Abstract
Natural killer (NK) cells are considered crucial for the elimination of emerging tumor cells. Effector NK-cell functions are controlled by interactions of inhibitory and activating killer-cell immunoglobulin-like receptors (KIRs) on NK cells with human leukocyte antigen (HLA) class I ligands on target cells. KIR and HLA are highly polymorphic genetic systems segregating independently, creating a great diversity in KIR/HLA gene profiles in different individuals. There is an increasing evidence supporting the relevance of KIR and HLA ligand gene background for the occurrence and outcome of certain cancers. However, the data are still controversial and the mechanisms of receptor-ligand mediated NK-cell action remain unclear. Here, the main characteristics and functions of KIRs and their HLA class I ligands are reviewed. In addition, we review the HLA and KIR correlations with different hematological malignancies and discuss our current understanding of the biological significance and mechanisms underlying these associations.
Keywords: NK cells, Killer immunoglobulin-like receptors, HLA ligands, Hematological malignancies
NK cells as a major player in innate immunity
Natural killer cells (NK) are a bone marrow-derived lymphoid population, constituting 10–20 % of all circulating lymphocytes [1]. They have important immunoregulatory roles in the natural immune response and protection against virus-infected and tumor-transformed cells [1, 2]. Recently, by grouping all innate lymphocytes in the family of “innate lymphoid cells—ILCs”, NK cells are assigned as “cytotoxic” ILCs [3, 4]. In many respects they are similar to cytotoxic T lymphocytes and may be considered as their innate counterparts. However, while cytotoxic T lymphocyte (CTL) responses require presentation of peptides by MHC class I molecules on antigen-presenting cells, the recognition of targets (stressed or transformed cells) by NK cells is not necessarily associated with HLA molecules, and therefore, it was assumed that NK cell-mediated cytotoxicity is not MHC restricted. However, more recent studies on NK cell receptors, their specific ligands, signal transduction mechanisms and functional activity have led to a reevaluation of this concept. Today’s understanding of NK-mediated cytotoxicity is based on the “missing self” hypothesis proposed by Kärre et al. [5] which explains the activation of NK cells through a simple mechanism: the lack of MHC class I expression on the target cells. According to this concept, NK and CTL are two complementary arms in the effector immune response, in which both cell types are MHC-dependent but act through different mechanisms. Unlike T and B cells, NK cells do not have receptors for specific recognition of foreign antigens. Their ability to distinguish autologous MHC class I or class I-like molecules is due to a unique class of NK-cell receptors with inhibitory and activating functions [6]. The condition for NK-cell tolerance assumes that each NK cell must carry at least one functional dominant inhibitory receptor for self-MHC molecules, protecting from autoreactivity under normal circumstances, proven by studies at clonal and polyclonal levels [7, 8]. Currently, it is accepted that NK cells recognize virus infected, neoplastic or allogeneic cells with altered expression of the corresponding MHC class I ligand [9]. The model of “missing self” does not fully explain the resistance of some normal cells with low/missing expression of MHC molecules to attack by NK cells and the mechanisms of NK-cell activation [10]. An alternative idea was proposed for “induced-self recognition”. According to this hypothesis, NK cells possess activating receptors recognizing molecules (ligands) that are not expressed by most normal cells but are displayed by infected or transformed cells, as confirmed in subsequent studies [11, 12]. Similar to the adaptive immune system, the innate immune system is able to differentiate “self” from “non-self” (foreign) or impaired self by finely regulated mechanisms involving cell surface receptors and MHC molecules, and the balance between activating and inhibitory receptors [12].
Currently, various surface receptors regulating NK-cell function known in humans are divided into the following groups: type I integral proteins with immunoglobulin-like domains (killer-cell immunoglobulin-like receptors—KIRs; immunoglobulin-like transcripts—ILTs), type II integral proteins with lectin-like domains (NKG2D, CD94/NKG2) and natural cytotoxicity receptors (NCR) [13–18]. The effector function of all these receptors is the result of complex interactions and the balance between activating and inhibitory signals driven by their specific binding to the corresponding ligands.
NK-cell receptors from the immunoglobulin superfamily
In humans, the major receptor type involved in NK-cell-mediated HLA class I recognition is encoded by a family of genes spanning 150 Kb within the leukocyte receptor complex located on chromosome 19q13.4 and designated “killer-cell immunoglobulin-like receptor” genes [14]. Currently KIR gene family includes 15 loci (KIR2DL1, KIR2DL2/L3, KIR2DL4, KIR2DL5A, KIR2DL5B, KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS4, KIR2DS5, KIR3DL1/S1, KIR3DL2, KIR3DL3 and two pseudogenes, KIR2DP1 and KIR3DP1) [15]. Each KIR gene has between 15 (KIR2DS3) and 112 (KIR3LD2) alleles [15] which further contributes to the genetic variety of these receptors. It is not yet clear if some of the originally described KIR genes are alleles of the same gene. At present, family segregation analysis and genomic sequencing revealed two pairs of KIR genes, KIR2DL2/KIR2DL3 and KIR3DL1/KIR3DS1 considered to segregates as alleles [16, 19, 20].
KIR gene products are type I transmembrane glycoproteins expressed on the surface of the majority of NK cells and some T-cell clones. According to the structural characteristics of the encoded proteins corresponding to their functions, there are two functionally different groups of receptors: inhibitory (iKIRs) and activating (aKIRs). KIRs are composed of two (2D) or three (3D) extracellular Ig-like domains and a cytoplasmic part that varies in length and accordingly is designated long (L) or short (S) tailed [15]. The length of the intracytoplasmic part determines the function of the appropriate receptor with some exceptions and particularly KIR2DL4. The presence of a long cytoplasmic tail with one or two immunoreceptor tyrosine-based inhibitory motifs (ITIMs) that bind to phosphatases SHP allows transduction of inhibitory signals and determines their inhibitory function in cellular cytotoxicity. Receptors with short cytoplasmic tails do not possess ITIMs but have a positively charged amino acid residue, lysine, in the transmembrane domain allowing their association with activating adapter proteins, such as DAP12, and defining them as activating receptors [21]. One KIR with a long cytoplasmic tail, KIR2DL4, containing an ITIM motif on the basis of which it was classified into the group of inhibitory receptors, binds to the activation motif of FcεRIγ and thus seems to transmit activating signals as well [22, 23].
The KIR genes in humans form two haplotypes, A and B, that differ in the number and types of genes [24]. Group A haplotypes contain up to 7 genes (KIR2DL1, KIR2DL3, KIR3DL1, KIR3DL2, KIR3DL3, KIR2DL4, KIR2DS4) most of them encoding receptors with inhibitory capacity, and KIR2DS4 and KIR2DL4 as the only activating KIRs. Additionally, they are characterized by small variations in the number of gene contained, but these also have significant allelic polymorphism. It is interesting to note that many A haplotypes carry null KIR2DS4 and KIR2DL4 variants that are not expressed on the cell surface [23, 25]. In fact, such haplotypes should not have functional activating KIRs. Group B haplotypes are characterized by a great diversity in both the number and the allelic variations of the genes. In addition to the framework genes (KIR2DL4, KIR3DL2, KIR3DL3), KIRs have one or more B haplotype-specific activating (KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS5, KIR3DS1) and inhibitory (KIR2DL2, KIR2DL5A, KIR2DL5B) genes. KIR haplotype zygosity determines the number of inhibitory and activating genes of an individual. Concerning KIR gene expression, it should be taken into account that it is the result of a complex process related to different factors. First, expression depends on the inherited KIR gene content and haplotypes. Second, the proportion of KIR-positive NK cells and the cell surface density of a given receptor are modulated by KIR allelic polymorphism which is well illustrated by expression patterns of KIR3DL1 allotypes. Using DX9 antibody staining, Gumperz et al. [26] demonstrated high expression of KIR3DL1*001, *002, *003 and *008, low surface levels of KIR3DL1*005, *006 and *007, and no expression of KIR3DL1*004 alleles. In addition, the correlation between KIR genotype polymorphism and phenotype variations within individuals was further compounded at the population level. Yawata et al. [27] showed that five common KIR3DL1 allotypes in Japanese have differences in their cellular expression with the following order of magnitude: KIR3DL1*005 < *007 < *001 < *020 < *01502, and a gene dose effect was also seen. In contrast, the non-functional 3DL1*004 allele common in Caucasians, Africans and South Asians is not represented in the Japanese population [27–30]. It should also be considered that the KIR2DS4 variant with deletion (KIR2DS4del) and many KIR2DL4 allele gene products are also not displayed on the cell surface [25, 31]. When cognate MHC class I KIR ligands are present, the amount of NK cells expressing the appropriate KIR is increased, while the frequency of NK cells displaying mismatched KIRs is decreased [27]. Finally, NK cells may express one or several KIR genes and each individual cell clone express only some of the inherited KIR genes on a relatively stochastic principle. This may lead to a structurally and functionally distinct NK-cell repertoire. Important features of KIR distribution patterns are their stable expression maintained upon expansion of mature NK-cell clones and also dynamics related to the acquisition of new receptors over time [7, 32]. This clonally restricted KIR expression pattern is partly due to gene silencing by DNA methylation [33, 34]. Although KIR2DL4 expression is also controlled by DNA methylation, it seems to be an exception compared to other receptors. Unlike the allele-specific random distribution patterns of all other KIRs, KIR2DL4 is biallelically expressed by most or all NK cells [35–37].
KIR ligands
The function of NK cells is modulated by interactions of their specific receptors with corresponding ligands expressed on target cells. HLA-C molecules are the main MHC class I ligands influencing the NK-cell activity by binding to inhibitory receptors with two immunoglobulin domains (KIR2D). The HLA-C recognition is restricted by two epitopes in the molecule defined by a dimorphism at positions 77 and 80 in the α1 helix of the heavy chain of MHC class I. Group 1 HLA-C (C1) ligands include allele-encoded molecules with serine and asparagine (Ser77 and Asn80), while group 2 HLA-C (C2) ligands are characterized by allele-encoded molecules with asparagine and lysine (Asn77 and Lys80) at these positions [38]. Group 1 epitopes bind specifically to the inhibitory KIR2DL2/3, while C2 are ligands for inhibitory KIR2DL1 [39, 40]. Recent data from in vitro studies suggest that KIR2DL2/2DL3 may also bind certain HLA-C2 epitopes (C*05:01, C*02:02, C*04:01) and some HLA-B allotypes that share polymorphisms with HLA-C (HLA-B*46:01, B*73:01) but with very low affinity [41, 42].
There is a hierarchy in the strength of inhibitory signals transmitted after interactions between KIRs and their cognate HLA-C ligands. The binding of C2 ligands with KIR2DL1 results in a strong inhibition, whereas C1 interaction with KIR2DL2 and KIR2DL3 generates intermediate and weak inhibitory signals, respectively [41, 43]. HLA-B allotypes with Bw4 motif at position 77–83 are ligands for the inhibitory KIR3DL1 [44, 45]. The binding affinity is more potent for HLA-Bw4 molecules having amino acid isoleucine at position 80 (Ile80) compared to those with threonine (Thr80) in the same position [44, 46]. Studies have shown that KIR3DL1 recognizes HLA-Bw4 alleles with the exception of B*13:01 and B*13:02, and some HLA-A alleles with Bw4 motifs (A*23:01, A*24:02 and A*32:01) [47, 48]. There is an evidence showing that polymorphisms in KIRs influence the recognition of HLA molecules, and vice versa; polymorphisms in HLA epitopes affect the binding to their specific receptors. For example, the KIR3DL1*005 allele recognizes HLA-A*24:02 [48] and KIR3DL1*002 is a stronger inhibitory receptor for Bw4 ligands in comparison with KIR3DL1*007 [49]. Moreover, KIR3DL1 binding to B*51:01 provides a strong inhibitory signal, followed by B*38:01 and B*58:01, while the receptor binding with B*27:05 generates weak inhibition [50]. Although HLA-B*27:05 appears to be a weak ligand for KIR3DL1 allotypes, its interaction with KIR3DL1*002 is much stronger compared to KIR3DL1*001 and the opposite is true for B*58:01 [50]. KIR3DL2 recognizes specifically HLA-A3, HLA-A11 [51–53]. Data on the KIR2DL4 ligands are still controversial. Several studies provided evidence that KIR2DL4 binds nonclassical MHC class I HLA-G molecules, but other researchers failed to detect such interaction [54–58].
The ligand specificity for activating KIRs is not completely determined. The activating receptor KIR2DS1 shares common sequences in extracellular domains with the corresponding inhibitory analog KIR2DL1, and therefore it was assumed that KIR2DS1 binds to the same HLA ligand specificities as KIR2DL1 but with lower affinity [59–61]. This observation is supported by the finding of weak interaction between KIR2DS1 and HLA-C2 allotypes [62]. Experimental data suggest that KIR2DS4 reacts with group C2 HLA-Cw4 allele-encoded molecules (HLA-C*05:01, *16:01) and HLA-A*11:02, and possibly with non-HLA ligands on melanoma cells [63–65]. The activating KIR3DS1 shares more than 95 % homology in the extracellular domain with its inhibitory counterpart KIR3DL1 and may also be implicated in Bw4 recognition [29, 66]. Although there is no direct evidence for interactions of KIR3DS1 with Bw4 allotype, some studies provide evidence that HLA-B*27:05 is a potential ligand for KIR3DS1 [67–69]. The ligand specificity of activating KIR2DS2, KIR2DS3 and KIR2DS5 remains unknown, but some authors suggest that they recognize non-HLA ligands [70–73]. Concerning KIR2DS2, selected publications reveal that it binds group 1 HLA-C allele-encoded products [42] and HLA-A*11 [74], but other studies failed to demonstrate interactions between KIR2DS2 and HLA molecules (reviewed in [75]).
It should also be pointed out that self-MHC class I ligands are involved in the educational process during NK-cell development being referred to as “arming” or “licensing” [76–79]. This adaptation of NK reactivity through finely regulated processes of KIR–HLA ligand interactions aims to ensure the self-tolerance and functional competence of NK cells [77, 80, 81]. In addition, NK-cell education can be tuned in a quantitative dynamic way. Thus, NK cells expressing iKIRs recognizing self-MHC molecules will be “educated” to stronger inhibition against autologous targets [82]. On the other hand, such NK cells will be capable of more potent cytotoxic responses when stimulated through activating receptors in comparison with NK cells expressing one of these lacking a self-recognizing iKIR (“unlicensed” NK cells) [83].
The fact that KIR, which is located on chromosome 19q13.4 and HLA, which is on chromosome 6q21.3, is highly polymorphic gene systems inherited independently, leads to a great diversity in both the number and type of KIR/HLA pairs in different individuals. NK-cell receptor-ligand recognition can vary at many different levels: KIR allelic polymorphism, KIR haplotype, HLA-B and C ligand polymorphism and variation in the clonal expression of genes. The proportion between activating (2DS, 3DS) and inhibitory (2DL, 3DL) receptors, the binding strength with the corresponding ligands and the balance between the generated signals, determine the behavior of NK cells in health and disease.
KIR genes and susceptibility to hematological malignancies
Similarly to other neoplastic diseases, the exact etiopathogenic mechanisms triggering the transformation of “normal” hematopoietic cells in blast proliferation has not been elucidated. The expansion of neoplastic cells and development of a hematologic malignancy are highly dependent on ability or failure of the immune system to attack blast cells. The anti-tumor potential of NK cells, originally established in “in vitro” studies, suggests their essential role in the anti-leukemic immune response [84, 85]. Important questions remain regarding whether or not immune genetic factors such as inherited KIRs, HLA ligands and KIR/HLA ligand profiles may influence the susceptibility to hemoblastosis. Evidence for associations of the underlying KIRs and KIR ligand repertoire with anti-leukemic effects of NK cells was first provided in allogeneic hematopoietic stem cell transplantation (HSCT) [86]. Experience to date has demonstrated that NK-receptor incompatibility in the graft-versus-host direction (recipient with missing HLA class I ligand for donor inhibitory KIRs) is associated with lower incidence of leukemia relapse and graft-versus-host disease [86–90], leading to improved survival and prognosis in transplant patients [91–93]. As a result, investigations have been focused on the search for associations between KIR and HLA ligand polymorphism with the development and progression of hematological malignancies. Activating KIR2DS3 was reported as a protective factor in three independent studies dealing with acute lymphoblastic leukemia (ALL) [94], acute myeloid leukemia (AML) [95] and chronic myeloid leukemia (CML) [96]. In a disease association study of individuals from the Bulgarian population with three subgroups of hemoblastosis (ALL, AML and CLL), we have demonstrated a lower frequency of KIR3DS1 in AML patients compared to the healthy control group (17.6 vs. 47.6 %, p = 0.02; OR 0.2, 95 % CI 0.07–0.9) [97]. Our data are supported by the trend toward association of KIR3DS1 with reduced risk of AML observed in Iranian patients [95]. However, studies in Han Chinese [98], Thai [99] and Belgian [100] patients did not establish significant relationship between KIR3DS1 and AML. Family studies on patients with Hodgkin’s lymphoma show a dominant protective effect of the activating KIR3DS1 and/or KIR2DS1 for disease development and progression, which is more strongly pronounced in EBV-positive patients [101]. Another activating KIR was also found to have a protective effect for leukemia development—KIR2DS2 in CML [102]. It is interesting to note that in allogeneic HSCT, a higher survival rate is associated with donor positivity for the activating KIR3DS1 [103] and KIR2DS2 [104] due to a reduced risk of disease recurrence. In contrast to the abovementioned observations, a higher frequency of the activating KIR2DS4 among patients with CML that was expected to be associated with higher anti-tumor activity is reported by Zhang et al. [94]. At the population level, the KIR2DS4del variant encoding functionally inactive receptor is more common. However, in the study mentioned above, no segregation analysis was performed and the impact of allele variants could not be assessed. Giebel et al. [105] have reported that there is a higher frequency of KIR2DS4del(+)/2DS4norm(−) in CML, demonstrating that at the gene level KIR2DS4 may be associated with increased risk for hematologic cancers, but at the allelic and functional levels this seems not to be the case. Based on the abovementioned observations, it could be assumed that the dominant prevalence of activating KIR genes may be the genetic basis for high NK-cell cytotoxicity and effective anti-tumor defense of the organism. On the other hand, it could be suggested that increased frequency of inhibitory KIRs may contribute to escape of malignant cells from immune attack. However, the above hypothesis cannot explain the decreased frequency of inhibitory KIR2DL5 logically associated with a lower inhibitory NK-cell capacity, as established by two independent research groups: Karabon et al. [96] in B-cell chronic lymphocytic leukemia (B-CLL) and Besson et al. [101] for Hodgkin’s disease. Our data also showed reduced incidence of KIR2DL5A variants in AML patients (17.6 vs. 47.7 %, p = 0.02, OR 0.2, 95 % CI 0.06–0.9) compared to healthy controls [97]. On the other hand, Du et al. [106] demonstrated that certain KIR2DL5 alleles are associated with KIR genotypes containing a lower number of activating receptors. Therefore, the low frequency of KIR2DL5A in AML and B-CLL may be considered as an indicator of lower activation rather than reduced inhibition. In contrast, it was reported that NK-cell leukemia with large granular lymphocyte (LGL) morphology was associated with an increased frequency of inhibitory KIR2DL5A and 2DL5B [107]. Recently, the presence of KIR2DL5A was found to correlate positively with minimal residual disease in ALL pediatric patients [108]. Interestingly, an anti-leukemic effect of this receptor was indirectly proven through clinically significant reduced risk of AML relapse after allogeneic stem cell transplantation from KIR2DL5A donors [103]. Other observations which do not support the concept of decreased activation related to lower incidence of KIR2DL2 were reported by Middleton et al. [102] and Verheyden et al. [100] who found higher frequency of KIR2DS2 in patients with leukemia (combined group of AML, CML, ALL and CLL). In both cases, it was assumed that the findings may be related to the well-documented strong positive linkage disequilibrium between KIR2DL2 and KIR2DS2 on the one hand and to not fully clarified ligand specificity of the activating receptor—KIR2DS2 on the other hand, making the interpretation of the results difficult. Additionally, Middleton et al. [102] suggested a possible association of the reduced KIR2DL2 frequency with lower incidence of KIRB haplotypes and correspondingly higher frequency of KIRAA which are preferentially inhibitory. The data for KIR genetic associations with the development of ALL in children are also conflicting. Babor et al. [109], for example, did not find any association of ALL in pediatric patients with any of the known inhibitory or activating KIRs, while Almalte et al. [110] established a significant reduction in the risk of childhood ALL with increasing cumulative sum of the activating KIR genes.
When assessing the impact of KIR genes as immunogenetic factors involved in hemoblastogenesis, it is of essential importance to consider the context of their related inheritance in haplo- and genotypes, which in turn defines the ratio between inhibitory and activating receptors. Data in support of this come from significant associations of two specific KIR phenotypes, AB1 and AB9 including all inhibitory and the lowest number of activating KIRs, with acute and chronic myeloid leukemia, respectively [100]. Thus, Verheyden et al. [100] were the first to demonstrate associations of higher number of inhibitory KIRs with leukemia. More recent studies have found a tendency for prevalence of inhibitory over activating KIR genes in patients with AML [95]. Similarly, Karabon et al. [96] demonstrated significantly increased frequency of genotypes with dominant inhibitory potential determined by the ratio between activating and inhibitory KIRs in B-CLL. Using a model of analysis analogous to that of Karabon et al. [96], we found two profiles that differentiate leukemic patients from healthy controls—higher frequency of KIR profile with value of 0.57 (4 aKIRs/7 iKIRs) and lower incidence of KIR ratio 0.66 (4 aKIRs/6 iKIRs) (Table 1). It is noteworthy that both KIR genotypes consist of equal numbers of activating but different numbers of inhibitory KIRs, which implies a stronger contribution of inhibitory activity to the risk of disease, compared to the protective KIR genotype. Additionally, KIR genotypes analysis based on the number of activating KIR genes revealed that KIR profiles with lower NK activating potential (composed of two activating KIRs) may be a risk for leukemia development [97]. The most recent findings demonstrated that KIR gene profiles may be predictive for treatment-free remission of CML patients. Thus, KIR A haplotype homozygosity and decreased inhibitory KIR2DL2 gene frequency were associated with achievement of complete molecular remission after tyrosine kinase inhibitor therapy, while the Bx haplotype may be indicative for leukemia relapse [111, 112]. In contrast, an evident beneficial survival effect was noticed in AML patients transplanted from unrelated donors bearing KIR B haplotypes in comparison with A haplotype homozygous donors [87, 88, 113]. Based on this finding, Cooley et al. [88] developed a donor selection algorithm that could be applied as a strategy for improving transplant outcome. The donor KIR B-content groups may be now easily defined as “best”, “better” or “neutral” using IPD-KIR [15].
Table 1.
KIR genotypes in combined group of patients with ALL, AML and CML and randomly selected healthy controls from the Bulgarian population
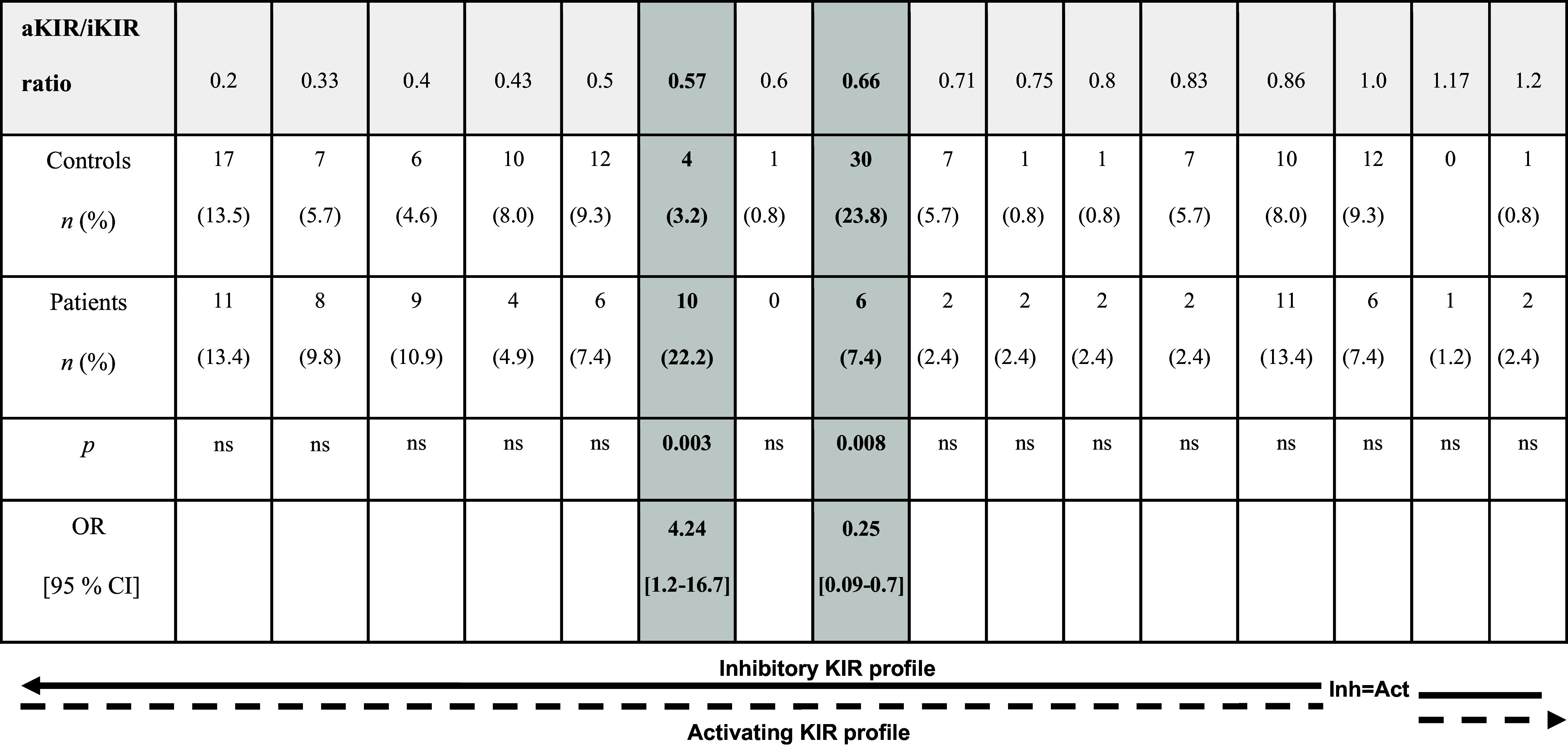
The genotypes were determined as ratios between the number of activating (aKIR) and inhibitory (iKIR) receptors [96]
Inhibitory KIR genes were counted, including framework KIR genes (KIR3DL2, 3DL3), and at least one other inhibitory KIR gene: KIR2DL1, 2DL2, 2DL3, 2DL5, 3DL1. Therefore, the minimum number of inhibitory KIR genes was 3 and the maximum was 7 (all inhibitory KIR genes present). From activating KIR genes, the deleted form of KIR2DS4 was excluded while KIR2DL4 was assigned as activating, although its function is still controversial
ALL acute lymphoblastic leukemia, AML acute myeloid leukemia, CLL chronic lymphocytic leukemia, CML chronic myeloid leukemia
De Smith et al. [114] demonstrated ethnic differences in the susceptibility to hemoblastosis, presented by significant associations between KIR A/A genotype, and between numbers of inhibitory and activating KIR genes as well as with childhood ALL in Hispanic but not in non-Hispanic patients. It is intriguing to hypothesize that an important factor in the etiology of ALL may be due to population-specific genetic variants in genes controlling the immune response. However, the authors suggest that the typical findings for Hispanic pediatric ALL could be related to environmental factors interacting with KIRs (e.g., types of infection) rather than to allelic differences between Hispanics and non-Hispanic whites.
Unlike the data on leukemia, studies in Lebanese patients with folicular lymphoma [115] and Hodgkin’s disease [116], and diffuse large B-cell lymphoma in Thai patients [99] showed no associations between KIR genotypes and lymphomas. Some data demonstrate that the effect of KIR genetic factors is not restricted to a specific nosological entity and is relevant for the development of a heterogeneous group of malignant diseases (Table 2). However, the results of some studies provide evidence for a weak anti-leukemic effects of NK cells against lymphoblasts compared to myeloblasts [117, 118]. Compiling available literature, it can be summarized that more reports support the hypothesis for dominant prevalence of inhibitory KIR gene profiles or the low frequency of activating ones as genetic prerequisites for decreased levels of innate immune protection against hematological malignancies. Similar data reported by different independent researchers are consistent with this opinion, e.g., predisposing associations of KIR haplo- and genotypes, expected to mediate predominantly inhibitory activity [95–97, 100]. The above statement is also supported by observations of protective effects of activating KIR3DS1 [95, 97, 101] and KIR2DS3 [94–96] in different types of leukemia. However, other findings have indicated protective effects of genes coding for inhibitory receptors KIR2DL5 [96, 101] and KIR2DL2 [102] or higher risks associated with activating receptors like KIR2DS2 [100]. Moreover, some KIRs are found with the opposite associations, for example KIR2DS2 as predisposing to [100] or protective for [102] leukemia. Together, these data show that KIRs as crucial receptors involved in the regulation of NK activity are essential for the innate anti-leukemia immune defense, but the exact impact of genetically determined KIR patterns remains incompletely understood.
Table 2.
KIR and KIR HLA ligand genetic associations in hematological malignancies
| Disease | Association | Observation | Potential NK cell effect | References |
|---|---|---|---|---|
| Leukemia | 2DL2 and 2DL2/2DL3 | Risk | ↑Inhibition | [100] |
| 2DS2 | Risk | nd | [100] | |
| AB phenotype (iKIRs > aKIRs) | Risk | ↑ Inhibition | [100] | |
| 3DL1/3DL1 without HLA-Bw4 | Protection | ↓ Inhibition | [100] | |
| HLA-Bw4 homozygous | Risk | ↑ Inhibition | [102] | |
| KIRs ratio 0.57 (4 aKIRs/7 iKIRs) | Risk | ↑ Inhibition | [97] | |
| KIRs ratio 0.66 (4 aKIRs/6 iKIRs) | Protection | ↓ Inhibition | [97] | |
| HLA-C1 homozygous | Risk | ↑ Inhibition | [99] | |
| HLA-C2 | Protection | ↓ Inhibition | [99] | |
| CLL | 2DS3 (female) | Protection | ↑ Activation | [96] |
| HLA-Bw4(Thr80) | Protection | ↓ Inhibition | [96] | |
| iKIRs > aKIRs | Risk | ↑ Inhibition | [96] | |
| 3DS1/HLA-Bw4 | Protection | ↑ Activation | [96] | |
| HLA-Bw4/Bw6 | Risk | ↑ Inhibition | [100] | |
| 3DL1/3DL1 with HLA-Bw4 | Risk | ↑ Inhibition | [100] | |
| ML | 2DL2/2DL3 and HLA-C1 | Risk | ↑ Inhibition | [119] |
| 2DL3/2DL3 and HLA-C1 | Protection | ↓ Inhibition | [119] | |
| 2DL2/HLA-C1 | Risk | ↑ Inhibition | [119] | |
| 2DS2/HLA-C1 | Risk | nd | [119] | |
| AML | AB1 phenotype | Risk | ↑ Inhibition | [100] |
| 2DS3 | Protection | ↑ Activation | [95] | |
| 3DS1 | Protection | ↑ Activation | [95, 97] | |
| iKIRs > aKIRs | Protection | ↑ Inhibition | [95] | |
| KIR2DL5A | Protection | nd | [97] | |
| CML | AB9 phenotype | Risk | ↑ Inhibition | [100] |
| 2DS4del +/2DS4norm- | Risk | ↑ Inhibition | [105] | |
| 2DS4 | Risk | nd | [102] | |
| HLA-Bw4 homozygous | Risk | ↑ Inhibition | [102] | |
| 2DL2/HLA-C1 | Protection | nd | [102] | |
| 2DS2/HLA-C1 | Protection | ↑ Activation | [102] | |
| ALL | 2DS3 | Protection | ↑Activation | [94] |
| KIR A/A haplotype (Hispanic children) | Risk | ↑ Inhibition | [114] | |
| HLA-Bw4 homozygous (non-Hispanic children) | Risk | ↑ Inhibition | [114] | |
| NK cell leukemia | KIR2DL5A and 2DL5B | Risk | ↑ Inhibition | [107] |
| Hodgkin disease | 3DS1, 2DS1 | Protection | ↑Activation | [103] |
| DLBCL | KIR3DL1/HLA-Bw4(Thr80) | Protection | nd | [99] |
ALL acute lymphoblastic leukemia, AML acute myeloid leukemia, CLL chronic lymphocytic leukemia, CML chronic myeloid leukemia, ML myeloid leukemia, DLBCL diffuse large B-cell lymphoma, nd not determined
KIR HLA class I ligands in hematological malignancies
HLA-C allele-encoded molecules are the main ligands for KIRs, but most studies failed to demonstrate associations between HLA-ligand genotypes and hematological malignancies [100, 119, 120]. Yet lower incidence of HLA-C2 and higher incidence of HLA-C1C1 was observed in Thai patients with AML [99]. It is worth mentioning the study of Karabon et al. [96], which establishes an association between HLA-C2 carriage and a decreased progression-free survival (PFS) in patients with B-CLL. In addition, among the HLA-C2-positive patients, a significantly lower probability of PFS was noted when the putative activation receptor for this ligand group, KIR2DS1, was missing. Contradictory results are reported for HLA-Bw4 ligands. Reduced incidence of HLA-Bw4 specificities has been established in patients with B-CLL [96] as a result of lower frequency of HLA-B allele-encoded molecules with Bw4 (Thr80) epitopes that bind with low affinity to the cognate inhibitory receptor. This correlation is statistically more significant for female patients. Homozygosity for HLA-Bw4 allele-encoded molecules is strongly associated with increased risk of ALL in non-Hispanic pediatric patients but not in Hispanic children [114]. Middleton et al. [102] also reported homozygosity for HLA-Bw4 ligands was an independent risk factor for the development of CML. Although HLA-B Bw6 allele-encoded molecules are not ligands for KIRs, their frequency should be taken into consideration. The lack of HLA-Bw6 in an individual may be interpreted in the context of the presence of two HLA-B Bw4 allele-encoded molecules (HLA-B Bw4/Bw4). Supporting the significance of HLA-Bw6 in hematological malignancies are the data of Middleton et al. [102], demonstrating a protective role of this subgroup of specificities for leukemia development, particularly for AML and CML. We also found a lower incidence of HLA-Bw6 allotypes in patients with myeloid leukemia compared to the control group, and the difference was more noticeable for CML [97]. The reduced frequency of HLA-Bw6 among our patients may be considered as an indication of more individuals with one or two HLA-B Bw4 alleles in their genotype. These ligands generate stronger inhibitory signals by binding to their cognate inhibitory receptor, KIR3DL1, and therefore maintain the NK cells in a state of hyporesponsiveness. HLA-B Bw6 allele-encoded molecules have no functional significance for KIRs, and their protective role may be associated with lower NK-cell inhibition and more effective anti-tumor protection. Additionally, the downregulation of HLA class I expression in leukemia is more common for HLA-Bw6 antigens [120]. It could be expected that in heterozygous patients (carriers of HLA-B Bw6/Bw4), the malignant cells will evade immune system detection by two mechanisms—escape from T-cell cytotoxicity by reduced/lost HLA-Bw6 expression and simultaneously evasion of NK-cell attack by increased HLA-Bw4/KIR3DL1-mediated inhibition.
The contradictory and irreproducible results reported by independent research groups make it hard to draw firm conclusions for KIR HLA ligand associations with hematological malignant diseases. Therefore, at present, the presence of HLA-Bw4 homozygosity as a risk factor for leukemia occurrence can only be considered possible.
Impact of inherited KIR/HLA class I ligand gene combinations in hematological malignancies
Effector NK functions are controlled not only by the expression of KIRs on NK cell surface and their HLA class I ligands on target cells, but also by the functional receptor-ligand combinations. In this context, a protective effect of the KIR2DS2/HLA-C1 pair associated with higher NK-cell cytotoxicity was established in CML [102]. On the other hand, the presence of HLA-C2 in absence of the activating KIR2DS1 was an indicator for more severe course of B-CLL [96]. Verheyden et al. [104] found a higher percentage of KIR2DL2/KIR2DL3 and heterozygous HLA-C1-positive individuals among patients with acute and chronic myeloid leukemia. Such combinations can lead to stronger inhibition due to higher binding affinity of KIR2DL2/KIR2DL3 compared to KIR2DL3 alone with their cognate HLA-C1 ligands. This idea is supported by the lower frequency of homozygous individuals for KIR2DL3 and HLA-C1 ligands [2DL3/L3(+)/HLA-C1C1(+)] among the same patient cohort that may be considered as a precondition for higher NK anti-tumor activity and protective effects. In patients with chronic lymphocytic leukemia, an increased incidence of KIR3DL1/HLA-Bw4 combinations mainly due to a prevalence of KIR3DL1 homozygous individuals [3DL1/3DL1/HLA-Bw4(+)] was observed [100]. Significant associations between the presence of inhibitory KIR3DL1/HLA-Bw4 combination and increased risk of leukemia were found in non-Hispanic white children with ALL as well [114]. Interesting data from Vejbaesya et al. [99] support a protective effect of KIR3DL1/HLA-Bw4(Thr80) carriage regarding the risk of diffuse large B-cell lymphoma (DLBCL). Karabon et al. [96] associated the presence of HLA-Bw4 along with the activation analog of KIR3DL1 [KIR3DS1(+)/HLA-Bw4(+)] not only with protection against B-CLL but also with a reduced risk of leukemia progression. The beneficial role of this KIR/ligand combination was indirectly confirmed by a decreased incidence of relapse observed in HSCT between KIR3DL1(+)/3DS1(+) donors and HLA-Bw4(+) recipients, probably due to a potential graft-versus-leukemia effect of alloreactive NK cells [121].
Despite the data on lymphoid neoplasia, Tao et al. [98] found no associations of KIR3DS1/Bw4 and KIR3DS1/Bw4-80I pairs with acute myeloid leukemia. Nearman et al. [122] did not observe a relationship between specific KIR/HLA profiles and cytotoxic T-cell LGL. However, they found significant mismatches between KIR3DL2 and KIR2DS1, and their respective HLA-A3, HLA-A11 and HLA-C2 ligands, but the biological significance of these results is difficult to explain.
Based on the data from disease association studies, we have attempted to integrate in a schematic representation the potential impact of KIRs and HLA ligands in the pathogenesis and resistance to blood cancers (Fig. 1). Overall, our current knowledge of KIR/ligand polymorphism in hematologic malignancies outlines two trends—higher levels of KIR-mediated inhibition and high levels of KIR-mediated activation. Inhibitory KIR phenotypes and KIR/ligand combinations may favor the escape of aberrant cells from immune control. Regardless of the loss of HLA expression by tumor cells, the downregulation may not affect all KIR ligands, which will not influence the inhibitory potential of NK cells. In support of this idea, the decreased expression of HLA class I molecules and mainly of Bw6 group in a heterogeneous group of patients with leukemias is established [120]. Leukemia cells with reduced/absent HLA-Bw6 expression but preserved expression of HLA-Bw4 molecules can evade from both CTL and NK cells. Moreover, in acute lymphoproliferative diseases, the loss of HLA class I expression is infrequent [123], and the inhibitory KIR-ligand combinations may facilitate the escape of leukemic cells from NK-cell immune surveillance. Activation KIR genotypes may have opposite effects in various malignancies depending on whether or not the infection is involved in tumor pathogenesis. Probably for malignant diseases with infectious pathogenesis (e.g., EBV-induced non-Hodgkin lymphoma, such as DLBCL), activating KIR genotypes may represent a genetic risk profile, while the inhibitory KIR genotypes may be relatively protective.
Fig. 1.
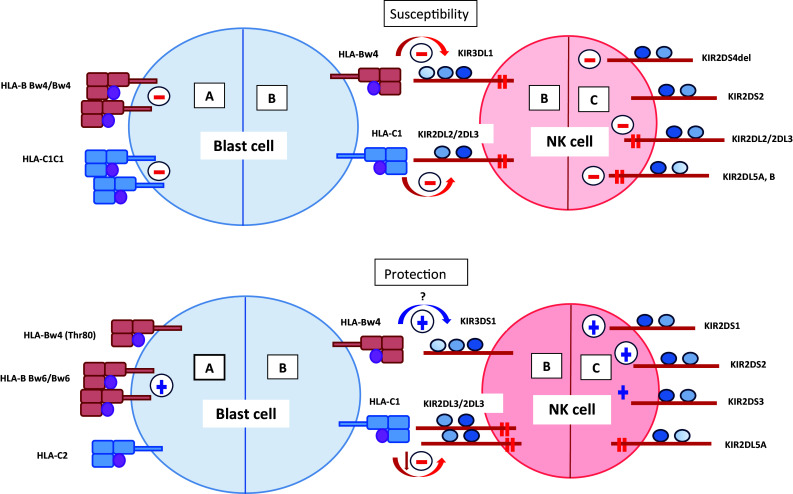
The most common KIR and HLA ligand associations observed in hematological malignancies. The figure shows possible associations of KIR HLA ligands alone (A), KIR/HLA ligand combinations (B) and KIRs alone (C) with susceptibility to (upper side) or protection (lower side) from hematological malignancies. Homozygosity for HLA-B Bw4 and HLA-C1 group allele-encoded molecules on target blast cells (A), or presence of inhibitory receptors and non-expressed form of KIR2DS4 (2DS4del) on NK cells (C) are a prerequisite for inhibition of NK cell reactivity (red minus). In addition, the interactions between inhibitory KIRs and their cognate HLA class I ligands (B) send strong signals resulting in no-killing of blasts. In associations related to protection, the presence of low affinity HLA-Bw4(Thr80) alleles and HLA-C2 ligands, and non-KIR ligands HLA-B Bw6 specificities (A) may be related to decreased inhibition and high activation (blue plus), respectively. Predominance of activating NK-cell receptors (C) and the interaction between activating KIR3DS1 and their putative (?) cognate HLA-Bw4 ligands (B) may generate activation signals, leading to elimination of neoplastic cells. Additionally, the weaker KIR2DL3/2DL3-HLA-C1 interaction (B) may result in decreased NK cell inhibition that can be overcome by activating signals, leading to a lysis of emerging malignant cells
Concluding remarks
There is growing evidence for associations of KIRs and KIR/ligand polymorphisms with the susceptibility and progression of malignant diseases, including hematologic neoplasia. However, the exact mechanisms of action of activating and inhibitory receptors remain unclear and no studies have been performed to assess the clinical consequences of KIR allelic diversity. Based on the available genetic data, several different models explaining the interactions between KIR and HLA genetic systems in NK-cell recognition of neoplastic cells have been proposed. We are in a period of intense development for immunotherapies against cancer aimed at effectively and safely modulating anti-tumor immune responses. The increasing knowledge of KIRs and their ligands illustrates the therapeutic potential of NK cells and paves the way for their application in cancer biotherapy (reviewed in [124–126]). Adoptive transfer of ex vivo bioprocessed NK cells for treatment of different malignancies is under extensive development and evaluation. This approach is considered in addition to the effect of KIR-ligand mismatch between donor and recipient to improve the clinical outcome of HSCT. Progress is also being made with administration of monoclonal antibodies or bispecific and trispecific antibody derivates, designated killer engagers, in order to boost antibody-dependent cellular cytotoxicity. Blocking inhibitory KIRs is another antibody-based therapeutic option aiming to increase NK-cell-mediated killing. Genetic manipulations oriented to overexpression of activating receptors, silencing of inhibitory receptors or transduction of NK cells with chimeric receptors are promising new approaches. The obtained therapeutic results are encouraging not only in experimental models but also in clinical trials. The future will undoubtedly provide new ways for using NK cells in cancer immunotherapy. This makes further studies on the inhibitory and activating NK-cell pathways important in order to determine the precise mechanisms for escaping innate immune surveillance used by different types of malignant cells and application of this knowledge in clinical practice.
Abbreviations
- aKIRs
Activating killer-cell immunoglobulin-like receptors
- ALL
Acute lymphoid leukemia
- AML
Acute myeloid leukemia
- CLL
Chronic lymphoid leukemia
- CML
Chronic myeloid leukemia
- CTL
Cytotoxic T lymphocytes
- DLBCL
Diffuse large B-cell lymphoma
- HLA
Human leukocyte antigen
- HSCT
Hematopoietic stem cell transplantation
- iKIRs
Inhibitory killer-cell immunoglobulin-like receptors
- ILCs
Innate lymphoid cells
- ILTs
Immunoglobulin-like transcripts
- ITIMs
Immunoreceptor tyrosine-based inhibitory motifs
- KIR2DS4del
KIR2DS4 variant with deletion
- KIRs
Killer-cell immunoglobulin-like receptors
- LGL
Large granular lymphocytes
- MHC
Major histocompatibility complex
- ML
Myeloid leukemia
- NCR
Natural cytotoxicity receptors
- NK cells
Natural killer cells
- PFS
Progression-free survival
Compliance with ethical standards
Conflict of interest
The authors have no conflict of interest to declare.
Footnotes
This article is part of the Symposium-in-Writing “Natural killer cells, ageing and cancer”, a series of papers published in Cancer Immunology, Immunotherapy.
References
- 1.Trinchieri G. Biology of natural killer cells. Adv Immunol. 1989;47:187–376. doi: 10.1016/S0065-2776(08)60664-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biron CA. Activation and function of natural killer cell responses during viral infections. Curr Opin Immunol. 1997;9:24–34. doi: 10.1016/S0952-7915(97)80155-0. [DOI] [PubMed] [Google Scholar]
- 3.Diefenbach A, Colonna M, Koyasu S. Development, differentiation, and diversity of innate lymphoid cells. Immunity. 2014;41(3):354–365. doi: 10.1016/j.immuni.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eberl G, Colonna M, Di Santo JP, McKenzie AN. Innate lymphoid cells: a new paradigm in immunology. Science. 2015;348(6237):aaa6566. doi: 10.1126/science.aaa6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kärre K, Ljunggren HG, Piontek G, Kiessling R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature. 1986;319:675–678. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- 6.Moretta L, Biassoni R, Bottiono C, Mingari MC, Moratta A. Natural killer cells: a mystery no more. Scand J Immunol. 2002;55:229–232. doi: 10.1046/j.1365-3083.2002.01055.x. [DOI] [PubMed] [Google Scholar]
- 7.Valiante N, Uhrberg M, Shilling H, Lienert-Weidenbach K, Arnett KL, D’Andrea A, Phillips JH, Lanier LL, Parham P. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7:739–751. doi: 10.1016/S1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- 8.Raulet D, Held W, Correa I, Dorfman J, Wu MF, Corral L. Specificity, tolerance and developmental regulation of natural killer cells defined by expression of class I-specific Ly49 receptors. Immunol Rev. 1997;155:41–52. doi: 10.1111/j.1600-065X.1997.tb00938.x. [DOI] [PubMed] [Google Scholar]
- 9.Ljunggren HG, Karre K. In search of the “missing self”: MHC molecules and NK cell recognition. Immunol Today. 1990;11:237–244. doi: 10.1016/0167-5699(90)90097-S. [DOI] [PubMed] [Google Scholar]
- 10.Liao N, Bix M, Zijlstra M, Jaenisch R, Raulet D. MHC class I deficiency susceptibility to natural killer (NK) cell and impaired NK activity. Science. 1991;253:199–202. doi: 10.1126/science.1853205. [DOI] [PubMed] [Google Scholar]
- 11.Diefenbach A, Raulet DH. Strategies for target cell recognition by natural killer cells. Immunol Rev. 2001;181:170–184. doi: 10.1034/j.1600-065X.2001.1810114.x. [DOI] [PubMed] [Google Scholar]
- 12.Lanier L. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 13.Bottino C, Biassoni R, Milo R, Moretta L, Morreta A. The human natural cytotoxic receptors (NCR) that induce HLA class I-independent NK cell triggering. Hum Immunol. 2000;61:1–6. doi: 10.1016/S0198-8859(99)00162-7. [DOI] [PubMed] [Google Scholar]
- 14.Martin AM, Freitas EM, Witt CS, Christiansen FT. The genomic organization and evolution of the natural killer immunoglobulin like receptor (KIR) gene cluster. Immunogenetics. 2000;51:268–280. doi: 10.1007/s002510050620. [DOI] [PubMed] [Google Scholar]
- 15.IPD-KIR: http://www.ebi.ac.uk/ipd/kir; Release 2.6.1, 17 February 2015. Accessed 08 Jan 2016
- 16.Wilson MJ, Torkar M, Haude A, Milne S, Jones T, Sheer D, Beck S, Trowsdale J. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci USA. 2000;97:4778–4783. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.López-Botet M, Pérez-Villar JJ, Carretero M, Rodríguez A, Melero I, Bellón T, Llano M, Navarro F. Structure and function of the CD94 C-type lectin receptor complex involved in recognition of HLA class I molecules. Immunol Rev. 1997;155:165–174. doi: 10.1111/j.1600-065X.1997.tb00949.x. [DOI] [PubMed] [Google Scholar]
- 18.Vales-Gomez M, Reyburn H, Erskine R, Lopez-Botet M, Strominger J. Kinetics and peptide dependency of the binding of the inhibitory NK receptor CD94/NKG2-A and the activating receptor CD94/NKG2-C to HLA-E. EMBO J. 1999;18:4250–4260. doi: 10.1093/emboj/18.15.4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gardiner CM, Guethlein LA, Shilling HG, Pando M, Carr WH, Rajalingam R, Vilches C, Parham P. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J Immunol. 2001;166:2992–3001. doi: 10.4049/jimmunol.166.5.2992. [DOI] [PubMed] [Google Scholar]
- 20.Parham P, Norman PJ, Abi-Rached L, Guethlein LA. Variable NK cell receptors exemplified by human KIR3DL1/S1. J Immunol. 2011;187(1):11–19. doi: 10.4049/jimmunol.0902332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lanier LL, Corliss BC, Wu J, Leong C, Phillips JH. Immunoreceptor DAP12 bearing a tyrosine-based activation motif is involved in activating NK cells. Nature. 1998;391:703–707. doi: 10.1038/35642. [DOI] [PubMed] [Google Scholar]
- 22.Kikuchi-Maki A, Catina T, Cambpbell K. Cutting edge: KIR2DL4 transduces signals into human NK cells through association with the Fc receptor gamma protein. J Immunol. 2005;174:3859–3863. doi: 10.4049/jimmunol.174.7.3859. [DOI] [PubMed] [Google Scholar]
- 23.Rajagopalan S, Fu J, Long E. Cutting edge: induction of IFN-γ production but not cytotoxicity by the killer cell Ig-like receptor KIR2DL4 (CD158d) in resting NK cells. J Immunol. 2001;167:1877–1881. doi: 10.4049/jimmunol.167.4.1877. [DOI] [PubMed] [Google Scholar]
- 24.Uhrberg M, Valiante N, Shum B, Shilling H, Lienert-Weidenbach K, Corliss B, Tyan D, Lanier LL, Parham P. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–763. doi: 10.1016/S1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 25.Steffens U, Vyas Y, Dupont B, Selvakumar A. Nucleotide and amino acid sequence alignment for human killer cell inhibitory receptors. Tissue Antigens. 1998;51:398–413. doi: 10.1111/j.1399-0039.1998.tb02981.x. [DOI] [PubMed] [Google Scholar]
- 26.Gumperz JE, Valiante NM, Parham P, Lanier LL, Tyan D. Heterogeneous phenotypes of expression of the NKB1 natural killer cell class I receptor among individuals of different human histocompatibility leukocyte antigens types appear genetically regulated, but not linked to major histocompatibility complex haplotype. J Exp Med. 1996;183:1817–1827. doi: 10.1084/jem.183.4.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med. 2006;203:633–645. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shilling HG, Guethlein LA, Cheng NW, Gardiner CM, Rodriguez R, Tyan D, Parham P. Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J Immunol. 2002;168(5):2307–2315. doi: 10.4049/jimmunol.168.5.2307. [DOI] [PubMed] [Google Scholar]
- 29.Norman PJ, Abi-Rached L, Gendzekhadze K, Korbel D, Gleimer M, Rowley D, Bruno D, Carrington CV, Chandanayingyong D, Chang YH, Crespí C, Saruhan-Direskeneli G, Fraser PA, Hameed K, Kamkamidze G, Koram KA, Layrisse Z, Matamoros N, Milà J, Park MH, Pitchappan RM, Ramdath DD, Shiau MY, Stephens HA, Struik S, Verity DH, Vaughan RW, Tyan D, Davis RW, Riley EM, Ronaghi M, Parham P. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat Genet. 2007;39:1092–1099. doi: 10.1038/ng2111. [DOI] [PubMed] [Google Scholar]
- 30.Gagne K, Willem C, Legrand N, Djaoud Z, David G, Rettman P, Bressollette-Bodin C, Senitzer D, Esbelin J, Cesbron-Gautier A, Schneider T, Retière C. Both the nature of KIR3DL1 alleles and the KIR3DL1/S1 allele combination affect the KIR3DL1 NK-cell repertoire in the French population. Eur J Immunol. 2013;43(4):1085–1098. doi: 10.1002/eji.201243007. [DOI] [PubMed] [Google Scholar]
- 31.Parham P. The genetic and evolutionary balances in human NK cell receptor diversity. Semin Immunol. 2008;20:311–316. doi: 10.1016/j.smim.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller JS, McCullar V. Human natural killer cells with polyclonal lectin and immunoglobulin like receptors develop from single hematopoietic stem cells with preferential expression of NKG2A and KIR2DL2/L3/S2. Blood. 2001;98:705–713. doi: 10.1182/blood.V98.3.705. [DOI] [PubMed] [Google Scholar]
- 33.Santourlidis S, Trompeter HI, Weinhold S, Eisermann B, Meyer KL, Wernet P, Uhrberg M. Crucial role of DNA methylation in determination of clonally distributed killer cell Ig-like receptor expression patterns in NK cells. J Immunol. 2002;169:4253–4261. doi: 10.4049/jimmunol.169.8.4253. [DOI] [PubMed] [Google Scholar]
- 34.Chan HW, Kurago ZB, Stewart CA, Wilson MJ, Martin MP, Mace BE, Carrington M, Trowsdale J, Lutz CT. DNA methylation maintains allele-specific KIR gene expression in human natural killer cells. J Exp Med. 2003;197(2):245–255. doi: 10.1084/jem.20021127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Presnell SR, Zhang L, Chlebowy CN, Al-Attar A, Lutz CT. Differential transcription factor use by the KIR2DL4 promoter under constitutive and IL-2/15-treated conditions. J Immunol. 2012;188(9):4394–4404. doi: 10.4049/jimmunol.1103352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan HW, Miller JS, Moore MB, Lutz CT. Epigenetic control of highly homologous killer Ig-like receptor gene alleles. J Immunol. 2005;175:5966–5974. doi: 10.4049/jimmunol.175.9.5966. [DOI] [PubMed] [Google Scholar]
- 37.Li G, Weyand CM, Goronzy JJ. Epigenetic mechanisms of age dependent KIR2DL4 expression in T cells. J Leukoc Biol. 2008;84:824–834. doi: 10.1189/jlb.0807583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mandelboim O, Reyburn H, Vales-Gomez M, Pazmany L, Colonna M, Borsellino G, Strominger JL. Protection from lysis by natural killer cells of group 1 and 2 specificity is mediated by residue 80 in human histocompatibility leukocyte antigen C alleles and also occurs with empty major histocompatibility complex molecules. J Exp Med. 1996;184:913–922. doi: 10.1084/jem.184.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winter C, Long E. A single amino acid in the p58 killer cell inhibitory receptor controls the ability of natural killer cells to discriminate between the two groups of HLA-C allotypes. J Immunol. 1997;158:4026–4028. [PubMed] [Google Scholar]
- 40.Colonna M, Samaridis J. Cloning of immunoglobulin-superfamily members associated with HLA-C and HLA-B recognition by human natural killer cells. Science. 1995;268:405–408. doi: 10.1126/science.7716543. [DOI] [PubMed] [Google Scholar]
- 41.Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J Immunol. 2008;180:3969–3979. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- 42.David G, Djaoud Z, Willem C, Legrand N, et al. Large spectrum of HLA-C recognition by Killer Ig-like receptor (KIR)2DL2 and KIR2DL3 and restricted C1 specifity of KIR2DS2: dominant impact of KIR2DL2/KIR2DS2 and KIR2D NK cell repertoire formation. J Immunol. 2013;191(9):4778–4788. doi: 10.4049/jimmunol.1301580. [DOI] [PubMed] [Google Scholar]
- 43.Ahlenstiel G, Martin MP, Gao X, Carrington M, Rehermann B. Distinct KIR/HLA compound genotypes affect the kinetics of human antiviral natural killer cell responses. J Clin Invest. 2008;118:1017–1026. doi: 10.1172/JCI32400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cella M, Longo A, Ferrara GB, Strominger JL, Colonna M. NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J Exp Med. 1994;180:1235–1242. doi: 10.1084/jem.180.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gumperz J, Litwin V, Philips J, Lanier L, Parham P. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J Exp Med. 1995;181:1133–1144. doi: 10.1084/jem.181.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gumperz J, Barber L, Valiante N, Percival L, et al. Conserved and variable residues within the Bw4 motif of HLA-B make separable contributions to recognition by the NKB1 killer cell-inhibitory receptor. J Immunol. 1997;158:5237–5241. [PubMed] [Google Scholar]
- 47.Foley B, De Santis D, Van Beelen E, Lathbury L, et al. The reactivity of Bw4+ HLA-B and HLA-A alleles with KIR3DL1: implications for patient and donor suitability for haploidentical stem cell transplantations. Blood. 2008;112:435–443. doi: 10.1182/blood-2008-01-132902. [DOI] [PubMed] [Google Scholar]
- 48.Thananchai H, Gillespie G, Martin MP, Bashirova A, et al. Cutting edge: allele-specific and peptide-dependent interactions between KIR3DL1 and HLA-A and HLA-B. J Immunol. 2007;178:33–37. doi: 10.4049/jimmunol.178.1.33. [DOI] [PubMed] [Google Scholar]
- 49.Carr W, Pando M, Parham P. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J Immunol. 2005;175:5222–5229. doi: 10.4049/jimmunol.175.8.5222. [DOI] [PubMed] [Google Scholar]
- 50.O’Connor G, Guinan K, Cunningham R, Middleton D, Parham P, Cl Gardiner. Functional polymorphism of the KIR3DL1/S1 receptor on human NK cell. J Immunol. 2007;178:235–241. doi: 10.4049/jimmunol.178.1.235. [DOI] [PubMed] [Google Scholar]
- 51.King A, Loke Y, Chaouat G. NK cells and reproduction. Immunol Today. 1997;18:64–66. doi: 10.1016/S0167-5699(97)01001-3. [DOI] [PubMed] [Google Scholar]
- 52.Pende D, Biasson R, Cantoni C, Verdiani S, Falco M, di Donato C, Accame L, Bottino C, Moretta A, Moretta L. The natural killer cell receptor specific for HLA-A allotypes: a novel member of the p58/p70 family of inhibitory receptors that is characterized by three immunoglobulin-like domains and is expressed as a 140-kD disulphide-linked dimer. J Exp Med. 1996;184:505–518. doi: 10.1084/jem.184.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dohring C, Scheidegger D, Samaridis J, Cella M, Colonna M. A human killer inhibitory receptor specific for HLA-A1,2. J Immunol. 1996;156:3098–3101. [PubMed] [Google Scholar]
- 54.Ponte M, Cantoni C, Biassoni R, Tradori-Cappai A, Bentivoglio G, Vitale C, Bertone S, Moretta A, Moretta L, Mingari MC. Inhibitory receptors sensing HLA-G1 molecules in pregnancy: decidua-associated natural killer cells express LIR-1 and CD94/NKG2A and acquire p49, an HLA-G1-specific receptor. Proc Natl Acad Sci USA. 1999;96:5674–5679. doi: 10.1073/pnas.96.10.5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rajagopalan S, Long EO. A human histocompatibility leukocyte antigen (HLA)-G-specific receptor expressed on all natural killer cells. J Exp Med. 1999;189:1093–1100. doi: 10.1084/jem.189.7.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yan WH, Fan LA. Residues Met76 and Gln79 in HLA-G alpha1 domain involve in KIR2DL4 recognition. Cell Res. 2005;15:176–182. doi: 10.1038/sj.cr.7290283. [DOI] [PubMed] [Google Scholar]
- 57.Allan DS, Colonna M, Lanier LL, Churakova TD, Abrams JS, Ellis SA, McMichael AJ, Braud VM. Tetrameric complexes of human histocompatibility leukocyte antigen (HLA)-G bind to peripheral blood myelomonocytic cells. J Exp Med. 1999;189:1149–1156. doi: 10.1084/jem.189.7.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Le Page ME, Goodridge JP, John E, Christiansen FT, Witt CS. Killer Ig-like receptor 2DL4 does not mediate NK cell IFN-g responses to soluble HLA-G preparations. J Immunol. 2014;192:732–740. doi: 10.4049/jimmunol.1301748. [DOI] [PubMed] [Google Scholar]
- 59.Stewart C, Laugier-Anfossi F, Vely F, Saulquin X, Riedmuller J, Tisserant A, Gauthier L, Romagné F, Ferracci G, Arosa FA, Moretta A, Sun PD, Ugolini S, Vivier E. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc Natl Acad Sci USA. 2005;102:13224–13229. doi: 10.1073/pnas.0503594102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Foley B, De Santis D, Lathbury L, Christiansen Witt C. KIR2DS1-mediated activation overrides NKG2A-mediated inhibition in HLA-C2-negative individuals. Int Immunol. 2008;20:555–563. doi: 10.1093/intimm/dxn013. [DOI] [PubMed] [Google Scholar]
- 61.Chewning J, Gudme C, Hsu K, Selvakumar A, Dupont B. KIR2DS1-positive NK cells mediate alloresponse against the C2 HLA–KIR ligand group in vitro. J Immunol. 2007;179:854–868. doi: 10.4049/jimmunol.179.2.854. [DOI] [PubMed] [Google Scholar]
- 62.Biassoni R, Pessino A, Malaspina A, Cantoni C, Bottino C, Sivori S, Moretta L, Moretta A. Role of amino acid position 70 in the binding affinity of p50.1 and p58.1 receptors for HLA-Cw4 molecules. Eur J Immunol. 1997;27:3095–3099. doi: 10.1002/eji.1830271203. [DOI] [PubMed] [Google Scholar]
- 63.Graef T, Moesta AK, Norman PJ, Abi-Rached L, Vago L, Older Aguilar AM, Gleimer M, Hammond JA, Guethlein LA, Bushnell DA, Robinson PJ, Parham P. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C. J Exp Med. 2009;206:2557–2572. doi: 10.1084/jem.20091010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Katz G, Markel G, Mizrahi S, Arnon TI, Mandelboim O. Recognition of HLA-Cw4 but not HLA-Cw6 by the NK cell receptor killer cell Ig-like receptor two domain short tail number 4. J Immunol. 2001;166:7260–7267. doi: 10.4049/jimmunol.166.12.7260. [DOI] [PubMed] [Google Scholar]
- 65.Katz G, Gazit R, Arnon TI, Gonen-Gross T, Tarcic G, Markel G, Gruda R, Achdout H, Drize O, Merims S, Mandelboim O. MHC class I-independent recognition of NK-activating receptor KIR2DS4. J Immunol. 2004;173:1819–1825. doi: 10.4049/jimmunol.173.3.1819. [DOI] [PubMed] [Google Scholar]
- 66.Vales-Gomez M, Reuburn H, Mandelboim M, Strominger J. Kinetics of interaction of HLA-ligands with natural killer cell inhibitory receptors. Immunity. 1998;9:337–344. doi: 10.1016/S1074-7613(00)80616-0. [DOI] [PubMed] [Google Scholar]
- 67.Martin MP, Gao X, Lee JH, Nelson GW, Detels R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, Trowsdale J, Wilson M, O’Brien SJ, Carrington M. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 68.Alter G, Martin MP, Teigen N, Carr WH, Suscovich TJ, Schneidewind A, Streeck H, Waring M, Meier A, Brander C, Lifson JD, Allen TM, Carrington M, Altfeld M. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J Exp Med. 2007;204:3027–3036. doi: 10.1084/jem.20070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alter G, Rihn S, Walter K, Nolting A, Martin M, Rosenberg ES, Miller JS, Carrington M, Altfeld M. HLA class I subtype dependent expansion of KIR3DS1+ and KIR3DL1+ NK cells during acute human immunodeficiency virus type 1 infection. J Virol. 2009;83:6798–6805. doi: 10.1128/JVI.00256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vales-Gomez M, Reyburn HT, Erskine RA, Strominger J. Differential binding to HLA-C of p50-activating and p58-inhibitory natural killer cell receptors. Proc Natl Acad Sci USA. 1998;95:14326–14331. doi: 10.1073/pnas.95.24.14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol. 2002;20:217–251. doi: 10.1146/annurev.immunol.20.092501.134942. [DOI] [PubMed] [Google Scholar]
- 72.Della Chiesa M, Romeo E, Falco M, Balsamo M, Augugliaro R, Moretta L, Bottino C, Moretta A, Vitale M. Evidence that the KIR2DS5 gene codes for a surface receptor triggering natural killer cell function. Eur J Immunol. 2008;38:2284–2289. doi: 10.1002/eji.200838434. [DOI] [PubMed] [Google Scholar]
- 73.VandenBussche CJ, Mulrooney TJ, Frazier WR, Dakshanamurthy S, Hurley CK. Dramatically reduced surface expression of NK cell receptor KIR2DS3 is attributed to multiple residues throughout the molecule. Genes Immun. 2009;10:162–173. doi: 10.1038/gene.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu J, Xiao Z, Ko HL, Shen M, Ren EC. Activating killer cell immunoglobulin-like receptor 2DS2 binds to HLA-A*11. Proc Natl Acad Sci USA. 2014;111(7):2662–2667. doi: 10.1073/pnas.1322052111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moesta AK, Parham P. Diverse functionality among human NK cell receptors for the C1 epitope of HLA-C: KIR2DS2, KIR2DL2, and KIR2DL3. Front Immunol. 2012;3:336. doi: 10.3389/fimmu.2012.00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Elliott JM, Yokoyama WM. Unifying concepts of MHC-dependent natural killer cell education. Trends Immunol. 2011;32:364–372. doi: 10.1016/j.it.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Raulet DH, Vance RE. Self-tolerance of natural killer cells. Nat Rev Immunol. 2006;6:520–531. doi: 10.1038/nri1863. [DOI] [PubMed] [Google Scholar]
- 78.Kim S, Poursine-Laurent J, Truscott SM, Lybarger L, Song YJ, French AR, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436:709–713. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- 79.Nash WT, Teoh J, Wei H, Gamache A, Brown MG. Know thyself: NK-cell inhibitory receptors prompt self-tolerance, education, and viral control. Front Immunol. 2014;5:175. doi: 10.3389/fimmu.2014.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Watzl C, Urlaub D, Fasbender F, Claus M. Natural killer cell regulation—beyond the receptors. F1000Prime Rep. 2014;6:87. doi: 10.12703/P6-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yokoyama WM, Kim S. Licensing of natural killer cells by self-major histocompatibility complex class I. Immunol Rev. 2006;214:143–145. doi: 10.1111/j.1600-065X.2006.00458.x. [DOI] [PubMed] [Google Scholar]
- 82.Brodin P, Lakshmikanth T, Johansson S, Kärre K, Höglund P. The strength of inhibitory input during education quantitatively tunes the functional responsiveness of individual natural killer cells. Blood. 2009;113:2434–2441. doi: 10.1182/blood-2008-05-156836. [DOI] [PubMed] [Google Scholar]
- 83.Joncker NT, Fernandez NC, Treiner E, Vivier E, Raulet DH. NK cell responsiveness is tuned commensurate with the number of inhibitory receptors for self-MHC class I: the rheostat model. J Immunol. 2009;182:4572–4580. doi: 10.4049/jimmunol.0803900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ciccone E, Pende D, Viale O, Di Donato C, Tripodi G, Orengo AM, Guardiola J, Moretta A, Moretta L. Evidence of a natural killer (NK) cell repertoire for (allo) antigen recognition: definition of five distinct NK-determined allospecificities in humans. J Exp Med. 1992;175:709–718. doi: 10.1084/jem.175.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lotzova E, Savary CA, Herberman RB. Inhibition of clonogenic growth of fresh leukemia cells by unstimulated and IL-2 stimulated NK cells of normal donors. Leuk Res. 1987;11:1059–1066. doi: 10.1016/0145-2126(87)90158-5. [DOI] [PubMed] [Google Scholar]
- 86.Ruggeri L, Capanni M, Urbani E, Perruccio K, Shlomchik WD, Tosti A, Posati S, Rogaia D, Frassoni F, Aversa F, Martelli MF, Velardi A. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. 2002;295:2097–2100. doi: 10.1126/science.1068440. [DOI] [PubMed] [Google Scholar]
- 87.Cooley S, Trachtenberg E, Bergemann TL, Saeteurn K, Klein J, Le CT, Marsh S, Guethlein LA, Parham P, Miller JS, Weisdorf DJ. Donors with group B KIR haplotypes improve relapse-free survival after unrelated hematopoietic cell transplantation for acute myelogenous leukemia. Blood. 2009;113:726–732. doi: 10.1182/blood-2008-07-171926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cooley S, Weisdorf DJ, Guethlein LA, Klein JP, Wang T, Le CT, Marsh S, Guethlein LA, Parham P, Miller JS. Donor selection for natural killer cell receptor genes leads to superior survival after unrelated transplantation for acute myelogenous leukemia. Blood. 2010;116:2411–2419. doi: 10.1182/blood-2010-05-283051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Giebel S, Locatelli F, Lamparelli T, Velardi A, Davies S, Frumento G, Maccario R, Bonetti F, Wojnar J, Martinetti M, Frassoni F, Giorgiani G, Bacigalupo A, Holowiecki J. Survival advantage with KIR ligand incompatibility in hematopoietic stem cell transplantation from unrelated donors. Blood. 2003;102:814–819. doi: 10.1182/blood-2003-01-0091. [DOI] [PubMed] [Google Scholar]
- 90.Ruggeri L, Capanni M, Martelli MF, Velardi A. Cellular therapy: exploting NK cell alloreactivity in transplantation. Curr Opin Hematol. 2001;8:355–359. doi: 10.1097/00062752-200111000-00007. [DOI] [PubMed] [Google Scholar]
- 91.Moretta A, Bottino C, Mingari MC, Biassoni R, Moretta L. What is a natural killer cell? Nat Immunol. 2002;3:6–8. doi: 10.1038/ni0102-6. [DOI] [PubMed] [Google Scholar]
- 92.Moretta A, Locatelli F, Moretta L. Human NK cells: from HLA class I- specific killer IG-like receptors to the therapy of acute leukemia. Immunol Rev. 2008;224:58–69. doi: 10.1111/j.1600-065X.2008.00651.x. [DOI] [PubMed] [Google Scholar]
- 93.Schellekens J, Gagne K, Marsh SG. Natural killer cells and killer-cell immunoglobulin-like receptor polymorphisms: their role in hematopoietic stem cell transplantation. Methods Mol Biol. 2014;1109:139–158. doi: 10.1007/978-1-4614-9437-9_9. [DOI] [PubMed] [Google Scholar]
- 94.Zhang Y, Wang B, Ye S, Liu S, Shen T, Teng Y, Qi J. Killer cell immunoglobulin-like receptor gene polymorphisms in patients with leukemia: possible association with susceptibility to the disease. Leuk Res. 2010;34:55–58. doi: 10.1016/j.leukres.2009.04.022. [DOI] [PubMed] [Google Scholar]
- 95.Shahsavar F, Tajik N, Entezami K, Fallah Rajabzadeh M, Asadifar B, Alimoghaddam K, Ostadali Dahaghi M, Jalali A, Ghashghaie A, Ghavamzadeh A. KIR2DS3 is associated with protection against acute myeloid leukemia. Iran J Immunol. 2010;7:8–17. [PubMed] [Google Scholar]
- 96.Karabon L, Jedynak A, Giebel S, Wołowiec D, Kielbinski M, Woszczyk D, Kapelko-Slowik K, Kuliczkowski K, Frydecka I. KIR/HLA gene combinations influence susceptibility to B-cell chronic lymphocytic leukemia and the clinical course of disease. Tissue Antigens. 2011;78:129–138. doi: 10.1111/j.1399-0039.2011.01721.x. [DOI] [PubMed] [Google Scholar]
- 97.Varbanova V (2013) NK cells receptors (KIRs) and HLA ligands—role of gene polymorphism for malignancies. Dissertation, Medical University, Sofia, Bulgaria
- 98.Tao S, He Y, Zhang W, Wang W, He J, Han Z, Chen N, Dong L, He J, Zhu F, Lv H. Comparison of the KIR3DS1/Bw4 distribution in Chinese healthy and acute myloid leukemia individuals. Hum Immunol. 2015;76:79–82. doi: 10.1016/j.humimm.2015.01.024. [DOI] [PubMed] [Google Scholar]
- 99.Vejbaesya S, Sae-Tam P, Khuhapinant A, Srinak D. Killer cell immunoglobulin-like receptors in Thai patients with leukemia and diffuse large B-cell lymphoma. Hum Immunol. 2014;75:673–676. doi: 10.1016/j.humimm.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 100.Verheyden S, Bernier M, Damanet C. Identification of natural killer cell receptor phenotypes associated with leukemia. Leukemia. 2004;18:2002–2007. doi: 10.1038/sj.leu.2403525. [DOI] [PubMed] [Google Scholar]
- 101.Besson C, Roetync S, Williams F, Orsi L, Amiel C, Lependeven C, Antoni G, Hemine O, Brice P, Ferme C, Carde P, Canioni D, Briere J, Raphael M, Nicolas J, Clavel J, Middleton D, Viver E, Abel L. Association of killer cell immunoglobulin-like receptor genes with Hodgkin lymphoma in a family study. PLoS ONE. 2007;2:e406. doi: 10.1371/journal.pone.0000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Middleton D, Diler A, Meenagh A, Sleator C, Gourraud P. Killer immunoglobulin-like receptors (KIR2DL2 and/or KIR2DS2) in presence of their ligand (HLA-C1 group) protect against chronic myeloid leukaemia. Tissue Antigens. 2009;73:553–560. doi: 10.1111/j.1399-0039.2009.01235.x. [DOI] [PubMed] [Google Scholar]
- 103.Stringaris K, Adams S, Uribe M, Eniafe R, Wu CO, Savani BN, Barrett AJ. Donor KIR genes 2DL5A, 2DS1 and 3DS1 are associated with a reduced rate of leukemia relapse after HLA-identical sibling stem cell transplantation for acute myeloid leukemia but not other hematologic malignancies. Biol Blood Marrow Transpl. 2010;16:1257–1264. doi: 10.1016/j.bbmt.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Verheyden S, Scots R, Duquet W, Demanet C. A defined donor activating natural killer cell receptor genotype protects leukemia relapse after related HLA-identical hematopoietic stem cell transplantation. Leukemia. 2005;19:1446–1451. doi: 10.1038/sj.leu.2403839. [DOI] [PubMed] [Google Scholar]
- 105.Giebel S, Nowak I, Wojnar J, Krawczyk-Kulis M, Holowiecki J, Kyrcz-Krzemien S, Kusnierczyk P. Association of KIR2DS4 and its variant KIR1D with leukemia. Leukemia. 2008;22:2129–2130. doi: 10.1038/leu.2008.108. [DOI] [PubMed] [Google Scholar]
- 106.Du Z, Sharma SK, Spellman S, Reed EF, Rajalingam R. KIR2DL5 alleles mark certain combination of activating KIR genes. Genes Immun. 2008;9:470–480. doi: 10.1038/gene.2008.39. [DOI] [PubMed] [Google Scholar]
- 107.Scquizzato E, Teramo A, Miorin M, Facco M, Piazza F, Noventa F, Trentin L, Agostini C, Zambello R, Semenzato G. Genotypic evaluation of killer immunoglobulin-like receptors in NK-type lymphoproliferative disease of granular lymphocytes. Leukemia. 2007;21:1060–1069. doi: 10.1038/sj.leu.2404634. [DOI] [PubMed] [Google Scholar]
- 108.Sullivan EM, Jeha S, Kang G, Cheng C, Rooney B, Holladay M, Bari R, Schell S, Tuggle M, Pui CH, Leung W. NK cell genotype and phenotype at diagnosis of acute lymphoblastic leukemia correlate with postinduction residual disease. Clin Cancer Res. 2014;20(23):5986–5994. doi: 10.1158/1078-0432.CCR-14-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Babor F, Manser A, Schonberg K, Enczmann J, Borkhardt A, Meisel R, Uhrberg M. Lack of association between KIR genes and acute lymphoblastic leukemia in children. Blood. 2012;120(13):2770–2772. doi: 10.1182/blood-2012-07-440495. [DOI] [PubMed] [Google Scholar]
- 110.Almalte Z, Samarani S, Iannello A, Debbeche O, Duval M, Infante-Rivard C, Amre DK, Sinnett D, Ahmad A. Novel associations between activating killer cell mmunoglobulin-like receptor genes and childhood leukemia. Blood. 2011;118(5):1323–1328. doi: 10.1182/blood-2010-10-313791. [DOI] [PubMed] [Google Scholar]
- 111.La Nasa G, Caocci G, Littera R, Atzeni S, Vacca A, Mulas O, Langiu M, Greco M, Orrù S, Orrù N, Floris A, Carcassi C. Homozygosity for killer immunoglobulin-like receptor haplotype A predicts complete molecular response to treatment with tyrosine kinase inhibitors in chronic myeloid leukemia patients. Exp Hematol. 2013;41(5):424–431. doi: 10.1016/j.exphem.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 112.Caocci G, Martino B, Greco M, Abruzzese E, Trawinska MM, Lai S, Ragatzu P, Galimberti S, Baratè C, Mulas O, Labate C, Littera R, Carcassi C, Passerini CG, La Nasa G. Killer immunoglobulin-like receptors can predict TKI treatment-free remission in chronic myeloid leukemia patients. Exp Hematol. 2015;43:1015–1018. doi: 10.1016/j.exphem.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 113.Impola U, Turpeinen H, Alakulppi N, Linjama T, Volin L, Niittyvuopio R, Partanen J, Koskela S. Donor Haplotype B of NK KIR Receptor Reduces the Relapse Risk in HLA-Identical Sibling Hematopoietic Stem Cell Transplantation of AML Patients. Front Immunol. 2014;5:405. doi: 10.3389/fimmu.2014.00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.De Smith AJ, Walsh KM, Ladner MB, Zhang S, Xiao C, Cohen F, Moore TB, Chokkalingam AP, Metayer C, Buffler PA, Trachtenberg EA, Wiemels JL. The role of KIR genes and their cognate HLA class I ligands in childhood acute lymphoblastic leukemia. Blood. 2014;123:2497–2503. doi: 10.1182/blood-2013-11-540625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Khalaf R, Hoteit R, Yazbek S, El Hajj N, Otrock Z, Khansa S, Sabbagh A, Shammaa D, Mahfouz R. Natural killer cell immunoglobulin-like receptor (KIR) genotypes in Follicular Lymphoma patients: results of a pilot study. Gene. 2013;525(1):136–140. doi: 10.1016/j.gene.2013.03.144. [DOI] [PubMed] [Google Scholar]
- 116.Hoteit R, Abboud M, Bazarbachi A, Salem Z, Shammaa D, Zaatari G, Mahfouz R. KIR genotype distribution among Lebanese patients with Hodgkin’s lymphoma. Meta Gene. 2015;4:57–63. doi: 10.1016/j.mgene.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nasrallah AG, Miale TD. Decreased natural killer cell activity in children with untreated acute leukemia. Cancer Res. 1983;43:5580–5585. [PubMed] [Google Scholar]
- 118.Ruggeri L, Mancusi A, Burchielli E, Capanni M, Carotti A, Aloisi T, Aversa F, Martelli MF, Velardi A. NK cell alloreactivity and allogeneic hematopoietic stem cell transplantation. Blood Cells Mol Dis. 2008;40(1):84–90. doi: 10.1016/j.bcmd.2007.06.029. [DOI] [PubMed] [Google Scholar]
- 119.Verheyden S, Demanet C. Susceptibility to myeloid and lymphoid leukemia is mediated by distinct inhibitory KIR–HLA ligand interactions. Leukemia. 2006;20:1437–1438. doi: 10.1038/sj.leu.2404279. [DOI] [PubMed] [Google Scholar]
- 120.Demanet C, Mulder A, Deneys V, Worshman M, Class F, Ferrone S. Down-regulation of HLA-A and HLA-Bw6, but not HLA-Bw4, allospecificities in leukemic cells: an escape mechanism from CTL and NK attack? Blood. 2004;103:3122–3130. doi: 10.1182/blood-2003-07-2500. [DOI] [PubMed] [Google Scholar]
- 121.Gagne K, Busson M, Bignon JD, Balère-Appert ML, Loiseau P, Dormoy A, Dubois V, Perrier P, Jollet I, Bois M, Masson D, Moine A, Absi L, Blaise D, Charron D, Raffoux C, ARS2000 FRM and FGM group Donor KIR3DL1/3DS1 gene and recipient Bw4 KIR ligand as prognostic markers for outcome in unrelated hematopoietic stem cell transplantation. Biol Blood Marrow Transpl. 2009;15(11):1366–1375. doi: 10.1016/j.bbmt.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 122.Nearman Z, Wlodarski M, Jankowska A, Howe E, Narvaez Y, Ball E, Maciejewski JP. Immunogenetic factors determining the evolution of T-cell large granular lymphocyte leukaemia and associated cytopenias. Br J Haematol. 2007;136:237–248. doi: 10.1111/j.1365-2141.2006.06429.x. [DOI] [PubMed] [Google Scholar]
- 123.Brouwer RE, van der Heiden P, Schreuder GM, Mulder A, Datema G, Anholts JD, Willemze R, Claas FH, Falkenburg JH. Loss or downregulation of HLA class I expression at the allelic level in acute leukemia is unfrequent but functionally relevant, and can be restored by interferon. Hum Immunol. 2002;63(3):200–210. doi: 10.1016/S0198-8859(01)00381-0. [DOI] [PubMed] [Google Scholar]
- 124.Cheng M, Chen Y, Xiao W, Sun R, Tian Z. NK cell-based immunotherapy for malignant diseases. Cell Mol Immunol. 2013;10(3):230–252. doi: 10.1038/cmi.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Leung W. Infusions of allogeneic natural killer cells as cancer therapy. Clin Cancer Res. 2014;20(13):3390–3400. doi: 10.1158/1078-0432.CCR-13-1766. [DOI] [PubMed] [Google Scholar]
- 126.Davis CT, Rizzieri D. Immunotherapeutic applications of NK cells. Pharmaceuticals (Basel) 2015;8(2):250–256. doi: 10.3390/ph8020250. [DOI] [PMC free article] [PubMed] [Google Scholar]


