Abstract
Tumor cells often evade killing by the complement system by overexpressing membrane-bound complement inhibitors. However, production of soluble complement inhibitors in cells other than hepatocytes was rarely reported. We screened several breast cancer cell lines for expression of soluble complement inhibitor, complement factor I (FI). We also analyzed local production of FI in tissue microarrays with tumors from 130 breast cancer patients by in situ hybridization and immunohistochemistry. We found expression of FI in breast adenocarcinoma cell line MDA-MB-468 and confirmed its functional activity. Expression of FI at mRNA and protein levels was also confirmed in tumor cells and tumor stroma, both in fibroblasts and infiltrating immune cells. Multivariate Cox regression analyses revealed that high expression of FI protein in tumor cells was correlated with significantly shorter cancer-specific survival (HR 2.8; 95 % CI 1.0–7.5; p = 0.048) and recurrence-free survival (HR 3.4; 95 % CI 1.5–7.4; p = 0.002). High FI expression was positively correlated with tumor size (p < 0.001), and Nottingham histological grade (p = 0.015) and associated with estrogen and progesterone receptor status (p = 0.03 and p = 0.009, respectively). Our data show that FI is expressed in breast cancer and is associated with unfavorable clinical outcome.
Electronic supplementary material
The online version of this article (doi:10.1007/s00262-015-1658-8) contains supplementary material, which is available to authorized users.
Keywords: Innate immunity, Complement system, Breast cancer, Tissue microarrays, Immunohistochemistry
Introduction
The complement system is a part of innate immunity and aims at elimination of “non-self” from our body. Such elimination is based on identification of molecular patterns that are atypical under physiological conditions and are recognized by a number of pattern-recognition molecules. Once activated, complement does not distinguish any more between self and non-self constituents and, therefore, additional protection of own cells and tissues from misguided complement attack is supported by endogenous complement inhibitors [1]. There is an increasing amount of evidence that both diminished and excessive activation of complement contributes to pathological processes including cancer. Tumor cells of different origin activate complement [2], and this event was long believed to act only in benefit of the host. This hypothesis was supported by a number of facts: (1) Tumor cells developed various strategies to counter-attack complement activation [3], (2) oncogenic viruses encode their own complement inhibitors [4], (3) tumor cells with knocked-down complement inhibitors grew slower as compared to control, mock-transfected cells, in mouse xenograft models [5]. However, new reports showed that tumor cells benefit from triggering complement activation due to C5a-dependent recruitment of myeloid-derived suppressor cells or induction of angiogenesis [6, 7]. Taken together, the role of complement in the pathogenesis of cancer is apparently more complex than initially believed and will likely differ between different types and stages of cancer.
Although membrane-bound complement inhibitors are abundant on the surface of various cell types, the production of soluble complement inhibitors was considered a rare phenomenon in cells other than hepatocytes [8]. The complement inhibitor factor I (FI) is a soluble serine protease with exceptionally narrow specificity. In the presence of appropriate cofactors, FI cleaves the early, activated complement components C3b and C4b to iC3b and iC4b, respectively. Neither iC3b nor iC4b can fuel the complement cascade any more. However, iC3b is recognized by complement receptors CR2 and CR3, which are present on epithelial cells as well as on a number of immune cells including B cells, follicular dendritic cells, tissue macrophages and mononuclear phagocytes, thymocytes and a subset of peripheral T cells [9]. Interaction of iC3b with its cellular receptors edits the immune response and influences local inflammation, which in turn may exert both pro- and antitumor effects [10]. While expression of FI was reported in a number of tumor cell lines including those originating from hepatoma [11], B cell leukemia [12], glioma [13], rhabdosarcoma [14] lung cancer [15] and cutaneous squamous cell carcinoma [16], only the latter study describes expression of FI by tumor cells of the skin in vivo and shows association of FI production and aggressive tumor phenotype [16]. Herein, we report local production of FI in breast cancer cells and associations between tumor-derived FI and clinical data obtained from 130 breast cancer patients.
Materials and methods
Cells
MDA-MB-231, MDA-MB-468, MCF-7 (all adenocarcinomas) and T47D (ductal carcinoma) breast cancer cell lines originally derived from metastatic sites were bought from American Type Culture Collection (ATCC) by Prof. Tommy Andersson (Lund University, Sweden), who generously shared these with us. Cells were frozen immediately after re-cultivation of the original aliquot, and all the experiments were performed on cultures originating from these secondary aliquots within no more than 5 passages. Cells were Mycoplasma negative and monthly tested for contamination with VenorGEM Classic kit (Minerva Biolabs). Clones of HEK293 cells expressing human full-length FI or transfected with empty vector were prepared as described in [17].
Purification of antibody against FI
Rabbit polyclonal PK9205 antibody against human FI was raised in-house and successively used before for FI detection [15, 18]. To employ this Ab for immunohistochemical FI staining, we purified it further by FI affinity chromatography. Recombinant FI (2.7 mg) produced as described in [19] was dialyzed and resuspended in 5 ml of 10 mM 3-(N-morpholino)propanesulfonic acid (MOPS) pH 7 and mixed with 1 ml of Affi-Gel 10 (Bio-Rad). After 4 h incubation at 4 °C, potential protein binding sites were blocked with addition of 100 μl of 1 M ethanolamine pH 8.0. The column was equilibrated with PBS, washed with the same buffer and eluted with 0.1 M glycine pH 2.5. The protein-containing eluate fractions were immediately neutralized with 1 M Tris, pH 8.0, pooled, dialyzed against PBS and concentrated.
qPCR
MDA-MB-231, MDA-MB-468, MCF-7 and T47D were grown to 80 % confluency in DMEM high-glucose medium supplemented with 10 % fetal calf serum (Gibco). RNA was isolated with the RNAeasy kit (Qiagen) according to the manufacturer’s instructions. cDNA was synthesized from 1 μg RNA by using 200 U Superscript III reverse transcriptase and 2.5 μM oligo(dT) primer (Life technologies). For the qPCR, 50 ng cDNA of each sample was used in triplicates. A primer (TaqMan probe set) specific for FI (Hs00989715_m1) and TaqMan gene expression master mix were bought from Applied Biosystems. Cyclophilin A, TATA box binding protein and hypoxanthine phosphoribosyltransferase 1 were used as reference genes. The relative expression of FI was calculated according to the ΔCt method by using the geometrical mean of the three reference genes [20].
Detection of FI by western blotting
Breast cancer cell lines were cultured in T25 flasks (Nunc) in DMEM high-glucose medium supplemented with 10 % fetal calf serum until 100 % confluency. Then, the cells were washed twice with PBS and 5 ml serum-free Optimum medium (Invitrogen) was added for 48 h. The medium was harvested and concentrated 50× with a concentrating device (Vivaspin) with molecular weight cutoff value of 10 kDa. The retentate was loaded onto a 10 % SDS–PAGE (30 μl per lane) and separated proteins were then transferred to a PVDF membrane. FI purified from plasma [21] was used as a positive control. The Western blot was developed with 3.5 μg/ml mouse monoclonal anti-FI antibody OX21, generously provided by Prof. R Sim (Oxford University) followed by goat anti-mouse F(ab)2 (Dako) diluted 1:1,000. Visualization of the bands was performed with Chemi Doc Visualization System (Bio-Rad) using Immobilon Western reagent (Millipore).
C3b/C4b degradation assay
The functionality of MDA-MB-468 cell-derived FI was analyzed by degradation of 125I-labeled C3b and C4b proteins, both purchased from Complement Technologies and iodinated by the chloramine-T method as described [22]. Conditioned or control OptiMEM medium (8 μl; concentrated 20×) or PBS buffer (negative control) was mixed with 10 μl of 0.25 mg/ml plasma-purified C4b-binding protein; C4BP [23] and trace amounts of iodinated C3b or C4b. As a positive control, 6 μg/ml of recombinant FI was added. 2 μl of FI function-blocking mAb #1 (Quidel, stock 1.1 mg/ml) was added to chosen samples to ensure the specificity of cleavage. The mixed ingredients in a total volume of 25 μl were incubated for 3 h at 37 °C, then reducing Laemmli buffer was added and samples were separated using 10 % SDS–PAGE. After electrophoresis, gels were dried, subjected to autoradiography (Fuji Film) and visualized in a Typhoon FLA 9500 phosphoimager (GE Healthcare).
Patient material and immunostaining of breast cancer tissue microarrays
We studied tissue microarrays (TMAs) constructed from tumor material obtained from female patients diagnosed with invasive breast cancer at Malmö University Hospital between 2001 and 2002. The cohort consisted originally of 179 patients and was described in detail previously [24]. Furthermore, we prepared sections from normal breast tissue that was removed for cosmetic purposes. Immunohistochemical data regarding hormone receptor status, Ki-67 and human epidermal growth factor receptor 2 (Her2) expression were available from previous studies on the same breast cancer patient cohort [25, 26]. Definitions of estrogen receptor (ER) and progesterone receptor (PR) negativity followed current Swedish clinical guidelines (<10 % positive nuclei). Similarly, Ki-67 status was assessed based on % of positive staining nuclei and dichotomized into ≤25 and >25 %. Her2 status was assessed by semiquantitative analysis according to a standard protocol (HercepTest; DakoCytomation) described in [27]. We grouped the specimens as weakly expressing (scores 0–2) and strongly expressing Her2 (score 3). Two 1.0-mm cores of tumor tissue were obtained from each tumor, and, in general, cores were taken from the peripheral aspect of the tumor, and necrotic tissue was avoided. TMA sections of 4 μm thickness were cut and automatically pretreated using the PT-link system (Dako; Agilent Technologies) and then stained in an Autostainer Plus (Dako) with the affinity-purified PK9205 polyclonal antibody (described above) at a final concentration of 0.25 μg/ml overnight. Subsequently, EnVision Flex HRP kit (Dako) was applied to the sections for 20 min followed by DAB reagent for visualization. Pellets of HEK293 freestyle cells transfected with human FI or vector only, treated in the same way as the TMAs, were used as negative and positive controls. Also, we included another negative control, in which the primary antibody was replaced with buffer. Mayer’s hematoxylin (Histolab) was used for counterstaining. After histopathological reevaluation, we eliminated detached or damaged specimens as well as those containing very little or no tumor cells. Finally, samples from 130 patients were used for scoring and statistical analyses. Ethical permission was obtained from the Lund University Regional Ethics Board, ref. no. 445/2007, whereby written consent was not required and patients were offered the option to opt out.
Scoring, evaluation and statistical analysis
The immunohistochemical staining was evaluated by an experienced pathologist and three scientists who were all blinded with regard to the clinical information. Since the staining within particular tissue sections was homogenous in the majority of cases, we did not analyze percentage of positive cells. Instead, we assessed the intensity of FI staining in tumor cells and tumor stroma separately and graded from 0 (negative), 1+ (weak), 2+ (moderate) to 3+ (strong). The final score was appointed to the highest intensity attainable in either of the two samples from the same patient. Original slides were scanned in Aperio ScanScope slide scanner and figures showing the representative stainings were prepared in the Image Scope software (Aperio) followed by Illustrator (Adobe). Kaplan–Meier analyses and Breslow test were used to illustrate and estimate the impact of high FI expression in cells and stroma on overall survival, cancer-specific survival (CSS) and recurrence-free survival (RFS). For Kaplan–Meier analyses, FI intensity was grouped into weak expression (score 0, 1 and 2) and strong expression (score 3). Cox regression proportional hazards models were used for estimation of hazard ratios (HR) for death from breast cancer or recurrence-free survival according to cellular and stromal FI expression in both uni- and multivariate analyses. Covariates, all with a p value < 0.05 in the univariate analysis, included in the multivariate analysis were: tumor size, histological grade, ER, Her2 and Ki-67 status (Ki-67 was only included for CSS since it was not significant in the univariate analysis for RFS). Differences in distributions of pathological and clinical parameters (age, tumor size, histological grade, lymph node status, hormone receptor status, Her2 and Ki-67) between different FI intensity scores in malignant cells and stroma were calculated using Spearman correlations or Mann–Whitney U tests, as indicated in Table 1. All statistical tests were two-sided and p < 0.05 was considered statistically significant. The calculations were preformed using SPSS Statistics version 20 (IBM).
Table 1.
Associations between FI expression and clinicopathologic features in primary breast cancer
| Factor | FI staining intensity in tumor cells | FI staining intensity in stroma | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | 0 | 1 | 2 | 3 | p value | n | 0 | 1 | 2 | 3 | p value | |
| All n (%) | 130 | 7 (5) | 42 (32) | 53 (41) | 28 (22) | 129 | 15 (12) | 44 (34) | 55 (43) | 15 (12) | ||
| Age | 130 | 0.143a | 129 | 0.507a | ||||||||
| Median (range) | 59 (47–81) | 64.5 (35–86) | 64 (34–97) | 73 (41–91) | 63 (35–91) | 64.5 (40–97) | 63 (34–91) | 75 (41–89) | ||||
| Size (tumor diameter, mm) | 130 | <0.001a | 129 | 0.405a | ||||||||
| Median (range) | 22 (11–28) | 19 (8–60) | 21 (8–80) | 27.5 (13–145) | 26 (11–60) | 22 (8–140) | 20 (8–145) | 21 (13–73) | ||||
| NHG | 0.015a | 0.326a | ||||||||||
| I (%) | 18 | 2 (11) | 9 (50) | 7 (39) | 0 (0) | 17 | 2 (12) | 6 (35) | 8 (47) | 1 (6) | ||
| II (%) | 58 | 2 (3) | 20 (35) | 23 (40) | 13 (22) | 58 | 6 (10) | 23 (40) | 24 (41) | 5 (9) | ||
| III (%) | 54 | 3 (6) | 13 (24) | 23 (43) | 15 (28) | 54 | 7 (13) | 15 (28) | 23 (43) | 9 (17) | ||
| Nodal status | 0.215a | 0.824a | ||||||||||
| 0 (%) | 65 | 4 (6) | 23 (35) | 26 (40) | 12 (19) | 65 | 8 (12) | 22 (34) | 26 (40) | 9 (14) | ||
| 1–3 (%) | 32 | 1 (3) | 12 (38) | 14 (44) | 5 (16) | 31 | 5 (16) | 12 (39) | 11 (36) | 3 (10) | ||
| 4 (%) | 21 | 1 (5) | 5 (24) | 7 (33) | 8 (38) | 21 | 2 (10) | 5 (24) | 11 (52) | 3 (14) | ||
| Missing | 12 | 12 | ||||||||||
| ER status | 0.030b | 0.004b | ||||||||||
| Negative (%) | 17 | 1 (6) | 2 (12) | 7 (41) | 7 (41) | 17 | 1 (6) | 1 (6) | 11 (65) | 4 (24) | ||
| Positive (%) | 113 | 6 (5) | 40 (35) | 46 (41) | 21 (19) | 112 | 14 (13) | 43 (38) | 44 (39) | 11 (10) | ||
| PR status | 0.009b | 0.020b | ||||||||||
| Negative (%) | 40 | 1 (3) | 9 (22) | 16 (40) | 14 (35) | 39 | 4 (10) | 8 (21) | 19 (49) | 8 (20) | ||
| Positive (%) | 90 | 6 (7) | 33 (37) | 37 (41) | 14 (16) | 90 | 11 (12) | 36 (40) | 36 (40) | 7 (8) | ||
| HER2c | 0.304b | 0.006b | ||||||||||
| Weak (%) | 118 | 6 (5) | 39 (33) | 49 (42) | 24 (20) | 117 | 15 (13) | 40 (34) | 52 (44) | 10 (9) | ||
| Strong (%) | 9 | 0 (0) | 3 (33) | 2 (22) | 4 (44) | 9 | 0 (0) | 2 (22) | 2 (22) | 5 (56) | ||
| Missing | 3 | 3 | ||||||||||
| Ki-67 | 0.516b | 0.137b | ||||||||||
| ≤25 % | 54 | 4 (7) | 15 (28) | 24 (44) | 11 (20) | 53 | 6 (11) | 23 (43) | 19 (36) | 5 (9) | ||
| >25 % | 59 | 2 (3) | 18 (31) | 23 (39) | 16 (27) | 59 | 7 (12) | 15 (25) | 29 (49) | 8 (14) | ||
| Missing | 17 | 17 | ||||||||||
aSpearman correlation, 2-tailed p value
bMann–Whitney U test for comparison of median, 2-tailed exact p value
cWeak (score 0–2), strong (score 3)
In situ hybridization
Paraffin-embedded slides containing breast cancer tissue or cell pellets were baked at 60 °C for 1 h and deparaffinized in xylene followed by absolute ethanol. Subsequently, detection of FI-specific RNA as well as PPIB housekeeping gene RNA (positive control) and bacterial DapB RNA (negative control) was performed with RNAscope 2.0 BROWN assay (Advanced Cell Diagnostics) [28], according to the manufacturer’s instructions. Original slides were scanned with Aperio ScanScope slide scanner. In our experimental conditions, hematoxilin counterstaining did not turn entirely blue and intracellular material remained purple/gray. Therefore, to better distinguish between counterstaining (purple) and signal from RNA probes (brown), we used Adobe Photoshop picture enhancement options, which included diminishing vibrance to −100 % followed by an increase of saturation to +100 %.
Results
Breast cancer cell lines produce functional FI
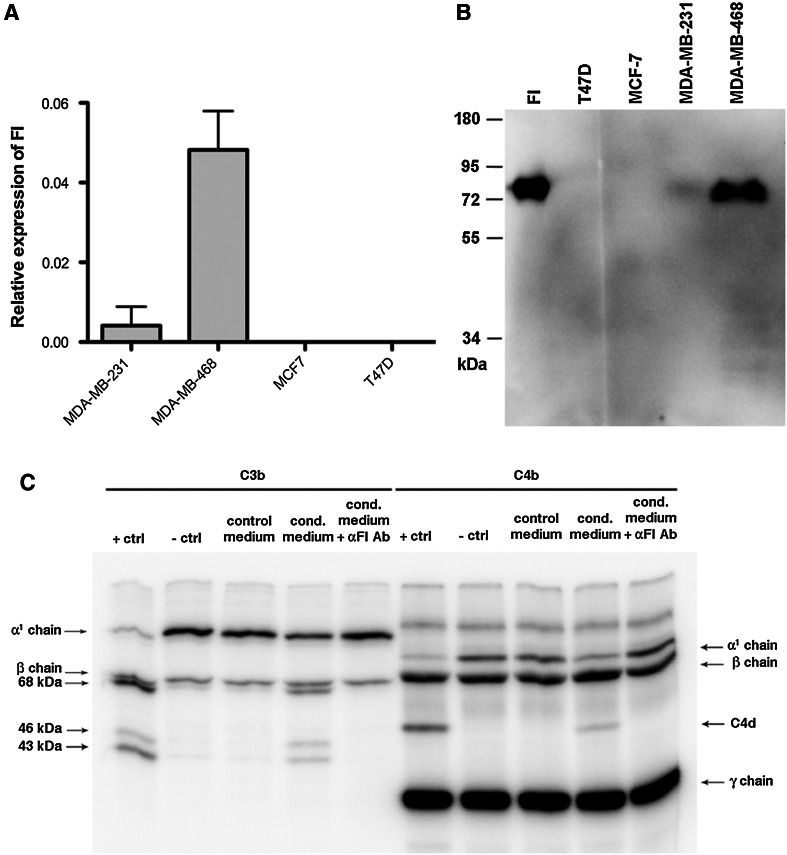
We analyzed expression of FI in four breast cancer cell lines: MDA-MB-231, MDA-MB-468, MCF-7 and T47D by qPCR (Fig. 1a). Two cell lines, MDA-MB-231 and MDA-MB-468 showed significant expression. In order to verify whether the qPCR results corresponded to FI expression at the protein level, we analyzed samples of concentrated, serum-free media conditioned by breast cancer cells using Western blotting. The clear band detected in medium conditioned by MDA-MB-468 showed the same molecular weight as the one in plasma-purified FI sample, whereas no band was detected in medium conditioned by T47D and MCF-7 cells. Very faint band was detected in the sample of medium conditioned by MDA-MB-231 cells (Fig. 1b). To determine whether FI produced by MDA-MB-468 cells retained its specific activity, we performed a degradation assay, which tested the ability of cell-derived FI to cleave its natural ligands C3b and C4b in the presence of a cofactor C4BP [29] (Fig. 1c). When MDA-MB-468 cells-conditioned, serum-free medium was used as a source of FI, the same pattern of C3b and C4b cleavage was revealed as when recombinant FI was applied. To further ensure that the observed cleavage pattern was solely due to FI activity and not due to the activity of other proteases, we used a function-blocking anti-FI mAb. When added to MDA-MB-468 cells-conditioned medium, it completely prevented cleavage of C3b and C4b.
Fig. 1.
Expression of FI in breast cancer cell lines. a FI expression in breast cancer cell lines was assessed on mRNA level by qPCR and related to three housekeeping genes. b Production of FI on protein level was confirmed by Western blotting performed on serum-free medium conditioned by breast cancer cells. c Functional activity of FI produced by MDA-MB-468 cells was shown using C3b/C4b degradation assay in which incubation of C3b or C4b with FI cofactor C4BP and conditioned medium resulted in appearance of degradations products of C3b and C4b. For both C3b and C4b, degradation was abolished when FI blocking antibody was added to the conditioned medium
FI is produced locally within breast cancer tissue
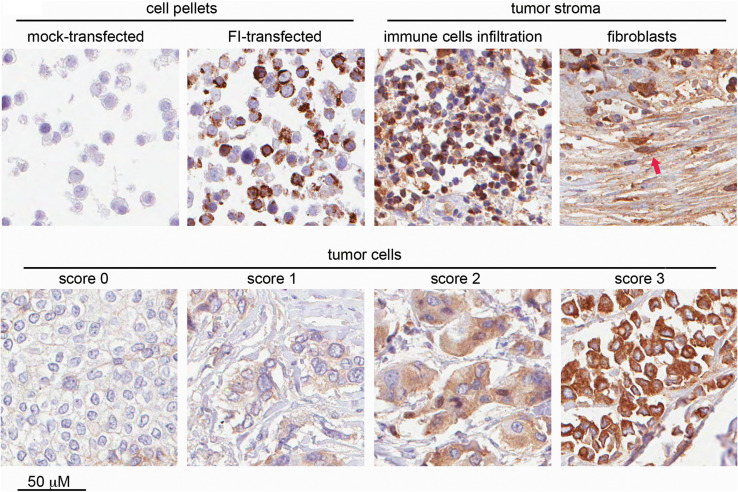
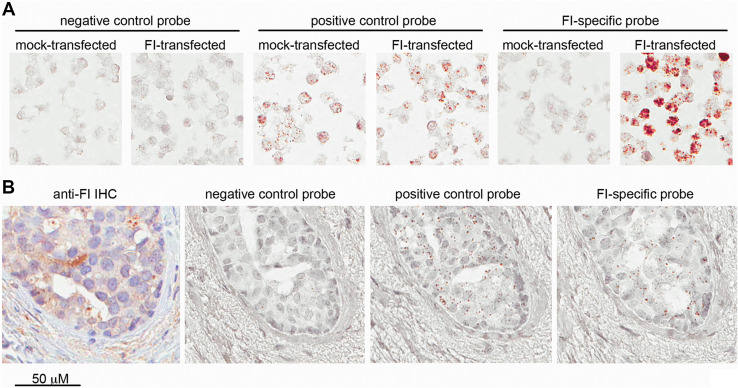
We aimed to examine whether FI expression is restricted to certain breast cancer cell lines or whether it also occurs in breast cancer tissue in vivo. For that purpose, we performed immunohistochemical staining with an affinity-purified, polyclonal anti-FI antibody. Validation of this method was performed on HEK293 cells transfected with human, full-length FI or empty vector. Cell pellets were formalin-fixed, paraffin-embedded and passed though all steps of immunohistochemical staining as routinely applied for tissue specimens. We found staining conditions where mock-transfected cells remained colorless and FI-transfected cells were strongly stained in their cytosolic compartment (Fig. 2, upper left panel), as expected. Using the same conditions, we stained TMAs from 130 breast cancer patients and found cytosolic staining of different intensity not only in breast cancer cells (Fig. 2, bottom panel), but also in tumor stroma including infiltrating immune cells and fibroblasts (Fig. 2, upper right panel). Furthermore, we analyzed TMAs of normal, healthy breast tissue from 37 women and detected a degree of expression of FI in ductal epithelial cells and stroma (supplementary Fig.1). However, the percentage of the specimens with the high expression of FI in cells was lower compared with tumor tissue (see details in Table 1 and supplementary Fig.1). Given the fact that FI is abundant in blood (the contents of small blood vessel in the tissue were accordingly strongly positive), we wanted to distinguish local production from diffusion and retention from the bloodstream. For this purpose, we employed the RNAscope technique, which we first validated on HEK293 cell pellets (Fig. 3a). Further experiments on TMAs revealed expression of FI-specific mRNA in malignant breast cancer cells in situ (Fig. 3b).
Fig. 2.
Immunohistochemical staining of FI-expressing cells and breast cancer tissue. Upper left panels represent pellets of HEK293 cells transfected with empty vector or full-length human FI. The same conditions were used for staining of breast cancer TMAs, which showed FI presence in stroma (infiltrating immune cells and fibroblasts pointed out by red arrow; upper right part) and breast cancer tumor cells (bottom panel). Examples of scoring of FI intensity in tumor cells are given in the bottom panel
Fig. 3.
Detection of FI-specific RNA by RNAscope technique. a Validation of RNA probes on HEK293 cells transfected with empty vector or full-length human FI followed by paraffin embedding and treatment identical to breast cancer tissue. b Representative photographs of consecutive slides of breast cancer tissue stained with negative control RNA probe, positive control RNA probe and FI-specific probe. The same area of tumor tissue stained for FI with immunohistochemistry is given at the left side
FI in tumor tissue is associated with clinicopathological variables
We assessed the intensity of FI staining from score 0 to 3 and performed separate analysis for tumor cells and tumor stroma including infiltrating cells (Table 1). The obtained values were associated with available clinical data: age, tumor size, Nottingham histological grade (NHG), lymph node metastases, ER, PR, Her2 and Ki-67. We also set out for separate assessment of FI staining in inflammatory cells, but we have not found any correlations or associations with clinicopathological parameters. There was a statistically significant positive correlation between tumor size and FI staining intensity in tumor cells (p < 0.001) but not in stroma (p = 0.405). Analyses of tumor diameter medians showed that breast cancer cells expressing the highest levels of FI (score 3) formed the biggest tumors in vivo. Tumor-specific (p = 0.015), but not stromal (p = 0.326), FI expression was also associated with NHG. Since there were virtually no tumors sharing FI score 3 and the lowest possible NHG score means that good differentiation, tubule formation and low mitotic rates, the trend toward more advanced NHG within tumors expressing more FI was observed. We observed an association between tumor cell-specific and stromal FI expression with ER status (p = 0.030 and p = 0.004, respectively). Distribution of ER-positive tumors followed the distribution of all tumors within the FI expression grades, whereas the majority of ER-negative tumors were associated with high expression of FI (score 2 and 3). Similar statistically significant association was observed for PR status with p values 0.009 and 0.020, respectively. More PR-negative tumors were grouped in high FI grades comparing to PR-positive tumors. Stromal but not tumor-specific FI expression was significantly associated with Her2 status (p = 0.006); however, the total number of Her2-positive tumors was low thus questioning the confidence of our observation. FI expression was not associated with proliferation as determined by Ki-67 expression.
High expression of FI in tumor cells is associated with poor cancer-associated survival and recurrence-free survival
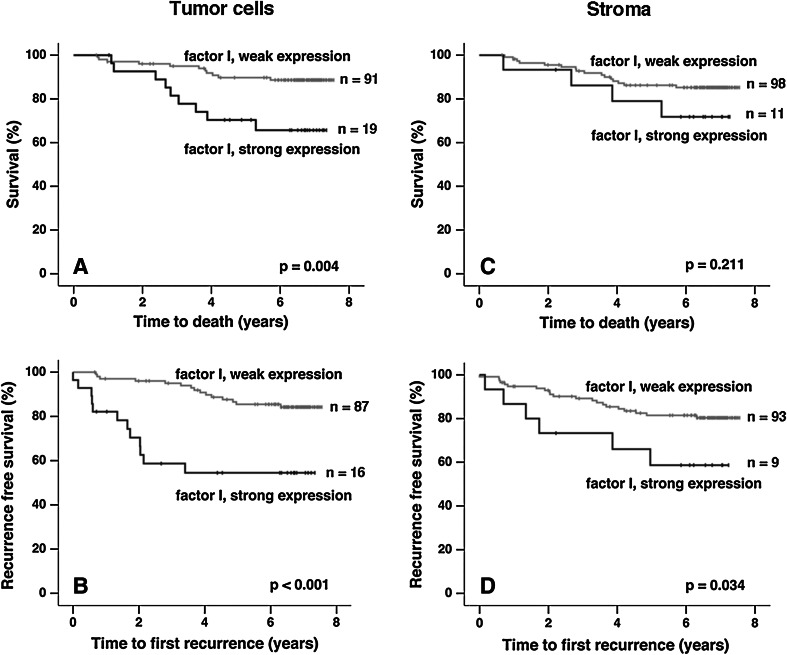
For survival analyses, a dichotomized variable was created, whereby tumors of FI intensity scores 0, 1 and 2 were denoted as having weak expression and tumors of intensity score 3 were considered as having strong FI expression. The rationale for this was a marked difference in FI staining intensity between score 2 and score 3 (see Fig. 2) and the fact that survival curves for patients with scores 0, 1 and 2 looked very similar (not shown). Kaplan–Meier analyses of cancer-specific survival (CSS) and recurrence-free survival (RFS) revealed statistically significant differences between patients bearing tumors with weak and strong expression of FI (p values 0.004 and < 0.001, respectively) (Fig. 4). Also, FI expression in stroma associated with RFS, but the p value was of lower confidence (p = 0.034) comparing to FI expression in tumor cells. Overall survival (not shown) in which all cases of death were taken into account was associated with FI expression in tumor cells (p = 0.02) but not in stroma (p = 0.383). Next, uni- and multivariate Cox regression analyses of CSS and RFS in relation to FI expression were performed using the same dichotomized variables as in the Kaplan–Meier analyses. The result revealed that strong expression of FI in tumor cells but not in stroma was an independent marker for CSS (p = 0.048; HR 2.8; 95 % CI 1.0–7.5) and RFS (p = 0.002; HR 3.4; 95 % CI 1.5–7.4) when the variables described in Table 2 were included. However, upon inclusion of nodal stage in the analysis, FI expression in tumor cells remained significant for RFS (p = 0.019; HR 2.8; 95 % CI 1.2–6.4) but not for CSS (p = 0.123; HR 2.5; 95 % CI 0.8–8.0). Details of Cox analyses for FI expression in tumor cells are reported in Table 2. Regarding FI expression in stroma, univariate Cox analysis resulted in no or weak statistical associations with CSS (p = 0.207; HR 2.0; 95 % CI 0.7–6.1) and RFS (p = 0.045; HR 2.5; 95 % CI 1.0–6.3).
Fig. 4.
Effect of weak versus strong FI expression on survival and recurrence. Kaplan–Meier estimates of cancer-specific survival (a, c) and recurrence-free survival (b, d) for weak (scores 0, 1, and 2) versus strong (score 3) FI expression in tumor cells and stroma. High expression of FI in tumor cells was significantly associated with shorter cancer-specific and recurrence-free survival. For high FI expression in stroma, only association with shorter recurrence-free survival was statistical significant
Table 2.
Univariate and multivariate Cox regression analyses of the impact on breast cancer-specific survival and recurrence-free survival for different patient and tumor characteristics
| Variable | Univariate | Multivariate | ||||||
|---|---|---|---|---|---|---|---|---|
| n | HR | 95 % CI | p | n | HR | 95 % CI | p | |
| Survival (CSS) | ||||||||
| FI (weak vs. strong) | 129 | 3.4 | 1.4–8.2 | 0.006 | 111 | 2.8 | 1.0–7.5 | 0.048 |
| ER (negative vs. positive) | 144 | 0.2 | 0.1–0.5 | 0.001 | 111 | 0.9 | 0.3–2.7 | 0.823 |
| HER2 (weak vs. strong) | 139 | 7.7 | 3.0–19.8 | <0.001 | 111 | 5.8 | 1.7–20.0 | 0.006 |
| NHG (1, 2, 3) | 144 | 5.1 | 2.0–13.0 | <0.001 | 111 | 1.1 | 0.3–4.0 | 0.923 |
| Size (≤20 vs. >20 mm) | 144 | 3.0 | 1.2–7.8 | 0.020 | 111 | 3.3 | 0.9–12.8 | 0.080 |
| Ki-67 (≤25 vs. >25 %) | 123 | 4.2 | 1.4–12.5 | 0.010 | 111 | 3.0 | 0.7–12.6 | 0.137 |
| Recurrence-free survival | ||||||||
| FI (weak vs. strong) | 130 | 4.4 | 2.0–9.4 | <0.001 | 127 | 3.4 | 1.5–7.4 | 0.002 |
| ER (negative vs. positive) | 144 | 0.3 | 0.1–0.6 | <0.001 | 127 | 0.8 | 0.3–2.0 | 0.577 |
| HER2 (weak vs. strong) | 139 | 7.1 | 3.0–16.9 | <0.001 | 127 | 5.7 | 2.3–14.5 | <0.001 |
| NHG (1, 2, 3) | 144 | 3.2 | 1.6–6.2 | <0.001 | 127 | 1.6 | 0.7–3.4 | 0.287 |
| Size (<20 vs. >20 mm) | 144 | 3.1 | 1.4–7.1 | 0.006 | 127 | 2.3 | 1.0–5.7 | 0.067 |
FI score was dichotomized into weak (value 0, 1, 2) and strong (3)
HR hazard ratio, CI confidence interval
Discussion
Our study contributes to the ongoing discussion about the exact role of complement in tumorigenesis. Although there are many reports supporting the theory of a dual role of complement in tumor growth (reviewed in [10]), only minor attention was devoted to soluble complement inhibitors. Of the existing studies, the majority relates to the main soluble inhibitor of the alternative complement pathway, factor H (FH). Experimental data show that FH expressed by tumor cells may influence tumor growth, as demonstrated in mouse xenograft experiments [5] and suggested by immunohistochemical staining of tumor tissue obtained from patients [30]. Interestingly, for our study, FH is one of the main cofactors required for the activity of FI. Of note, there is only one study showing FI production by tumor cells in vivo in cutaneous squamous cell carcinoma [16] where there was association of FI staining intensity with aggressive tumor phenotype [16]. Previously, we reported that non-small cell lung cancer (NSCLC) cell lines secrete several soluble complement inhibitors: FI and its cofactors FH and C4BP [15]. Although NSCLC cells strongly express membrane-bound complement inhibitors, we showed that addition of FI and its cofactors provides additional protection beyond the level attainable for membrane-bound inhibitors only [15]. Another interesting observation was that NSCLC cells seemed to purposely give up their protection from complement under hypoxic condition and this phenomenon remains in sharp contrast to endothelial cells [31]. These results underline the importance of soluble complement inhibitors as diffusible compounds, which may be used by tumor cells to actively control complement activation in their local microenvironment.
The expression of FI may face difficulties in cells other than hepatocytes due to insufficient cellular machinery for posttranslational processing, resulting in secretion of a precursor devoid of proteolytic activity [32]. Herein, we showed for the first time that breast cancer cell lines produce and secrete fully functional FI. This finding stimulated us to analyze whether FI is also expressed in breast cancer tissue in vivo and whether its expression associates with clinical outcome. Interestingly, we detected varying degrees of FI staining in ductal epithelial cells as well as in surrounding stroma already in the healthy breast tissue. Importantly, we found that this local expression varies in cancer development and that a high expression of FI in breast cancer cells correlates to a more aggressive phenotype including lower differentiation of tumor cells and larger tumor size. We also observed majority of ER and PR receptor-negative tumors to be distributed within high grades of FI expression, as compared to general FI grade distribution. Loss of ER and PR by breast cancer cells is an attribute of the basal molecular subtype, which is associated with poor prognosis [33] and demands harsh adjuvant chemotherapy. These associations may be reflected in the significantly shorter survival observed for patients with tumors expressing high levels of FI. Notably, our results regarding FI expression in breast cancer remain in line with the evidence from cell lines. Two out of four breast cancer cell lines identified in our study to express FI, namely MDA-MB-468 and MDA-MB-231 and these cell lines do not express ER and PR. Cell lines lacking FI expression, MCF-7 cells are ER positive, and T47D cells express both ER and PR.
There are several hypotheses why local production of soluble complement inhibitors may be important for tumor cells. In spite of the fact that these proteins are ubiquitous in the bloodstream (FI: 35 μg/ml, FH: 500 μg/ml, C4BP: 250 μg/ml), tumor vasculature is often malformed causing blood flow stagnation [34]. Reduced diffusion would also affect retention of complement components but neighboring cells may produce them instead. For example, adipocytes (usually adjacent to breast tissue) express three complement proteins: C3, factor B and protein D, which are sufficient for activation of the alternative complement pathway. Our current results are in agreement with findings showing that local production of FH favors tumor growth [5, 30]. Noteworthily, FI is a more universal inhibitor since it inactivates not only the alternative, but also the classical and lectin complement pathways. Another important observation from our study is the FI expression in tumor stroma, implying that not only tumor cells, but also tumor-associated fibroblasts and infiltrating immune cells may actively regulate local complement activation within tumor tissue. We revealed that the list of FI-expressing cells is longer than initially believed and a systematic study on FI expression is needed to clarify this issue.
Beside an obvious pro-tumor activity of endogenous complement inhibitors, there are also reports showing tumor-sustaining events driven by complement activation, such as mobilization of immune suppressor cells [6] and angiogenesis [7]. The reason why human immunohistochemical studies has so far only confirmed pro-tumorigenic effects of soluble complement inhibitors may be related to the fact that these studies enable insight only into outgrown tumors but miss the very first stages of tumor formation. It is highly possible that the same mechanisms, which protect outgrown tumors from the complement attack, must be suspended at the earlier stages of tumorigenesis. Keeping that in mind, we conclude that FI supports breast cancer at the certain stage when the tumor is palpable. However, one must be careful with extrapolation of our findings to the whole sequence of tumor growth. Conversion of surface-bound C3b into iC3b, an opsonin recognized more readily by CR3 expressing cells including phagocytes, T cells and NK cells, is another consequence of FI activity, yet unfavorable for tumor cells. However, inhibition of complement-mediated cell lysis by FI may be physiologically more relevant than activation of complement-dependent cell cytotoxicity (CDCC) by iC3b due to different accessibility of tumor cells for soluble components and CDCC effector cells. Moreover, iC3b stimulated CDCC may need additional priming, as reviewed in [35].
In conclusion, the results from this study demonstrate, for the first time, that FI is expressed in breast cancer cells in vivo and its expression correlates to clinical outcome. We not only noticed correlation of FI expression with tumor size and NHG or association with steroid receptor status, but also revealed the prognostic value of FI in tumor cells. When the metastases (nodal stage) are present, an obvious and exceptionally strong prognostic marker for survival and recurrence in breast cancer is excluded, tumor-specific FI expression acts as an independent marker for RFS and CSS. Even if nodal status is included, tumor-derived FI still independently predicted RFS in the current cohort. These findings underline the importance of soluble complement inhibitors in tumor progression.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We are grateful to Dr. Linda Werner-Hartman for help with statistical analyses, to Prof. Tommy Andersson for sharing breast cancer cell lines, Viktor Fransson for technical help with purification of FI antibodies and to Dr. Sara Nilsson for producing recombinant FI used for affinity purification of antibodies. This work was supported by the Swedish Cancer Society (to Anna Blom), the Swedish Research Council grants (K2012-66X-14928-09-5 to Anna Blom and K2010-80X-21514-01-4 to Marcin Okroj), grant for clinical research (ALF, to Anna Blom) and the Foundations of Österlund (to Anna Blom), Greta and Johan Kock (to Anna Blom), Malmo Cancer (to Marcin Okroj and Anna Blom), Tore Nilsson (to Marcin Okroj), Knut and Alice Wallenberg (to Anna Blom) and Inga-Britt and Arne Lundberg (to Anna Blom), as well as grants from the Skåne University Hospital (to Anna Blom and Marcin Okroj).
Conflict of interest
Authors declare no conflict of interest.
Abbreviations
- ATCC
American Type Culture Collection
- CDCC
Complement-dependent cell cytotoxicity
- CI
Confidence interval
- CSS
Cancer-specific survival
- C4BP
C4b-binding protein
- ER
Estrogen receptor
- FH
Complement factor H
- FI
Complement factor I
- Her2
Epidermal growth factor receptor 2
- HR
Hazard ratio
- MOPS
3-(N-morpholino)propanesulfonic acid
- NHG
Nottingham histological grade
- NSCLC
Non-small cell lung cancer
- PR
Progesterone receptor
- PVDF
Polyvinylidene difluoride
- RFS
Recurrence-free survival
- SDS–PAGE
Sodium dodecyl sulfate, polyacrylamide gel electrophoresis
- TMA
Tissue microarray
References
- 1.Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat Immunol. 2010;11:785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Okroj M, Osterborg A, Blom AM. Effector mechanisms of anti-CD20 monoclonal antibodies in B cell malignancies. Cancer Treat Rev. 2013;39:632–639. doi: 10.1016/j.ctrv.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 3.Jurianz K, Ziegler S, Garcia-Schuler H, Kraus S, Bohana-Kashtan O, Fishelson Z, Kirschfink M. Complement resistance of tumor cells: basal and induced mechanisms. Mol Immunol. 1999;36:929–939. doi: 10.1016/S0161-5890(99)00115-7. [DOI] [PubMed] [Google Scholar]
- 4.Mark L, Lee WH, Spiller OB, Proctor D, Blackbourn DJ, Villoutreix BO, Blom AM. The Kaposi’s sarcoma-associated herpesvirus complement control protein mimics human molecular mechanisms for inhibition of the complement system. J Biol Chem. 2004;279:45093–45101. doi: 10.1074/jbc.M407558200. [DOI] [PubMed] [Google Scholar]
- 5.Ajona D, Hsu YF, Corrales L, Montuenga LM, Pio R. Down-regulation of human complement factor H sensitizes non-small cell lung cancer cells to complement attack and reduces in vivo tumor growth. J Immunol. 2007;178:5991–5998. doi: 10.4049/jimmunol.178.9.5991. [DOI] [PubMed] [Google Scholar]
- 6.Markiewski MM, DeAngelis RA, Benencia F, Ricklin-Lichtsteiner SK, Koutoulaki A, Gerard C, Coukos G, Lambris JD. Modulation of the antitumor immune response by complement. Nat Immunol. 2008;9:1225–1235. doi: 10.1038/ni.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corrales L, Ajona D, Rafail S, Lasarte JJ, Riezu-Boj JI, Lambris JD, Rouzaut A, Pajares MJ, Montuenga LM, Pio R. Anaphylatoxin c5a creates a favorable microenvironment for lung cancer progression. J Immunol. 2012;189:4674–4683. doi: 10.4049/jimmunol.1201654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlaf G, Demberg T, Beisel N, Schieferdecker HL, Gotze O. Expression and regulation of complement factors H and I in rat and human cells: some critical notes. Mol Immunol. 2001;38:231–239. doi: 10.1016/S0161-5890(01)00045-1. [DOI] [PubMed] [Google Scholar]
- 9.Holers VM. Complement and its receptors: new insights into human disease. Ann Rev Immunol. 2014;32:433–459. doi: 10.1146/annurev-immunol-032713-120154. [DOI] [PubMed] [Google Scholar]
- 10.Stover C. Dual role of complement in tumour growth and metastasis (review) Int J Mol Med. 2010;25:307–313. doi: 10.3892/ijmm_00000346. [DOI] [PubMed] [Google Scholar]
- 11.Minta JO, Fung M, Paramaswara B. Transcriptional and post-transcriptional regulation of complement factor I (CFI) gene expression in Hep G2 cells by interleukin-6. Biochim Biophys Acta. 1998;1442:286–295. doi: 10.1016/S0167-4781(98)00189-4. [DOI] [PubMed] [Google Scholar]
- 12.Vetvicka V, Reed W, Hoover ML, Ross GD. Complement factors H and I synthesized by B cell lines function to generate a growth factor activity from C3. J Immunol. 1993;150:4052–4060. [PubMed] [Google Scholar]
- 13.Gasque P, Julen N, Ischenko AM, Picot C, Mauger C, Chauzy C, Ripoche J, Fontaine M. Expression of complement components of the alternative pathway by glioma cell lines. J Immunol. 1992;149:1381–1387. [PubMed] [Google Scholar]
- 14.Legoedec J, Gasque P, Jeanne JF, Fontaine M. Expression of the complement alternative pathway by human myoblasts in vitro: biosynthesis of C3, factor B, factor H and factor I. Eur J Immunol. 1995;25:3460–3466. doi: 10.1002/eji.1830251238. [DOI] [PubMed] [Google Scholar]
- 15.Okroj M, Hsu YF, Ajona D, Pio R, Blom AM. Non-small cell lung cancer cells produce a functional set of complement factor I and its soluble cofactors. Mol Immunol. 2008;45:169–179. doi: 10.1016/j.molimm.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 16.Riihila P, Nissinen L, Farshchian M, Kivisaari A, Ala-Aho R, Kallajoki M, Grenman R, Meri S, Peltonen S, Peltonen J, Kahari VM. Complement factor I promotes progression of cutaneous squamous cell carcinoma. J Invest Dermatol. 2015;135:579–588. doi: 10.1038/jid.2014.376. [DOI] [PubMed] [Google Scholar]
- 17.Nilsson SC, Karpman D, Vaziri-Sani F, Kristoffersson AC, Salomon R, Provot F, Fremeaux-Bacchi V, Trouw LA, Blom AM. A mutation in factor I that is associated with atypical hemolytic uremic syndrome does not affect the function of factor I in complement regulation. Mol Immunol. 2007;44:1835–1844. doi: 10.1016/j.molimm.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 18.Malm S, Jusko M, Eick S, Potempa J, Riesbeck K, Blom AM. Acquisition of complement inhibitor serine protease factor I and its cofactors C4b-binding protein and factor H by Prevotella intermedia. PLoS ONE. 2012;7:e34852. doi: 10.1371/journal.pone.0034852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nilsson SC, Kalchishkova N, Trouw LA, Fremeaux-Bacchi V, Villoutreix BO, Blom AM. Mutations in complement factor I as found in atypical hemolytic uremic syndrome lead to either altered secretion or altered function of factor I. Eur J Immunol. 2010;40:172–185. doi: 10.1002/eji.200939280. [DOI] [PubMed] [Google Scholar]
- 20.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nilsson SC, Blom AM. Purification and functional characterization of factor I. Methods Mol Biol. 2014;1100:177–188. doi: 10.1007/978-1-62703-724-2_15. [DOI] [PubMed] [Google Scholar]
- 22.Greenwood FC, Hunter WM, Glover JS. The preparation of I-131-labelled human growth hormone of high specific radioactivity. Biochem J. 1963;89:114–123. doi: 10.1042/bj0890114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohlin FC, Blom AM. Purification and functional characterization of C4b-binding protein (C4BP) Methods Mol Biol. 2014;1100:169–176. doi: 10.1007/978-1-62703-724-2_14. [DOI] [PubMed] [Google Scholar]
- 24.Helczynska K, Larsson AM, Mengelbier LH, Bridges E, Fredlund E, Borgquist S, Landberg G, Pahlman S, Jirstrom K. Hypoxia-inducible factor-2alpha correlates to distant recurrence and poor outcome in invasive breast cancer. Cancer Res. 2008;68:9212–9220. doi: 10.1158/0008-5472.CAN-08-1135. [DOI] [PubMed] [Google Scholar]
- 25.Svensson KJ, Christianson HC, Kucharzewska P, Fagerstrom V, Lundstedt L, Borgquist S, Jirstrom K, Belting M. Chondroitin sulfate expression predicts poor outcome in breast cancer. Int J Oncol. 2011;39:1421–1428. doi: 10.3892/ijo.2011.1164. [DOI] [PubMed] [Google Scholar]
- 26.Borgquist S, Holm C, Stendahl M, Anagnostaki L, Landberg G, Jirstrom K. Oestrogen receptors alpha and beta show different associations to clinicopathological parameters and their co-expression might predict a better response to endocrine treatment in breast cancer. J Clin Pathol. 2008;61:197–203. doi: 10.1136/jcp.2006.040378. [DOI] [PubMed] [Google Scholar]
- 27.Ryden L, Jirstrom K, Bendahl PO, Ferno M, Nordenskjold B, Stal O, Thorstenson S, Jonsson PE, Landberg G. Tumor-specific expression of vascular endothelial growth factor receptor 2 but not vascular endothelial growth factor or human epidermal growth factor receptor 2 is associated with impaired response to adjuvant tamoxifen in premenopausal breast cancer. J Clin Oncol. 2005;23:4695–4704. doi: 10.1200/JCO.2005.08.126. [DOI] [PubMed] [Google Scholar]
- 28.Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, Wu X, Vo HT, Ma XJ, Luo Y. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blom AM, Kask L, Dahlbäck B. CCP1-4 of the C4b-binding protein a-chain are required for factor I mediated cleavage of C3b. Mol Immunol. 2003;39:547–556. doi: 10.1016/S0161-5890(02)00213-4. [DOI] [PubMed] [Google Scholar]
- 30.Cui T, Chen Y, Knösel T, Yang L, Zöller K, Galler K, Berndt A, Mihlan M, Zipfel PF, Petersen I. Human complement factor H is a novel diagnostic marker for lung adenocarcinoma. Int J Oncol. 2011;39:161–168. doi: 10.3892/ijo.2011.1010. [DOI] [PubMed] [Google Scholar]
- 31.Okroj M, Corrales L, Stokowska A, Pio R, Blom AM. Hypoxia increases susceptibility of non-small cell lung cancer cells to complement attack. Cancer Immunol Immunother. 2009;58:1771–1880. doi: 10.1007/s00262-009-0685-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ullman CG, Chamberlain D, Ansari A, Emery VC, Haris PI, Sim RB, Perkins SJ. Human complement factor I: its expression by insect cells and its biochemical and structural characterisation. Mol Immunol. 1998;35:503–512. doi: 10.1016/S0161-5890(98)00052-2. [DOI] [PubMed] [Google Scholar]
- 33.O’Brien KM, Cole SR, Tse CK, Perou CM, Carey LA, Foulkes WD, Dressler LG, Geradts J, Millikan RC. Intrinsic breast tumor subtypes, race, and long-term survival in the Carolina Breast Cancer Study. Clin Cancer Res. 2010;16:6100–6110. doi: 10.1158/1078-0432.CCR-10-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munn LL. Aberrant vascular architecture in tumors and its importance in drug-based therapies. Drug Discov Today. 2003;8:396–403. doi: 10.1016/S1359-6446(03)02686-2. [DOI] [PubMed] [Google Scholar]
- 35.Ross GD, Vĕtvicka V. CR3 (CD11b, CD18): a phagocyte and NK cell membrane receptor with multiple ligand specificities and functions. Clin Exp Immunol. 1993;92:181–184. doi: 10.1111/j.1365-2249.1993.tb03377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.