Abstract
Rubella virus (RV) virions contain two glycosylated membrane proteins, E1 and E2, that exist as a heterodimer and form the viral spike complexes on the virion surface. Formation of an E1-E2 heterodimer is required for transport of E1 out of the endoplasmic reticulum lumen to the Golgi apparatus and plasma membrane. To investigate the nature of the E1-E2 interaction, we have introduced mutations in the internal hydrophobic region (residues 81 to 109) of E1. Substitution of serine at Cys82 (mutant C82S) or deletion of this hydrophobic domain (mutant dt) of E1 resulted in a disruption of the E1 conformation that ultimately affected E1-E2 heterodimer formation and cell surface expression of both E1 and E2. Substitution of either aspartic acid at Gly93 (G93D) or glycine at Pro104 (P104G) was found to impair neither E1-E2 heterodimer formation nor the transport of E1 and E2 to the cell surface. Fusion of RV-infected cells is induced by a brief treatment at a pH below 6.0. To test whether this internal hydrophobic domain is involved in the membrane fusion activity of RV, transformed BHK cell lines expressing either wild-type or mutant spike proteins were exposed to an acidic pH and polykaryon formation was measured. No fusion activity was observed in the C82S, dt, and G93D mutants; however, the wild type and the P104G mutant exhibited fusogenic activities, with greater than 60% and 20 to 40% of the cells being fused, respectively, at pH 4.8. These results suggest that it is likely that the region of E1 between amino acids 81 and 109 is involved in the membrane fusion activity of RV and that it may be important for the interaction of that protein with E2 to form the E1-E2 heterodimer.
Rubella virus (RV), the etiological agent of German measles, is a small enveloped RNA virus that belongs to the togavirus family (31) and bears similarities to the prototype alphaviruses in terms of its genomic organization and strategy for viral gene expression (11). RV virions contain two glycosylated envelope proteins (E1 [58 kDa] and E2 [42 to 47 kDa]), located on the virion surface (32), that exist as heterodimers in the viral spike complexes (3, 49). E1, the major target antigen of RV, contains virus-neutralizing and hemagglutinin epitopes (6). The biological role of E2 is not well defined, although it has been reported to possess strain-specific epitopes and possibly a neutralizing domain (9). The RV structural proteins are synthesized from a subgenomic RNA as a polyprotein precursor that is cleaved to form the viral polypeptides C, E2, and E1 (33). The coordinate and individual expression of the C, E2, and E1 proteins shows that E2 is necessary for the cell surface expression of E1 and that E1 is not required for exit of E2 from the endoplasmic reticulum (ER) and its passage through the Golgi complex to the cell surface (16). However, E1 seems to increase the rate of transport of E2 to the cell surface (16). In the absence of E2, E1 is arrested in a post-ER, pre-Golgi complex compartment (18) and is only transported to the cell surface in the presence of E2 (16). Since the association of E1 and E2 is a hydrophobic interaction (3, 16), it is likely that E2 interacts specifically with an E1 hydrophobic domain to form the E1-E2 complexes that are transport competent.
Although RV appears to be similar to alphaviruses in terms of its structure as well as its structural-protein expression, its replication cycle kinetics are different. Cells infected with alphaviruses generally reach maximum rates of virus production 4 to 8 h after infection (22). RV, in contrast, has a latent period of more than 12 h, and peak virus production is reached between 24 and 48 h postinfection (13). The maturation and intracellular transport of alphavirus structural proteins have been extensively studied (27, 47, 48, 51). Compared to that of alphaviruses, RV glycoprotein processing occurs relatively slowly and the transport of glycoproteins E2 and E1 to the plasma membrane is inefficient (16). Like Semliki Forest virus (SFV), RV infects cells via endocytosis and an acid-triggered fusion step (1, 3, 26, 45). The reported acid-induced conformational changes in RV E1 and E2 and the removal of E2 by proteolytic digestion of RV particles, resulting in E1 particles that can still bind to liposomes (23), suggest that RV E1 plays a dominant role in membrane fusion in the acidic endosomal compartment, similar to that of SFV E1 (24, 25, 30), and may contain a noncleavable fusion peptide in the internal region of E1.
Inspection of the amino acid sequence of RV E1 revealed that there is an internal hydrophobic domain within E1, 29 amino acids long and 82 residues from the E1 NH2 (Fig. 1). We speculated that this internal hydrophobic domain is involved in E1-E2 interaction as well as in cell fusion. We initiated experiments to determine the role of this E1 hydrophobic region in the E1-E2 interaction and fusogenic activity of BHK cells expressing RV antigens. In vitro mutagenesis was used to introduce four mutations into the hydrophobic region of E1. BHK cells were transfected with recombinant plasmids, and stable transformed BHK cell lines expressing wild-type or mutant spike proteins were isolated via methotrexate selection (38). Here we present evidence that the internal hydrophobic domain of RV E1 plays a major role in E1-E2 interaction and membrane fusion of RV.
FIG. 1.
Schematic diagram of RV cDNAs and the hydrophobic domain of the RV E1 glycoprotein. Respective portions of the C, E2, and E1 genes are indicated above the constructs. Intergene borders are marked by vertical lines extending through the cDNAs. The translation start site of the capsid protein is utilized in all constructs. Signal peptides are indicated by open boxes, and the transmembrane regions are shown as closed boxes. The hydrophobic domain of E1 is depicted as an open box 81 amino acid residues from the N terminus of E1. Restriction endonuclease sites are abbreviated as follows: E, EcoRI; H, HindIII; B, BamHI; and N, NcoI. The bar represents approximately 500 nucleotides. Beneath the arrows are the single-amino-acid changes and the deletion (dt) mutation. 24S, the cDNA containing all three structural protein genes of RV; E2E1, the cDNA containing the E1 and E2 genes.
MATERIALS AND METHODS
Plasmid construction and mutagenesis.
RV cDNAs encoding the polyprotein precursor for all three structural proteins (24S) (7) and the E2E1 polyprotein precursor (E2E1) (16) are shown in Fig. 1. The cDNAs were subcloned into the SmaI site of the transfer vector pNUT (34) under the control of the metallothionein 1 promoter. The construction of plasmids pNUT-24S and pNUT-E2E1 has been previously described (38). Plasmid pNUT-E2 was constructed by insertion of a blunted EcoRI-HindIII fragment from plasmid pCMV5-E2 (15) into the SmaI site of the pNUT vector.
Mutations (substitution of serine at Cys82 of E1 [C82S] and deletion of the hydrophobic domain [residues 81 to 109] of E1 [dt]) were introduced by oligonucleotide-directed mutagenesis on a uracil-containing single-stranded DNA template (28). M13mp18-3′E2E1 was constructed by insertion of the EcoRI-HindIII fragment from p3′E2E1 (14) into M13mp18 vector which had been digested with the EcoRI and HindIII restriction enzymes. Mutagenic oligonucleotides were pCAGAAGGTCGACGCGCGCT (mutated bases are underlined) and pGTGGTACTGCTTCTAGCGCTGTGTGCCATT, respectively, for the C82S and dt mutants. To create an E2E1 cDNA containing an introduced mutation in E1, the NcoI fragment from pCMV5-E2 (15), encoding the N-terminal 202 amino acid residues of E2 and a portion of C (8 and 54 amino acid residues at the N and C termini of the C protein, respectively) (Fig. 1), was inserted into the NcoI site of the mutant plasmid. The resulting cDNA was subcloned into the SmaI site of the pNUT vector. The identities of the recombinant plasmids were confirmed by DNA sequencing and restriction analysis.
Generation of mutations at residues Gly93 and Pro104 was carried out with a Quik Change site-directed mutagenesis kit (Stratagene) with mutagenic oligonucleotides pGCCTACTCCTCTGACGGGTACGCGCAG-3′ and pCTGCGCGTACCCGTCAGAGGAGTAGG-3′ for the Gly93-to-aspartic acid substitution (G93D) and pCCTCTTACTTCAACGGTGGCGGCAGCTAC and pGTAGCTGCCGCCACCGTTGAAGTAAGAGG for alteration of Pro104 to glycine (P104G) (mutated bases are underlined). The cDNA used in the mutagenesis was pSPT19 E2E1 (39), and the cycling reactions consisted of 16 cycles of 95°C for 30 s, 55°C for 1 min, and 68°C for 14 min. The resulting E2E1 cDNA containing the introduced mutation was subcloned into the SmaI site of the pNUT vector. The identity of the introduced mutation was confirmed by DNA sequencing.
Generation of stable transformed BHK cell lines.
BHKtk− cells (46) grown in Dulbecco modified Eagle medium (DMEM) with 5% fetal bovine serum were transfected with recombinant plasmids by the calcium phosphate method (12). After 24 h, the medium was changed to DMEM containing 500 μM methotrexate. The presence of dihydrofolate reductase cDNA driven by the simian virus 40 early promoter in the pNUT vector (34) permits the selection of transfected BHK cells in the presence of high concentration of methotrexate (38). After 10 days in the selection medium, methotrexate-resistant colonies were picked and screened for the expression of RV structural proteins by Western blot analysis (44). The resultant cell lines were named BHK-E2E1(wt), BHK-E2E1(C82S), BHK-E2E1(dt), BHK-E2E1(G93D), and BHK-E2E1(P104G).
Metabolic labelling, biotinylation, immunoprecipitation, and endoglycosidase digestion.
Transformed BHK cells, grown as monolayers (in 35-mm-diameter petri dishes) in growth medium (DMEM containing 2.5% fetal bovine serum), were induced with zinc sulfate (40 μM) for 16 h. Induced BHK cells were labelled with 100 μCi of [35S]methionine for 30 min and chased with 1 mM unlabelled methionine for various time periods. Cell surface biotinylation was carried out as described by Duffus et al. (10). Briefly, labelled cells were placed on ice, washed twice with phosphate-buffered saline (PBS), and incubated with 0.5 ml of sulfosuccinimidobiotin (0.5 mg/ml in PBS) for 15 min on ice. The biotinylation reaction was repeated once with fresh reagent. The reaction was stopped by washing with cold 10 mM glycine containing 10 mM Tris-HCl (pH 7.4) and 150 mM NaCl. The cells were lysed with lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 2 mM EDTA, and 1% Triton X-100), and spike proteins were immunoprecipitated with human anti-RV serum. The biotinylated spike proteins were recovered from streptavidin-agarose and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (29). Immunoprecipitation of cellular spike proteins was performed as described by Wahlberg et al. (47). Briefly, at the end of the pulse or chase period, the monolayers were washed twice with PBS and lysed with buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 2 mM EDTA, 1% Nonidet P-40 (NP-40), and 74 μM antipain dihydrochloride (Boehringer Mannheim). RV antiserum-coated protein A-Sepharose was washed with lysis buffer and incubated with 35S-labelled cellular lysate overnight at 4°C. The beads were washed twice with washing buffer containing 10 mM Tris-HCl (pH 7.4), 150 mM NaCl, 2 mM EDTA, and 0.2% NP-40 and once with 10 mM Tris-HCl (pH 7.4). RV antigen was eluted into 100 mM sodium citrate (pH 5.5) containing 0.15% SDS and 1 mM phenylmethylsulfonyl fluoride at 100°C for 4 min and analyzed by SDS-PAGE (29). Some immunoprecipitates were digested with endoglycosidase H (5 mU/50 μl) as described previously (16).
Fusion assay.
BHK cells expressing either wild-type or mutant RV spike proteins were cultured in six-well plates. After ZnSO4 induction for 16 h, the cells were washed once with fusion medium (DMEM without bicarbonate, containing 0.2% bovine serum albumin, 10 mM HEPES, and 10 mM morpholinoethanesulfonic acid; pH 7.0) and then incubated for 20 min at 37°C with fusion medium adjusted to a pH of 4.0 to 7.0 as required. After exposure to the desired pH, the fusion medium was replaced with growth medium of pH 7.0 and the cells were incubated for an additional 4 h at 37°C to allow both maximal RV spike protein expression in the newly fused cells and morphological reorganization of the cell nuclei. Polykaryons containing more than five nuclei were counted under a phase-contrast microscope. Fusion activity was presented as the percentage of fused cells, which is the ratio of the mean total number of polykaryons counted in five random fields to the mean total number of unfused cells and polykaryons.
Localization of spike protein by immunofluorescence.
Transformed BHK cells were seeded onto poly-l-lysine-coated coverslips. After ZnSO4 induction, the cells were washed three times with PBS containing 0.7 mM CaCl2 and 0.3 mM MgCl2, fixed for 20 min at room temperature in 2% formaldehyde–PBS, and then washed with PBS. Some cells were permeabilized with 0.075% NP-40–PBS for 30 min prior to being blocked with bovine serum albumin (1% in PBS) for localization of intracellular RV antigen (16). The cells were then treated with mouse monoclonal antibodies against E1 (1:50) or E2 (1:50) followed by incubation with fluorescein- or rhodamine-conjugated secondary antibodies (Kirkegaard and Perry Laboratories).
Sucrose velocity gradients.
Samples (500 μl) were layered onto 11.5-ml linear gradients of 5 to 20% sucrose in 50 mM Tris-HCl (pH 7.5)–150 mM NaCl–2 mM EDTA–0.1% NP-40 and centrifuged for 30 h at 40,000 rpm in a Beckman SW41 rotor. Fractions (0.5 ml) were collected from the bottom of the tube.
RESULTS
Expression of mutant proteins.
To determine whether the internal hydrophobic region (residues 81 to 109) is involved in E1-E2 interaction and cell-cell fusion, four mutants (C82S, dt, G93D, and P104G) were constructed by oligonucleotide-directed mutagenesis. Since the antigenic structure of E1 is dependent on N-linked glycosylation and intramolecular disulfide bonding (37), the Cys82 residue in the hydrophobic region was selected as a target for studying E1-E2 interaction. The substitution of aspartic acid at Gly93 was based on the assumption that if the E1 internal hydrophobic region interacts directly with the host cell membrane as a prerequisite of or during the fusion event, the introduction of a charged residue may inhibit the ability of E1 to induce membrane fusion. The internal fusion domains have been postulated to contain helix-breaking residues near their centers (50); therefore, proline 104 was changed to glycine.
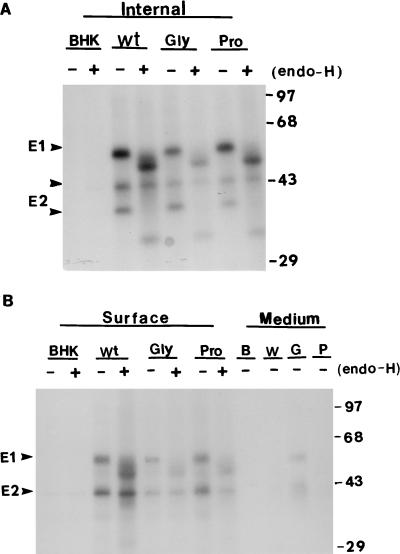
Because the processing of RV glycoprotein occurs relatively slowly and transport of E1 and E2 to the plasma membrane is inefficient in transfected COS cells (16), stably transformed BHK cell lines expressing mutant proteins were isolated by transfection of BHK cells with recombinant plasmids as described in Materials and Methods. Metabolic labelling and immunoprecipitation were used to evaluate the expressed spike proteins. Transformed BHK cells were incubated with growth medium containing 40 μM zinc sulfate for 16 h to induce the expression of RV structural proteins from the metallothionein promoter (34). Induced transformed BHK cells were labelled with [35S]methionine for 30 min and then chased with 1 mM unlabelled methionine for 2 h. Labelled intracellular RV proteins were immunoprecipitated with human anti-RV serum and treated with endo-β-N-acetylglycosaminidase H (endo-H) to monitor the processing of N-linked sugars (43). In the cell lysates of the wild type and three of the mutants (C82S, G93D, and P104G) were found protein species corresponding to E1 (57 kDa) and E2 (39 kDa) while the intracellular E1 dt (54 kDa) and E2 (39 kDa) of the dt mutants were immunoprecipitated (Fig. 2A). Deletion of 29 amino acids of E1 resulted in the loss of approximately 3 kDa of mass from E1. No RV-specific proteins of the size of E1 and E2 were detected in control BHK cells (Fig. 2A). Digestion with endo-H reduced the sizes of E1, E1 dt, and E2 to 51, 48, and 32 kDa, respectively (Fig. 2A), indicating the presence of immature high-mannose N-linked sugars on these proteins within the compartments of the ER and the medial Golgi apparatus. The levels of E1 and E2 proteins expressed by both the C82S and dt mutants were greatly reduced compared to those of the wild type and the G93D and P104G mutants (Fig. 2A).
FIG. 2.
Expression of mutant proteins. Induced BHK cells were labelled with [35S]methionine for 30 min and chased with 1 mM unlabelled methionine for 2 h. Intracellular RV antigens were isolated by immunoprecipitation with human anti-RV serum (A) or with anti-E1 monoclonal antibody (B), followed by SDS-PAGE and fluorography. Portions of the immunoprecipitates were (+) or were not (−) digested with endo-H. The positions of apparent molecular mass markers are shown at the right (in kilodaltons). BHK, BHK cells; Wt, BHK-E2E1(wt); Cys, BHK-E2E1(C82S); dt, BHK-E2E1(dt); Gly, BHK-E2E1(G93D); Pro, BHK-E2E1)(P104G); Human, human anti-RV serum; Mab-E1, monoclonal antibody against RV E1.
It is interesting that in the C82S and dt mutants the ratio of the amount of E1 immunoprecipitated to that of immunoprecipitated E2 was smaller than the E1/E2 ratios in the wild type and the G93D and P104G mutants (Fig. 2A). To investigate whether the introduced mutations in the C82S and dt mutants affect the E1 structure recognized by antibodies, we performed immunoprecipitation in nonionic detergent, using a conformation-sensitive anti-E1 monoclonal antibody (6). As expected, E1 was immunoprecipitated only from the lysates of the wild type and the G93D and P104G mutants, not from those of the C82S and dt mutants (Fig. 2B), indicating that the antigenic structure in C82S and dt mutants has been altered by the mutation introduced in E1.
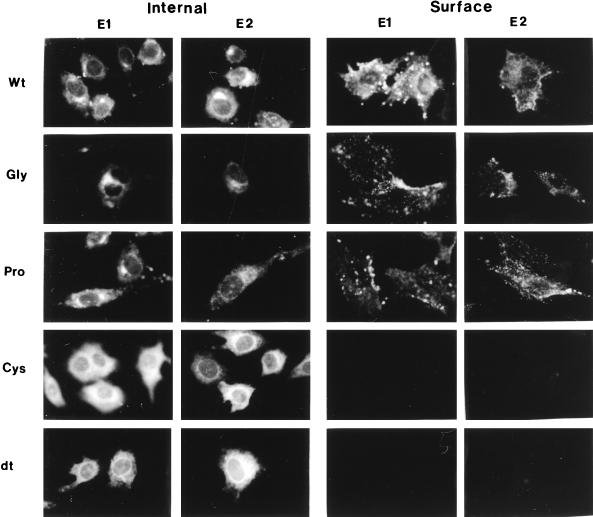
To determine whether mutant proteins reached the plasma membrane, cell surface expression of mutant proteins was determined by indirect immunofluorescence with RV anti-E1 and anti-E2 monoclonal antibodies. Transformed BHK cells were seeded onto poly-l-lysine-coated coverslips, induced with ZnSO4, and processed for detection of the presence of cell surface RV antigen in live unpermeabilized cells (16). We observed that the wild type and the G93D and P104G mutants displayed strong cell surface staining, as evidenced by the binding of the anti-E1 and anti-E2 monoclonal antibodies (Fig. 3). No cell surface staining was observed in the C82S and dt mutants (Fig. 3), indicating that E2 and E1 from these mutants did not reach the cell surface. The permeabilized cells of the wild type and the G93D and P104G mutants exhibited strong fluorescence in a juxtanuclear region likely corresponding to the Golgi apparatus (Fig. 3). In contrast, RV antigen in the C82S and dt mutants was found mostly in a reticular distribution not corresponding to the Golgi region (Fig. 3), indicating that RV antigen in the C82S and dt mutants accumulated in the ER. These results indicate that the processing and intracellular transport of E1 and E2 in the C82S and dt mutants were greatly affected by the mutation introduced in E1.
FIG. 3.
Localization of RV spike proteins by immunofluorescence. Induced BHK cells were fixed with 2% formaldehyde–PBS, incubated with monoclonal antibodies against E1 (1:50) or E2 (1:50), and then incubated with fluorescein isothiocyanate-conjugated anti-mouse immunoglobulin G. Wt, BHK-E2E1; Gly, BHK-E2E1 (G93D); Pro, BHK-E2E1(P104G); Cys, BHK-E2E1(C82S); dt, BHK-E2E1(dt). Magnifications, ×400.
To quantitate the cell surface E1 and E2 in the G93D and P104G mutants, induced BHK cells were labelled with [35S]methionine for 30 min and chased with excess unlabelled methionine for 2 or 8 h. The cells were derivatized with sulfosuccinimidobiotin and then lysed (10). RV spike proteins were precipitated with human anti-RV serum and separated into surface and internal proteins by streptavidin binding. In the absence of a chase, no cell surface antigen was detected in the wild type or the mutants (data not shown). After a 2-h chase, only a small fraction (10%) of the total RV antigen was detected at the cell surfaces of the wild type and both mutants (data not shown). After an 8-h chase, about 40% of the RV antigen reached the cell surface (Fig. 4) and the proportion of E1 and E2 in G93D mutant that was derivatized with biotin was less than that of the wild type (Fig. 4B). This appeared to be due to the release of E1 and E2 into the medium during the chase period, possibly due to proteolytic degradation (Fig. 4B). Quantitation of the protein bands by scintillation counting indicated that the amounts of E1 and E2 expressed at the cell surfaces of the G93D and P104G mutants were 45 and 82%, respectively, of that of the wild type (100%). The majority of the cell surface E2 existed as an endo-H-resistant 43-kDa form that contained both N- and O-linked sugars (36, 40). The cell surface E1 of the wild type and both mutants was partially endo-H resistant (Fig. 4B), whereas greater proportions of the internal spike protein E1 and E2 were endo-H sensitive (Fig. 4A).
FIG. 4.
Cell surface expression of wild-type and mutant proteins. Induced BHK cells were labelled with [35S]methionine for 30 min and chased with 1 mM unlabelled methionine for 2 or 8 h. Cell surface RV antigens were derivatized with sulfosuccinimidobiotin and isolated as described in Materials and Methods. The virus spike proteins were precipitated and separated into internal (A) and surface (B) proteins by streptavidin binding. Portions of the immunoprecipitates were (+) or were not (−) digested with endo-H. The positions of apparent molecular mass markers are shown at the right (in kilodaltons). Internal, intracellular antigens; surface, cell surface antigens; medium, RV antigens released into the culture medium; Wt, W, BHK-E2E1; Gly, G, BHK-E2E1(G93D); Pro, P, BHK-E2E1(P104G).
We have shown previously that E1 secreted from insect cells was partially resistant to endo-H digestion (42) whereas E1 secreted from CHO cells was endo-H resistant (20). There are three N-linked glycosylation sites in RV E1 (17). It is likely that one or more of the three high-mannose N-linked oligosaccharide chains on E1 were processed to a mature form in the medial Golgi apparatus before reaching the cell surface and that N glycosylation, but not processing of N-linked oligosaccharide, is required for cell surface expression of E1 in BHK cells.
Taken together, our results indicate that substitution of serine at Cys82 or deletion of the hydrophobic domain of E1 completely impaired the transport of E1 and E2 to the plasma membrane. In contrast, the mutations introduced in the G93D and P104G mutants did not affect the cell surface expression of E1 and E2, although the expression level was lower than that of the wild type.
Glycan processing and intracellular stability of mutant proteins.
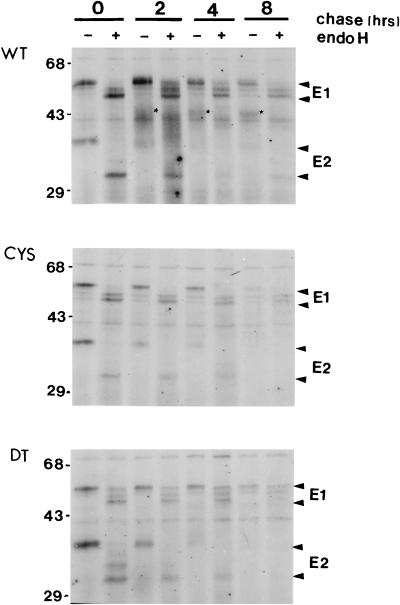
To study the processing and stability of the C82S and dt mutant proteins, pulse-chase experiments were carried out. In the wild type, after 30 min of pulse-labelling, E2 was found predominantly as a 39-kDa form, and removal of high-mannose glycans by digestion with endo-H reduced the molecular size to 31 kDa (Fig. 5, top panel). Approximately 20, 40, and 60% of E2 was further processed to complex-type glycans (16) (indicated by asterisks in Fig. 5, top panel). In contrast, E2 from the mutants was not processed to complex-type glycans and was not detected after the 8-h chase period (Fig. 5, middle and bottom panels). It is likely that the unassociated E2 in the C82S and dt mutants was either degraded in the ER or transported to the cell surface by itself.
FIG. 5.
Time course of glycan processing of wild-type and mutant proteins. Induced transformed BHK cells were pulse-labelled with [35S]methionine for 30 min and chased for various periods of time as indicated. Some immunoprecipitates were digested with endo-H for 8 h (+); others were not (−). E1 and E2 arrowheads indicate the E1 and E2 protein species in the absence or presence of endo-H treatment. The processing of E2 is indicated by the asterisks. The positions of molecular size markers are shown on the left (in kilodaltons). WT, BHK-E2E1; CYS, BHK-E2E1(C82S); DT, BHK-E2E1(dt).
Following a 30-min pulse-labelling, an RV-specific protein the size of E1 (57 kDa in the wild type and the C82S mutant; 54 kDa in the dt mutant) was detected in both the wild type and the mutants. Digestion with endo-H reduced the size of E1 to 53 and 51 kDa, indicating the presence of high-mannose sugars on the protein (Fig. 5). The presence of the 53-kDa endo-H digestion product just above the 51-kDa E1 indicates that a fraction of E1 acquired complex sugar moieties. Heterogeneous processing of E1 glycans was observed after increasing the length of the chase period (Fig. 5). In the wild type, after an 8-h chase, about 40% of the E1 was partially endo-H resistant (Fig. 5, top panel). The processing of E1 by the mutants was similar to that by the wild type, although the level of expression was only about 10 to 20% of the wild-type level (Fig. 5, middle and bottom panels). In the dt mutant, a fraction of 54-kDa E1 was still present after endo-H digestion (Fig. 5, bottom panel). It is possible that in the absence of the hydrophobic peptide, E1 in the dt mutant became misfolded and the misfolded E1 was more resistant to endo-H digestion.
Effect of mutation on E1 conformation.
RV E1 and E2 are rich in cysteine residues and contain intramolecular disulfide bridges that are important in maintenance of the proper protein folding (49). RV E1 and E2 show increased mobility in SDS-PAGE gels if disulfide bond reduction is omitted (49). This difference in mobility is probably due to the presence of intrachain disulfide bonds in E1 and E2. To determine whether a mutation would affect the conformation of E1, Western blot analysis was used to detect the binding of E1 to human anti-RV serum and monoclonal antibodies against E1 or E2 under reducing and nonreducing conditions. Under reducing conditions, the binding of antibodies to the wild-type E1 and their binding to mutant E1 proteins were similar (Fig. 6). In contrast, under nonreducing conditions, E1 proteins from the wild type and the G93D and P104G mutants retained their antibody binding activities, whereas the E1 proteins from the C82S and dt mutants lost most of their ability to bind the human anti-RV serum and anti-E1 monoclonal antibody (Fig. 6) and no E1-E2 heterodimer was detected (note asterisks in Fig. 6). As expected, E2 binding activity was not affected under nonreducing conditions in either the wild type or the mutants (Fig. 6). This decrease in E1 antibody binding activity of the C82S and dt mutants is not due to the formation of E1-E1 or E1-E2 oligomers under nonreducing conditions, since no higher-molecular-weight protein species were observed (Fig. 6). Thus, the antibodies appear to detect conformational changes with respect to intrachain disulfide bonding in the C82S and dt mutants. This result is consistent with our data on the immunoprecipitation of RV antigen with a conformation-specific anti-E1 monoclonal antibody (Fig. 2B).
FIG. 6.
Immunoblot analysis of E1 mutant proteins. Monolayers of induced BHK cells were lysed in lysis buffer containing 10 mM iodoacetamide to prevent the formation of disulfide bonds. The protein samples were denatured in the presence (+β-Me) or absence (−β-Me) of 2-mercaptoethanol, separated by SDS-PAGE, and transferred to nitrocellulose membranes for immunoblot analysis. RV antigens were detected by using human anti-RV serum (Human), anti-E1 monoclonal antibody exhibiting hemagglutination-inhibiting activity (Mab-E1), and anti-E2 monoclonal antibody (Mab-E2). The positions of the apparent molecular mass standards are indicated on the right (in kilodaltons). E1-E2 heterodimer is indicated by an asterisk. Wt, BHK-E2E1; Cys, BHK-E2E1(C82S); dt, BHK-E2E1(dt); Gly, BHK-E2E1(G93D); Pro, BHK-E2E1(P104G).
E1-E2 heterodimer interaction.
Since correct E1-E2 oligomerization appears to be required for efficient transport of these two proteins through the Golgi apparatus and to the plasma membrane (3), alteration of the E1 conformation may affect the E1-E2 heterodimer interaction. Biochemical studies have demonstrated that RV particles can be solubilized by nonionic detergent into an E1-E2 heterodimer structure and nucleocapsid (32). This E1-E2 noncovalent protein complex is not dissociated by NP-40 lysis buffer. Therefore, the formation of intracellular E1-E2 dimers can be detected by sedimentation analysis in sucrose gradients. To determine whether the mutations in E1 affect the interaction between E1 and E2, induced BHK cells were solubilized in lysis buffer containing 1% NP-40 and cellular lysates were fractionated on isokinetic gradients (3). Gradient fractions were analyzed by SDS-PAGE under nonreducing conditions. RV antigens transferred to membranes were detected with human anti-RV serum. Two distinct E1 peaks were observed in the wild type and in both the G93D and P104G mutants (Fig. 7, top three panels). The slower-migrating peak (fractions 15 to 19) was the monomeric E1, and the faster-sedimenting peak (fractions 7 to 10) contained E1-E2 heterodimers as well as E1 oligomers. Baron and Forsell (3) have reported that RV E1 migrates as two peaks in sucrose velocity gradients. The lighter peak is the E1 monomer, while the heavier peak is the E1 oligomer, which is larger than the E1-E2 heterodimer. Therefore, it is likely that the fractions between 11 and 14 contained mostly E1-E2 heterodimer. This interpretation is further supported by the band intensity of undissociated E1-E2 in the Western blots (Fig. 7, top three panels). In the Cys and dt mutants, only a small fraction of E1 and E2 was found as a heterodimer. Most of E2 was detected as E2 oligomers (fractions 7 to 13), and no oligomeric E1 was observed (Fig. 7, bottom two panels). It appears that alteration of the conformation of E1 in the C82S and dt mutants resulted in prevention of E1-E2 interaction.
FIG. 7.
Sucrose velocity analysis of E2 and E1 from cellular lysates. Induced BHK cells were lysed in lysis buffer, and the lysates were spun to remove the nuclei. Each lysate was analyzed on a 5 to 20% linear sucrose gradient as described in Materials and Methods. Fractions from each gradient were analyzed by immunoblotting. E2 and E1 were detected with human anti-RV serum (1:100 dilution). Sedimentation is from right to left. The positions of the apparent molecular mass standards are indicated on the right (in kilodaltons). Wt, BHK-E2E1; Gly, BHK-E2E1(G93D); Pro, BHK-E2E1(P104G); Cys, BHK-E2E1(C82S); dt, BHK-E2E1(dt).
Cell-cell fusion activities of mutants.
Katow and Sugiura (23) suggested that RV E1 plays a vital role in membrane fusion in the acidic endosomal compartment. Bernasconi et al. (5) reported that RV E1 containing a glycosylphosphatidylinositol (GPI) anchor was transported to the cell surface, where it retained the hemadsorption activity characteristic of the wild-type E1-E2 heterodimer. To examine whether E1 is the fusogenic protein of RV, transformed BHK cell lines expressing RV E2 (BHK-E2) were isolated and used in the fusion assay in parallel with the wild type, BHK-E2E1. Induced BHK cells were treated with fusion medium at pH 5.0, 6.0, or 7.0 for 20 min at 37°C. The cells were washed with growth medium (pH 7.0) and incubated with the same medium at 37°C for an additional 3 to 4 h. The polykaryons formed were viewed under a phase-contrast microscope. The wild type, BHK-E2E1, showed extensive polykaryon formation at both pH 5.0 and 6.0 but not at pH 7.0 (Fig. 8A). The majority of polykaryons contained 20 to 50 nuclei at pH 5.0 and 5 to 20 nuclei at pH 6.0 (Fig. 8A). No syncytium formation was observed in the induced BHK-E2 cells at any pH tested (data not shown). The lack of detectable cell fusion observed in BHK-E2 cells is not due to limited cell surface expression, since we have shown previously that RV E2 can reach the cell surface by itself (16) and the expression level in transformed BHK cells is five times higher than that of the transfected COS cells (data not shown). Taken together, these results indicate that E1 contains the fusogenic domain of RV, consistent with the finding that RV E1 particles can bind to liposomes in the absence of E2 (23).
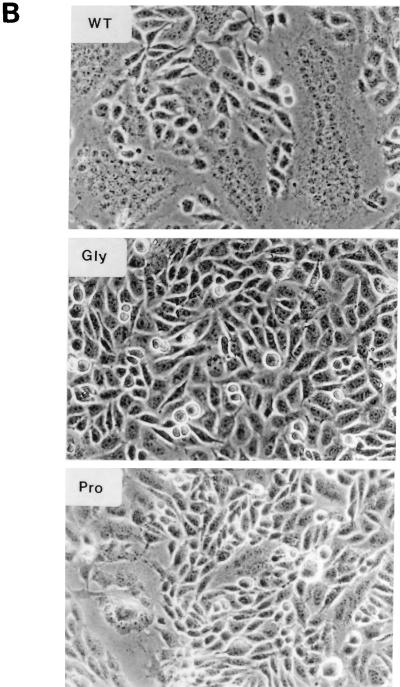
FIG. 8.
Syncytium formation in cells expressing wild-type or mutant proteins. (A) pH dependence of polykaryon formation. BHK-E2E1 (WT) and BHK cells were induced with ZnSO4 for 16 h, exposed to fusion medium at pH 5.0, 6.0, or 7.0 for 20 min, incubated in regular growth medium for 4 h, and photographed. (B) Polykaryon formation by mutant proteins. Induced BHK cells were exposed to fusion medium at pH 5.0 for 20 min, incubated in regular growth medium for 4 h, fixed, and photographed. WT, BHK-E2E1; Gly, BHK-E2E1(G93D); Pro, BHK-E2E1(P104G).
To examine whether the internal hydrophobic domain of E1 is involved in membrane fusion, the pH dependence of fusion activity of the mutants was investigated. Induced BHK cells were treated with fusion medium over the pH range of 4.0 to 7.0, and polykaryons formed were quantitated. Since the extent of syncytium formation in the cultures is dependent on both the number of cells expressing RV antigen and the amount of RV antigen on the cell surface, the results presented in Table 1 are the averages of data from four separate experiments. The fusion activity is presented as the percentage of cell fused in the culture for each mutant at the pH indicated. As expected, no fusion activity was observed in the C82S and dt mutants, since RV antigen was not expressed on the cell surface in these mutants (Table 1). For the G93D mutant, no fusogenic activity was detected, although occasionally a cell fusion rate of less than 0.5% was observed at pH 5 to 4.5. We found that the wild type and the P104G mutant had a broad pH range for fusion, with maximum fusogenic activity at around pH 4.8 and corresponding fusion rates of greater than 60% and between 20 and 40%, respectively (Table 1 and Fig. 8B). The average polykaryon sizes were 20 to 30 nuclei for the wild type and 5 to 10 nuclei for the P104G mutant (Fig. 8B). The decrease in fusion activity of the P104G mutant and the block in cell fusion observed in the G93D mutant are not due to the limited amount of spike protein at the cell surface. We have assayed the fusogenic activity of wild-type BHK cells induced at various ZnSO4 concentrations in order to determine the minimal level of cell surface spike protein required for fusion. We found that in the absence of ZnSO4 induction (10% cell surface antigen, compared to 100% at 40 μM ZnSO4), cell fusion rates of 5 to 10% were observed, while at 10 μM ZnSO4 induction (25% cell surface antigen), more than 30% of cells showed polykaryon formation at pH 4.8 (data not shown). Therefore, it is likely that this hydrophobic region of E1 represents the fusion domain of RV.
TABLE 1.
Fusion activities of the wild-type and the mutant cell lines at different pHs
| Cell linea | Fusion activityb at pH:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 7.0 | 6.5 | 6.0 | 5.6 | 5.3 | 5.0 | 4.8 | 4.5 | 4.2 | |
| wt | − | − | + | + | ++ | +++ | ++++ | +++ | ++ |
| Cys | − | − | − | − | − | − | − | − | − |
| dt | − | − | − | − | − | − | − | − | − |
| Gly | − | − | − | − | − | − | − | − | − |
| Pro | − | − | − | + | + | ++ | ++ | + | + |
wt, BHK-E2E1; Cys, BHK-E2E1(C82S); dt, BHK-E2E1(dt); Gly, BHK-E2E1(G93D); Pro, BHK-E2E1(P104G).
The percentage of fused cells for each mutant is represented as follows: −, no fusion or less than 5% fused cells; +, 5 to 20% fused cells; ++, 20 to 40% fused cells; +++, 40 to 60% fused cells; and ++++, greater than 60% fused cells. Values are averages of data from four separate experiments.
DISCUSSION
RV envelope glycoproteins E1 and E2 are targeted to the Golgi complex as heterodimers. It has been suggested that heterodimerization of E1 with E2 is required for the former protein to be correctly folded and transported to the Golgi complex (4, 19), where the complex accumulates due to a Golgi retention signal in the membrane-spanning domain of E2 (21). However, RV E1 can be rendered transport competent in the absence of E2 by the addition of a GPI anchor to the C terminus of the E1 ectodomain (5). Addition of a GPI anchor has been used for studying the role of individual domains of several membrane proteins in oligomerization and transport (8, 41) and has been shown to influence folding (2). Thus, it is conceivable that E1 retention is mediated by the binding of E1 to another protein, perhaps via a sequence(s) in the ectodomain, and this interaction may be prevented by oligomerization with E2 or fusion to a GPI anchor.
Our results showed that the internal hydrophobic domain (residues 81 to 109) in RV E1 plays a major role in the formation of the E1-E2 heterodimer and in the low-pH-induced cell fusion. We have shown previously that the antigenic structure of E1 is dependent on both N-linked glycans and intramolecular disulfide bonding (37). It is likely that substitution of serine at Cys82 disturbed the intramolecular disulfide bridges in E1 that are critical in maintaining the conformation of E1 for E1-E2 interaction and cell surface transport. The defect in dimerization and transport in the deletion mutant may imply that the E1 hydrophobic domain indirectly interacts with E2. However, we believe that the transport defect is more likely due to an overall disruption of the conformation of E1 in the absence of this hydrophobic domain. It is possible that the E1 hydrophobic domain is normally masked in the ER by association of E1 and E2, in order for transport to proceed further. In the absence of this hydrophobic moiety, E1 became misfolded or bound to ER proteins, such as the resident ER protein Bip (35).
Our results with G93D and P104G mutants demonstrated that the hydrophobic domain of E1 is closely involved in RV membrane fusion. This interpretation is further supported by our recent experiments using virus-like particles and an infectious clone containing a G93D or P104G mutation. We have shown previously that C protein of RV mediates the assembly of RV spike glycoproteins into virus-like particles (38). We found that virus-like particles containing either G93D or P104G mutations were produced as efficiently as the wild type (unpublished results). In the G93D mutant, the release of spike proteins into the medium was not observed in the presence of C protein. We have also carried out the fusion assay using BHK cells producing G93D or P104G virus-like particles; the results obtained are in agreement with those reported here. The other evidence supporting our interpretation lies in experiments in which defective G93D virus was produced from an RV infectious RNA transcript containing the G93D mutation. Further studies on G93D and P104G viruses to define the steps in fusion processes that are affected by these mutations are in progress.
A variety of mutations affecting the fusion activity of SFV have been reported (30). Although mutations at many positions affect the pH dependence of fusion, only a mutation (Gly91 to Asp) in the hydrophobic domain of SFV E1 was found to completely block fusion (30). It is interesting that in RV E1, substitution of aspartic acid at Gly93 also resulted in complete blockage of cell fusion.
The binding and fusion process mediated by the spike proteins in the envelope of the virus particle usually involve a series of conformational changes in these proteins. In the case of SFV, conformational changes in E1 and E2 on exposure to a pH that induces fusion have been investigated by using protease digestion and monoclonal antibody assays (24, 25). Under the acidic conditions of the endosomes, the E1-E2 heterodimer of SFV dissociates and an NP-40-resistant E1 trimer is formed (48). Due to the lack of conformation-specific monoclonal antibodies available for RV, little is known about the low-pH-mediated fusion process of RV structural proteins. Although RV E2 is transported to the plasma membrane in the absence of E1 (16) and does not have fusion activity, the involvement of RV E2 in the fusion process cannot be ruled out. It is possible that conformational changes in RV E2 are necessary for activation of E1 acid sensitivity. Unfortunately, the E2 and E1 subunits are correctly folded and transported only as a heterodimer. No direct evidence for this mechanism is yet available. It is also possible that a maturation step occurring late in the exocytic pathway is required to confer acid sensitivity on newly synthesized E1.
ACKNOWLEDGMENTS
This work was supported by a grant from the Medical Research Council of Canada. Zhiyong Qiu was the recipient of a British Columbia’s Children’s Hospital Foundation postdoctoral fellowship. Shirley Gillam is an investigator of British Columbia’s Children’s Hospital Foundation.
REFERENCES
- 1.Anderson R G W, Orci L. A view of acidic intracellular compartments. J Cell Biol. 1988;106:539–543. doi: 10.1083/jcb.106.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barboni E, Rivero B P, George A J T, Martin S R, Renouf D V, Hounsell E F, Barber P C, Morris R J. The glycosylphosphatidylinositol anchor affects the conformation of Thy-I protein. J Cell Sci. 1995;108:487–497. doi: 10.1242/jcs.108.2.487. [DOI] [PubMed] [Google Scholar]
- 3.Baron M, Forsell K. Oligomerization of the structural proteins of rubella virus. Virology. 1991;185:811–819. doi: 10.1016/0042-6822(91)90552-m. [DOI] [PubMed] [Google Scholar]
- 4.Baron M D, Ebel T, Suomalainen M. Intracellular transport of rubella virus structural proteins expressed from cloned cDNA. J Gen Virol. 1992;73:1073–1086. doi: 10.1099/0022-1317-73-5-1073. [DOI] [PubMed] [Google Scholar]
- 5.Bernasconi E, Fasel N, Wittek R. Cell surface expression of a functional rubella virus E1 glycoprotein by addition of a GPI anchor. J Cell Sci. 1996;109:1195–1201. doi: 10.1242/jcs.109.6.1195. [DOI] [PubMed] [Google Scholar]
- 6.Chaye H, Chong P, Tripet B, Brush B, Gillam S. Localization of the virus neutralizing and hemagglutinin epitopes of E1 glycoprotein of rubella virus. Virology. 1992;189:483–492. doi: 10.1016/0042-6822(92)90572-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark D M, Loo T W, Hui I, Chong P, Gillam S. Nucleotide sequence and in vitro expression of rubella virus 24S subgenomic mRNA encoding the structural proteins E1, E2 and C. Nucleic Acids Res. 1987;15:3041–3057. doi: 10.1093/nar/15.7.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crise B, Ruusala A, Zagouras P, Shaw A, Rose J K. Oligomerization of glycolipid-anchored and soluble forms of the vesicular stomatis virus glycoprotein. J Virol. 1989;63:5328–5333. doi: 10.1128/jvi.63.12.5328-5333.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorsett, P. H., D. C. Miller, K. Y. Green, and F. I. Byrd. 1985. Structure and function of the rubella virus proteins. Rev. Infect. Dis. 7(Suppl. I):S150–S156. [DOI] [PubMed]
- 10.Duffus W A, Levy-Mintz P, Klimjack M R, Kielian M. Mutations in the putative fusion peptide of Semliki Forest virus affect spike protein oligomerization and virus assembly. J Virol. 1995;69:2471–2479. doi: 10.1128/jvi.69.4.2471-2479.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frey T K. Molecular biology of rubella virus. Adv Virus Res. 1994;44:69–160. doi: 10.1016/S0065-3527(08)60328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hemphill M L, Forng R, Abernathy E S, Frey T K. Time course of virus-specific macromolecular synthesis during rubella virus infection in Vero cells. Virology. 1988;162:65–75. doi: 10.1016/0042-6822(88)90395-9. [DOI] [PubMed] [Google Scholar]
- 14.Hobman T C, Shukin R, Gillam S. Translocation of rubella virus glycoprotein E1 into the endoplasmic reticulum. J Virol. 1988;62:4259–4264. doi: 10.1128/jvi.62.11.4259-4264.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hobman T C, Gillam S. In vitro and in vivo expression of rubella virus E2 glycoprotein: the signal peptide is located in the C-terminal region of capsid protein. Virology. 1989;173:241–250. doi: 10.1016/0042-6822(89)90240-7. [DOI] [PubMed] [Google Scholar]
- 16.Hobman T C, Lundström M L, Gillam S. Processing and intracellular transport of rubella virus structural proteins in COS cells. Virology. 1990;178:122–133. doi: 10.1016/0042-6822(90)90385-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hobman T C, Qiu Z, Chaye H, Gillam S. Analysis of rubella virus E1 glycosylation mutants expressed in COS cells. Virology. 1991;181:768–772. doi: 10.1016/0042-6822(91)90915-x. [DOI] [PubMed] [Google Scholar]
- 18.Hobman T C, Woodward L, Farquhar M G. The rubella virus E1 glycoprotein is arrested in a novel post-ER, pre-Golgi compartment. J Cell Biol. 1992;118:795–811. doi: 10.1083/jcb.118.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hobman T C, Woodward L, Farquhar M G. The rubella virus E2 and E1 spike glycoproteins are targeted to the Golgi complex. J Cell Biol. 1993;121:269–281. doi: 10.1083/jcb.121.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hobman T C, Seto N, Gillam S. Expression of soluble forms of rubella virus glycoproteins in mammalian cells. Virus Res. 1994;31:277–289. doi: 10.1016/0168-1702(94)90022-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hobman T C, Woodward L, Farquhar M G. Targeting of a heterodimeric membrane protein complex to the Golgi: rubella virus E2 glycoprotein contains a transmembrane Golgi retention signal. Mol Biol Cell. 1995;6:7–20. doi: 10.1091/mbc.6.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kääriäinen L, Soderlund H. Structure and replication of alphaviruses. Curr Top Microbiol Immunol. 1978;82:15–69. doi: 10.1007/978-3-642-46388-4_2. [DOI] [PubMed] [Google Scholar]
- 23.Katow S, Sugiura A. Low pH-induced conformational changes of rubella virus envelope proteins. J Gen Virol. 1988;69:2797–2807. doi: 10.1099/0022-1317-69-11-2797. [DOI] [PubMed] [Google Scholar]
- 24.Kielian M, Helenius A. pH-induced alterations in the fusogenic spike protein of Semliki Forest virus. J Cell Biol. 1985;101:2284–2291. doi: 10.1083/jcb.101.6.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kielian M, Jungerwirth S, Sayad K U, DeCandido S. Biosynthesis, maturation, and acid activation of the Semliki Forest virus fusion protein. J Virol. 1990;64:4614–4624. doi: 10.1128/jvi.64.10.4614-4624.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobayashi N. Hemolytic activity of rubella virus. Virology. 1978;89:6610–6612. doi: 10.1016/0042-6822(78)90202-7. [DOI] [PubMed] [Google Scholar]
- 27.Kondor-Koch C, Burke B, Garoff H. Expression of Semliki Forest virus proteins from cloned complementary DNA. 1. The fusion activity of the spike glycoproteins. J Cell Biol. 1983;97:644–651. doi: 10.1083/jcb.97.3.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kunkel T, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection methods. Methods Enzymol. 1987;154:367–387. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Levy-Mintz P, Kielian M. Mutagenesis of the putative fusion domain of the Semliki Forest virus spike protein. J Virol. 1991;65:4292–4300. doi: 10.1128/jvi.65.8.4292-4300.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matthews R E F. Classification and nomenclature of viruses. Third report of the International Committee on Taxonomy of Viruses. Intervirology. 1982;17:1–199. doi: 10.1159/000149278. [DOI] [PubMed] [Google Scholar]
- 32.Oker-Blom C, Kalkkinen N, Kääriäinen L, Pettersson R F. Rubella virus contains one capsid protein and three envelope glycoproteins, E1, E2a, and E2b. J Virol. 1983;46:964–973. doi: 10.1128/jvi.46.3.964-973.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oker-Blom C. The gene order for rubella virus structural proteins is NH2-C-E2-E1-COOH. J Virol. 1984;51:354–358. doi: 10.1128/jvi.51.2.354-358.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palmiter R D, Behringer R R, Quaife C J, Maxwell F, Maxwell I H, Brinster R I. Cell lineage ablation in transgenic mice by cell-specific expression of toxic gene. Cell. 1987;50:435–443. doi: 10.1016/0092-8674(87)90497-1. [DOI] [PubMed] [Google Scholar]
- 35.Pelham H R B. Speculation on the functions of the major heat shock and glucose-regulated proteins. Cell. 1986;46:959–961. doi: 10.1016/0092-8674(86)90693-8. [DOI] [PubMed] [Google Scholar]
- 36.Qiu Z, Hobman T C, McDonald H L, Seto N O L, Gillam S. Role of N-linked oligosaccharides in processing and intracellular transport of E2 glycoprotein of rubella virus. J Virol. 1992;66:3514–3521. doi: 10.1128/jvi.66.6.3514-3521.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qiu Z, Tufaro F, Gillam S. The influence of N-linked glycosylation on the antigenicity and immunogenicity of rubella virus E1 glycoprotein. Virology. 1992;190:876–881. doi: 10.1016/0042-6822(92)90929-j. [DOI] [PubMed] [Google Scholar]
- 38.Qiu Z, Ou D, Wu H, Hobman T C, Gillam S. Expression and characterization of virus-like particles containing rubella virus structural proteins. J Virol. 1994;68:4086–4091. doi: 10.1128/jvi.68.6.4086-4091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qiu Z, McDonald H L, Chen J, Hobman T C, Gillam S. Mutational analysis of the arginine residues in the E2-E1 junction region on the proteolytic processing of the polyprotein precursor of rubella virus. Virology. 1994;200:821–825. doi: 10.1006/viro.1994.1250. [DOI] [PubMed] [Google Scholar]
- 40.Qiu Z, Tufaro F, Gillam S. Brefeldin A and monensin arrest cell surface expression of membrane glycoproteins and release of rubella virus. J Gen Virol. 1995;76:855–863. doi: 10.1099/0022-1317-76-4-855. [DOI] [PubMed] [Google Scholar]
- 41.Salzwedel K, Johnston P B, Roberts S J, Dubay J W, Hunter E. Expression and characterization of glycophospholipid-anchored human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 1993;67:5279–5288. doi: 10.1128/jvi.67.9.5279-5288.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seto N L, Ou D, Gillam S. Expression and characterization of secreted forms of rubella virus E2 glycoprotein in insect cells. Virology. 1995;206:736–741. doi: 10.1016/s0042-6822(95)80098-0. [DOI] [PubMed] [Google Scholar]
- 43.Tarentino A L, Maley F. Purification and properties of an endo-β-N-acetylglucosaminidase from Streptomyces griseus. J Biol Chem. 1974;249:811–817. [PubMed] [Google Scholar]
- 44.Towbin H, Staehelin Y, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Väänänen P, Kääriäinen L. Fusion and haemolysis of erthrocytes caused by three togaviruses: Semliki Forest, Sindbis and rubella. J Gen Virol. 1980;46:467–475. doi: 10.1099/0022-1317-46-2-467. [DOI] [PubMed] [Google Scholar]
- 46.Waechter D E, Baserga R. Effect of methylation on expression of microinjected genes. Proc Natl Acad Sci USA. 1982;79:1106–1110. doi: 10.1073/pnas.79.4.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wahlberg J M, Boere W A M, Garoff H. The heterodimeric association between the membrane proteins of Semliki Forest virus changes its sensitivity to low pH during virus maturation. J Virol. 1989;63:4991–4997. doi: 10.1128/jvi.63.12.4991-4997.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wahlberg J M, Bron R, Wilschut J, Garoff H. Membrane fusion of Semliki Forest virus involves homotrimers of the fusion protein. J Virol. 1992;66:7309–7318. doi: 10.1128/jvi.66.12.7309-7318.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waxham M W, Wolinsky J S. Immunochemical identification of rubella virus hemagglutinin. Virology. 1983;126:194–203. doi: 10.1016/0042-6822(83)90471-3. [DOI] [PubMed] [Google Scholar]
- 50.White J M. Membrane fusion. Science. 1992;258:917–924. doi: 10.1126/science.1439803. [DOI] [PubMed] [Google Scholar]
- 51.Zhao H, Garoff H. Role of cell surface spikes in alphavirus budding. J Virol. 1992;66:7089–7095. doi: 10.1128/jvi.66.12.7089-7095.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]