Abstract
Cytotoxic T lymphocytes (CTL) can kill Hodgkin’s lymphoma (HL) cells, and CTL have been used for the treatment of Epstein-Barr virus (EBV)-positive HL. For patients with EBV-negative HL, this strategy cannot be employed and alternative target structures have to be defined. In order to establish a system for the stimulation of HL-reactive T cells, we used dendritic cells (DC) as antigen-presenting cells for autologous T cells and transfected these DC with RNA from established HL cell lines. After stimulation of peripheral blood mononuclear cells (PBMC) with RNA-transfected DC, we analyzed the reactivity of primed PBMC by interferon gamma enzyme-linked immunospot. Our results suggest the presence of antigens with expression in HL cell lines and recognition of these antigens in combination with DC-derived human leukocyte antigen molecules. By the analysis of Gene Expression Omnibus microarray data sets from HL cell lines and primary HL samples in comparison with testis and other normal tissues, we identified HL-associated cancer testis antigens (CTA) including the preferentially expressed antigen in melanoma (PRAME). After stimulation of PBMC with RNA-transfected DC, we detected PRAME-reactive T cells. PRAME and other HL-associated CTA might be targets for HL-specific immune therapy or for the monitoring of HL-directed immune responses.
Electronic supplementary material
The online version of this article (doi:10.1007/s00262-012-1239-z) contains supplementary material, which is available to authorized users.
Keywords: Hodgkin’s lymphoma, Gene expression, Cancer/testis antigens, ZBTB32, PRAME
Introduction
Hodgkin’s lymphoma (HL) is a lymphoproliferative disorder of unknown etiology. In most cases, a B-cell origin of the tumor cells has been suggested by molecular markers, but cases with expression of T-cell markers have as well been described [1]. In addition, HL cells exhibit the phenotype of professional antigen-presenting cells [2]. Regardless of the phenotype, HL cell lines have a characteristic gene expression profile that allows to discriminate these cells from other hematopoietic cells [3] Today more than 90% of HL patients are cured by a combination of radiotherapy and chemotherapy [4]. However, a significant number of patients cannot be cured with current therapy regimes. Even allogeneic stem cell transplantation has not led to satisfying results, because a significant proportion of the patients relapsed or died as a consequence of toxicity. It was suggested that reduced-intensity conditioning might improve the prognosis [5]. In addition, a remarkable graft-versus-lymphoma effect was demonstrated [5]. However, a high proportion of patients who had been treated with donor lymphocyte infusion after allogeneic transplantation developed severe graft-versus-host disease, and some patients died as a consequence of this complication. Taken together, these and other data suggest that HL cells can be targeted by immunocompetent cells, but that the specificity of this strategy is not sufficiently high. Therefore, the induction of T cells with specificity for HL-associated antigens might be an interesting strategy. In the case of Epstein-Barr virus (EBV)-positive HL, EBV-encoded antigens can be used as target structures for such T cells [6]. For patients with EBV-negative HL, alternative target structures have to be defined. In general, two strategies can be used for the induction of tumor-specific T cells. Firstly, peptides derived from known tumor antigens can be used for the stimulation of T cells. For this strategy, it is necessary to know which antigens are expressed in the tumor cells and which peptides can be presented by the major histocompatibility complex (MHC) antigens of the individual patient. Alternatively, proteins or ribonucleic acids (RNA) can be isolated from tumor cells and used for the stimulation of T cells. For the stimulation of T cells with specificity for pre-selected peptides as well as for the induction of tumor-specific T cells by the use of RNA-transfected antigen-presenting cells, dendritic cells (DC) are highly potent, because DC are able to stimulate naïve T cells [7]. The number of tumor cells in affected lymph nodes from HL patients is very low, and the establishment of cell lines is difficult. Therefore, it is not possible to prepare fully autologous vaccines for each HL patient. In other tumor models, the use of allogeneic vaccines based on permanently growing allogeneic tumor cell lines has been suggested [8–10]. In this context, clinical trials have demonstrated safety and immunogenicity of such vaccines [9, 10]. In addition to genetically modified allogeneic tumor cells, DC-based vaccination strategies have been used. These strategies include DC after transfection with RNA from allogeneic tumor cells [10]. Even after transfection of RNA from autologous tumor cells, DC-stimulated T cells are not always specific for antigens encoded by the transfected RNAs [11]. Nevertheless, in clinical studies, neither the induction of autoimmunity nor other severe side effects have been observed.
In a previous study, we have characterized gene expression and chemotherapy resistance of established HL cell lines [12]. For all tested cytostatic drugs, HL cell line L-540 showed the highest sensitivity, whereas L-1236-cells showed the lowest sensitivity. We identified several genes that are differentially expressed between resistant and sensitive cells. Resistant cells showed an increased expression of genes with known association to apoptosis inhibition (e.g., CD40) or chemotherapy resistance (e.g., MARCKS). In addition, we observed high expression of co-stimulatory molecules (e.g., CD80) on the surface of resistant cells. The expression of co-stimulatory molecules suggests that immunological targeting of tumor cells might be an interesting option for patients with resistant HL. Therefore, we established a system for the stimulation of HL-reactive T cells.
Materials and methods
Cells and cell culture
Sarcoma cell lines SK-N-MC [13] and TE-671 [14] as well as HL cell lines L-1236 [15], L-540 [16], L-428 [17], HDLM-2 [17] and KM-H2 [18] were obtained from the Deutsche Sammlung für Mikroorganismen und Zellkulturen (Braunschweig, Germany). Ewing tumor cell line A673 [19] was obtained from the American Type Culture Collection (Manassas, VA, USA). T2 cells [20] were a kind gift from Dr. A. B. Reske-Kunz (Mainz, Germany). All cells were cultured in RPMI1640 medium (Invitrogen, Karlsruhe, Germany) supplemented with 10% fetal calf serum and penicillin/streptomycin. In some experiments, cells were treated with 5 μM 5′-azacytidine (Sigma, Heidelberg, Germany) for 2 weeks. Peripheral blood mononuclear cells (PBMC), CD3-, CD4-, CD8-, CD14-, CD15-, CD16,- CD19-, and CD56-positive cells from healthy donors were isolated as described [21] with informed consent and approval by the ethics committee of the Medical Faculty of the Martin-Luther-University Halle-Wittenberg. Dendritic cells were generated from CD14-positive PBMC or the plastic adherent fraction of PBMC using standard methods [22]. Human leukocyte antigen (HLA) haplotypes from tumor cell lines and PBMC donors were determined by molecular HLA typing. In addition, serological typing was used for the analysis of HLA class I from PBMC donors. HLA haplotypes of the used cell lines are summarized in Table 1.
Table 1.
HLA types of the used cell lines and PBMC donors
| Cells | Cell type | HLA-A | HLA-B | HLA-Cw | HLA-DRB1 | HLA-DQB1 |
|---|---|---|---|---|---|---|
| L-428 | HL | 03 | 35 | 04 | 12 | 03 |
| L-540 | HL | 03, 11 | 51 | 02, 15 | 04, 11 | 03 |
| L-1236 | HL | 02 | 51 | 02 | 14 | 05 |
| KM-H2 | HL | 11, 24 | 15, 52 | 04, 12 | 04, 11 | 03 |
| HDLM-2 | HL | 01, 02 | 08, 44 | 05, 07 | 13, 15 | 06 |
| TE-671 | RMS | 01 | 37 | 06 | 03 | 02 |
| A673 | EFT | 01, 02 | 07 | 07 | 04, 15 | 03, 06 |
| SK-N-MC | EFT | 01, 25 | 08 | 07 | 03, 15 | 02, 06 |
| Donor 1 | PBMC | 02, 24 | 15 | 03, 07 | 04, 11 | 03 |
| Donor 7 | PBMC | 01, 02 | 08, 15 | 04, 07 | 01, 11 | 03, 05 |
| Donor 8 | PBMC | 01, 03 | 08, 35 | 04, 07 | 03, 11 | 03 |
Presented are HLA types of the tumor cell lines used in this study (HL Hodgkin’s lymphoma cell lines, RMS cell line from rhabdomyosarcoma, EFT Ewing family tumor cell lines). In addition, HLA types from three PBMC donors are presented
Reverse transcription polymerase chain reaction and sequence analysis
RNA from cultured cells and PBMC was isolated by using Trizol reagent (Invitrogen) following the manufacturer’s protocol. In addition, RNA from spleen, kidney and testis was obtained from Becton–Dickinson (Heidelberg, Germany). After reverse transcription of 2 μg of RNA, conventional and quantitative polymerase chain reaction (PCR) was performed as described elsewhere [12, 23]. The following primer combinations have been used: actin beta (ACTB): 5′-ggc atc gtg atg gac tcc g-3′ and 5′-gct gga agg tgg aca gcg a-3′; glyceraldehyde-3-phosphate dehydrogenase (GAPDH): 5′-cca tgg aga agg ctg ggg-3′ and 5′-caa agt tgt cat gga tga cc-3′; PRAME: 5′-gct gtg ctt gat gga ctt ga-3′ and 5′-aag gtg ggt agc ttc cag gt-3′; and zinc finger and BTB (bric-a-brac, tramtrack, broad-complex) domain containing 32 (ZBTB32): 5′-aga tgt tgc aca agc act cg-3′ and 5′-tgg ggg tgc tat ggt aga ag-3′. Sequencing of PRAME was performed as described [24] by using primers: 5′-aaa ctc gag cgt ggc aac aag tga-3′ and 5′-aaa ctc gag tgc aca tcc tgg ctt-3′. Sequence analysis was performed with BLAST [25].
DNA microarray analysis and identification of HL-associated CTA
DNA microarray analysis of HL cell lines using Affymetrix HG-U133A microarrays was performed as described [8, 12]. For the identification of CTA, data sets from HL cell lines (GEO data set GSE26325) [12] were analyzed together with data from normal tissues (GEO data set GSE2361) [26]. Additional Affymetrix HG-U133Plus2.0 microarray data files from HL samples, testis and other normal tissues were obtained from the GEO database [27]. Primary data analysis was performed with Expression Console (Affymetrix, Santa Clara, USA) applying the Microarray Suite 5 algorithm. All microarray data sets were scaled to the same target intensity of 500. Data visualization and cluster analysis were performed with Genesis [28]. For the identification of putative cancer/testis antigens, we developed a short analysis program “MAFilter.” This program allows the filtering of tab-separated text files (e.g., exported from the Affymetrix Expression Console) on the basis of programmable statistical criteria. The program is available online from the homepage of the authors (http://steingrube-home.de/software_MAFilter.html). For the identification of HL-associated CTA, an algorithm based on Wilks’ lambda score (WLS) [29] was applied. The microarray database used for the identification of HL-associated CTA was constructed using the following data sets: 37 data sets from naïve B cells, memory B cells, centrocytes, centroblasts, plasma cells and micro-dissected classical HL (GSE12453); 164 data sets from different normal tissues, selected from GSE7307 and GSE3526; 4 data sets from micro-dissected classical HL (GSE14879); and additional data sets from established HL cell lines from data sets GSE20011, GSE25986 and GSE12427 [30–35].
T-cell stimulation
Mature DC (2 × 106) were transfected with RNA (50 μg/ml) from tumor cell lines by electroporation (square wave, 500 V, 0.5 ms, 0.2 ml cuvette) using a GenePulser Xcell (Bio-Rad, Munich, Germany). Transfected DC were used for the stimulation of PBMC or isolated CD8-positive T cells from the same donor. Alternatively, PBMC were stimulated with mitomycin C (50 μg/ml)-treated T2 cells that had been pulsed with HLA-A2-binding peptides from CTA. Peptides corresponding to PRAME100-108 (VLDGLDVLL: = VLD), PRAME300-309 (ALYVDSLFFL: = ALY), PRAME142-151 (SLYSFPEPEA: = SLY) and membrane-associated phosphatidic acid-selective phospholipase A1-beta (LIPI) (LDYTDAKFV : = LDY) were synthesized by WITA (Telto, Germany). Successful stimulation of PBMC was monitored by pentamer staining, interferon gamma enzyme-linked immunospot (ELISPOT) or lactate dehydrogenase assay (Roche, Mannheim, Germany), according to manufacturer’s instructions. Spots from ELISPOT analysis were enumerated automatically using the KS Elispot system (Carl Zeiss, Jena, Germany). VLD/HLA-A2-pentamers and ALY/HLA-A2-pentamers were purchased from ProImmune (Oxford, UK). Additional antibodies and ELISPOT reagents were purchased from Becton–Dickinson (Heidelberg, Germany). Multicolor flow cytometry was performed by using BD Oncomark (T-cell receptor (TCR)α/β-fluorescein isothiocyanate (FITC)/TCRγ/δ-phycoerythrin (PE)/CD3-PerCP-Cy5.3) or TriTEST (CD4-FITC, CD8-PE and CD3-PerCP) reagents (Becton–Dickinson). Flow cytometry was performed essentially as described [36].
Results
T-cell stimulation by HL-RNA-loaded DC
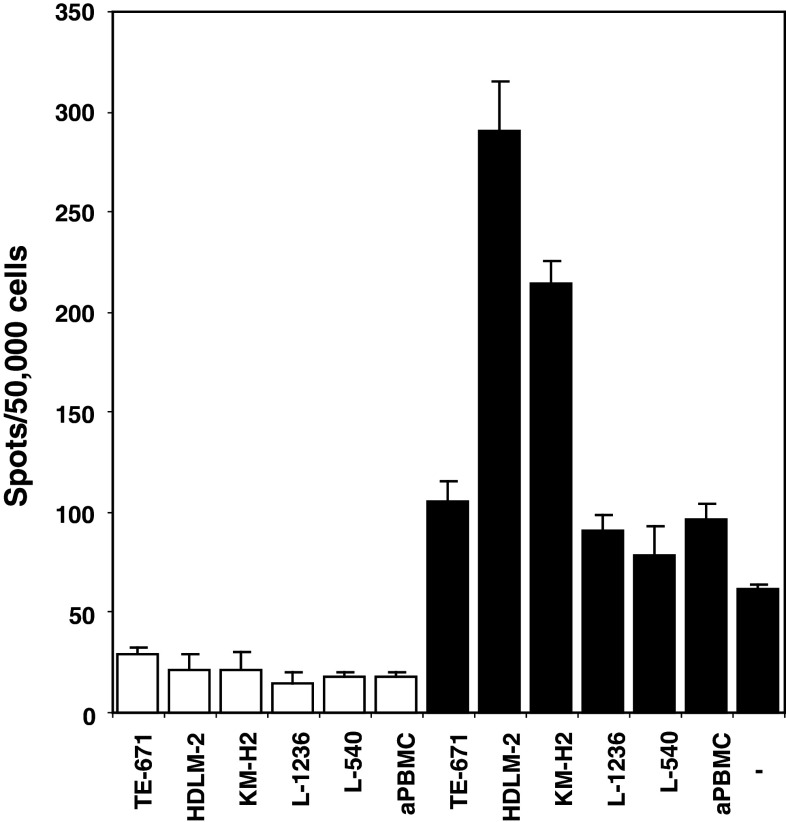
In order to establish a system for the stimulation of HL-reactive T cells, we used DC as antigen-presenting cells for autologous T cells and transfected these DC with RNA from established HL cell lines. After stimulation of PBMC with HL-RNA-transfected DC, we observed reactivity of primed PBMC against different HL cell lines (supplementary Table 1). As was expected in the presence of different HLA haplotypes, reactivity against different HL cell lines and cross-reactivity with unrelated tumor cell lines varied from donor to donor. Most donors exhibit an increased reactivity against HL cell lines with a more pronounced B-cell phenotype (L-1236 and KM-H2), but other cell lines were also recognized. After transfection of RNA from L-540 cells, one donor did not react with L-540 cells but showed some reactivity against other HL cell lines. When we transfected DC (HLA-A*02,24; B*15; Cw*03,07; DRB1*04,11; DQB1*03) with RNA from HL cell line HDLM-2 (HLA-A*01,02; B*08,44; Cw*05,07; DRB1*13,15; DQB1*06) and used these DC for priming of the PBMC from the same DC-donor, we observed that the stimulated cells reacted with HDLM-2 cells but not with irrelevant tumor cells (TE-671 sarcoma cells; HLA-A*01; B*37; Cw*06; DRB1*03; DQB1*02) or autologous PBMC (Fig. 1). Interestingly, these HDLM-2-primed cells also reacted with HL cell line KM-H2 (HLA-A*11,24; B*52,15; Cw*04,12; DRB1*04,11; DQB1*03) but not with HL cell lines L-1236 (HLA-A*02; B*51; Cw*02; DRB1*14; DQB1*05) nor with L-540 (HLA-A*03,11; B*51; Cw*02,15; DRB1*04,11; DQB1*03). Because HDLM-2 cells and KM-H2 cells have completely different MHC haplotypes, and KM-H2 cells share 6/10 MHC molecules with the used PBMC, these observations suggest that HL-specific antigens, expressed in both HL cell lines and presented in the context of the MHC molecules of both HL cell lines, led to the T-cell response. Similar results were obtained with other donors with different HLA haplotypes (supplementary Table 1).
Fig. 1.
ELISPOT analysis of PBMC after stimulation with HDLM-2 RNA-transfected dendritic cells. Dendritic cells were transfected with total RNA from HL cell line HDLM-2 and used as antigen-presenting cells for PBMC from the same donor. On day 7, primed PBMC were re-stimulated with RNA-transfected DC. Six days after re-stimulation, the reactivity of stimulated cells against indicated cell lines was assessed by interferon gamma ELISPOT. Stimulatory cells were incubated in the absence (open bars) or presence (closed bars) of primed PBMC (50.000 cells per well). Fresh autologous PBMC (aPBMC) and medium without stimulatory cells (-) served as controls. Spots were automatically detected by a KS Elispot system (Carl Zeiss). Presented are means and standard deviations from triplicates
Identification of putative HL-associated antigens
In order to identify possible candidates for antigens that are recognized by HL-reactive T cells, we analyzed microarray data from established HL cell lines in comparison with a panel of normal tissues. We used the same database that was used previously for the identification of chemotherapy resistance factors [12]. Due to cancer/testis antigens (CTA) are an important group of antigens, we focused our analysis on the identification of CTA in HL cells. By using the MAFilter program, we used the following algorithm for the identification of CTA: the data sets in the microarray database were grouped into (a) data sets from HL cell lines, (b) data sets from testis and (c) data sets from other normal tissues. Firstly, the database was filtered for a high quotient 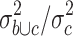 of the variance
of the variance 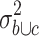 in the merged group b and c and the variance
in the merged group b and c and the variance 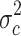 in group c alone. The 2,228 probe sets (10% of all probe sets) with highest quotient were used for further analysis. These probe sets were filtered for a high quotient
in group c alone. The 2,228 probe sets (10% of all probe sets) with highest quotient were used for further analysis. These probe sets were filtered for a high quotient 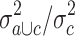 of the variance
of the variance 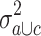 in group a and c and the variance
in group a and c and the variance  . The 223 probe sets (1% of all probe sets) with highest quotient were further filtered for significant (p < 0.01) differences between HL samples and normal tissues by applying Student’s t test. 115 probe sets passed the filter (supplementary Table 2). Among these probe sets, we found few known CTA, for example, the outer dense fiber of sperm protein 2 (ODF2) [37]. In addition, we found new genes with CTA-like expression pattern. The probe set with highest WLS in the comparison of HL and normal tissues corresponds to the zinc finger and BTB (bric-a-brac, tramtrack, broad-complex) domain containing 32 (ZBTB32) protein (supplementary Table 2). RT-PCR proved expression of ZBTB32 in HL cell lines and testis but not other normal tissues (Fig. 2a). Quantitative PCR indicated expression of ZBTB32 in isolated B cells but not in other hematopoietic cells (supplementary Figure 2). We extended our analysis by using a second microarray database. This database was constructed on the basis of Affymetrix HG_U133Plus2.0 microarray data sets (see “Materials and methods” section) and has several advantages: the number of probe sets on this array platform is higher and covers a higher percentage of the human genome, the database contains more samples from normal testis, the database contains samples from normal B cells, and finally, the database contains not only data sets from HL cell lines but also data sets from primary micro-dissected HL cells. For the identification of CTA in this database, we followed a similar strategy as described above. In a first step, the data sets in the database were grouped into (a1) data sets from micro-dissected HL cells, (a2) data sets from established HL cell lines, (b) data sets from testis and (c) data sets from other normal tissues. The database was filtered for a high quotient
. The 223 probe sets (1% of all probe sets) with highest quotient were further filtered for significant (p < 0.01) differences between HL samples and normal tissues by applying Student’s t test. 115 probe sets passed the filter (supplementary Table 2). Among these probe sets, we found few known CTA, for example, the outer dense fiber of sperm protein 2 (ODF2) [37]. In addition, we found new genes with CTA-like expression pattern. The probe set with highest WLS in the comparison of HL and normal tissues corresponds to the zinc finger and BTB (bric-a-brac, tramtrack, broad-complex) domain containing 32 (ZBTB32) protein (supplementary Table 2). RT-PCR proved expression of ZBTB32 in HL cell lines and testis but not other normal tissues (Fig. 2a). Quantitative PCR indicated expression of ZBTB32 in isolated B cells but not in other hematopoietic cells (supplementary Figure 2). We extended our analysis by using a second microarray database. This database was constructed on the basis of Affymetrix HG_U133Plus2.0 microarray data sets (see “Materials and methods” section) and has several advantages: the number of probe sets on this array platform is higher and covers a higher percentage of the human genome, the database contains more samples from normal testis, the database contains samples from normal B cells, and finally, the database contains not only data sets from HL cell lines but also data sets from primary micro-dissected HL cells. For the identification of CTA in this database, we followed a similar strategy as described above. In a first step, the data sets in the database were grouped into (a1) data sets from micro-dissected HL cells, (a2) data sets from established HL cell lines, (b) data sets from testis and (c) data sets from other normal tissues. The database was filtered for a high quotient 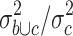 of the variance
of the variance 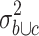 in the merged group b and c and the variance
in the merged group b and c and the variance 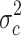 in group c alone. The 5,468 probe sets (10%) with highest quotient were used for further analysis. Among these probe sets, we selected those with a high quotient
in group c alone. The 5,468 probe sets (10%) with highest quotient were used for further analysis. Among these probe sets, we selected those with a high quotient 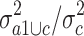 of the variance
of the variance 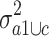 in group a1 and c and the variance
in group a1 and c and the variance 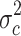 . The 547 (1%) probe sets with highest quotient were then filtered for a high quotient
. The 547 (1%) probe sets with highest quotient were then filtered for a high quotient 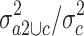 of the variance
of the variance 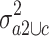 in group a2 and c and the variance
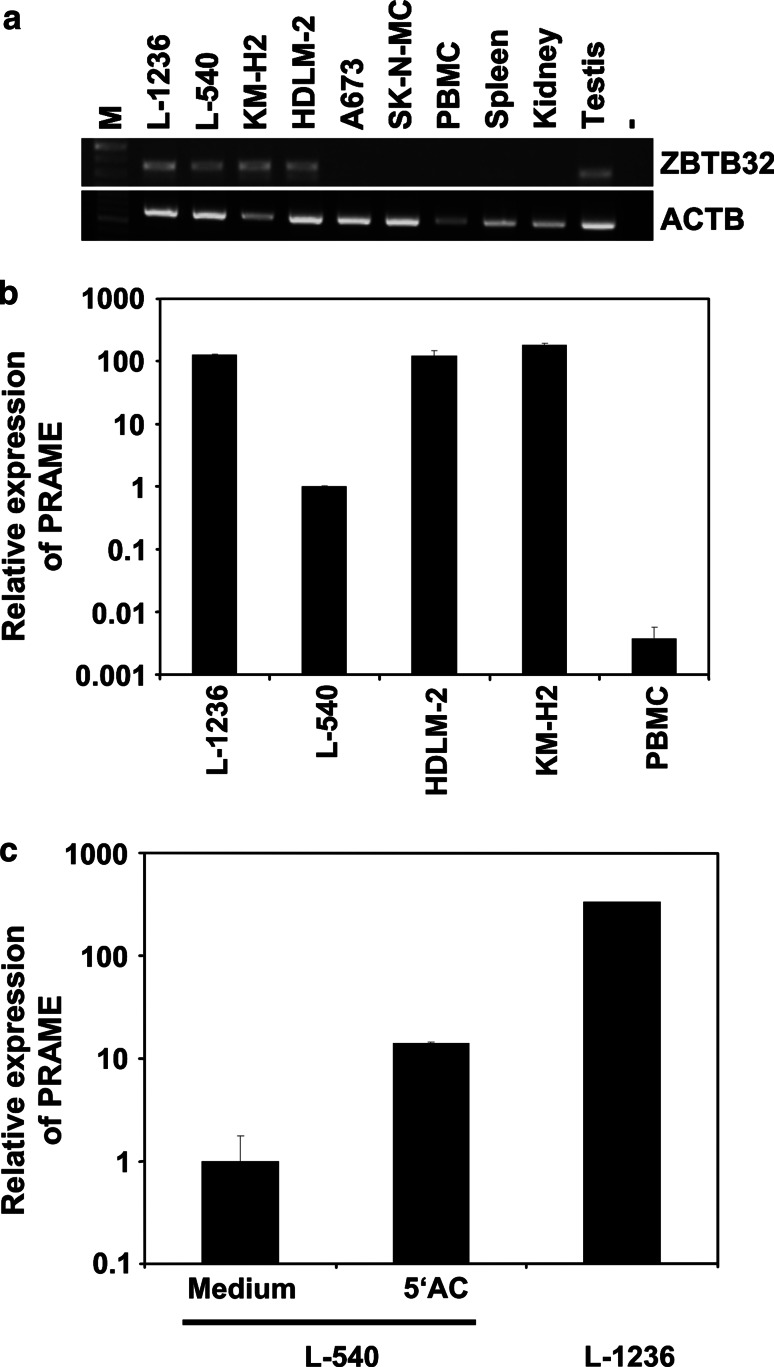
in group a2 and c and the variance 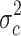 . Finally, the 273 probe sets with highest signal intensities in testis and HL samples were filtered for significant (p < 0.01) differences between HL samples (group a1 and a2) and data sets from normal B cells in the database by applying Student’s t test. All calculations and variance filtering were performed with the MAFilter program. A total of 198 probe sets passed the filtering procedure (supplementary Table 3). We found CTA with known expression in HL, for example, cancer/testis antigen 45 (CT45) [38] or preferentially expressed antigen in melanoma (PRAME) [12, 39]. Quantitative RT-PCR proved high expression of PRAME in HL cell lines (Fig. 2b). The expression of PRAME in cells with relatively low expression (L-540) was increased after treatment with 5′-azacytidine (Fig. 2c and supplementary Figure 3). Other examples for HL-associated antigens are illustrated in Fig. 3. In addition to new genes with CTA-like expression pattern (e.g., the histone H2AFB1), some antigens showed high expression in HL samples and testis but also expression in a restricted panel of other normal tissues. An example for this class of antigens was ZBTB32 that shows high signal intensities in normal memory B cells (Fig. 3). In cluster analyses with these probe sets as data points, HL and testis clustered together (Fig. 4). Similar results were obtained by using the probe sets from the HG_U133A analysis as data points (supplementary Figure 1). Among the micro-dissected HL cells, a subgroup with a higher similarity with testis and established HL cell lines was present (Fig. 4). We sequenced the cDNA of PRAME from HL cell lines L-1236, L-428, L-540, KM-H2 and HDLM-2. Sequences have been submitted to GenBank (accession numbers JQ511981, JQ511982, JQ511983, JQ511984 and JQ511985). All cell lines contained the wild-type sequence. The PRAME sequence from cell line HDLM-2 differed from the other cell lines at positions 7 and 47 (supplementary Figure 4). Both differences are known single-nucleotide polymorphisms (rs41310248 and rs2266988). For the polymorphism rs41310248 (L47M), frequency data are not available. Frequencies of the polymorphism rs2266988 (W7R) in HL cell lines were not significantly different from the frequencies (for Utah residents with ancestry from northern and western Europe) of the HAPMAP database [40] judged by a χ2 test (p > 0.06). Known immunodominant epitopes [41–44] were not affected by these polymorphisms (supplementary Figure 4).
. Finally, the 273 probe sets with highest signal intensities in testis and HL samples were filtered for significant (p < 0.01) differences between HL samples (group a1 and a2) and data sets from normal B cells in the database by applying Student’s t test. All calculations and variance filtering were performed with the MAFilter program. A total of 198 probe sets passed the filtering procedure (supplementary Table 3). We found CTA with known expression in HL, for example, cancer/testis antigen 45 (CT45) [38] or preferentially expressed antigen in melanoma (PRAME) [12, 39]. Quantitative RT-PCR proved high expression of PRAME in HL cell lines (Fig. 2b). The expression of PRAME in cells with relatively low expression (L-540) was increased after treatment with 5′-azacytidine (Fig. 2c and supplementary Figure 3). Other examples for HL-associated antigens are illustrated in Fig. 3. In addition to new genes with CTA-like expression pattern (e.g., the histone H2AFB1), some antigens showed high expression in HL samples and testis but also expression in a restricted panel of other normal tissues. An example for this class of antigens was ZBTB32 that shows high signal intensities in normal memory B cells (Fig. 3). In cluster analyses with these probe sets as data points, HL and testis clustered together (Fig. 4). Similar results were obtained by using the probe sets from the HG_U133A analysis as data points (supplementary Figure 1). Among the micro-dissected HL cells, a subgroup with a higher similarity with testis and established HL cell lines was present (Fig. 4). We sequenced the cDNA of PRAME from HL cell lines L-1236, L-428, L-540, KM-H2 and HDLM-2. Sequences have been submitted to GenBank (accession numbers JQ511981, JQ511982, JQ511983, JQ511984 and JQ511985). All cell lines contained the wild-type sequence. The PRAME sequence from cell line HDLM-2 differed from the other cell lines at positions 7 and 47 (supplementary Figure 4). Both differences are known single-nucleotide polymorphisms (rs41310248 and rs2266988). For the polymorphism rs41310248 (L47M), frequency data are not available. Frequencies of the polymorphism rs2266988 (W7R) in HL cell lines were not significantly different from the frequencies (for Utah residents with ancestry from northern and western Europe) of the HAPMAP database [40] judged by a χ2 test (p > 0.06). Known immunodominant epitopes [41–44] were not affected by these polymorphisms (supplementary Figure 4).
Fig. 2.
Detection of CTA in HL cell lines by PCR. a Expression of ZBTB32 was analyzed by RT-PCR in HL cell lines (L-1236, L-540, KM-H2 and HDLM-2), irrelevant tumor cell lines (A673 and SK-N-MC) and a panel of normal tissues. Actin beta (ACTB) was used as housekeeping control. b quantitative RT-PCR was used for the quantification of PRAME transcripts in HL cell lines. For the calculation of relative expression values, ACTB was used as housekeeping control and expression in L-540 cells was set as 1. Presented are means and standard deviations from triplicates (HL cell lines). PBMCs were used as control. Presented are means and standard deviations from 8 experiments (PBMC samples from 4 independent donors were analyzed in duplicates). c quantitative RT-PCR was used for the quantification of PRAME transcripts in HL cells. L-540 cells were cultured for 2 weeks in the presence or in the absence of 5 μM 5′-azacytidine. For calculation of relative expression values, glyceraldehyde-3-phosphate dehydrogenase was used as housekeeping control and expression in untreated L-540 cells was set as 1. Presented are means and standard deviation from three independent experiments
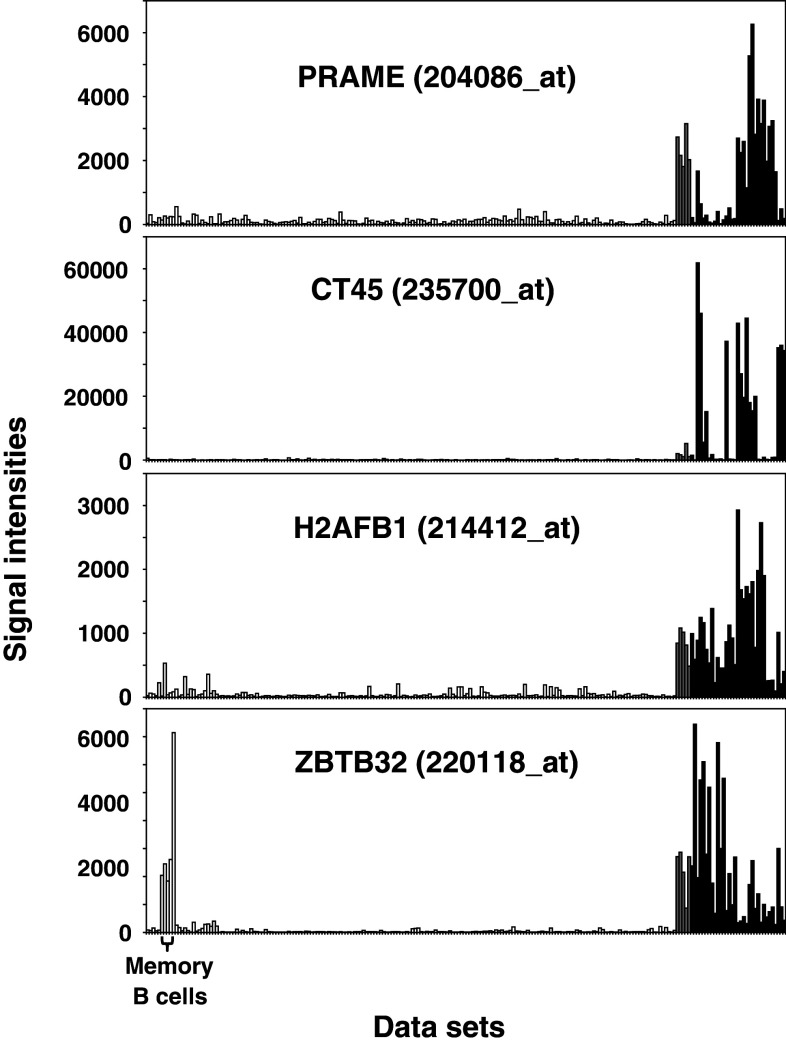
Fig. 3.
Expression patterns of putative HL-associated CTA. Presented are examples of signal intensities from probe sets of identified HL-associated CTA in normal tissues (open bars), testis (gray bars) and HL samples (black bars). The following GEO data sets have been used: GSE12453, GSE7307, GSE3526, GSE14879, GSE20011, GSE25986 and GSE12427. PRAME, CT54 and H2AFB1 are expressed nearly exclusively in testis and HL samples, whereas ZBTB32 shows high signal intensities in memory B cells
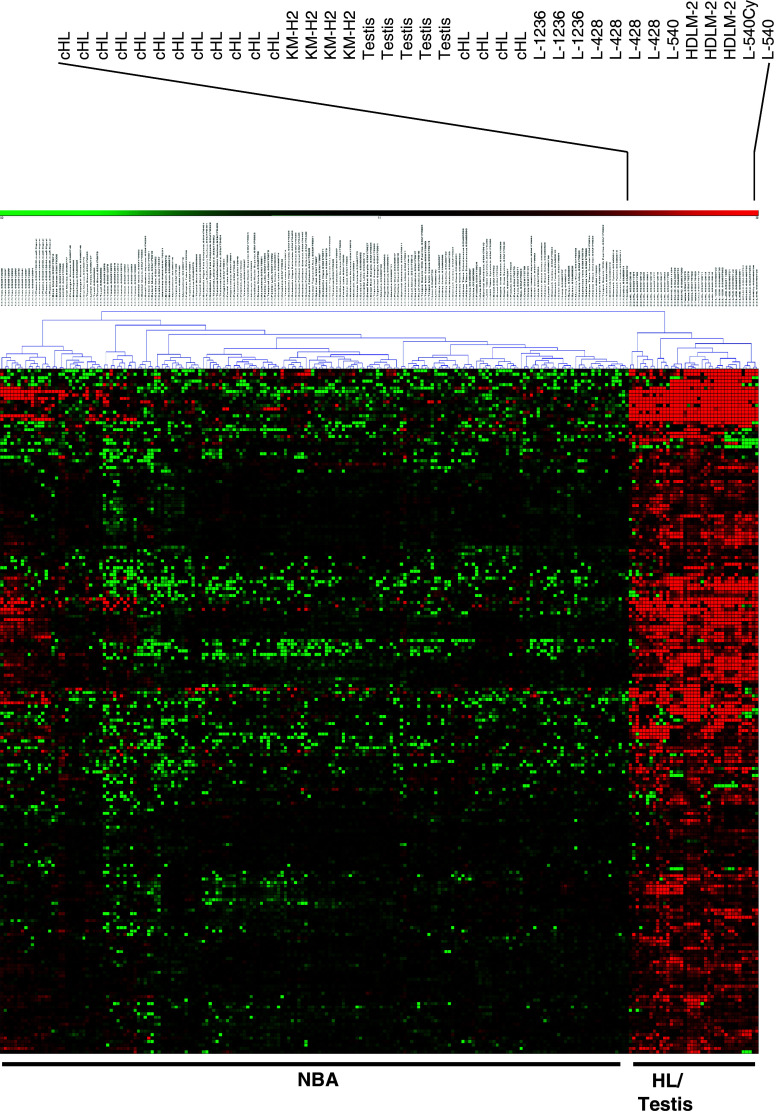
Fig. 4.
Expression of putative CTA in HL cells (HG_U133Plus2.0 analysis). Putative HL-associated CTA were identified by using MAFilter software as described in the text. The following GEO data sets have been used: GSE12453, GSE7307, GSE3526, GSE14879, GSE20011, GSE25986 and GSE12427. Presented is a cluster analysis with the identified probe sets (log2-transformed and median-centered signal intensities, absolute Manhattan distance and complete linkage clustering). Cluster analysis was performed with Genesis. The positions from samples from HL cell lines, micro-dissected HL cells (cHL), testes and other normal samples (NBA) are indicated
HL-RNA-transfected DC prime T cells for recognition of cancer/testis antigens
We asked whether PBMC after stimulation with RNA-transfected DC might recognize HL-associated CTA. Flow cytometric analysis of PBMC after stimulation with transfected DC showed the presence of variable numbers of TCRαβ-positive, CD8-positive and CD4-positive T cells and very low numbers of NK cells and TCRγδ-positive T cells (supplementary Figure 5). Increased expression of activation markers HLA-DR (data not shown) and CD69 (supplementary Figure 5) was observed on the surface of both CD4-positive and CD8-positive cells. By using two PRAME/HLA-A2 pentamers (corresponding to PRAME peptides ALY and VLD), we found increased frequencies of T cells binding these pentamers after in vitro stimulation of HLA-A2-positive donors with HL-RNA-transfected DC compared to cells stimulated with mock-transfected DC (supplementary Table 4; Fig. 5). Vice versa, PBMC that had been stimulated previously with PRAME peptide-pulsed HLA-A2-positive T2 cells showed, in addition to variable alloreactivity, a response not only against these cells but also against HLA-A2-positive L-1236 cells (Table 2). In all experiments, the number of cells reacting against PRAME peptide-pulsed T2 cells increased after stimulation with PRAME peptide-pulsed T2 cells, whereas the reactivity was low without primary stimulation or after primary stimulation with un-pulsed T2 cells. Similarly, reactivity against L-1236 cells increased in 4/5 donors after stimulation with PRAME peptide-pulsed T2 cells. Three of these donors had also a higher reactivity against PRAME peptide-pulsed T2 cells than against T2 cells without peptide pulse, suggesting peptide specificity (Table 1). Similar results were obtained in cytotoxicity assay (supplementary Figure 6). After priming with PRAME peptides, the PBMC showed PRAME specificity. No such specificity was observed after priming with an irrelevant peptide with similar HLA-A2 stabilization activity.
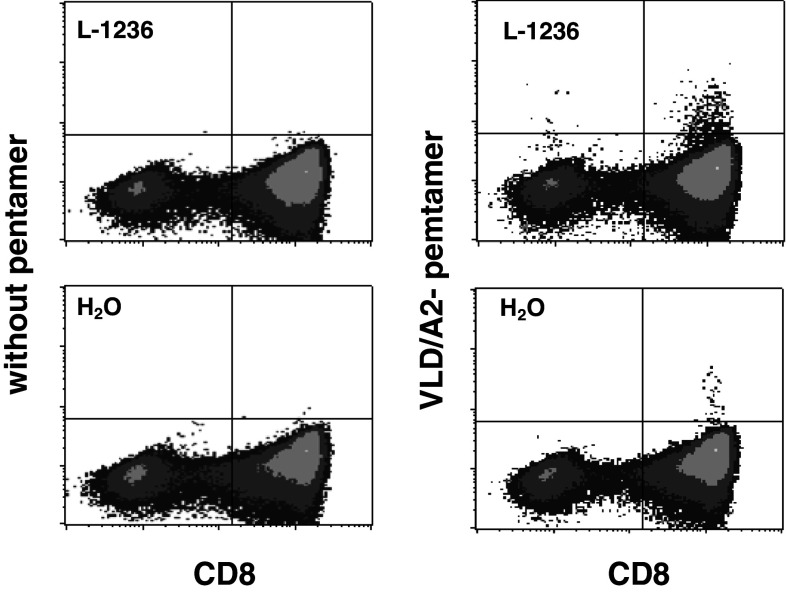
Fig. 5.
Characterization of T cells after DC stimulation in vitro. PBMC (5 × 106) from an HLA-A2-positive donor were primed with DC (1 × 106) that had been transfected with total RNA from cell line L-1236. After one re-stimulation, PRAME-specific cells were analyzed by pentamer staining with VLD/HLA-A2 pentamers. PBMC after stimulation with mock-transfected DC (H2O) were used as control
Table 2.
PRAME-stimulated PBMC react with L-1236 HL cells
| Primary stimulation | Stimulatory cells in ELISPOT | Donor 1 | Donor 2 | Donor 3 | Donor 4 | Donor 5 |
|---|---|---|---|---|---|---|
| None | None | 0 | 0 | 0 | 24 | – |
| None | T2 | 26 | 31 | 27 | 34 | – |
| None | T2-SLY (PRAME) | 46 | 66 | 26 | 33 | – |
| None | L-1236 | 80 | 31 | 16 | 38 | – |
| T2 | None | 0 | 0 | – | – | 24 |
| T2 | T2 | 0 | 36 | – | – | 110 |
| T2 | T2-SLY (PRAME) | 0 | 119 | – | – | – |
| T2 | L-1236 | 0 | 22 | – | – | 137 |
| T2-SLY | None | 39 | 0 | 23 | 222 | 60 |
| T2-SLY | T2 | 288 | 344 | 278 | 290 | – |
| T2-SLY | T2-LDY (LIPI) | – | – | – | – | 150 |
| T2-SLY | T2-SLY (PRAME) | 283 | 538 | 390 | 362 | 160 |
| T2-SLY | L-1236 | 190 | 294 | 237 | 295 | 107 |
| T2-ALY | None | – | – | – | – | 46 |
| T2-ALY | T2-LDY (LIPI) | – | – | – | – | 190 |
| T2-ALY | T2-ALY (PRAME) | – | – | – | – | 248 |
| T2-ALY | L-1236 | – | – | – | – | 246 |
| T2-VLD | None | – | – | – | – | 62 |
| T2-VLD | T2-LDY (LIPI) | – | – | – | – | 222 |
| T2-VLD | T2-VLD (PRAME) | – | – | – | – | 203 |
| T2-VLD | L-1236 | – | – | – | – | 215 |
HLA-A2-positive PBMC were primed with T2 cells or with T2 cells that had been pulsed with 100 μg/ml of PRAME-derived peptides SLY, ALY and VLD, respectively (primary stimulation). Reactivity of stimulated cells with T2 cells and L-1236 cells was assessed by interferon gamma ELISPOT. For this end, T2 cells had been pulsed with PRAME-derived peptides (T2-SLY, T2-ALY and T2-VLD), peptides derived by the irrelevant antigen LIPI (T2-LDY) or left un-pulsed (T2). Presented are means from triplicates after background subtraction. – not done
Discussion
Tumor antigens have been identified as targets for naturally occurring CTL responses in patients with varying malignancies. Tumor specificity of such antigens seems to be the rule [45]. However, the majority of known tumor antigens are not specific for tumor cells but are additionally expressed at least in subsets of normal cells. Cancer/testis antigens are one important group of these tumor-associated antigens. Not all CTA can be detected by DNA microarray analysis [23]. Nevertheless, comparative microarray analysis can be used for the identification of genes with CTA-like expression pattern. Interestingly, the algorithm presented in the present manuscript did not only allow the identification of genes with a conventional CTA expression pattern (e.g., PRAME or CT45) but also the identification of genes with a mixed CTA/differentiation antigen pattern, for example, ZBTB32. Differentiation antigens serve as targets for CTL responses. Differentiation antigens with exclusive expression in hematopoietic cells are particularly promising for the development of passive or adoptive immunotherapy in combination with allogeneic hematopoietic stem cell transplantation (aHSC). Tolerance to such antigens can be circumvented by the use of T cells with restriction to nonself MHC molecules [46]. In agreement with our microarray and PCR results, Northern blot analysis of ZBTB32 showed nearly exclusive expression in testis [47]. It has been recently shown that the transcriptional repressor function of ZBTB32 was partially inhibited by interaction of ZBTB32 with the Rec protein from the human endogenous retrovirus (HERV) HERV-K [48]. Interestingly, HERV activity has been observed not only in testicular cancer but also in HL cells [49]. Further investigations are necessary to elucidate which genes are repressed in memory B cells and/or HL cell lines by ZBTB32 and whether HERV activity in HL cells interfere with the transcriptional repression by ZBTB32.
Antigens that would be optimally suited for immunotherapy should be those involved in proliferation, survival or abnormal differentiation of the tumor cells, because loss of such antigens by immunoselection [50] is unlikely. PRAME promises to be a good candidate for such an antigen, because this antigen inhibits differentiation of hematopoietic cells [51]. Recently, a method for the efficient generation of PRAME-specific T cells from tumor patients and healthy donors has been described [43]. Importantly, the investigators describe the successful generation of PRAME-specific T cells from patients with HL [43].
Our observation of increased expression of PRAME after treatment of HL cells with 5′-azacytidine is in agreement with epigenetic regulation of PRAME. Such regulation has been seen in other malignant cells [52]. Re-expression of PRAME in tumor cells after treatment with methylation-blocking agents might increase recognition by PRAME-specific cells. In our in vitro model, we found no increased recognition of 5′-azacytidine-treated L-540 cells (supplementary Table 1). However, it is unclear whether the HLA molecules expressed on L-540 cells are able to present PRAME peptides. The low reactivity with this cell line that we observed for most donors might reflect the low expression of co-stimulatory molecules on this HL cell line with a T-cell phenotype [12].
In addition to peptide vaccination strategies, tumor-derived material can be used for the preparation of tumor antigen-loaded professional antigen-presenting cells. The advantage of such strategies is that they can be employed irrespective of the patient’s HLA haplotype. In addition, the simultaneous presence of multiple tumor-derived antigens in such vaccines renders immune escape by loss of antigen expression unlikely. The low number of tumor cells together with the difficulty to establish tumor cell lines hampers the adaptation of this strategy for the treatment of HL. Established allogeneic cell lines might allow overcoming these problems. As seen in other tumor models, the gene expression profile of HL cell lines and primary HL cells is not identical. However, microarray analysis indicates a high similarity of cell lines and micro-dissected cells with regard to expression of putative CTA. Our data suggest that DC after loading with RNA from established HL cell lines are able to stimulate T-cell responses against HL-associated tumor antigens, for example, PRAME. Similar responses against PRAME have been observed in an RNA-DC vaccine model for neuro-ectodermal tumors [53]. Antigens recognized on allogeneic tumor cells consist of a mixture of tumor-specific antigens and alloantigens. As transfected tumor RNA can also encode HLA molecules from the tumor cells, these molecules might prime T cells against alloantigens in an allo-HLA restricted manner. Therefore, the response against HL cells after priming with HL-RNA-transfected DC might include a strong alloreactive component. The optimal antigen sources and preparation (RNA from one cell line, assemblages from different cell lines; addition of adjuvant; RNA isolation and stabilization procedure) have to be determined. T cells with specificity for antigens with known presence in the vaccine (e.g., PRAME) might be useful for monitoring tumor-specific immune responses after vaccination with such antigen composition.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported by grants from Peter-Escher-Foundation for children with cancer.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Drexler HG, Minowada J. Hodgkin’s disease derived cell lines: a review. Hum Cell. 1992;5:42–53. [PubMed] [Google Scholar]
- 2.Sahin U, Neumann F, Tureci O, Schmits R, Perez F, Pfreundschuh M. Hodgkin and Reed-Sternberg cell-associated autoantigen CLIP-170/restin is a marker for dendritic cells and is involved in the trafficking of macropinosomes to the cytoskeleton, supporting a function-based concept of Hodgkin and Reed-Sternberg cells. Blood. 2002;100:4139–4145. doi: 10.1182/blood.V100.12.4139. [DOI] [PubMed] [Google Scholar]
- 3.Schwering I, Bräuninger A, Klein U, Jungnickel B, Tinguely M, Diehl V, et al. Loss of the B-lineage-specific gene expression program in Hodgkin and Reed-Sternberg cells of Hodgkin lymphoma. Blood. 2003;101:1505–1512. doi: 10.1182/blood-2002-03-0839. [DOI] [PubMed] [Google Scholar]
- 4.Körholz D, Claviez A, Hasenclever D, Kluge R, Hirsch W, Kamprad F, et al. The concept of the GPOH-HD 2003 therapy study for pediatric Hodgkin’s disease: evolution in the tradition of the DAL/GPOH studies. Klin Padiatr. 2004;216:150–156. doi: 10.1055/s-2004-822627. [DOI] [PubMed] [Google Scholar]
- 5.Peggs KS, Hunter A, Chopra R, Parker A, Mahendra P, Milligan D, et al. Clinical evidence of a graft-versus-Hodgkin’s-lymphoma effect after reduced-intensity allogeneic transplantation. Lancet. 2005;365:1934–1941. doi: 10.1016/S0140-6736(05)66659-7. [DOI] [PubMed] [Google Scholar]
- 6.Kennedy-Nasser AA, Bollard CM, Rooney CM. Adoptive immunotherapy for Hodgkin’s lymphoma. Int J Hematol. 2006;83:385–390. doi: 10.1532/IJH97.06107. [DOI] [PubMed] [Google Scholar]
- 7.Grunebach F, Muller MR, Brossart P. New developments in dendritic cell-based vaccinations: RNA translated into clinics. Cancer Immunol Immunother. 2005;54:517–525. doi: 10.1007/s00262-004-0605-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Staege MS, Hansen G, Baersch G, Burdach S. Functional and molecular characterization of interleukin-2 transgenic Ewing tumor cells for in vivo immunotherapy. Pediatr Blood Cancer. 2004;43:23–34. doi: 10.1002/pbc.20013. [DOI] [PubMed] [Google Scholar]
- 9.Buchner A, Pohla H, Willimsky G, Frankenberger B, Frank R, Baur-Melnyk A, et al. Phase 1 trial of allogeneic gene-modified tumor cell vaccine RCC-26/CD80/IL-2 in patients with metastatic renal cell carcinoma. Hum Gene Ther. 2010;21:285–297. doi: 10.1089/hum.2008.192. [DOI] [PubMed] [Google Scholar]
- 10.Mu LJ, Kyte JA, Kvalheim G, Aamdal S, Dueland S, Hauser M, et al. Immunotherapy with allotumour mRNA-transfected dendritic cells in androgen-resistant prostate cancer patients. Br J Cancer. 2005;93:749–756. doi: 10.1038/sj.bjc.6602761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kyte JA, Kvalheim G, Lislerud K, thor Straten P, Dueland S, Aamdal S, et al. T cell responses in melanoma patients after vaccination with tumor-mRNA transfected dendritic cells. Cancer Immunol Immunother. 2007;56:659–675. doi: 10.1007/s00262-006-0222-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Staege MS, Banning-Eichenseer U, Weissflog G, Volkmer I, Burdach S, Richter G, et al. Gene expression profiles of Hodgkin’s lymphoma cell lines with different sensitivity to cytotoxic drugs. Exp Hematol. 2008;36:886–896. doi: 10.1016/j.exphem.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 13.Biedler JL, Helson L, Spengler BA. Morphology and growth, tumorigenicity, and cytogenetics of human neuroblastoma cells in continuous culture. Cancer Res. 1973;33:2643–2652. [PubMed] [Google Scholar]
- 14.McAllister RM, Isaacs H, Rongey R, Peer M, Au W, Soukup SW, et al. Establishment of a human medulloblastoma cell line. Int J Cancer. 1977;20:206–212. doi: 10.1002/ijc.2910200207. [DOI] [PubMed] [Google Scholar]
- 15.Wolf J, Kapp U, Bohlen H, Kornacker M, Schoch C, Stahl B, et al. Peripheral blood mononuclear cells of a patient with advanced Hodgkin’s lymphoma give rise to permanently growing Hodgkin-Reed Sternberg cells. Blood. 1996;87:3418–3428. [PubMed] [Google Scholar]
- 16.Diehl V, Kirchner HH, Schaadt M, Fonatsch C, Stein H, Gerdes J, et al. Hodgkin’s disease: establishment and characterization of four in vitro cell lines. J Cancer Res Clin Oncol. 1981;101:111–124. doi: 10.1007/BF00405072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drexler HG, Gaedicke G, Lok MS, Diehl V, Minowada J. Hodgkin’s disease derived cell lines HDLM-2 and L-428: comparison of morphology, immunological and isoenzyme profiles. Leuk Res. 1986;10:487–500. doi: 10.1016/0145-2126(86)90084-6. [DOI] [PubMed] [Google Scholar]
- 18.Kamesaki H, Fukuhara S, Tatsumi E, Uchino H, Yamabe H, Miwa H, et al. Cytochemical, immunologic, chromosomal, and molecular genetic analysis of a novel cell line derived from Hodgkin’s disease. Blood. 1986;68:285–292. [PubMed] [Google Scholar]
- 19.Giard DJ, Aaronson SA, Todaro GJ, Arnstein P, Kersey JH, Dosik H, et al. In vitro cultivation of human tumors: establishment of cell lines derived from a series of solid tumors. J Natl Cancer Inst. 1973;51:1417–1423. doi: 10.1093/jnci/51.5.1417. [DOI] [PubMed] [Google Scholar]
- 20.Salcedo M, Momburg F, Hämmerling GJ, Ljunggren HG. Resistance to natural killer cell lysis conferred by TAP1/2 genes in human antigen-processing mutant cells. J Immunol. 1994;152:1702–1708. [PubMed] [Google Scholar]
- 21.Foell JL, Volkmer I, Giersberg C, Kornhuber M, Horneff G, Staege MS. Loss of detectability of Charcot-Leyden crystal protein transcripts in blood cells after treatment with dimethyl sulfoxide. J Immunol Methods. 2008;339:99–103. doi: 10.1016/j.jim.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 22.Meyer-Wentrup F, Burdach S. Efficacy of dendritic cell generation for clinical use: recovery and purity of monocytes and mature dendritic cells after immunomagnetic sorting or adherence selection of CD14+ starting populations. J Hematother Stem Cell Res. 2003;12:289–299. doi: 10.1089/152581603322023025. [DOI] [PubMed] [Google Scholar]
- 23.Foell JL, Hesse M, Volkmer I, Schmiedel BJ, Neumann I, Staege MS. Membrane-associated phospholipase A1 beta (LIPI) is an Ewing tumour-associated cancer/testis antigen. Pediatr Blood Cancer. 2008;51:228–234. doi: 10.1002/pbc.21602. [DOI] [PubMed] [Google Scholar]
- 24.Neumann I, Foell JL, Bremer M, Volkmer I, Korholz D, Burdach S, et al. Retinoic acid enhances sensitivity of neuroblastoma cells for imatinib mesylate. Pediatr Blood Cancer. 2010;55:464–470. doi: 10.1002/pbc.22603. [DOI] [PubMed] [Google Scholar]
- 25.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ge X, Yamamoto S, Tsutsumi S, Midorikawa Y, Ihara S, Wang SM, et al. Interpreting expression profiles of cancers by genome-wide survey of breadth of expression in normal tissues. Genomics. 2005;86:127–141. doi: 10.1016/j.ygeno.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 27.Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF et al. (2011) NCBI GEO: archive for functional genomics data sets–10 years on. Nucleic Acids Res 39 Database issue:D1005-1010 [DOI] [PMC free article] [PubMed]
- 28.Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- 29.Dangond F, Hwang D, Camelo S, Pasinelli P, Frosch MP, Stephanopoulos G, et al. Molecular signature of late-stage human ALS revealed by expression profiling of postmortem spinal cord gray matter. Physiol Genomics. 2004;16:229–239. doi: 10.1152/physiolgenomics.00087.2001. [DOI] [PubMed] [Google Scholar]
- 30.Brune V, Tiacci E, Pfeil I, Döring C, Eckerle S, van Noesel CJ, et al. Origin and pathogenesis of nodular lymphocyte-predominant Hodgkin lymphoma as revealed by global gene expression analysis. J Exp Med. 2008;205:2251–2268. doi: 10.1084/jem.20080809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roth RB, Hevezi P, Lee J, Willhite D, Lechner SM, Foster AC, et al. Gene expression analyses reveal molecular relationships among 20 regions of the human CNS. Neurogenetics. 2006;7:67–80. doi: 10.1007/s10048-006-0032-6. [DOI] [PubMed] [Google Scholar]
- 32.Eckerle S, Brune V, Döring C, Tiacci E, Bohle V, Sundström C, et al. Gene expression profiling of isolated tumour cells from anaplastic large cell lymphomas: insights into its cellular origin, pathogenesis and relation to Hodgkin lymphoma. Leukemia. 2009;23:2129–2138. doi: 10.1038/leu.2009.161. [DOI] [PubMed] [Google Scholar]
- 33.Köchert K, Ullrich K, Kreher S, Aster JC, Kitagawa M, Jöhrens K, et al. High-level expression of Mastermind-like 2 contributes to aberrant activation of the NOTCH signaling pathway in human lymphomas. Oncogene. 2010;30:1831–1840. doi: 10.1038/onc.2010.544. [DOI] [PubMed] [Google Scholar]
- 34.Steidl C, Shah SP, Woolcock BW, Rui L, Kawahara M, Farinha P, et al. MHC class II transactivator CIITA is a recurrent gene fusion partner in lymphoid cancers. Nature. 2011;471:377–381. doi: 10.1038/nature09754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu TY, Wu SJ, Huang MH, Lo FY, Tsai MH, Tsai CH, et al. EBV-positive Hodgkin lymphoma is associated with suppression of p21cip1/waf1 and a worse prognosis. Mol Cancer. 2010;9:32. doi: 10.1186/1476-4598-9-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoennscheidt C, Max D, Richter N, Staege MS. Expression of CD4 on Epstein-Barr virus-immortalized B cells. Scand J Immunol. 2009;70:216–225. doi: 10.1111/j.1365-3083.2009.02286.x. [DOI] [PubMed] [Google Scholar]
- 37.Ghafouri-Fard S, Abbasi A, Moslehi H, Faramarzi N, Taba Taba Vakili S, Mobasheri MB, et al. Elevated expression levels of testis-specific genes TEX101 and SPATA19 in basal cell carcinoma and their correlation with clinical and pathological features. Br J Dermatol. 2010;162:772–779. doi: 10.1111/j.1365-2133.2009.09568.x. [DOI] [PubMed] [Google Scholar]
- 38.Chen YT, Chadburn A, Lee P, Hsu M, Ritter E, Chiu A, et al. Expression of cancer testis antigen CT45 in classical Hodgkin lymphoma and other B-cell lymphomas. Proc Natl Acad Sci USA. 2010;107:3093–3098. doi: 10.1073/pnas.0915050107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willenbrock K, Küppers R, Renné C, Brune V, Eckerle S, Weidmann E, et al. Common features and differences in the transcriptome of large cell anaplastic lymphoma and classical Hodgkin’s lymphoma. Haematologica. 2006;91:596–604. [PubMed] [Google Scholar]
- 40.Thorisson GA, Smith AV, Krishnan L, Stein LD. The international HapMap project Web site. Genome Res. 2005;15:1592–1593. doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ikeda H, Lethé B, Lehmann F, van Baren N, Baurain JF, de Smet C, et al. Characterization of an antigen that is recognized on a melanoma showing partial HLA loss by CTL expressing an NK inhibitory receptor. Immunity. 1997;6:199–208. doi: 10.1016/S1074-7613(00)80426-4. [DOI] [PubMed] [Google Scholar]
- 42.Kessler JH, Beekman NJ, Bres-Vloemans SA, Verdijk P, van Veelen PA, Kloosterman-Joosten AM, et al. Efficient identification of novel HLA-A(*)0201-presented cytotoxic T lymphocyte epitopes in the widely expressed tumor antigen PRAME by proteasome-mediated digestion analysis. J Exp Med. 2001;193:73–88. doi: 10.1084/jem.193.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quintarelli C, Dotti G, Hasan ST, De Angelis B, Hoyos V, Errichiello S, et al. High-avidity cytotoxic-T-lymphocytes specific for a new preferentially expressed antigen of melanoma (PRAME)-derived peptide can target leukemic- and leukemic-precursor cells. Blood. 2011;117:3353–3362. doi: 10.1182/blood-2010-08-300376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawahara M, Hori T, Matsubara Y, Okawa K, Uchiyama T. Identification of HLA class I-restricted tumor-associated antigens in adult T cell leukemia cells by mass spectrometric analysis. Exp Hematol. 2006;34:1496–1504. doi: 10.1016/j.exphem.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 45.Parmiani G, De Filippo A, Novellino L, Castelli C. Unique human tumor antigens: immunobiology and use in clinical trials. J Immunol. 2007;178:1975–1979. doi: 10.4049/jimmunol.178.4.1975. [DOI] [PubMed] [Google Scholar]
- 46.Gao L, Downs AM, Stauss HJ. Immunotherapy with CTL restricted by nonself MHC. Methods Mol Med. 2005;109:215–228. doi: 10.1385/1-59259-862-5:215. [DOI] [PubMed] [Google Scholar]
- 47.Lin W, Lai CH, Tang CJ, Huang CJ, Tang TK. Identification and gene structure of a novel human PLZF-related transcription factor gene, TZFP. Biochem Biophys Res Commun. 1999;264:789–795. doi: 10.1006/bbrc.1999.1594. [DOI] [PubMed] [Google Scholar]
- 48.Kaufmann S, Sauter M, Schmitt M, Baumert B, Best B, Boese A, et al. Human endogenous retrovirus protein Rec interacts with the testicular zinc-finger protein and androgen receptor. J Gen Virol. 2010;91:1494–1502. doi: 10.1099/vir.0.014241-0. [DOI] [PubMed] [Google Scholar]
- 49.Lamprecht B, Walter K, Kreher S, Kumar R, Hummel M, Lenze D, et al. Derepression of an endogenous long terminal repeat activates the CSF1R proto-oncogene in human lymphoma. Nat Med. 2010;16:571–579. doi: 10.1038/nm.2129. [DOI] [PubMed] [Google Scholar]
- 50.Khong HT, Wang QJ, Rosenberg SA. Identification of multiple antigens recognized by tumor-infiltrating lymphocytes from a single patient: tumor escape by antigen loss and loss of MHC expression. J Immunother. 2004;27:184–190. doi: 10.1097/00002371-200405000-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oehler VG, Guthrie KA, Cummings CL, Sabo K, Wood BL, Gooley T, et al. The preferentially expressed antigen in melanoma (PRAME) inhibits myeloid differentiation in normal hematopoietic and leukemic progenitor cells. Blood. 2009;114:3299–3308. doi: 10.1182/blood-2008-07-170282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ortmann CA, Eisele L, Nückel H, Klein-Hitpass L, Führer A, Dührsen U, et al. Aberrant hypomethylation of the cancer-testis antigen PRAME correlates with PRAME expression in acute myeloid leukemia. Ann Hematol. 2008;87:809–818. doi: 10.1007/s00277-008-0514-8. [DOI] [PubMed] [Google Scholar]
- 53.Morandi F, Chiesa S, Bocca P, Millo E, Salis A, Solari M, et al. Tumor mRNA-transfected dendritic cells stimulate the generation of CTL that recognize neuroblastoma-associated antigens and kill tumor cells: immunotherapeutic implications. Neoplasia. 2006;8:833–842. doi: 10.1593/neo.06415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.