylera F, Boston M, et al. Focused antibody response in plasma cell-infiltrated non-medullary (NOS) breast cancers. Breast Cancer Res Treat. 2007;104:129–144. doi: 10.1007/s10549-006-9409-3. [DOI] [PubMed] [Google Scholar]
- 7.Hansen MH, Nielsen HV, Ditzel HJ. Translocation of an intracellular antigen to the surface of medullary breast cancer cells early in apoptosis allows for an antigen-driven antibody response elicited by tumor-infiltrating B cells. J Immunol. 2002;169:2701–2711. doi: 10.4049/jimmunol.169.5.2701. [DOI] [PubMed] [Google Scholar]
- 8.Kotlan B, Simsa P, Teillaud J-L, et al. Novel ganglioside antigen identified by B cells in human medullary breast carcinomas: the proof of principle concerning the tumor-infiltrating B lymphocytes. J Immunol. 2005;175:2278–2285. doi: 10.4049/jimmunol.175.4.2278. [DOI] [PubMed] [Google Scholar]
- 9.Pavoni E, Monteriù G, Santapaola D, et al. Tumor-infiltrating B lymphocytes as an efficient source of highly specific immunoglobulins recognizing tumor cells. BMC Biotechnol. 2007;7:70. doi: 10.1186/1472-6750-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rothe A, Klimka A, Tur MK, et al. Construction of phage display libraries from reactive lymph nodes of breast carcinoma patients and selection for specifically binding human single chain Fv on cell lines. Int J Mol Med. 2004;14:729–735. doi: 10.3892/ijmm.14.4.729. [DOI] [PubMed] [Google Scholar]
- 11.Belimezi MM, Papanastassiou D, Merkouri E, et al. Growth inhibition of breast cancer cell lines overexpressing Her2/neu by a novel internalized fully human Fab antibody fragment. Cancer Immunol Immunother. 2006;55:1091–1099. doi: 10.1007/s00262-005-0100-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ayat H, Burrone OR, Sadghizadeh M, et al. Isolation of scFv antibody fragments against HER2 and CEA tumor antigens from combinatorial antibody libraries derived from cancer patients. Biologicals. 2013;41(6):345–354. doi: 10.1016/j.biologicals.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 13.Krag DN, Anderson SJ, Julian TB, et al. Sentinel-lymph-node resection compared with conventional axillary-lymph-node dissection in clinically node-negative patients with breast cancer: overall survival findings from the NSABP B-32 randomised phase 3 trial. Lancet Oncol. 2010;11:927–933. doi: 10.1016/S1470-2045(10)70207-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Cicco C, Chinol M, Paganelli G. Intraoperative localization of the sentinel node in breast cancer: technical aspects of lymphoscintigraphic methods. Semin Surg Oncol. 1998;15:268–271. doi: 10.1002/(SICI)1098-2388(199812)15:4<268::AID-SSU16>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 15.Setiadi AF, Ray NC, Kohrt HE, et al. Quantitative, architectural analysis of immune cell subsets in tumor-draining lymph nodes from breast cancer patients and healthy lymph nodes. PLoS ONE. 2010;5:e12420. doi: 10.1371/journal.pone.0012420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang AY, Bhattacharya N, Mu J, et al. Spatial organization of dendritic cells within tumor draining lymph nodes impacts clinical outcome in breast cancer patients. J Transl Med. 2013;11:242. doi: 10.1186/1479-5876-11-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei S, Shreiner AB, Takeshita N, et al. Tumor-induced immune suppression of in vivo effector T-cell priming is mediated by the B7-H1/PD-1 axis and transforming growth factor beta. Cancer Res. 2008;68:5432–5438. doi: 10.1158/0008-5472.CAN-07-6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Q, Lao X, Pan Q, et al. Adoptive transfer of tumor reactive B cells confers host T cell immunity and tumor regression. Clin Cancer Res. 2011;17(15):4987–4995. doi: 10.1158/1078-0432.CCR-11-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zirakzadeh AA, Marits P, Sherif A, Winqvist O. Multiplex B cell characterization in blood, lymph nodes, and tumors from patients with malignancies. J Immunol. 2013;190:5847–5855. doi: 10.4049/jimmunol.1203279. [DOI] [PubMed] [Google Scholar]
- 20.Barbas CF, III, Burton DR, Scott JK, Silverman GJ. Phage display: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 21.Brochet X, Lefranc M-P, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehr R, Sternberg-Simon M, Michaeli M, Pickman Y. Models and methods for analysis of lymphocyte repertoire generation, development, selection and evolution. Immunol Lett. 2012;148:11–22. doi: 10.1016/j.imlet.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 23.Dixon WJ, Massey FJ. Introduction to statistical analysis. 4. New York: McGraw-Hill Science, Engineering & Mathematics; 1983. [Google Scholar]
- 24.Kao J, Salari K, Bocanegra M, et al. Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PLoS ONE. 2009;4:e6146. doi: 10.1371/journal.pone.0006146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shukla GS, Krag DN. A sensitive and rapid chemiluminescence ELISA for filamentous bacteriophages. J Immunoass Immunochem. 2005;26:89–95. doi: 10.1081/IAS-200051990. [DOI] [PubMed] [Google Scholar]
- 26.Sun Y, Shukla GS, Kennedy GG, et al. Biopanning phage-display libraries on small tissue sections captured by laser capture microdissection. J Biotech Res. 2009;1:55–63. [PMC free article] [PubMed] [Google Scholar]
- 27.Chang B, Casali P. The CDR1 sequences of a major proportion of human germline Ig VH genes are inherently susceptible to amino acid replacement. Immunol Today. 1994;15:367–373. doi: 10.1016/0167-5699(94)90175-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muramatsu M, Kinoshita K, Fagarasan S, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/S0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 29.Stavnezer J, Guikema JEJ, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thie H, Toleikis L, Li J, et al. Rise and fall of an anti-MUC1 specific antibody. PLoS ONE. 2011;6:e15921. doi: 10.1371/journal.pone.0015921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansen MH, Nielsen H, Ditzel HJ. The tumor-infiltrating B cell response in medullary breast cancer is oligoclonal and directed against the autoantigen actin exposed on the surface of apoptotic cancer cells. Proc Natl Acad Sci USA. 2001;98:12659–12664. doi: 10.1073/pnas.171460798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kotlan B, Simsa P, Foldi J, et al. Immunoglobulin repertoire of B lymphocytes infiltrating breast medullary carcinoma. Hum Antibodies. 2003;12:113–121. [PubMed] [Google Scholar]
- 33.Lavinder JJ, Wine Y, Giesecke C, et al. Identification and characterization of the constituent human serum antibodies elicited by vaccination. Proc Natl Acad Sci USA. 2014;111(6):2259–2264. doi: 10.1073/pnas.1317793111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.