Abstract
Aim
We compared the expression of genes related to inflammatory and cytotoxic functions between MSI and MSS (HLA-class I-negative and HLA-class I-positive) colorectal cancers (CRCs), seeking evidence of differences in inflammatory mediators and cytotoxic T-cell responses. Twenty-two CRCs were divided into three study groups as a function of HLA class I expression and MSI phenotype: 8 MSI tumours, 6 MSS/HLA− tumours and 6 MSS/HLA+ tumours (controls).
Findings
A first comparison between eight MSI and six MSS/HLA-positive (control) cancers, based on microarray analysis on an Affymetrix® HG-U133-Plus-PM plate, identified 1974 differentially expressed genes (P < 0.05). We grouped genes in Gene Ontology functional categories: apoptotic programme (72 genes, P = 5.5·10−3), leucocyte activation (43 genes, P = 1.8·10−5), T-cell activation (24 genes, P = 6.3·10−4), inflammatory response (40 genes, 2.3·10−2) and cytokine production (10 genes, P = 1.9·10−2). Real-time PCR and immunohistochemical evaluation were used to validate the data, finding that increased mRNA levels of pro-inflammatory cytokines and cytotoxic mediators were associated with greater infiltration by CD8+T lymphocytes in the MSI group (P < 0.001). Finally, HLA-class I-negative tumours were not grouped together but rather in accordance with features of the gene expression profile of MSI or MSS tumours. As expected, genes associated with antigen processing machinery and MHC class I molecules (TAP2, B2m) were downregulated in MSS/HLA-class I-negative CRCs (n = 6) in comparison to controls.
Conclusions
In conclusion, microarray and immunohistochemical data may be useful to comprehensively assess tumour–host interactions and differentiate MSI from MSS cancers. The two types of tumour, MSI/HLA-class I-negative and MSS/HLA-class I-negative, showed marked differences in the composition and intensity of infiltrating leucocytes, suggesting that their immune escape strategies involve distinct pathways.
Keywords: Colorectal cancers, Microsatellite instability, Human leucocyte antigen, Gene expression profile, Inflammatory response
Introduction
Microsatellite instability (MSI) is one of the main carcinogenic pathways and has been observed in 15% of colorectal cancers (CRC). This type of genetic instability consists of the accumulation of numerous somatic mutations during DNA replication (insertions and deletions of single nucleotides and single base substitutions) in repetitive DNA sequences (microsatellites) because of alterations in the DNA mismatch repair (MMR) system [1–3]. The microsatellite instability (MSI) phenotype is the hallmark of Lynch syndrome-associated cancers characterized by MMR gene germline mutations (predominantly hMLH1 or hMSH2), and it is also found in ~10–15% of sporadic CRCs, in which MMR gene promoter methylation is observed [3, 4]. Besides microsatellite instability, there is another carcinogenic pathway that promotes emergence of the remaining 85% of sporadic CRCs, i.e., chromosomal instability (CIN), characterized by numerical and structural chromosomal alterations, by the accumulation of mutations in oncogenes (e.g., K-ras) or tumour suppressor genes (p53) and, in most cases, by not being accompanied by CpG island metylator phenotype (CIMP) [4, 5].
Finally, cDNA microarray studies have demonstrated that microsatellite stable (MSS) and MSI colorectal cancers clearly differ in gene expression profiling, strongly supporting the different pathogeneses of these tumours [6, 7]. However, few studies have focused on the analysis of immune response genes, which may elucidate the distinct biological behaviours of the two cancer types [8].
Tumour development takes place despite activation of cytotoxic T lymphocytes (CTLs) in the response of the host immune system. In CRCs, it is possible to correlate the MSI phenotype with the strength of the anti-tumour immune response characterized by a high lymphocytic infiltration [9, 10]. In attempts to explain the high immunogenicity of this type of tumour, various studies have suggested that a defective DNA MMR system results in insertions and deletions at coding microsatellites in target genes that lead to the abundant generation of new immunogenic frameshift peptides (FSPs) that can be presented to CTLs [11, 12]. However, the biological meaning of the lymphocyte infiltrate in microsatellite instability tumours is controversial. Some authors propose that intra-epithelial lymphocytes (IELs) possess the characteristics of predominantly cytotoxic cells and release mediators of target cell death, whereas others argue that these infiltrates are secondary phenomena representing the proliferation of resident lamina propia lymphocytes with no immunological implications or biological role [8, 13]. Emergence of tumour escape variants due to immunoedition may explain the failure of CTLs to eliminate tumour cells [14]. In MSI cancers, a frequent mechanism to escape CTL immunosurveillance is the total loss of surface expression of HLA class I molecules and subsequent loss of tumour antigen presentation capacity [15]. Cells with HLA class I loss due to B2-microglobulin (B2m) gene mutations [16–19], especially in tumours with mutator phenotype (i.e. insertions and deletions leading to a change in the translational reading frame), would be immunoselected under immune pressure.
In a previous study, we observed that the characteristics and intensity of the infiltrate clearly differed as a function of the microsatellite instability phenotype but not the HLA expression on tumour cells [20]. In the present study, we used the same set of colorectal tumours to compare the expression of genes influencing inflammatory response and cytotoxic activity between MSI and MSS colorectal tumours in order to explain the above differences in leucocyte infiltration. We observed a differential expression of genes involved in the anti-tumour immune response that may elucidate the molecular cancer escape mechanisms.
Materials and methods
Histopathology and selection of tumour samples
We selected 20 colorectal carcinomas (CRCs) based on the immunohistochemical study of HLA class I expression and analysis of the MSI phenotype. The rational for dividing the groups of tumours is based on their immunogenicity for CD8+T cells, associated with HLA class I expression. Two groups consists of HLA-negative tumours (both MSI and MSS), while another group includes control tumours with genomic stability and no detectable alterations in HLA expression. Therefore, the three study groups were: 8 MSI tumours, including 7 HLA-class I-negative (MSI/HLA-negative) and 1 HLA-class I-positive (MSI/HLA-positive); 6 MSS tumours, all with total HLA class I loss (MSS/HLA-negative); and 6 MSS/HLA-positive tumours (control group), with no detectable alterations in HLA or B2m genes (MSS/HLA-positive). The clinical characteristics of the 20 patients are listed in Table 1.
Table 1.
Summary of main clinical and histological parameters of patient samples
| Samples | Age | Sex | TNM | Tumour location | Tumour stage | Tumour grade |
|---|---|---|---|---|---|---|
| MSI tumours | ||||||
| CRC-1 | 81 | Female | pT3N0M0 | Colorectal | IIA | IV |
| CRC-2 | 39 | Male | PT3N1M1 | Colorectal | IVA | II |
| CRC-3 | 73 | Female | pT4N1M0 | Colorectal | IIIB | III |
| CRC-4 | 45 | Male | pT2N0M0 | Colorectal | I | II |
| CRC-5 | 79 | Male | pT4N2M0 | Colorectal | IIIC | II |
| CRC-6 | 59 | Female | pT3N0M0 | Colorectal | IIA | II |
| CRC-7 | 72 | Female | pT3N2M0 | Colorectal | IIIB | III |
| CRC-8 | 82 | Female | – | Colorectal | – | III |
| MSS/HLA-negative tumours | ||||||
| CRC-9 | 64 | Female | pT3N1M0 | Colorectal | IIIB | II |
| CRC-10 | 71 | Female | pT3N0M0 | Colorectal | IIA | II |
| CRC-11 | 72 | Male | pT3N0M0 | Colorectal | IIA | II |
| CRC-12 | 70 | Male | pT3N2M0 | Colorectal | IIIB | III |
| CRC-13 | 66 | Male | pT3N0M0 | Colorectal | IIA | II |
| CRC-14 | 54 | Male | pT4N1Mx | Colorectal | IIIB | II |
| MSS/HLA-positive tumours (Conrols) | ||||||
| CRC-15 | 61 | – | pT3N0M0 | Colorectal | IIA | II |
| CRC-16 | 67 | Male | PT3N0M0 | Colorectal | IIA | II |
| CRC-17 | – | – | – | Colorectal | – | II |
| CRC-18 | 62 | Male | pT2N0M0 | Colorectal | I | II |
| CRC-19 | 70 | Female | pT3N2M1 | Colorectal | IVA | II |
| CRC-20 | 83 | Male | pT3N0M0 | Colorectal | IIA | II |
– Data not available
We analysed cryopreserved samples (Virgen de las Nieves University Hospital [VNUH] Tumour-Tissue Biobank) that were previously studied and provided by the Department of Pathology, VNUH, Granada (Spain). The diagnosis followed the WHO pathological classification and TNM staging criteria [21]. Clinical and pathological reports were available, and approval was obtained from the ethical investigation review board of our hospital.
Immunohistological analysis of HLA class I expression
Frozen tissue sections of 4–8 μm thickness were cut on a microtome-cryostat (Bright), allowed to dry at room temperature for 4–18 h, fixed in acetone at 4°C for 10 min and stored at −40°C until staining. Immunohistological techniques were performed with the Biotin-Streptavidin System (supersensitive Multilink HRP/DAB kit, BioGenex, The Hague, The Netherlands). The following mouse monoclonal antibodies (mAbs) were used to analyse HLA class I expression: W6/32 against HLA-A, B and C heavy chain/B2m complex (kind gift from Dr Bodmer, Tissue Antigen Laboratory, Imperial Cancer Research Fund Laboratories, London, UK); GRH-1, which recognizes free and HLA class I heavy chain-associated B2m chain [22]; and HC-10 against free heavy chain of HLA-B and C molecules [23]. The primary antibody was replaced with PBS for negative controls, in which no immunohistochemical staining was detected.
DNA isolation
Total DNA was obtained from tumour fragments microdissected (PALM Microlaser System, Olympus) from two frozen tissue sections (4–8 μm thick) using the REDExtract-N-Amp Tissue PCR extraction kit. Normal DNA was obtained from PBLs using the Quiagen DNA isolation kit (QIAamp Tissue Kit, Wetsbsurg, Leusden, the Netherlands).
MSI analysis
MSI status was determined according to the criteria of the National Cancer Institute workshop, using various panels of dinucleotide and mononucleotide repeat sequences and classifying tissue samples with MSI in ≥30–40% of tested markers as MSI and those with MSI in <30–40% of markers as MSS [3]. The MSI analysis conditions and markers used are reported elsewhere [20].
Microarray profiling
Total RNA was isolated from 10 serial cuts (30 μm thick) from a liquid nitrogen-preserved tumour specimen that was also used in the tumour diagnosis by pathologists. The RNA isolation kit used was RNeasy Mini Kit (Quiagen). All samples were found to have good quality mRNA, with little degradation and correct ratios of 28S:18S ribosomal peaks. The RNA Integrity Number (RIN), a measure of the integrity of total sample RNA, was calculated by using an algorithm that takes account of the entire electrophoretic trace. RIN values range from 1 (maximum degradation) to 10 (intact RNA). RIN values in our RNA samples ranged from 7 to 9.5. Biotinylated cRNA was synthesised from 200 ng of total RNA from each sample by using GeneChip 3′ IVT Express kit (Affymetrix), following the manufacturer’s recommendations. Oligonucleotide array hybridization and scanning were performed according to Affymetrix protocols. Gene expression profiles were determined by using Affymetrix HG-U133-Plus-PM Array Plate which contains 54,670 probe sets, including >33,000 well-characterized genes and UniGene clusters per sample.
Data pre-processing
Raw data were pre-processed by using the Robust MultiArrays Average (RMA) Bioconductor package RMA [24]. We removed control probes (102) and those with a background level hybridization signal and then filtered out the probes with no expression change across all samples, resulting in 9,695 probes that were used for further analysis.
Microarray analysis
Gene expression
As a first step in the analysis of pre-processed data, we identified the differentially expressed genes by means of the Partek Genomic Suite (Partek Inc. St Louis, MO, USA). Unsupervised hierarchical clustering of the samples was done by using Partek Genomic Suite software, with the euclidean distance as proximity measure and the average-linkage method as linkage criteria. We also applied principal components analysis (PCA), to simplify the analysis and visualization of the expression data sets. The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus [25] and are accessible through GEO Series accession number GSE27544 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE27544).
Gene ontology
Gene Ontology (GO) information was used to assign biological meaning to selected genes, [26, 27] using csbl.go, a recently published tool, implemented as an R package [28, 29]. Furthermore, after analysing the set of genes and their GO annotations, we computed the most enriched GO terms. P-values were obtained by applying Fisher’s exact test.
Pathways
The data sets of differentially expressed genes were uploaded into the Ingenuity Pathways Knowledge Base (IPKB) (IPA program) for the construction of networks and the analysis of biofunctions and biological pathways [30].
Microarray data validation
Real-time RT-PCR and immunohistochemical analysis were performed to validate the microarray data.
Real-time RT-PCR analysis
Total RNA was extracted from 10 new sections (30 μm thick) of each tumour specimen and from microdissected fragments using miRNeasy Mini Kit (Qiagen) according to the manufacturer’s recommendations. cDNA synthesis was performed with the iScript cDNA Synthesis Kit (Bio-Rad), using 1 μg RNA and following the manufacturer’s instructions. RT-PCR products were analysed for gene expression of four target genes identified in microarray analysis: B2m (RNA isolated from tumour microdissected fragments), MIP1α (CCL3), TGF1β and STAT1 (RNA isolated from 10 serial cuts of tumour specimen) by quantitative real-time RT-PCR. To control for variations in amounts of mRNA, we also tested glucose-6 phosphate dehydrogenase (G6PDH) as control gene. All reactions were performed in a LightCycler instrument using the LC-FastStart DNA Master SYBR Green I kit (Roche Diagnostics, Manheim, Germany) with the exception of G6PDH and B2m, for which FastStart DNA Master Plus HybProbe Kit (Roche Diagnostics, Manheim, Germany) was used. HouseKeeping Gene Set kit (Roche Diagnostics) was used for G6PDH amplification. B2m primers used in the analysis were: Fw 5′ TCAGGAAATTTGACTTTCCATTC 3′; Bw 5′ TTCTGGCCTGGAGGCTATC 3′ (TIB MOLBIOL, Berlin, Germany). Primers used for MIP1α (CCL3), TGF1β and STAT1 gene amplification reactions were part of the LightCycler-Primer Set (GmbH, Heidelberg, Germany). The final expression levels of target genes were given relative to the expression levels of G6PDH.
Real-time PCR data were expressed as median ± standard deviation. Normal distribution of results was confirmed by Kolmogorov–Smirnov test. The Student’s t test was used to analyse data in the validation experiment. Statistical significance was defined as P ≤ 0.05. SPSS version 15.0 (SPSS, Inc., Chicago, IL) was used for data analyses.
Immunohistological study
Immunohistological techniques were applied as specified above. For the staining of tumour infiltrates, we took consecutive cuts (4–8 μm thick) from the frozen tumour specimen used to obtain histological sections for RNA isolation. The following mouse mAbs were used to stain tumour infiltrates: GRT2 (anti-CD45), produced in our lab [31]; OKT3 (anti-CD3 hybridoma, ATCC, Teddington, UK), OKT8 (anti-CD8 hybridoma, ATCC), anti-CD4 (Clone RPA-T4, Becton–Dickinson Biosciences (BDB), San Jose, CA), anti-perforin (Clone δG9, BD Biosciences), anti-CD64 (Clone 10.1, BD Biosciences), anti-CD206 (macrophage mannose receptor; Clone 19.2, BD Biosciences), anti-CD163 (macrophage scavenger receptor; Clone Ber-Mac3, MBL, Woburn, MA) and anti-CD56 (Clone 123C3, Dako, Barcelona, Spain. A mouse monoclonal Ab (1D4.1) provided by Dr. Jaime Sancho (Department of Cellular Biology and Immunology, Instituto de Parasitología y Biomedicina, Granada, Spain) was used to stain the CD3 complex ζ-chain [32].
Two observers analysed tissue tumour infiltrates, defining the infiltrate for each mAb staining as: stromal (inflammatory infiltrate surrounding tumour mass), interstitial (in fibrous septa around glands), or intratumoral (inflammatory cells in close contact with neoplastic epithelial cells). Each stromal, interstitial and intratumoral infiltration was scored as: 0 (Absent), + (Low), ++ (Moderate) and +++ (High).
In the statistical analysis, for each studied tumour, we calculated the overall infiltration score (0–9) for each marker (CD45, CD3, CD8, CD64, CD163, CD206, CD247 [CD3 complex ζ-chain] and perforin) separately, as the sum of the scores for stromal, interstitial and intratumoral infiltration. After the normal distribution of the data was confirmed by means of the Kolmogorov–Smirnov test, one-way ANOVA and Tukey post hoc tests were used to compare the means for each marker. P ≤ 0.05 was considered significant. SPSS version 15.0 (SPSS, Inc., Chicago, IL) was used for the data analyses.
Results
Gene expression profiles
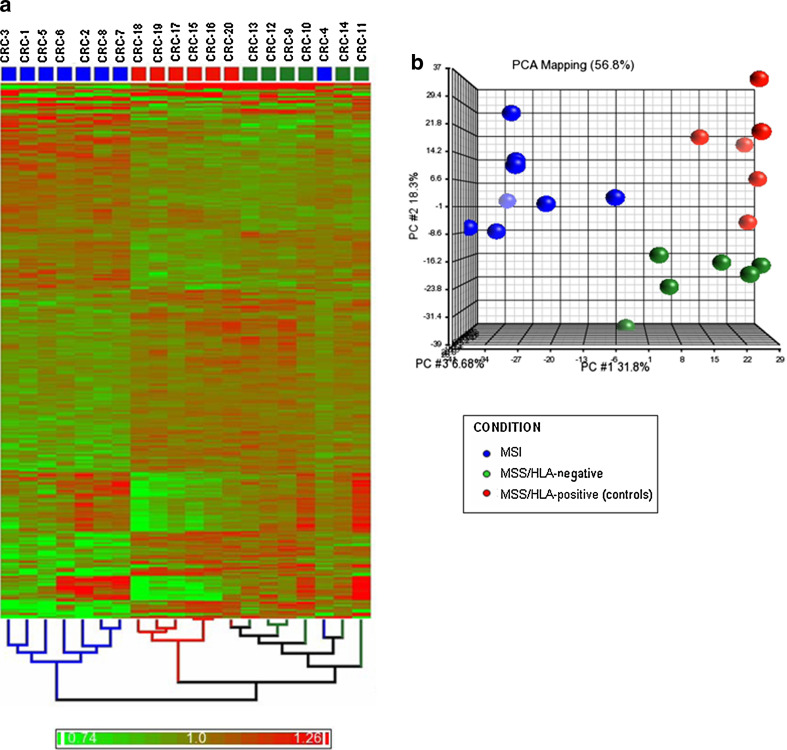
Parket Genomic Suite software was used for unsupervised hierarchical clustering analysis from the significantly differentially expressed sequences. As expected, the samples tended to cluster together in accordance with their experimental conditions. Figure 1a depicts the hierarchical clustering for 2,057 sequences differentially expressed across all groups (P < 0.05), showing that the control group and MSS/HLA-negative samples are more closely related and have similar gene expression profiles, whereas MSI samples, except for the CRC-4 tumour, clearly differ from the rest, as confirmed by PCA of the significant sequences. Patient groups can be distinguished into a linearly separable gene expression data space (Fig. 1b).
Fig. 1.
Bidimensional-hierarchical clustering and PCA of MSI, MSS/HLA-negative and MSS/HLA-positive (control) cancers, using 2,057 significantly differentiated sequences (P < 0.05). a Two-way hierarchical clustering in which samples (columns) and array targets (rows) were ordered. Red, upregulated; green, downregulated. MSI tumours, except for the CRC-4 sample, were clearly differentiated from MSS CRCs (HLA negative and HLA positive), which shared similar expression profiles. b Discriminating genes were used to generate a three-dimensional (from 2,057 dimensional plot) plot of the data. The cumulative proportions of the variance captured by each principal component are (1) principal component axis, 31.8%; (2) principal component axis, 18.3%; and (3) principal component axis, 6.68%. PCA- based multidimensional scaling visualization separated MSI, MSS/HLA-negative and MSS/HLA-positive (controls) samples into linearly separable 2,057-gene expression data spaces
The statistical analysis revealed 1974 and 1070 sequences corresponding to genes differentially expressed in MSI versus MSS/HLA-positive and in MSS/HLA-negative versus MSS/HLA-positive tumours, respectively. Comparison of MSI and MSS/HLA-positive groups revealed upregulation of 964 genes and downregulation of 1,010 genes in the former.
Gene function analysis
GO functions associated with the differentially expressed genes in the MSI versus control (MSS/HLA-positive) experiment were: apoptotic program (72 genes, P = 5.5·10−3), leucocyte activation (43 genes, P = 1.8·10−5), T-cell activation (24 genes, P = 6.3·10−4), chemotaxis (24 genes, P = 9.8·10−3), inflammatory response (40 genes, P = 2.3·10−2) or cytokine production (10 genes, P = 1.9·10−2). Among the last three groups, we mainly focused on the genes involved in the inflammatory response and cytotoxic activity.
Differentially expressed immune response genes in MSI tumours versus MSS/HLA-class I-positive (controls) tumours
Fifty-two genes associated with inflammatory and cytotoxic functions were differentially expressed (P < 0.05) in MSI tumours versus controls (Tables 2 and 3). Most of these genes were upregulated (fold change ranging from 1.4 to 6.79) in MSI tumours, (Fig. 2a), and only genes involved in anti-inflammatory pathways (TGFBR2, GPX2) and eosinophil chemotaxis (CCL11) were downregulated.
Table 2.
List of inflammatory- and macrophage markers-related gene-specific probes differentially expressed (P < 0.05) between MSI and MSS/HLA-positive (control) colorectal cancers
| Probe ID | Entrez gene | Fold change | P-value | Gene symbol | Gene name |
|---|---|---|---|---|---|
| Cytokine, chemokine and inflammation-related molecules | |||||
| 205114_PM_s_at | 414162 | 5.09 | 0 | CCL3 | Chemokine (C–C motif) ligand 3 |
| 210133_PM_at | 6356 | −6.15 | 0.01 | CCL11 | Chemokine (C–C motif) ligand 11 |
| 32128_PM_at | 6362 | 6.79 | 0.01 | CCL18 | Chemokine (C–C motif) ligand 18 |
| 223454_PM_at | 58191 | 1.92 | 0 | CXCL16 | Chemokine (C-X-C motif) ligand 16 |
| 207375_PM_s_at | 3601 | 1.47 | 0.03 | IL15RA | Interleukin 15 receptor, alpha |
| 206026_PM_s_at | 7130 | 4.79 | 0 | TNFAIP6 | Tumour necrosis factor, alpha-induced protein 6 |
| 209969_PM_s_at | 6772 | 2.09 | 0.02 | STAT1 | Signal transducer and activator of transcription 1, 91 kDa |
| 203010_PM_at | 6776 | 1.76 | 0 | STAT5A | Signal transducer and activator of transcription 5A |
| 209875_PM_s_at | 6696 | 21.15 | 0 | SPP1 | Secreted phosphoprotein 1 |
| 209949_PM_at | 4688 | 3.24 | 0 | NCF2 | Neutrophil cytosolic factor 2 |
| 203574_PM_at | 4783 | 1.46 | 0.04 | NFIL3 | Nuclear factor, interleukin 3 regulated |
| 209732_PM_at | 9976 | 2 | 0.02 | CLEC2B | C-type lectin domain family 2, member B |
| 202638_PM_s_at | 3383 | 2.36 | 0 | ICAM1 | Intercellular adhesion molecule 1 |
| 202803_PM_s_at | 3689 | 2.49 | 0.02 | ITGB2 | Integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| 205786_PM_s_at | 3684 | 4.02 | 0 | ITGAM | Integrin, alpha M (complement component 3 receptor 3 subunit) |
| 214578_PM_s_at | 6093 | 1.39 | 0.03 | ROCK1 | Rho-associated, coiled-coil containing protein kinase 1 |
| 228176_PM_at | 1903 | 2.9 | 0 | S1PR3 | Sphingosine-1-phosphate receptor 3 |
| 211661_PM_x_at | 5724 | 1.98 | 0.01 | PTAFR | Platelet-activating factor receptor |
| 204446_PM_s_at | 240 | 1.82 | 0.03 | ALOX5 | Arachidonate 5-lipoxygenase |
| 202831_PM_at | 2877 | −1.8 | 0.01 | GPX2 | Glutathione peroxidase 2 (gastrointestinal) |
| 208944_PM_at | 7048 | −1.55 | 0.03 | TGFBR2 | Transforming growth factor, beta receptor II (70/80 kDa) |
| Macrophage markers | |||||
| 203561_PM_at | 2212 | 3.32 | 0 | FCGR2A | Fc fragment of IgG, low affinity IIa, receptor (CD32) |
| 210889_PM_s_at | 2213 | 2.76 | 0.01 | FCGR2B | Fc fragment of IgG, low affinity IIb, receptor (CD32) |
| 210992_PM_x_at | 9103 | 2.63 | 0.01 | FCGR2C | Fc fragment of IgG, low affinity IIc, receptor for (CD32) |
| 216950_PM_s_at | 100132417///2209 | 3.79 | 0.01 | FCGR1A///FCGR1C | Fc fragment of IgG, high affinity Ia///Ic, receptor (CD64) |
| 210895_PM_s_at | 942 | 2.92 | 0.02 | CD86 | CD86 molecule |
| 201743_PM_at | 929 | 2.31 | 0.01 | CD14 | CD14 molecule |
| 212722_PM_s_at | 23210 | 1.38 | 0.04 | JMJD6 | Jumonji domain containing 6 |
| 221698_PM_s_at | 64581 | 2.57 | 0.01 | CLEC7A | C-type lectin domain family 7, member A |
| 203645_PM_s_at | 9332 | 4.56 | 0 | CD163 | CD163 molecule |
| 203946_PM_s_at | 384 | 3.14 | 0.02 | ARG 2 | Arginase, type II |
| 204924_PM_at | 7097 | 2.71 | 0 | TLR2 | Toll-like receptor 2 |
| 229560_PM_at | 51311 | 2.95 | 0.01 | TLR8 | Toll-like receptor 8 |
| 206584_PM_at | 23643 | 2.48 | 0.02 | LY96 | Lymphocyte antigen 96 |
| 228234_PM_at | 35376 | 3.12 | 0 | TICAM2 | Toll-like receptor adaptor molecule 2 |
Table 3.
List of T lymphocyte and cytotoxicity markers-related gene-specific probes differentially expressed (P < 0.05) between MSI and MSS/HLA-positive (control) colorectal cancers
| Probe ID | Entrez gene | Fold change | P-value | Gene symbol | Gene name |
|---|---|---|---|---|---|
| 217143_PM_s_at | 6955///6964 | 2.7 | 0.05 | TRA@///TRD@ | T-cell receptor alpha locus///T-cell receptor delta locus |
| 210031_PM_at | 919 | 2.6 | 0.05 | CD247 | CD247 molecule |
| 205269_PM_at | 3937 | 2.46 | 0.02 | LCP2 | Lymphocyte cytosolic protein 2 (SH2 domain containing leucocyte protein of 76 kDa) |
| 216033_PM_s_at | 2534 | 2.95 | 0 | FYN | FYN oncogene related to SRC, FGR, YES |
| 36564_PM_at | 127544 | 1.55 | 0.03 | RNF19B | Ring finger protein 19B |
| 203416_PM_at | 63 | 2.32 | 0.03 | CD53 | CD53 molecule |
| 205988_PM_at | 8832 | 2.49 | 0.01 | CD84 | CD84 molecule |
| 204158_PM_s_at | 10312 | 1.46 | 0.04 | TCIRG1 | T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 subunit A3 |
| 212809_PM_at | 84901 | 1.56 | 0 | NFATC2IP | Nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 |
| 206907_PM_at | 8744 | 3.81 | 0.03 | TNFSF9 | Tumour necrosis factor (ligand) superfamily, member 9 |
| 223501_PM_at | 10673 | 2.56 | 0.01 | TNFSF13B | Tumour necrosis factor (ligand) superfamily, member 13b |
| 226545_PM_at | 135228 | 5.61 | 0 | CD109 | CD109 molecule |
| 37145_PM_at | 10578 | 4.62 | 0.01 | GNLY | Granulysin |
| 214617_PM_at | 5551 | 3.02 | 0.02 | PRF1 | Perforin 1 (pore forming protein) |
| 238542_PM_at | 80324 | 4.99 | 0.02 | ULBP2 | UL16-binding protein 2 |
| 205904_PM_at | 4276 | 1.82 | 0.01 | MICA | MHC class I polypeptide-related sequence A |
| 206247_PM_at | 4277 | 2.4 | 0.01 | MICB | MHC class I polypeptide-related sequence B |
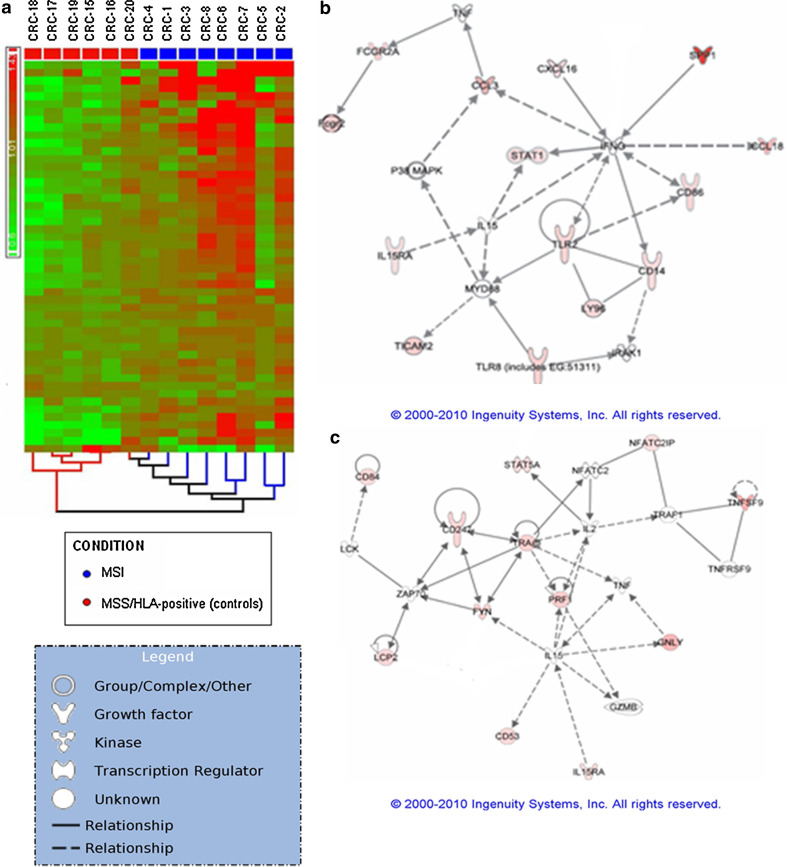
Fig. 2.
a Bidimensional-hierarchical clustering of MSI and MSS/HLA-positive (control) cancers, using 52 significantly differentiated sequences (P < 0.05). Red, upregulated; green, downregulated. Discriminator sequences, related to inflammatory and cytotoxic functions, were mainly upregulated in MSI tumours. b, c Ingenuity pathways analysis (IPA) of 26 differentially expressed genes in MSI CRCs versus MSS control groups that were qualified as networks. a This network centred on various macrophage markers, cytokines and chemokines. All differentially expressed genes were upregulated (red). Increased gene expression of MIP-1-α (CCL3) but not STAT1 was confirmed by RT-PCR analysis (Table 4). b This network centred on T lymphocyte activation via TCR and cytotoxicity, with 12 molecules from the data set of differentially expressed genes that were upregulated
Genes related to inflammation (cytokines and chemokines)
Among the upregulated differentially expressed genes (Table 2), we found genes encoding macrophage- and T lymphocyte-attractant chemokines and cytokine receptors (CCL3, CCL18, CXCL16, IL15RA), molecules related to cytokine-signalling pathways (TNFAIP6, STAT1, STAT5A, NFIL3, SPP1), proteins implicated in the extravasation of leucocytes from blood to tissues (ICAM1, ITGB2, ITGAM, ROCK1) and molecules that contribute to inflammation (CLEC2B, S1PR3, PTAFR, ALOX5).
Macrophage-related genes
We also observed several upregulated macrophage marker genes (Table 2), including genes for Fc Receptors (FCGR2A, FCGR2B, FCGR1A…), PAMP receptor (CLEC7A), scavenger receptor related to M2 macrophages (CD163), co-activator molecule (CD86), TLRs and proteins implicated in TLR signalling pathway (CD14, TLR2, TLR8, LY96, TICAM2) and molecules involved in macrophage activation (JMJD6) (Fig. 2b).
Lymphocyte activation and cytotoxicity-related genes
We found overexpressed gene sequences related to T lymphocytes (Table 3; Fig. 2c), including those encoding for: molecules that participate in signal transduction through the TCR (CD53, α chain of TCR, ξ chain of CD3 (CD247), LCP2, FYN, NFATC2IP); proteins involved in cell activation and proliferation (TCIRG1, CD84); cytolytic activity markers (RNF19B, TNFSF9, GNLY, PRF1); and a protein expressed in activated T cells that negatively regulates the TGFβ pathway (CD109).
A further gene encoding a negative regulator of immune response, Arginase II (ARG2), was upregulated in MSI tumours versus controls (Table 2).
NKG2D-ligand-encoding genes (MICA, MICB, ULBP2) were also upregulated in MSI tumour cells versus controls, implying that MSI tumour cells expressing these stress molecules may be susceptible to the cytotoxic activity of T or NK cells through their NKG2D receptors (Table 3).
Immune response-related genes differentially expressed in MSS/HLA-class I-negative versus MSS/HLA-positive tumours (controls)
Genes differentially expressed between MSS/HLA-negative tumours and controls were related to: cell adhesion (64 genes, P = 5.8·10−7), immune response (45 genes, P = 0.01), phagocytosis (9 genes, P = 2·10−3), chemotaxis (17 genes, P = 3.4·10−3) and negative regulation of apoptosis (29 genes, P = 6.6·10−3). The MSS/HLA-negative group showed upregulation versus controls of genes encoding: monokines and other chemokines (CCL3, CCL4, CCL8, fold change >4), macrophage markers (FCGR1A, FCGR2A, FCGR2B, FCGR2C,CD14, TLR2, TLR8, CLEC7A, MRC1, CD163) (fold change ranging from 2 to 5.26), other molecules related to TLR pathway (LY96, fold change = 2.86; TICAM2, fold change = 2.2), and antigen-presenting cell (CD86, fold change = 3.28; IFI30, fold change = 2.25) markers, transcription factors detected in nuclei of monocytes and granulocytes (MNDA, fold change = 3.12) or involved in monocyte/macrophage differentiation (MAFB, fold change = 2.07), neutrophil-related markers (NCF2, fold change = 2.84; SPARC, fold change = 1.91), adhesion-related molecules (SKAP2, fold change = 3; ITGAM, fold change = 4.53; ICAM1, fold change = 2.12; ROCK1, fold change = 1.35), complement-related molecules (C1QB, fold change = 2.83; C3AR1, fold change = 3.06), negative regulators of immune response (VSIG4, fold change = 5.2; PPP3CB, fold change = 1.3; PLA2G7, fold change = 3.13) and molecules involved in the TGFβ pathway (TGFB1, fold change = 1.83; TGFB3, fold change = 3.37) and the downregulation of genes related to antigen processing machinery and MHC class I molecules (TAP2, fold change = −1.56; ERAP1, fold change = −2.22; B2m, fold change = −2.44).
Microarray data validation
Real-time PCR analysis and immunohistochemistry were performed to validate microarray results. Four statistically significant genes were selected for these experiments: STAT1 and MIP1α (CCL3) gene sequences were upregulated in MSI tumours compared with MSS/HLA-positive (control) group, and TGF1β and B2m genes were up- and downregulated, respectively, in MSS/HLA-negative tumour group versus controls. Real-time PCR confirmed these findings (P < 0.05), with the exception of STAT1 gene expression, which did not significantly differ between MSI tumours and controls (Table 4).
Table 4.
Summarized results of quantitative real-time RT-PCR analysis in CRC samples
| Ratioa MSI tumours | Ratioa MSS/HLA-negative tumours | Ratioa MSS/HLA-positive tumours | P-value | |
|---|---|---|---|---|
| B2m | 1.99 | 108.6 | 0.05 | |
| STAT1 | 10.5 | 37.2 | 0.16 | |
| MIP1α (CCL3) | 6.018 | 0.418 | 0.04 | |
| TGFβ1 | 2.3·10−2 | 7·10−4 | 0.02 |
Expression results of B2m, MIP1α (CCL3) and TGFβ1 genes by Real-Time RT-PCR confirmed the data obtained in microarray analysis (P ≤ 0.05), excluding STAT1 gene expression, for which no significant differences were observed
aMean values of target gene mRNA copy numbers normalized against G6PDH mRNA copy numbers
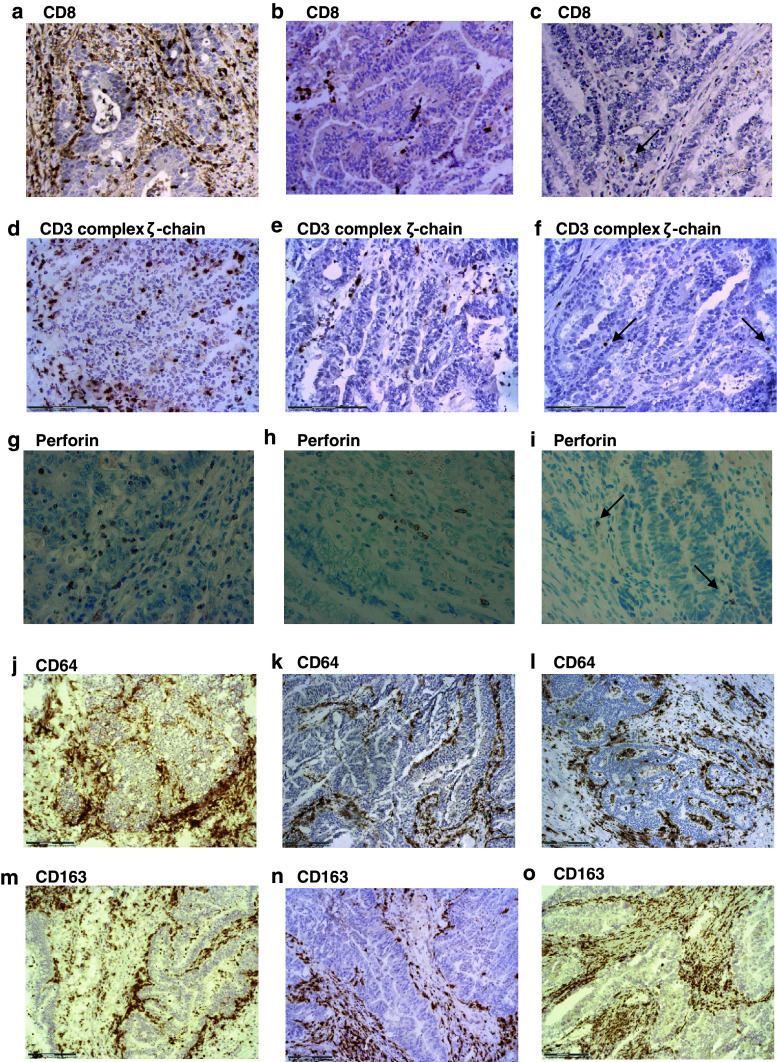
The microarray analysis results were also confirmed by immunohistochemistry, measuring the expression of two proteins encoded by discriminator genes identified in the MSI/control comparison: CD3 complex ζ-chain and perforin (Table 3). Expression of these proteins was higher in MSI tumours than in control CRCs (Fig. 3; Table 5). Expression of the selected proteins was closely related to the presence of cytotoxic CD8+T cells in the tissue. We observed a high infiltration by these cytotoxic CD8+T lymphocytes in MSI tumours (Fig. 3a; Table 5), which is also compatible with the upregulation of other genes expressed by these cells and identified in the MSI/control comparison (GNLY, α chain of TCR, RNF19B, TNFSF9, LCP2, FYN, NFATC2IP, TCIRG1, etc. [fold change > 1.46]) (Table 3). MSI tumours showed the highest leucocyte infiltration, represented mainly by CD8+T lymphocytes, whereas the leucocyte infiltration was lower in MSS/HLA-negative samples, with low or absent CD8+ cells (Fig. 3a, c) [20]. MSS/HLA-positive tumours (controls) largely showed moderate/high leucocyte infiltration, with a low presence of CD8+ cells in comparison to MSI tumours (Fig. 3b).
Fig. 3.
Immunohistological results of microarray data validation in CRC tissue samples. a MSI tumour (CRC-6), b MSS/HLA-positive (control) tumour (CRC-19) and c MSS/HLA-negative tumour (CRC-14). A higher infiltration by CD8+ cells was observed in MSI tumour versus control CRC, whereas no CD8+ cells were observed in MSS/HLA-negative CRC. d Elevated expression of CD3 complex ζ-chain in MSI tumour (CRC-1) compared with e Control tumour (CRC-19) and f MSS/HLA-negative sample (CRC-14). Perforin staining in paraffin-embedded samples was higher in g MSI tumours (CRC-6) than in h Control tumours (CRC-16) and i MSS/HLA-negative CRCs (CRC-11). No differences were observed in infiltration by M1 (CD64) and M2 (CD163) macrophages in CRC samples. j, m MSI tumour (CRC-1); k, n MSS/HLA-positive sample (CRC-19); l MSS/HLA-negative CRCs (CRC-11) and o (CRC-12)
Table 5.
Summarized results of immunohistochemical study of leucocyte infiltration in CRC samples
| Markers | MSI tumours | MSS/HLA-negative tumours | MSS/HLA-positive tumours (controls) | P-value |
|---|---|---|---|---|
| CD45 | ++/+++a | +/++ | +/++ | <0.001 |
| CD3 | ++/+++a | + | + | 0.001 |
| CD8 | ++a | 0/+ | + | <0.001 |
| CD247 (ζchain) | ++a | 0/+ | + | <0.001 |
| Perforin | ++a | 0/+ | + | <0.001 |
| CD64 | ++ | +/++ | ++ | 0.63 |
| CD163 | +/++ | +/++ | ++ | 0.51 |
| CD206 | ++ | +/++ | +/++ | 0.84 |
The scoring system and statistical analysis used are reported in “Materials and methods”
In the table, the score represents intensity of infiltration: +++ (high), ++ (moderate); + (low); 0 (absent). One-way ANOVA test was used to compare the mean of each marker between the three CRC groups: We observed significant differences in infiltration by total leucocytes (CD45) and T (CD3) lymphocytes, represented by CD8+ cells, between CRC groups. CD247 and perforin expression were higher in MSI tumours than in MSS CRCs. No significant differences were observed in infiltration by classical or M1 macrophages (CD64) and by immunosuppressive or M2 macrophages (CD163 and CD206) among tumour groups
aAfter using the Tukey post hoc test, significant differences were found between MSI and control groups (CD45, P = 0.001; CD3, P = 0.001; CD8, P = 0.001; CD247, P = 0.001 and perforin, P = 0.001) and between MSI and MSS/HLA-negative tumour groups (CD45, P < 0.001; CD3, P = 0.003; CD8, P < 0.001, CD247, P < 0.001 and perforin, P = 0.001). There was no significant differences in infiltration by CD45 (P = 0.85), CD3 (P = 0.88), or CD8 (P = 0.18) cells and in CD247 (P = 0.8) or perforin (P = 0.1) expression between MSS/HLA-negative and control tumours
The density of M1 (CD64) macrophage infiltration was similar among the three CRC groups (Fig. 3j–l). M2 macrophages (CD163, CD206) were detected in all selected CRC samples, with no major differences in infiltration pattern among tumour groups (Fig. 3m–o; Table 5). No significant differences in infiltration density were found between M1 and M2 macrophages in any CRC group (Fig. 3j–o). The microarray data showed significant differences in the expression of genes related to M1 (FCGR2A, FCGR2B, FCGR1A, CD86, CD14, TLR2, TLR8 (fold change > 2.31)) and M2 (CD163, ARG2 (fold change > 3.14)) macrophages between MSI tumours and controls, but these differences were not reflected in the immunohistochemistry results (Table 5). This discrepancy may be due to the semi-quantitative nature of immunohistochemical analysis and the intense infiltration by macrophages, which accumulated mainly in interstitial and stromal tissue areas. Accordingly, further Real-Time PCR studies are warranted to confirm the microarray data.
Discussion
In this study, we compared the genome-wide gene expression microarray analysis profile between HLA-class I-negative tumours (MSI and MSS) and HLA-class I-positive tumours (controls). A main finding was that the two groups of HLA-class I-negative tumours (MSI and MSS) differed in the number of differentially expressed genes related to local anti-tumour immune reactivity, which was confirmed by hierarchical cluster analysis.
Only a small number of genes were differentially expressed (P < 0.05) between MSS-HLA-class I-positive and MSS/HLA-class I-negative tumours, including the upregulation of molecules involved in TGFβ pathway (TGFB1, TGFB3) and, as expected, the downregulation of genes related to antigen processing machinery and MHC class I molecules (TAP2, ERAP1, B2m) in the HLA-I-negative group versus controls.
Among the 9,695 genes analysed, the expression of 2,057 significantly differed among all of the groups. Samples were non-randomly enriched in various categories of biological processes, including leucocyte activation, T-cell activation, inflammatory response or cytokine production, all of which showed a significantly higher expression in MSI versus MSS (control) cancers. Apoptosis was another GO category that discriminated between these cancers (P = 5.5·10−3), identifying 72 genes with pro- or anti-apoptotic functions that were over- or under-expressed in MSI cancers. This observation highlights the complex interaction between pro- and anti-apoptotic agents in MSI tumours, which requires further investigation.
A marked difference between MSI tumours and controls was also found for 52 genes involved in inflammatory responses against cancer (Tables 2 and 3), including the overexpression (in MSI) of genes related to immune response intensity and cytotoxic cell activity. The observed pattern of gene expression might be considered as a biomarker for MSI cancers.
Ingenuity pathway analysis (IPA) showed that the mechanisms underling the generation of variants with defective HLA class I expression differed according to the MSI/MSS phenotype (Figs. 2b, c). For instance, 40 genes in the inflammatory and cytotoxic signalling pathway were upregulated (P = 0.02) in MSI versus MSS/HLA-positive tumours, suggesting the importance of this pathway in the MSI groups.
The immunohistochemical data showed higher infiltration by CD8+T lymphocytes with activated phenotype in MSI tumours versus controls (Fig. 3a, b). In agreement with these findings, the microarray data evidenced overexpression in MSI tumours of genes encoding lymphocyte markers related to: TCR-mediated signal transduction and lymphocyte activation, proliferation and cytotoxic activity. The high infiltration of CD8+ lymphocytes in MSI tumours may result from the release of specific chemokines. In this context, cytokines detected in MSI tumours were reported to favour recruitment of cytotoxic T cells and increase Th1 responses and upregulation of antigenic peptides potentially recognizable by CD8+T lymphocytes [8, 33]. In the present study, we identified a higher expression of genes encoding macrophage- and T lymphocyte-attractant chemokines and cytokines, molecules related to cytokine-signalling pathways and proteins implicated in the extravasation of leucocytes from blood to tissues. However, MSI cancers showed a markedly decreased expression of TGFBR2 gene versus MSS controls (Table 2). MSI analysis revealed that 6 out of 8 MSI carcinomas showed instability in the microsatellite marker of the coding region in TGFBR2 gene [20], confirming that this gene is frequently affected by the MSI pathway due to mutational inactivation [34]. However, we detected no appreciable differences in the intensity of infiltration and gene expression profiles between TGFBRII wild type tumours (CRC-2 and CRC-6) and the remaining MSI cancers with instability in this microsatellite marker.
In MSI carcinomas, the overexpression of M1 markers involved in inflammation and Th1 responses was more frequent than the overexpression of M2 markers related to inhibitory functions. However, despite this finding, no major differences were observed by immunohistochemistry between M1 (CD64) and M2 (CD163) polarized macrophage populations in MSI tumours (Fig. 3j–o).
The above findings suggest that a combined analysis of microarray and immunohistochemical data might be useful to comprehensively assess tumour–host interactions. Thus, our data suggest that the immune microenvironment of MSI tumours favours immune-mediated tumour rejection, in line with previous suggestions of the protective role of immune infiltrates in colorectal tumours [35–38].
However, the outgrowth of MSI CRCs, despite the presence of a dense lymphocytic infiltration, is probably due to several mechanisms that interfere with the efficacy of the host immune response in vivo [39–41]. We found two possible mechanisms for tumour immune evasion in the MSI tumours studied: first, total loss of HLA class I molecules on tumour cell surface, mainly attributable to the accumulation of somatic mutations in the B2m gene and cell clonal expansion [20]; second, inhibition of T-cell responses by factors associated with the presence of M2 macrophages. For instance, we found increased gene expression levels of ARG2 (fold change = 3.14) and VSIG4 (V-set and immunoglobulin domain containing 4, fold change = 4.08) in MSI tumours compared with controls [42, 43].
We believe that molecular inactivation due to somatic mutation of the B2m gene may play a critical role in MSI tumour development, whereas this mutation was less frequent in the present MSS cancers. Interestingly, the presence of CTLs in MSI may be a necessary but not sufficient condition for tumour rejection. In fact, the high infiltration of CD8+ lymphocytes in MSI tumours persists after immunoedition (immune selection of B2m-deficient cells), probably recruited or locally expanded by the elevated expression of specific chemokines in the tumour microenvironment.
Finally, genes encoding NKG2D ligands (MICA, MICB and ULBP2) were found to be overexpressed in MSI tumours compared with controls, which, together with the total loss of HLA class I expression, could promote the cytotoxic activity of NK cells and CD8+ NKG2D+T lymphocytes. Although the absence or downregulation of HLA class I renders cells susceptible to NK cells, we found only scant CD56+ cells in most of the tumours, with no differences as a function of HLA class I cell surface expression, in agreement with previous reports [44, 45].
In summary, immune-mediated tumour rejection may be favoured by the characteristics of MSI tumours, including the abundant generation of new immunogenic frameshift peptides presented to CTLs and the dense infiltration with CTLs. Nevertheless, changes in the intrinsic phenotype of cancer cells (e.g. total HLA class loss) and the participation of immune regulatory mechanisms can lead to tumour immune escape. Finally, our results also strongly suggest a divergence in the mechanisms that drive immune escape in HLA-I-negative MSI and MSS tumours.
Acknowledgments
The authors thank Eva García, Antonia Moreno and Ana Isabel Rodríguez for technical assistance. They are also grateful to the Tumour-Tissue Biobank of Virgen de las Nieves University Hospital for providing study samples. This investigation was partially supported by grants from the Fondo de Investigaciones Sanitarias (08/0528), Red Genómica del Cáncer (RETICRD 06/020), Consejería de Salud de la Junta de Andalucía (PI-0080-2010), Consejería de Innovación, Ciencia y Empresa de la Junta de Andalucía (P08-TIC-4299), Dirección General de Investigación y Gestión del Plan Nacional I + D+i (TIN2009-13489), Proyecto de Investigación de Excelencia (CTS-3952, CVI-4740 and P06/-CTS-02200) and Plan Andaluz de Investigación (PAI, Group CTS) and from the European Searchable Tumour Cell Line Database (ESTDAB) project, contract No. QLRI-CT-2001-01325 (http://www.ebi.ac.uk/estdab), the European Network for the identification and validation of antigens and biomarkers in cancer and their application in clinical tumour immunology (ENACT) project (European community LSHC-CT-2004-503306) and the Cancer Immunotherapy project (European community OJ 2004/c158,18234).
Conflict of interest
The authors declare that they have no conflict of interests.
Abbreviations
- APM
Antigen processing machinery
- CRC
Colorectal cancer
- HLA
Human leucocyte antigen
- MHC
Major histocompatibility complex
- MMR
Mismatch repair
- MSI
Microsatellite instability
- MSS
Microsatellite stability
References
- 1.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 2.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 3.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 4.Imai K, Yamamoto H. Carcinogenesis and microsatellite instability: the interrelationship between genetics and epigenetics. Carcinogenesis. 2008;29:673–680. doi: 10.1093/carcin/bgm228. [DOI] [PubMed] [Google Scholar]
- 5.Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50:113–130. doi: 10.1111/j.1365-2559.2006.02549.x. [DOI] [PubMed] [Google Scholar]
- 6.Bertucci F, Salas S, Eysteries S, Nasser V, Finetti P, Ginestier C, Charafe-Jauffret E, Loriod B, Bachelart L, Montfort J, Victorero G, Viret F, et al. Gene expression profiling of colon cancer by DNA microarrays and correlation with histoclinical parameters. Oncogene. 2004;23:1377–1391. doi: 10.1038/sj.onc.1207262. [DOI] [PubMed] [Google Scholar]
- 7.Lanza G, Ferracin M, Gafà R, Veronese A, Spizzo R, Pichiorri F, Liu CG, Calin GA, Croce CM, Negrini M. mRNA/microRNA gene expression profile in microsatellite unstable colorectal cancer. Mol Cancer. 2007;6:54. doi: 10.1186/1476-4598-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banerjea A, Ahmed S, Hands RE, Huang F, Han X, Shaw PM, Feakins R, Bustin SA, Dorudi S. Colorectal cancers with microsatellite instability display mRNA expression signatures characteristic of increased immunogenicity. Mol Cancer. 2004;3:21. doi: 10.1186/1476-4598-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dolcetti R, Viel A, Doglioni C, Russo A, Guidoboni M, Capozzi E, Vecchiato N, Macrì E, Fornasarig M, Boiocchi M. High prevalence of activated intraepithelial cytotoxic T lymphocytes and increased neoplastic cell apoptosis in colorectal carcinomas with microsatellite instability. Am J Pathol. 1999;154:1805–1813. doi: 10.1016/S0002-9440(10)65436-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smyrk TC, Watson P, Kaul K, Lynch HT. Tumor-infiltrating lymphocytes are a marker for microsatellite instability in colorectal carcinoma. Cancer. 2001;91:2417–2422. doi: 10.1002/1097-0142(20010615)91:12<2417::AID-CNCR1276>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 11.Linnebacher M, Gebert J, Rudy W, Woerner S, Yuan YP, Bork P, von Knebel Doeberitz M. Frameshift peptide-derived T-cell epitopes: a source of novel tumor-specific antigens. Int J Cancer. 2001;93:6–11. doi: 10.1002/ijc.1298. [DOI] [PubMed] [Google Scholar]
- 12.Saeterdal I, Bjørheim J, Lislerud K, Gjertsen MK, Bukholm IK, Olsen OC, Nesland JM, Eriksen JA, Møller M, Lindblom A, Gaudernack G. Frameshift-mutation-derived peptides as tumor-specific antigens in inherited and spontaneous colorectal cancer. Proc Natl Acad Sci USA. 2001;98:13255–13260. doi: 10.1073/pnas.231326898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michael-Robinson JM, Biemer-Huttmann A, Purdie DM, Walsh MD, Simms LA, Biden KG, Young JP, Leggett BA, Jass JR, Radford-Smith GL. Tumour infiltrating lymphocytes and apoptosis are independent features in colorectal cancer stratified according to microsatellite instability status. Gut. 2001;48:360–366. doi: 10.1136/gut.48.3.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunn GP, Old LJ, Schreiber RD. The immunobiology of cancer immunosurveillance and immunoediting. Immunity. 2004;21:137–148.2. doi: 10.1016/j.immuni.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 15.Seliger B, Cabrera T, Garrido F, Ferrone S. HLA class I antigen abnormalities and immune escape by malignant cells. Semin Cancer Biol. 2002;12:3–13. doi: 10.1006/scbi.2001.0404. [DOI] [PubMed] [Google Scholar]
- 16.Kloor M, Michel S, Buckowitz B, Rüschoff J, Büttner R, Holinski-Feder E, Dippold W, Wagner R, Tariverdian M, Benner A, Schwitalle Y, Kuchenbuch B, et al. Beta2-microglobulin mutations in microsatellite unstable colorectal tumors. Int J Cancer. 2007;121:454–458. doi: 10.1002/ijc.22691. [DOI] [PubMed] [Google Scholar]
- 17.Bicknell DC, Kaklamanis L, Hampson R, Bodmer WF, Karran P. Selection for beta 2-microglobulin mutation in mismatch repair-defective colorectal carcinomas. Curr Biol. 1996;6:1695–1697. doi: 10.1016/S0960-9822(02)70795-1. [DOI] [PubMed] [Google Scholar]
- 18.Yamamoto H, Yamashita K, Perucho M. Somatic mutation of the beta2-microglobulin gene associates with unfavorable prognosis in gastrointestinal cancer of the microsatellite mutator phenotype. Gastroenterology. 2001;120:1565–1567. doi: 10.1053/gast.2001.24497. [DOI] [PubMed] [Google Scholar]
- 19.Cabrera CM, Jiménez P, Cabrera T, Esparza C, Ruiz-Cabello F, Garrido F. Total loss of MHC class I in colorectal tumors can be explained by two molecular pathways: beta2-microglobulin inactivation in MSI-positive tumors and LMP7/TAP2 downregulation in MSI-negative tumors. Tissue Antigens. 2003;61:211–219. doi: 10.1034/j.1399-0039.2003.00020.x. [DOI] [PubMed] [Google Scholar]
- 20.Bernal M, Concha A, Sáenz-López P, Rodríguez AI, Cabrera T, Garrido F, Ruiz-Cabello F. Leukocyte infiltrate in gastrointestinal adenocarcinomas is strongly associated with tumor microsatellite instability but not with tumor immunogenicity. Cancer Immunol Immunother. 2011;60:869–882. doi: 10.1007/s00262-011-0999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sobin L, Gospodarowiaz M, Wittekind CH. TNM classification of malignant tumours UICC. 7. Oxford: Wiley-Blackwell; 2009. [Google Scholar]
- 22.López Nevot MA, Cabrera T, de la Higuera B, Ruiz-Cabello F, Garrido F. Obtención y caracterización de anticuerpos monoclonales frente a leucemias humanas. Inmunología. 1986;5:51–59. [Google Scholar]
- 23.Stam NJ, Spits H, Ploegh HL. Monoclonal antibodies raised against denatured HLA-B locus heavy chains permit biochemical characterization of certain HLA-C locus products. J Immunol. 1986;137:2299–2306. [PubMed] [Google Scholar]
- 24.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson PN, Wollstein A, Bohme U, Beattie B. Ontologizing gene-expression microarray data: characterizing clusters with gene ontology. Bioinformatics. 2004;20:979–981. doi: 10.1093/bioinformatics/bth040. [DOI] [PubMed] [Google Scholar]
- 27.Cheng J, Sun S, Tracy A, Hubbell E, Morris J, Valmeekam V, Kimbrough A, Cline MS, Liu G, Shigeta R, Kulp D, Siani-Rose MA. NetAffx gene ontology mining tool: a visual approach for microarray data analysis. Bioinformatics. 2004;20:1462–1463. doi: 10.1093/bioinformatics/bth087. [DOI] [PubMed] [Google Scholar]
- 28.Ovaska K, Laakso M, Hautaniemi S. Fast gene ontology based clustering for microarray experiments. BioData Min. 2008;1:1–11. doi: 10.1186/1756-0381-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Resnik P (1995) Using information content to evaluate semantic similarity in a taxonomy In: Smith Y (ed) Proceedings of the 14th international joint conference on artificial intelligence, Montreal, pp 448–453
- 30.Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK, Miller-Graziano C, Moldawer LL, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 31.Huelin C, Gonzalez M, Pedrinaci S, de la Higuera B, Piris MA, San Miguel J, Ruiz-Cabello F, Garrido F. Distribution of the CD45R antigen in the maturation of lymphoid and myeloid series. The CD45R negative phenotype is a constant finding in T CD4 positive lymphoproliferative disorders. Br J Haematol. 1988;69:173–179. doi: 10.1111/j.1365-2141.1988.tb07619.x. [DOI] [PubMed] [Google Scholar]
- 32.Sancho J, Franco R, Chatila T, Hall C, Terhorst C. The T cell receptor associated CD3-epsilon protein is phosphorylated upon T cell activation in the two tyrosine residues of a conserved signal transduction motif. Eur J Immunol. 1993;23:1636–1642. doi: 10.1002/eji.1830230736. [DOI] [PubMed] [Google Scholar]
- 33.Kloor M, Michel S, von Knebel Doeberitz M. Immune evasion of microsatellite unstable colorectal cancers. Int J Cancer. 2010;127:1001–1010. doi: 10.1002/ijc.25283. [DOI] [PubMed] [Google Scholar]
- 34.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan RS, Zborowska E, Kinzler KW, Vogelstein B, et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 35.Pagès F, Kirilovsky A, Mlecnik B, Asslaber M, Tosolini M, Bindea G, Lagorce C, Wind P, Marliot F, Bruneval P, Zatloukal K, Trajanoski Z, Berger A, Fridman WH, Galon J. In situ cytotoxic and memory T cells predict outcome in patients with early-stage colorectal cancer. J Clin Oncol. 2009;27:5944–5951. doi: 10.1200/JCO.2008.19.6147. [DOI] [PubMed] [Google Scholar]
- 36.Pagès F, Galon J, Dieu-Nosjean MC, Tartour E, Sautès-Fridman C, Fridman WH. Immune infiltration in human tumors: a prognostic factor that should not be ignored. Oncogene. 2010;29:1093–1102. doi: 10.1038/onc.2009.416. [DOI] [PubMed] [Google Scholar]
- 37.Buckowitz A, Knaebel HP, Benner A, Bläker H, Gebert J, Kienle P, von Knebel Doeberitz M, Kloor M. Microsatellite instability in colorectal cancer is associated with local lymphocyte infiltration and low frequency of distant metastases. Br J Cancer. 2005;92:1746–1753. doi: 10.1038/sj.bjc.6602534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pagès C, Tosolini M, Camus M, Berger A, Wind P, Zinzindohoué F, Bruneval P, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 39.Garrido F, Ruiz-Cabello F, Cabrera T, et al. Implications for immunosurveillance of altered HLA class I phenotypes in human tumours. Immunol Today. 2006;18:89–95. doi: 10.1016/S0167-5699(96)10075-X. [DOI] [PubMed] [Google Scholar]
- 40.Drake CG, Jaffee E, Pardoll DM. Mechanisms of immune evasion by tumors. Adv Immunol. 2006;90:51–81. doi: 10.1016/S0065-2776(06)90002-9. [DOI] [PubMed] [Google Scholar]
- 41.Michel S, Benner A, Tariverdian M, Wentzensen N, Hoefler P, Pommerencke T, Grabe N, von Knebel Doeberitz M, Kloor M. High density of FOXP3-positive T cells infiltrating colorectal cancers with microsatellite instability. Br J Cancer. 2008;99:1867–1873. doi: 10.1038/sj.bjc.6604756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Norian LA, Rodriguez PC, O’Mara LA, Zabaleta J, Ochoa AC, Cella M, Allen PM. Tumor-infiltrating regulatory dendritic cells inhibit CD8+T cell function via l-arginine metabolism. Cancer Res. 2009;69:3086–3094. doi: 10.1158/0008-5472.CAN-08-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vogt L, Schmitz N, Kurrer MO, Bauer M, Hinton HI, Behnke S, Gatto D, Sebbel P, Beerli RR, Sonderegger I, Kopf M, Saudan P, et al. VSIG4, a B7 family related protein, is a negative regulator of T cell activation. J Clin Invest. 2006;116:2817–2826. doi: 10.1172/JCI25673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sandel MH, Speetjens FM, Menon AG, Albertsson PA, Basse PH, Hokland M, Nagelkerke JF, Tollenaar RA, van de Velde CJ, Kuppen PJ. Natural killer cells infiltrating colorectal cancer and MHC class I expression. Mol Immunol. 2005;42:541–546. doi: 10.1016/j.molimm.2004.07.039. [DOI] [PubMed] [Google Scholar]
- 45.Cozar JM, Canton J, Tallada M, Concha A, Cabrera T, Garrido F, Ruiz-Cabello Osuna F. Analysis of NK cells and chemokine receptors in tumor infiltrating CD4 T lymphocytes in human renal carcinomas. Cancer Immunol Immunother. 2005;54:858–866. doi: 10.1007/s00262-004-0646-1. [DOI] [PMC free article] [PubMed] [Google Scholar]