Abstract
Immune checkpoint molecules are highly relevant as potential prognostic markers and therapeutic targets in malignant diseases. HVEM belongs to the TNF receptor family and provides stimulatory as well as inhibitory signals depending on the ligand. Abnormal HVEM expression has been described in various malignancies, but the role in AML is unknown. Here we report extensive data on HVEM surface protein expression analyzed by flow cytometry on bone marrow leukemic cells of 169 AML patients at diagnosis. An independent cohort of 512 AML patients was analyzed for HVEM mRNA expression in bone marrow samples by Affymetrix microarrays. Consistently for both cohorts and methods, we show that HVEM was differentially expressed and that expression levels were associated with defined genetic markers. HVEM expression was lower in cases with FLT3-ITD (p = 0.001, p < 0.001), with mutations in NPM1 (p = 0.001, p < 0.001) or with the combination of NPM1 mutation and FLT3 wild type (p = 0.049, p = 0.050), while a biallelic mutation in CEBPA correlated positively with higher HVEM expression (p = 0.015, p < 0.001). In a differential gene expression analysis, we found 13 genes including HOXA9, MEIS1 and MN1 that were closely associated with HVEM expression. Besides, four gene sets closely linked to immunity were enriched in HVEM high samples. Finally, high expression of HVEM was associated with a trend toward longer relapse-free survival. The results of this study provide new information on the potential significance of HVEM in AML.
Electronic supplementary material
The online version of this article (doi:10.1007/s00262-015-1755-8) contains supplementary material, which is available to authorized users.
Keywords: Acute myeloid leukemia (AML), Herpesvirus entry mediator (HVEM), Costimulation, Immune checkpoint molecules, Immunophenotyping, Gene expression analysis
Introduction
Herpesvirus entry mediator (HVEM) was originally discovered as the entry route for herpes simplex virus, but has since been assigned to the growing number of immune checkpoint molecules. It is a member of the tumor necrosis factor receptor (TNFR) family and interacts with other TNFR (LIGHT, lymphotoxin-α) as well as Ig superfamily (BTLA, CD160) molecules, both in trans and in cis position [1]. Although it can provide stimulatory (via interaction with LIGHT or lymphotoxin-α) [2, 3] as well as inhibitory (via interaction with BTLA or CD160) [4] signals depending on the ligand, its overall inhibitory function seems to be dominant as demonstrated by studies with HVEM knockout mice [5]. Within normal cells, HVEM is highly expressed mainly on non-activated B and T lymphocytes, but can also be found at more moderate levels on a wide range of other hematopoietic (NK cells, monocytes, immature dendritic cells) as well as non-hematopoietic cells.
Initially, HVEM and its ligands were perceived as potential therapeutic targets in autoimmunity and transplant rejection [6]. However, more recently the focus shifted toward their significance in tumor immunology and immunotherapy. Abnormal expression has been described in B cell malignancies [7] and in malignant melanoma [8], but recently also in esophageal squamous cell cancer [9], hepatocellular carcinoma [10] and colorectal cancer [11]. In contrast, there is little knowledge about the role of the HVEM network in myeloid diseases such as acute myeloid leukemia (AML). Expression of HVEM was found in only one out of seven AML cell lines [7] and was generally described to be low in leukemias of myeloid origin [12]. Based on the analysis of seven AML cell lines, it was speculated that the expression might be higher on more mature (FAB type M5) compared to immature (FAB type M0–M2) leukemias [13]. Within the latter study, HVEM expression was also demonstrated on primary AML cells of eight bone marrow (BM) samples, albeit at rather low levels.
Here we report data based on flow cytometric HVEM surface expression analysis on CD33+ leukemic cells in BM samples of 169 AML patients at diagnosis. For an independent cohort of 512 AML patients, HVEM mRNA expression was analyzed by Affymetrix microarrays, also in BM samples from diagnosis. Expression levels were correlated with morphologic, cytogenetic and molecular characteristics of the disease and with the prognosis of the patients with respect to relapse-free survival. Besides, differential gene expression analysis and gene set enrichment analysis were performed to study whether HVEM expression was part of a larger functional context. The results of this study cast new light on the potential significance of this coinhibitory molecule as prognostic marker as well as therapeutic target in AML.
Subjects and methods
Patient cohorts and routine diagnostics
Surface expression analysis by flow cytometry was done on BM samples taken for routine diagnostics at the time of diagnosis from 169 AML patients, excluding APL. All patients of this cohort were included in the AML registry of the German AML Cooperative Group (AML-CG) and treated according to their treatment recommendations. Patient characteristics are presented in Table 1a. Expression of mRNA was analyzed by microarray on BM samples from an independent cohort of 512 patients at the time of diagnosis. All patients of this cohort were included in the AML-CG 99 trial (NCT00266136). Characteristics of these patients are summarized in Table 1b. For both cohorts, all patients gave written informed consent for scientific workup of the samples. Within clinical routine diagnostics, the samples were analyzed morphologically for cytogenetic and molecular (mutations in nucleophosmin = NPM1, internal tandem repeats in Fms-like tyrosine kinase 3 = FLT3, mutations in CCAAT/enhancer-binding protein alpha = CEBPA) aberrations. The AML specimens were classified into morphological subgroups (FAB criteria), except for cases of secondary AML (sAML). Additionally, patients were assigned to three different risk groups (favorable, intermediate, adverse) according to cytogenetics (refined Medical Research Council (MRC) criteria) [14].
Table 1.
Characteristics of patients used for protein analysis (a) and mRNA analysis (b)
| (a) Patients for protein analysis | n = 169 |
|---|---|
| Age | |
| Median (range) | 59 (20–92) |
| Gender, n (%) | |
| Male | 93 (55 %) |
| Female | 76 (45 %) |
| FAB, n (%) | |
| M0 | 7 (4 %) |
| M1 | 46 (27 %) |
| M2 | 25 (15 %) |
| M4 | 41 (24 %) |
| M5 | 15 (9 %) |
| M6 | 3 (2 %) |
| M7 | 1 (0.6 %) |
| Cytogenetics, n (%) | |
| t (8;21) | 4 (2 %) |
| inv(16)/t(16;16)(p13;q22) | 4 (2 %) |
| Normal karyotype | 90 (53 %) |
| t(9;11)(p21-22;q23) or t(11;19)(q23;p13) | 5 (3 %) |
| Abnormalities not classified as favorable or adverse | 20 (12 %) |
| Complex karyotype | 16 (9 %) |
| Other adverse risk abnormalities | 20 (12 %) |
| Cytogenetic risk group, n (%) | |
| Favorable | 8 (5 %) |
| Intermediate | 115 (68 %) |
| Adverse | 36 (21 %) |
| Mutations, n (%) | |
| NPM1 mut/FLT3 wt (normal karyotype) | 30 (18 %) |
| NPM1 mut/FLT3-ITD (normal karyotype) | 23 (14 %) |
| NPM1 wt/FLT3-ITD (normal karyotype) | 5 (3 %) |
| ELN, n (%) | |
| Favorable | 40 (24 %) |
| Intermediate | 78 (46 %) |
| Adverse | 40 (24 %) |
| (b) Patients for RNA analysis | n = 512 |
|---|---|
| Age | |
| Median (range) | 58 (18–85) |
| Gender, n (%) | |
| Male | 257 (50 %) |
| Female | 255 (50 %) |
| FAB, n (%) | |
| M0 | 21 (4 %) |
| M1 | 98 (19 %) |
| M2 | 127 (25 %) |
| M4 | 106 (21 %) |
| M5 | 56 (11 %) |
| M6 | 18 (4 %) |
| M7 | 3 (0.6 %) |
| Cytogenetics, n (%) | |
| t (8;21) | 30 (6 %) |
| inv(16)/t(16;16)(p13;q22) | 38 (7 %) |
| Normal Karyotype | 209 (41 %) |
| t(9;11)(p21-22;q23) or t(11;19)(q23;p13) | 22 (4 %) |
| Abnormalities not classified as favorable or adverse | 85 (17 %) |
| Complex karyotype | 69 (13 %) |
| Other adverse risk abnormalities | 53 (10 %) |
| Cytogenetic risk group, n (%) | |
| Favorable | 68 (13 %) |
| Intermediate | 312 (61 %) |
| Adverse | 126 (25 %) |
| Mutations, n (%) | |
| NPM1 mut/FLT3 wt (normal karyotype) | 54 (11 %) |
| NPM1 mut/FLT3-ITD (normal karyotype) | 54 (11 %) |
| NPM1 wt/FLT3-ITD (normal karyotype) | 27 (5 %) |
| ELN, n (%) | |
| Favorable | 143 (28 %) |
| Intermediate | 222 (43 %) |
| Adverse | 127 (25 %) |
HVEM protein surface expression analysis
BM samples of 169 AML patients at time of diagnosis were analyzed by flow cytometry for surface expression of HVEM (clone eBioHVEM-122, eBioscience, San Diego, CA, USA) on the CD33+/SSClow (clone D3HL60.251, Beckman-Coulter, Krefeld, Germany) leukemic cell population. Specific fluorescence intensity (SFI) was calculated by dividing the mean fluorescence intensity (MFI) of the HVEM antibody by the MFI of the respective isotype control.
PCR for HVEM mRNA expression analysis
For analysis of the correlation between surface protein and mRNA expression of HVEM, 14 patients of the protein analysis cohort were chosen for PCR analysis, based on the availability of material for mRNA extraction; 8 of the samples were selected from the low end of surface protein expression (SFI between 0.3 and 0.6) and 6 from the top end (SFI between 7.1 and 11.0). mRNA expression analysis was done by qPCR using the Taqman Assay of Life Technologies (Darmstadt, Germany). HVEM expression relative to ABL as housekeeping gene was determined and compared between both groups.
HVEM mRNA expression analysis by Affymetrix
BM samples of 512 patients at time of diagnosis were analyzed using Affymetrix U133A+ B and Affymetrix U133 Plus2.0 microarrays (Affymetrix, Santa Clara, CA) as published previously [15]. The microarray data have been deposited in the Gene Expression Omnibus with the accession number GSE37642. Expression data are presented as log2 throughout the study.
Differential gene expression analysis by LIMMA and GSEA
Gene expression data from the microarray analysis were evaluated with respect to differential gene expression for the sub-cohort of 275 patients within the mRNA cohort that was also used for analysis of relapse-free survival (see below). The Linear Models for Microarray Data (Limma) package [16] was used to compute differentially regulated genes based on HVEM high (n = 69) versus HVEM low (n = 206) groups. To balance gene expression data for sensitivity and specificity, we filtered genes for an adjusted p value of ≤0.001 and fold change (FC) ≥1. Gene set enrichment analysis (GSEA) was performed with GSEA software (BROAD Institute, Cambridge, MA) to assess significant changes in gene expression levels [17]. The GSEA was run with default settings and compared with “Signal2Noise” to the “c2.cp.kegg.v4.0” collection from the Molecular Signatures Database MsigDB (http://www.broadinstitute.org/gsea/msigdb/index.jsp) consisting of 186 gene sets from the KEGG pathway database (http://www.genome.jp/kegg/pathway.html).
Statistical analyses
All results of distribution analyses are presented in box-and-whisker plots. As interindividual values were not distributed normally, differences between groups were assessed using the nonparametric two-tailed Mann–Whitney U test. Relapse-free survival (RFS) was analyzed within the sub-cohorts of patients that achieved a CR and had clinical data available (n = 97 for protein, n = 275 for mRNA expression analysis). RFS time was measured from the date of diagnosis (protein analysis cohort) or the date of CR (mRNA analysis cohort) to the date of relapse or death. Patients not known to have any of these events at last follow-up or patients undergoing allogeneic stem cell transplantation (SCT) were censored, while patients with an allogeneic SCT in first CR were analyzed separately (n = 26 for protein, n = 31 for mRNA expression analysis). Survival was estimated according to the Kaplan–Meier method. The log-rank test was used to assess statistical significance. Cox regression was used to assess the association of HVEM expression level with RFS. p values < 0.05 were considered statistically significant. All analyses were calculated with PASW Statistics 21 (SPSS, Chicago, IL, USA).
Results
HVEM was differentially expressed between AML patients on protein and RNA level
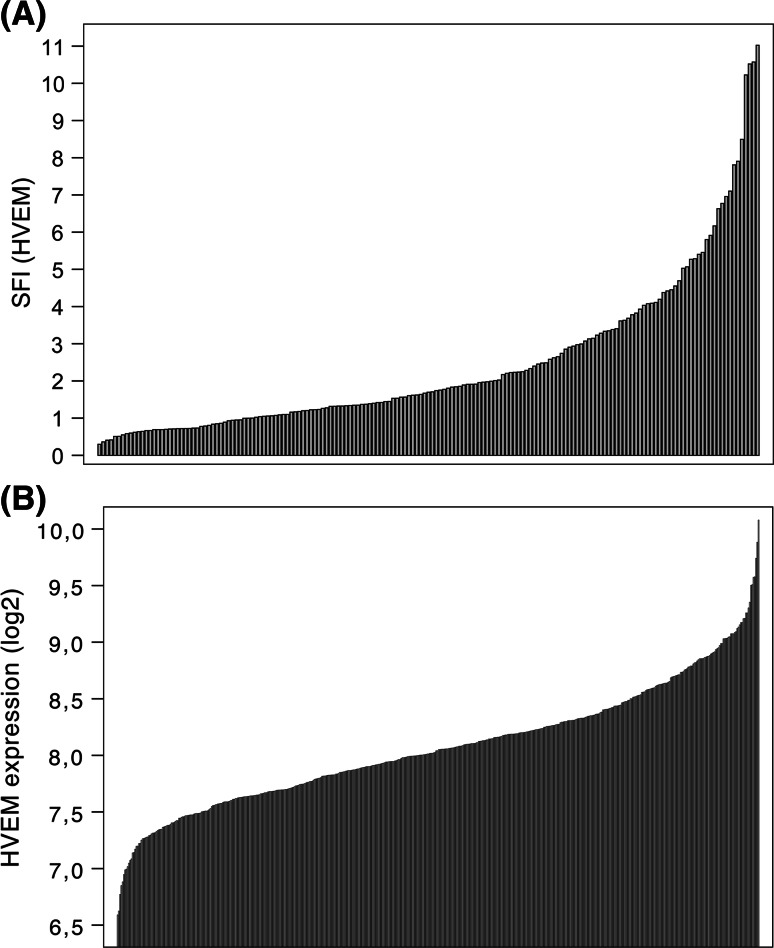
HVEM expression was analyzed for all patients of both cohorts (Fig. 1). Expression of HVEM on the protein level was quite variable within the 169 samples, with the SFI ranging from 0.3 to 11.0. The median SFI for all samples was 1.7, but almost a quarter of the samples (40 of 169) were completely negative for HVEM (SFI ≤ 1), while the highest quartile had an SFI of ≥3.2 (Fig. 1a).
Fig. 1.
Distribution of HVEM expression on leukemic cells of AML patients. HVEM surface protein (a) and mRNA (b) expression was measured in independent cohorts for 169 and 512 patient samples, respectively. Distribution of expression levels is shown
Expression of HVEM on the mRNA level similarly proved to be variable between 512 patients. Expression levels ranged from 6.6 to 10.1, with a median of 8.0, and the highest quartile had an expression level of ≥8.4 (Fig. 1b).
Correlation between mRNA and protein expression of HVEM was tested for 14 samples of the protein analysis cohort. HVEM mRNA expression relative to ABL as housekeeping gene was compared between samples with low and high HVEM protein expression (Supplementary Fig. 1). Median relative HVEM expression in the low SFI group was 22.2 %, while it was 92.7 % in the high SFI group (p = 0.02).
HVEM expression was independent of sex, age and AML morphology
For both cohorts and methods, there was no evidence for an association with the sex of the patient or the age at diagnosis (data not shown). When correlating protein surface expression levels with FAB subgroups (Supplementary Fig. 2A), we could not confirm the hypothesis that it is higher on monocytoid leukemias as represented by AML M5 (n = 15; median SFI 1.9; p = 0.556 vs. all other cases) or AML M4/5 (n = 56; median SFI 1.8; p = 0.377 vs. all other cases). Instead, we found slightly increased expression in cases of AML M2 (n = 25; median SFI 2.3; p = 0.035).
Similarly, when correlating mRNA expression levels with FAB subgroups (Supplementary Fig. 2B), levels in AML M5 (n = 56; median expression 8.0; p = 0.219 vs. all other cases) or AML M4/5 (n = 162; median 8.0; p = 0.553 vs. all other cases) were not different from those of non-monocytoid leukemias. In cases of AML M6, expression was lower than the rest (n = 18; median 7.7; p = 0.010). Taken together, HVEM expression levels did not significantly and consistently differ between AMLs with different morphology.
HVEM expression was largely independent of cytogenetic risk classification
A potential association between HVEM expression levels and cytogenetic risk classification was analyzed (Supplementary Fig. 3). Within the cohort for protein surface expression analysis, cytogenetic risk classification according to the refined MRC criteria could be done for 159 patients, and HVEM expression was compared between the favorable, intermediate and adverse groups (n = 8, 115 and 36, respectively; see Supplementary Fig. 3A). We found that patients in the adverse risk group showed a significantly higher expression than those in the intermediate group (median SFI of 2.1 vs. 1.4; p = 0.015) or than those in the favorable and intermediate group combined (median SFI of 2.1 vs. 1.5; p = 0.023).
This finding, however, could not be reproduced in the mRNA expression analysis. Cytogenetic risk classification could be done for 506 patients (n = 68, 312 and 126, respectively; see Supplementary Fig. 3B). Median expression in the adverse risk group was 8.1, which was neither higher than that in the intermediate group (p = 0.454) nor in the favorable and intermediate group combined (p = 0.936).
HVEM expression was associated with several molecular markers
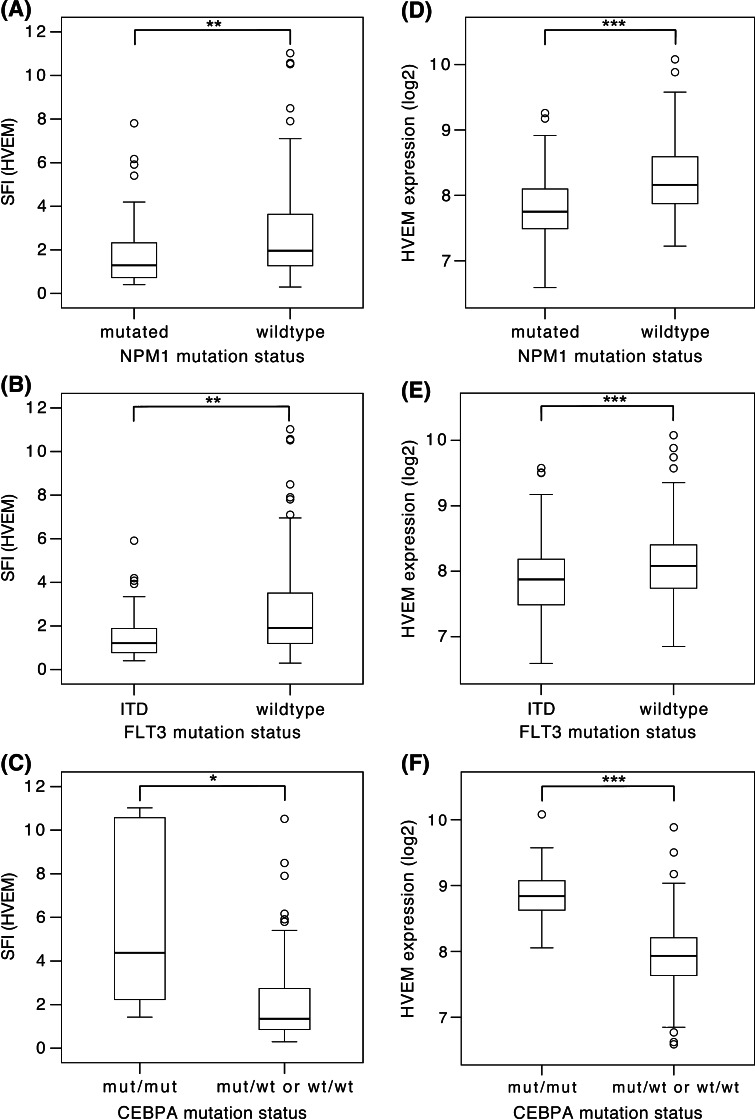
Next, HVEM expression intensity was correlated with single molecular aberrations in the leukemic cells (Fig. 2). Within the cohort for protein surface expression analysis, the mutation status was known for 166 patients in case of NPM1 (68 mutated and 98 wild type), for 165 patients in case of FLT3-ITD (46 ITD and 119 wild type) and for 88 patients in case of CEBPA (5 biallelic mutations and 83 monoallelic mutations or wild type). We found a significantly lower expression of HVEM in cases with a mutation in NPM1 (median SFI of 1.3 vs. 2.0; p = 0.001; Fig. 2a) and in cases of FLT3-ITD (median SFI of 1.2 vs. 1.9; p = 0.001; Fig. 2b), but considerably higher expression in cases with biallelic mutation in CEBPA (median SFI of 4.4 vs. 1.3; p = 0.015; Fig. 2c). When the analysis was reduced to the cases with normal karyotype, these differences still held true for NPM1 (n = 53 and 36; median SFI of 1.3 vs. 2.6; p = 0.003) and CEBPA (n = 5 and 65; median SFI of 4.4 vs. 1.3; p = 0.008), but not for FLT3 (p = 0.285). The prognostically favorable subgroup of cases with NPM1 mut/FLT3 wt within the normal karyotype leukemias also showed significantly lower expression compared to all other cases with normal karyotype (n = 30 and 59; median SFI of 1.3 vs. 1.9; p = 0.049).
Fig. 2.
Association of HVEM expression with molecular markers. HVEM surface protein (a–c) and mRNA (d–f) expression levels were correlated with the mutational status of NPM1 (a, d), FLT3 (b, e) and CEBPA (c, f). Significant and consistent differences in expression levels were found for all three mutations. *p < 0.05; **p < 0.01; ***p < 0.001
All of these associations could be confirmed in the second patient cohort by mRNA expression analysis. The mutation status was known for 287 patients in case of NPM1 (116 mutated and 171 wild type), for 502 patients in case of FLT3 (124 ITD and 378 wild type) and for 184 patients in case of CEBPA (10 biallelic mutations and 174 monoallelic mutations or wild type). We found a significantly lower expression of HVEM in cases with a mutation in NPM1 (median expression of 7.8 vs. 8.2; p < 0.001; Fig. 2d) and in cases with an FLT3-ITD (median of 7.9 vs. 8.1; p < 0.001; Fig. 2e). Again, cases with a biallelic mutation in CEBPA showed a considerably higher HVEM expression (median of 8.8 vs. 7.9; p < 0.001; Fig. 2f). When the analysis was reduced to the cases with normal karyotype, these differences held true for all three molecular aberrations: NPM1 (n = 108 and 94; median of 7.8 vs. 8.2; p < 0.001), FLT-ITD3 (n = 82 and 127; median of 7.9 vs. 8.1; p < 0.001) and CEBPA (n = 10 and 174; median of 8.8 vs. 7.9; p < 0.001). The prognostically favorable subgroup of cases with NPM1 mut/FLT3 wt within the normal karyotype leukemias also showed significantly lower expression compared to all other cases with normal karyotype (n = 54 and 148; median of 7.8 vs. 8.0; p = 0.050).
High HVEM expression was significantly associated with several single genes implied in AML pathogenesis and with immunity-related pathways
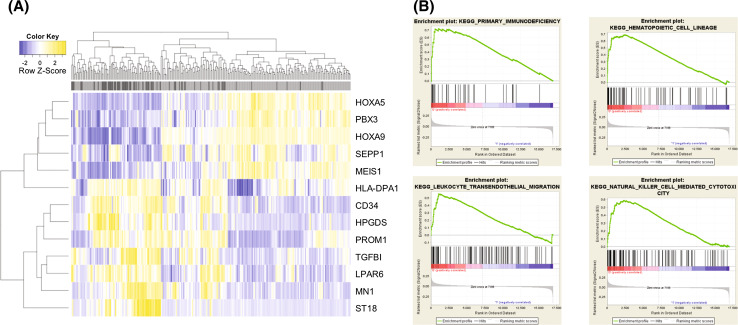
Gene expression data from the microarray analysis were studied for 275 patients within the mRNA expression analysis cohort with complete remission (CR) and clinical data available. Based on the expression data described above, we compared the smaller group with high HVEM expression (upper quartile; n = 69) to the larger group with low HVEM expression (lower three quartiles; n = 206). Limma analysis showed 953 differentially expressed probe sets (p < 0.05). Upregulated expression in the HVEM high group was seen in 578 probe sets, whereas 375 probe sets showed lower expression. Thirteen probe sets were highly significant at p ≤ 0.001 and fold change ≥1 (Supplementary Table 1). A heat map of the top 13 differentially expressed probe sets is shown in Fig. 3a. Remarkably, genes like HOXA9, MEIS1, HOXA5 and PBX3, which are already known to have implications in AML pathogenesis [18–20], showed significant downregulation, whereas MN1, a gene that is associated with inferior survival in AML [21], was significantly upregulated.
Fig. 3.
Association of HVEM expression with single genes implied in AML pathogenesis and with immunity-related pathways. Differential gene expression between the HVEM high and the HVEM low group was analyzed for 275 patients of the mRNA expression analysis cohort. a Limma analysis showed 13 probe sets with highly significant (p ≤ 0.001 and fold change ≥1) differences for single genes between HVEM high (dark gray) and HVEM low (light gray). A heat map of the top 13 differentially expressed probe sets is shown. b Gene set enrichment analysis resulted in four gene sets enriched in the HVEM high group at p < 0.05 and at a false discovery rate <0.25
To test for an association with gene sets, a GSEA analysis was performed. Eleven gene sets were enriched in the HVEM high group at p < 0.05. Four gene sets (primary immunodeficiency, hematopoietic cell lineage, leukocyte transendothelial migration and natural killer cell-mediated cytotoxicity) were enriched at a false discovery rate < 0.25 (Supplementary Table 2 and Fig. 3B).
High HVEM expression was associated with a trend toward better RFS in the ELN intermediate risk groups or after allogeneic SCT
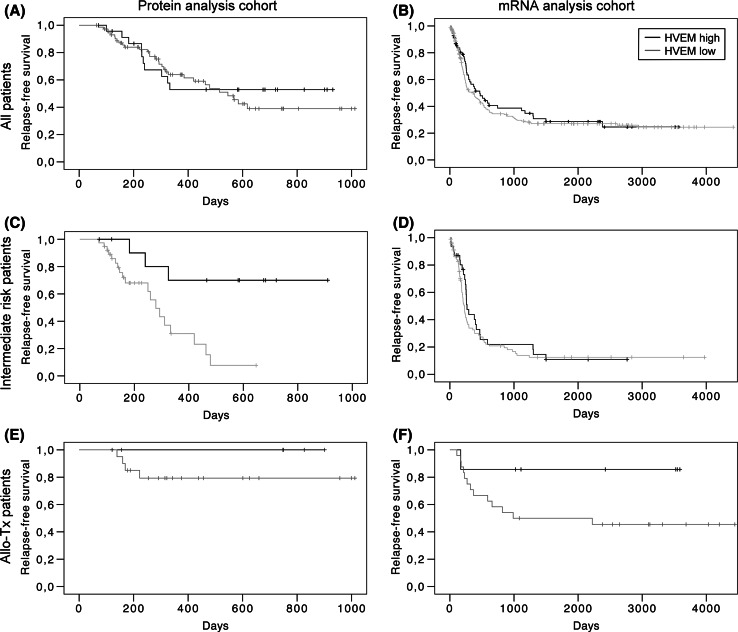
Finally, HVEM expression levels were correlated with the clinical course of the disease after CR (Fig. 4). Data from 97 and 275 patients, respectively, were available for analysis of RFS from both cohorts. Patients undergoing allogeneic SCT were censored at the time of transplantation. Based on the expression data described above, we compared the smaller group with high HVEM expression (upper quartile; n = 24 and n = 69) to the larger group with low HVEM expression (lower three quartiles; n = 73 and n = 206).
Fig. 4.
Association of HVEM expression with prognosis. Patients that achieved CR after induction chemotherapy were classified into two groups according to HVEM expression level (high: upper quartile; low: lower three quartiles). Relapse-free survival in both groups was compared for all evaluable patients (a, b), for all patients in the intermediate risk group according to ELN classification (c, d) and for all patients with allogeneic SCT in first CR (e, f), both within the protein analysis cohort (a, c, e) and the mRNA analysis cohort (b, d, f)
No difference in RFS was found between those two groups when all evaluable patients were included. For the surface protein analysis cohort, median RFS was 333 and 420 days, respectively (Fig. 4a); for the mRNA analysis cohort, median RFS was 467 and 377 days (Fig. 4b), but neither was statistically different between both groups. We then specified the analysis to the intermediate risk subgroup (intermediate I and intermediate II combined) of patients according to the European LeukemiaNet (ELN) criteria, which incorporate cytogenetic and molecular genetic data about the disease [22]. Within this subgroup of the surface protein analysis cohort (n = 50), high expression of HVEM correlated with significantly higher RFS (p = 0.005; Fig. 4c). Median RFS in the HVEMhigh group (n = 12) was not reached after 910 days, while it was 279 days in the HVEMlow group (n = 38). In contrast, no difference was seen in the favorable risk group (n = 8 and n = 24, respectively; median RFS of 515 and 584 days, respectively; p = 0.919); the number of patients in the adverse risk group was too low for a statistical analysis (n = 9). For the intermediate risk patients within the mRNA analysis cohort (n = 123), there was a trend toward higher RFS in the HVEM high group (n = 31; median RFS 267 days) versus the HVEM low group (n = 92; median RFS 226 days), but this was not statistically significant (p = 0.417; Fig. 4d).
Finally, we studied the impact of HVEM expression on RFS in patients who received an allogeneic SCT in first CR. Within the surface protein analysis cohort, 6 patients with high HVEM expression were compared to 20 patients with low HVEM expression. Median RFS was not reached in both groups, but there was a trend toward higher RFS in the HVEMhigh group (p = 0.330; Fig. 4e). This trend was stronger, although still not significant (p = 0.115; Fig. 4f) in the mRNA analysis cohort. Median RFS in the HVEM high group (n = 7) was not reached after 3588 days; on the other hand, it was 987 days in the HVEM low group (n = 24).
Taken together, high HVEM expression was associated with a favorable prognosis (statistically significant or by trend) in several analyses within both cohorts, particularly for the ELN intermediate risk subgroup and after allogeneic SCT.
Discussion
HVEM is an extraordinary immune checkpoint molecule in several respects: It interacts with members of both the TNFR as well as the Ig superfamily, both in trans and in cis [1]. Besides, it can provide stimulatory as well as inhibitory signals depending on the ligand. In the light of very promising clinical results with various immune checkpoint blockade antibodies (particularly anti-CTLA-4 and anti-PD1), HVEM and its ligands are increasingly discussed as additional important cancer immune evasion molecules and new targets to enhance anti-tumor immunity [23].
For esophageal squamous cell carcinoma, it has recently been shown that the level of HVEM expression on cancer cells was associated with the depth of tumor invasion, lymph node metastases and the number of tumor-infiltrating lymphocytes [9]. HVEM status (high vs. low) was identified as an independent prognostic marker. Consequently, it was demonstrated that HVEM gene silencing provided an anti-tumor effect, both in vitro and in vivo [9]. Similarly, the identification of prognostic subtypes by HVEM expression status as well as possibilities for novel immunotherapeutic strategies by targeting the HVEM pathway is discussed for other solid tumors [23].
Hematological tumors, such as AML, are especially prone to systemic immunotherapy, because leukemic cells in the blood and BM are not surrounded by a stroma barrier and are thus more easily accessible by the immune cells. Besides, initial cytotoxic chemotherapy often succeeds in achieving a status of hematological remission with minimal residual disease, in which immune responses can be particularly effective. Therefore, immunotherapeutic strategies are highly attractive for post-remission therapy of AML [24, 25]. One promising approach is the use of immunomodulatory antibodies directed against inhibitory molecules on AML blasts, thus unleashing preexisting immune responses. Expression of coinhibitory molecules on AML cells has been analyzed, and preclinical mouse experiments or early clinical trials have given hints to potential effects of a CTLA-4 antibody [26, 27], of PD-L1 or PD-L2 antibodies [28, 29] and of Tim-3/galectin-9 blockade [30].
In contrast, very little is known about the role of HVEM in AML. There has been no study quantifying HVEM surface protein expression on a larger number of patients. Nothing has been reported about an association of HVEM expression with cytogenetic or molecular aberrations or with other genes and gene sets. And finally, a potential prognostic impact of HVEM expression has not been analyzed.
In our study, we report data based on HVEM surface expression analysis of CD33+ leukemic cells and on HVEM mRNA expression analysis in BM samples of two large independent patient cohorts at diagnosis each. Both methods could be used in parallel, as we were able to prove a positive correlation between HVEM protein surface expression and HVEM mRNA expression. For the majority of comparisons, both datasets revealed remarkably similar results, although tested both in different cohorts and with different methodologies, strengthening the reliability of the association results. These data demonstrate for the first time that HVEM is differentially expressed on leukemic cells of AML patients. The expression of HVEM was shown to be lower in cases with an FLT3-ITD, with mutations in NPM1 or with the combination of mutated NPM1 and FLT3 wild type. In contrast, a biallelic mutation in CEBPA correlated positively with higher expression of HVEM.
To address the question whether HVEM expression is part of a larger functional context, we extended our gene expression analysis. A comparison of gene expression profiles between samples of the HVEM high and the HVEM low group revealed significant differences both on single transcript and on pathway level. Interestingly, several genes already known to have implications in AML pathogenesis (HOXA9, MEIS1, HOXA5 and PBX3) [18–20] showed significant downregulation in the HVEM high group, whereas MN1, a gene that is associated with inferior survival in AML [21], was significantly upregulated. Of the 186 gene sets from the KEGG pathway database that were analyzed, an enrichment of four pathways related to immunodeficiency, migration, cytotoxicity and hematopoietic cell lineage was revealed. Thus, our data suggest a significant association of HVEM expression in the pathogenesis of AML and immunoregulation.
Finally, high expression of HVEM was shown to be associated with a trend toward favorable prognosis in several analyses, particularly for the very heterogeneous group of intermediate risk patients and for patients after allogeneic SCT. This data suggest the notion that HVEM expression is a positive prognostic marker in AML.
The mechanism by which interactions of HVEM with its various ligands might potentially influence immune surveillance and relapse or survival is intriguing. It seems surprising at first that overexpression of a predominantly inhibitory molecule should correlate with better control of residual leukemic cells. However, it has recently been suggested that activated CD8+ T cells in the tumor microenvironment are required for the upregulation of immunosuppressive mechanisms (e.g., PD-L1, indoleamine-2,3-dioxygenase and regulatory T cells) [31]. Similarly, high expression of HVEM might be a reflection of an ongoing active immune reaction against the leukemia, which by itself could be prognostically favorable. Functional studies of HVEM interaction in the setting of AML in vitro and in vivo will be needed to reveal the functional basis for its prognostic impact and to figure out the potential of HVEM blockade for immunomodulatory therapy.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors would like to thank Maria Sauerland from the Institute for Biometrics and Clinical Research at the Westfälische Wilhelms-Universität Münster and all doctors of the participating centers for the recruitment of patients and help with acquisition of the clinical data; Elke Habben, Karin Hecht, Sabine Reinkunz and Ewelina Zientara for their excellent technical assistance; and the patients for providing the research samples used in this study. The work was supported by funds from BayImmuNet, the Bavarian Immunotherapy Network (http://www.bayimmunet.de), and by a Metiphys fellowship of the Medical Faculty of the Ludwig-Maximilian Universität München to Felix S. Lichtenegger.
Author contributions
Marion Subklewe was the principal investigator and takes primary responsibility for the paper. Thomas Büchner, Wolfgang E. Berdel, Bernhard J. Wörmann and Wolfgang Hiddemann were principle investigators of the AML-CG 99 trial and recruited the patients for this analysis. Karsten Spiekermann and Stephanie Schneider provided the morphological, cytogenetic and molecular characterization of the patients. Tobias Herold provided the mRNA expression data and their statistical analysis. Felix S. Lichtenegger, Isabell Kondla, Michael Krempasky and Anna L. Weber collected and analyzed the data. Felix S. Lichtenegger and Marion Subklewe coordinated the research and wrote the manuscript.
Abbreviations
- AML
Acute myeloid leukemia
- AML-CG
AML Cooperative Group
- BM
Bone marrow
- CEBPA
CCAAT/enhancer-binding protein alpha
- CR
Complete remission
- ELN
European LeukemiaNet
- FLT3
Fms-like tyrosine kinase 3
- GSEA
Gene set enrichment analysis
- HVEM
Herpesvirus entry mediator
- Limma
Linear Models for Microarray Data
- MFI
Mean fluorescence intensity
- MRC
Medical Research Council
- NPM1
Nucleophosmin
- OS
Overall survival
- RFS
Relapse-free survival
- sAML
Secondary AML
- SCT
Stem cell transplantation
- SFI
Specific fluorescence intensity
- TNFR
Tumor necrosis factor receptor
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interest.
References
- 1.Pasero C, Speiser DE, Derré L, Olive D. The HVEM network: new directions in targeting novel costimulatory/co-inhibitory molecules for cancer therapy. Curr Opin Pharmacol. 2012;12:478–485. doi: 10.1016/j.coph.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Morel Y, Truneh A, Sweet RW, Olive D, Costello RT. The TNF superfamily members LIGHT and CD154 (CD40 ligand) costimulate induction of dendritic cell maturation and elicit specific CTL activity. J Immunol. 2001;167:2479–2486. doi: 10.4049/jimmunol.167.5.2479. [DOI] [PubMed] [Google Scholar]
- 3.Duhen T, Pasero C, Mallet F, Barbarat B, Olive D, Costello RT. LIGHT costimulates CD40 triggering and induces immunoglobulin secretion; a novel key partner in T cell-dependent B cell terminal differentiation. Eur J Immunol. 2004;34:3534–3541. doi: 10.1002/eji.200425598. [DOI] [PubMed] [Google Scholar]
- 4.Murphy TL, Murphy KM. Slow down and survive: enigmatic immunoregulation by BTLA and HVEM. Annu Rev Immunol. 2010;28:389–411. doi: 10.1146/annurev-immunol-030409-101202. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Subudhi SK, Anders RA, Lo J, Sun Y, Blink S, Wang Y, Wang J, Liu X, Mink K, Degrandi D, Pfeffer K, Fu YX. The role of herpesvirus entry mediator as a negative regulator of T cell-mediated responses. J Clin Invest. 2005;115:711–717. doi: 10.1172/JCI200522982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.del Rio ML, Lucas CL, Buhler L, Rayat G, Rodriguez-Barbosa JI. HVEM/LIGHT/BTLA/CD160 cosignaling pathways as targets for immune regulation. J Leukoc Biol. 2010;87:223–235. doi: 10.1189/jlb.0809590. [DOI] [PubMed] [Google Scholar]
- 7.Costello RT, Mallet F, Barbarat B, De Colella JMS, Sainty D, Sweet RW, Truneh A, Olive D. Stimulation of non-Hodgkin’s lymphoma via HVEM: an alternate and safe way to increase Fas-induced apoptosis and improve tumor immunogenicity. Leukemia. 2003;17:2500–2507. doi: 10.1038/sj.leu.2403175. [DOI] [PubMed] [Google Scholar]
- 8.Derré L, Rivals JP, Jandus C, Pastor S, Rimoldi D, Romero P, Michielin O, Olive D, Speiser DE. BTLA mediates inhibition of human tumor-specific CD8+ T cells that can be partially reversed by vaccination. J Clin Invest. 2010;120:157–167. doi: 10.1172/JCI40070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Migita K, Sho M, Shimada K, Yasuda S, Yamato I, Takayama T, Matsumoto S, Wakatsuki K, Hotta K, Tanaka T, Ito M, Konishi N, Nakajima Y. Significant involvement of herpesvirus entry mediator in human esophageal squamous cell carcinoma. Cancer. 2014;120:808–817. doi: 10.1002/cncr.28491. [DOI] [PubMed] [Google Scholar]
- 10.Hokuto D, Sho M, Yamato I, Yasuda S, Obara S, Nomi T, Nakajima Y. Clinical impact of herpesvirus entry mediator expression in human hepatocellular carcinoma. Eur J Cancer. 2015;51:157–165. doi: 10.1016/j.ejca.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Inoue T, Sho M, Yasuda S, Nishiwada S, Nakamura S, Ueda T, Nishigori N, Kawasaki K, Obara S, Nakamoto T, Koyama F, Fujii H, Nakajima Y. HVEM expression contributes to tumor progression and prognosis in human colorectal cancer. Anticancer Res. 2015;35:1361–1367. [PubMed] [Google Scholar]
- 12.Pasero C, Barbarat B, Just-Landi S, Bernard A, Aurran-Schleinitz T, Rey J, Eldering E, Truneh A, Costello RT, Olive D. A role for HVEM, but not lymphotoxin-beta receptor, in LIGHT-induced tumor cell death and chemokine production. Eur J Immunol. 2009;39:2502–2514. doi: 10.1002/eji.200939069. [DOI] [PubMed] [Google Scholar]
- 13.Hobo W, Norde WJ, Schaap N, Fredrix H, Maas F, Schellens K, Falkenburg JH, Korman AJ, Olive D, van der Voort R, Dolstra H. B and T lymphocyte attenuator mediates inhibition of tumor-reactive CD8+ T cells in patients after allogeneic stem cell transplantation. J Immunol. 2012;189:39–49. doi: 10.4049/jimmunol.1102807. [DOI] [PubMed] [Google Scholar]
- 14.Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, Wheatley K, Harrison CJ, Burnett AK, National Cancer Research Institute Adult Leukaemia Working Group Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116:354–365. doi: 10.1182/blood-2009-11-254441. [DOI] [PubMed] [Google Scholar]
- 15.Li Z, Herold T, He C, Valk PJ, Chen P, Jurinovic V, Mansmann U, Radmacher MD, Maharry KS, Sun M, Yang X, Huang H, Jiang X, Sauerland MC, Büchner T, Hiddemann W, Elkahloun A, Neilly MB, Zhang Y, Larson RA, et al. Identification of a 24-gene prognostic signature that improves the European LeukemiaNet risk classification of acute myeloid leukemia: an international collaborative study. J Clin Oncol. 2013;31:1172–1181. doi: 10.1200/JCO.2012.44.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawrence HJ, Rozenfeld S, Cruz C, Matsukuma K, Kwong A, Kömüves L, Buchberg AM, Largman C. Frequent co-expression of the HOXA9 and MEIS1 homeobox genes in human myeloid leukemias. Leukemia. 1999;13:1993–1999. doi: 10.1038/sj.leu.2401578. [DOI] [PubMed] [Google Scholar]
- 19.Zhao P, Tan L, Ruan J, Wei XP, Zheng Y, Zheng LX, Jiang WQ, Fang WJ. Aberrant expression of HOXA5 and HOXA9 in AML. Asian Pac J Cancer Prev. 2015;16:3941–3944. doi: 10.7314/APJCP.2015.16.9.3941. [DOI] [PubMed] [Google Scholar]
- 20.Li Z, Zhang Z, Li Y, Arnovitz S, Chen P, Huang H, Jiang X, Hong GM, Kunjamma RB, Ren H, He C, Wang CZ, Elkahloun AG, Valk PJ, Döhner K, Neilly MB, Bullinger L, Delwel R, Löwenberg B, Liu PP, et al. PBX3 is an important cofactor of HOXA9 in leukemogenesis. Blood. 2013;121:1422–1431. doi: 10.1182/blood-2012-07-442004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Metzeler KH, Dufour A, Benthaus T, Hummel M, Sauerland MC, Heinecke A, Berdel WE, Büchner T, Wörmann B, Mansmann U, Braess J, Spiekermann K, Hiddemann W, Buske C, Bohlander SK. ERG expression is an independent prognostic factor and allows refined risk stratification in cytogenetically normal acute myeloid leukemia: a comprehensive analysis of ERG, MN1, and BAALC transcript levels using oligonucleotide microarrays. J Clin Oncol. 2009;27:5031–5038. doi: 10.1200/JCO.2008.20.5328. [DOI] [PubMed] [Google Scholar]
- 22.Döhner H, Estey EH, Amadori S, Appelbaum FR, Büchner T, Burnett AK, Dombret H, Fenaux P, Grimwade D, Larson RA, Lo-Coco F, Naoe T, Niederwieser D, Ossenkoppele GJ, Sanz MA, Sierra J, Tallman MS, Löwenberg B, Bloomfield CD, LeukemiaNet European. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115:453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 23.Pasero C, Olive D. Interfering with coinhibitory molecules: BTLA/HVEM as new targets to enhance anti-tumor immunity. Immunol Lett. 2013;151:71–75. doi: 10.1016/j.imlet.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 24.Lichtenegger FS, Schnorfeil FM, Hiddemann W, Subklewe M. Current strategies in immunotherapy for acute myeloid leukemia. Immunotherapy. 2013;5:63–78. doi: 10.2217/imt.12.145. [DOI] [PubMed] [Google Scholar]
- 25.Lichtenegger FS, Krupka C, Köhnke T, Subklewe M. Immunotherapy for acute myeloid leukemia. Semin Hematol. 2015;52:207–214. doi: 10.1053/j.seminhematol.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 26.Zhong RK, Loken M, Lane TA, Ball ED. CTLA-4 blockade by a human MAb enhances the capacity of AML-derived DC to induce T-cell responses against AML cells in an autologous culture system. Cytotherapy. 2006;8:3–12. doi: 10.1080/14653240500499507. [DOI] [PubMed] [Google Scholar]
- 27.Bashey A, Medina B, Corringham S, Pasek M, Carrier E, Vrooman L, Lowy I, Solomon SR, Morris LE, Holland HK, Mason JR, Alyea EP, Soiffer RJ, Ball ED. CTLA4 blockade with ipilimumab to treat relapse of malignancy after allogeneic hematopoietic cell transplantation. Blood. 2009;113:1581–1588. doi: 10.1182/blood-2008-07-168468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berger R, Rotem-Yehudar R, Slama G, Landes S, Kneller A, Leiba M, Koren-Michowitz M, Shimoni A, Nagler A. Phase I safety and pharmacokinetic study of CT-011, a humanized antibody interacting with PD-1, in patients with advanced hematologic malignancies. Clin Cancer Res. 2008;14:3044–3051. doi: 10.1158/1078-0432.CCR-07-4079. [DOI] [PubMed] [Google Scholar]
- 29.Zhou Q, Munger ME, Highfill SL, Tolar J, Weigel BJ, Riddle M, Sharpe AH, Vallera DA, Azuma M, Levine BL, June CH, Murphy WJ, Munn DH, Blazar BR. Program death-1 signaling and regulatory T cells collaborate to resist the function of adoptively transferred cytotoxic T lymphocytes in advanced acute myeloid leukemia. Blood. 2010;116:2484–2493. doi: 10.1182/blood-2010-03-275446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Q, Munger ME, Veenstra RG, Weigel BJ, Hirashima M, Munn DH, Murphy WJ, Azuma M, Anderson AC, Kuchroo VK, Blazar BR. Coexpression of Tim-3 and PD-1 identifies a CD8+ T-cell exhaustion phenotype in mice with disseminated acute myelogenous leukemia. Blood. 2011;117:4501–4510. doi: 10.1182/blood-2010-10-310425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spranger S, Spaapen RM, Zha Y, Williams J, Meng Y, Ha TT, Gajewski TF. Up-regulation of PD-L1, IDO, and tregs in the melanoma tumor microenvironment is driven by CD8+ T cells. Sci Transl Med. 2013;5:200ra116. doi: 10.1126/scitranslmed.3006504. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.