Abstract
Herpes simplex virus type 1 (HSV-1) DNA replication intermediates exist in a complex nonlinear structure that does not migrate into a pulsed-field gel. Genetic evidence suggests that the product of the UL12 gene, termed alkaline nuclease, plays a role in processing replication intermediates (R. Martinez, R. T. Sarisky, P. C. Weber, and S. K. Weller, J. Virol. 70:2075–2085, 1996). In this study we have tested the hypothesis that alkaline nuclease acts as a structure-specific resolvase. Cruciform structures generated with oligonucleotides were treated with purified alkaline nuclease; however, instead of being resolved into linear duplexes as would be expected of a resolvase activity, the artificial cruciforms were degraded. DNA replication intermediates were isolated from the well of a pulsed-field gel (“well DNA”) and treated with purified HSV-1 alkaline nuclease. Although alkaline nuclease can degrade virion DNA to completion, digestion of well DNA results in a smaller-than-unit-length product that migrates as a heterogeneous smear; this product is resistant to further digestion by alkaline nuclease. The smaller-than-unit-length products are representative of the entire HSV genome, indicating that alkaline nuclease is not inhibited at specific sequences. To further probe the structure of replicating DNA, well DNA was treated with various known nucleases; our results indicate that replicating DNA apparently contains no accessible double-stranded ends but does contain nicks and gaps. Our data suggest that UL12 functions at nicks and gaps in replicating DNA to correctly repair or process the replicating genome into a form suitable for encapsidation.
Herpes simplex virus type 1 (HSV-1) is an enveloped double-stranded DNA virus (59). The linear genome becomes circular upon infection (20), and the circular template is replicated into longer-than-unit-length head-to-tail concatemers (32). The packaging machinery then cleaves concatemeric DNA to monomeric units, which are packaged into preassembled capsids (74). DNA-containing capsids bud from the nucleus to the cytoplasm, where they acquire an envelope and mature glycoproteins (59).
Although the viral proteins required for DNA replication have been identified, the exact mechanism of replication remains unclear (reviewed in references 5 and 77). Despite suggestions that HSV-1 DNA replication proceeds by a rolling-circle mechanism (58, 67, 68), there is no direct evidence that larger-than-unit-length replication intermediates result solely from rolling-circle replication. HSV-1 DNA replication and recombination appear to be linked (2, 12, 13, 76), and the earliest replication products that can be observed have undergone genomic inversion (1, 45, 83). By analogy with bacteriophage T4, recombination may play a role in the generation of DNA replication intermediates. When cesium chloride-banded viral DNA is viewed under the electron microscope, a number of unusual structures are observed. Such structures include large tangled masses similar to those in replicating DNA from bacteriophage T4 (29), as well as Y-shaped structures and replication bubbles (33, 34, 66). Taken together, these results may indicate that recombination plays a role in the generation of viral replication intermediates.
Recent analysis by pulsed-field gel electrophoresis (PFGE) also suggests that simple rolling-circle replication cannot account for the complex structure of DNA replication intermediates. PFGE can resolve high-molecular-weight linear DNA up to 2,000 kb (75). DNA replication intermediates from cells infected with HSV-1, however, do not even migrate into the pulsed-field gel (1, 10, 45, 48, 61, 83). Although restriction enzymes that cut once per genome length can release linear molecules into the gel, the majority of replicating DNA remains in the well (at the gel origin). These results suggest that replicating DNA is not composed merely of linear concatemers or of large circular forms. Y- and X-shaped branches are found within this structure (62), and these branches may account for its inability to migrate in a pulsed-field gel. DNA replication intermediates may be topologically constrained as well (22, 54). In summary, replicating DNA exists in a complex nonlinear structure whose precise conformation remains unclear.
Since DNA packaged into capsids is linear, replication intermediates must presumably be processed from a complex to a linear form prior to or during cleavage and encapsidation. Viral mutants bearing mutations in the UL12 gene appear deficient in this processing step. The UL12 null mutant (AN-1) is severely compromised for overall growth, with production of progeny at 0.1 to 1% of the levels of the wild type (79). During AN-1 infection, DNA replication produces wild-type quantities of DNA, and full-length genomes are cleaved and packaged into capsids (63). A large number of abortive A capsids are produced, however, and few DNA-containing capsids are detectable in the cytoplasm (63). Alkaline nuclease is therefore required for efficient egress of DNA-containing capsids from the nucleus. The observation that subtle mutations that eliminate exonucleolytic activity in vitro also eliminate function in vivo suggests that the role of alkaline nuclease in vivo is probably a exonucleolytic one (21). Henderson et al. (23) recently reported that the endonucleolytic activity of UL12 is required in vivo; however, the assay system used in that study did not clearly differentiate between endo- and exonucleolytic activities. In any case, UL12 enzymatic activity is required for viral growth.
Cleavage of well DNA (DNA replication intermediates from the well of a pulsed-field gel) isolated from AN-1-infected cells with a restriction enzyme that cuts once per genome length releases no discrete bands into a pulsed-field gel (45). Replicating DNA from AN-1-infected cells is therefore more complex than that from cells infected with wild-type virus. We have proposed that alkaline nuclease is required for a genome maturation step, “processing” complex replication intermediates into a form suitable for encapsidation. Genomes which are cleaved and packaged in the absence of functional alkaline nuclease may retain some level of complexity, such as small branches or other modifications. We suggest that capsids that contain these complex unit-length molecules are unstable and unable to egress from the nucleus (45, 77).
Bacteriophage T4 gene 49 encodes an enzyme termed endonuclease VII, which resolves cruciform structures in replicating DNA into a linear form in order to ensure proper packaging (50). A gene 49 mutant accumulates highly branched, complex DNA (15, 35). In order to determine whether alkaline nuclease performs the analogous function of converting complex DNA into a linear form, we examined its activity in vitro. In addition, we digested well DNA with UL12 and other nucleases with defined specificities to probe the function of UL12 and the structure of well DNA itself. Various models for UL12 function are considered.
MATERIALS AND METHODS
Cells and viruses.
African green monkey kidney cells (Vero; American Type Culture Collection, Manassas, Va.) were propagated as described previously (78). The 6-5 and S22 cell lines, which are permissive for UL12 mutants, have previously been described (8, 63). The KOS strain of HSV-1 was used as the wild-type virus. The UL12-null mutant virus AN-1 has previously been described (79). The AD169 strain of human cytomegalovirus (HCMV) was used for the HCMV studies; this strain was used to infect human foreskin fibroblast (HFF) cells. Both virus and cells were kindly provided by John Shanley, University of Connecticut Health Center.
Proteins.
Recombinant UL12 protein and two UL12 mutant proteins, D340E and G336A/S338A, were purified to >95% homogeneity from Sf9 insect cells infected with recombinant baculoviruses as described previously (21). Purified wild-type UL12 exhibits a specific activity of 2 U/μg; a unit is defined as the amount of enzyme required to render 1 μg of DNA soluble in 1 min. Exonuclease V (ATP-dependent exonuclease) from Micrococcus luteus was obtained from U.S. Biochemicals (Cleveland, Ohio). S1 nuclease was obtained from GIBCO (Gaithersburg, Md.). Staphylococcal nuclease and DNase I were obtained from Pharmacia (Piscataway, N.J.). T4 endonuclease VII (T4 Endo VII) (39) was kindly provided by Börries Kemper (Institut fur Genetik der Universitat zu Koln, Cologne, Germany).
Resolvase assay.
Artificial branched structures were prepared as described previously (4). Briefly, five 60-mer partially complementary oligonucleotides were provided by National Biosciences, Plymouth, Minn., and were termed 4X12-1 (5′-GACGCTGCCGAA TTCTACCAGTGC CTTGCTAGGACA TCTTTGCCCACC TGCAGGTTCACC C-3′), 4X12-2 (5′-TGGGTGAACCTG CAGGTGGGCAAA GATGTCCTAGCA ATGTAATCGTCA AGCTTTATGCCG TT-3′), 4X12-3 (5′-CAACGGCATAAA GCTTGACGATTA CATTGCTAGGAC ATGCTGTCTAGA GGATCCGACTAT CGA-3′), 4X12-4 (5′-ATCGATAGTCGG ATCCTCTAGACA GCATGTCCTAGC AAGGCACTGGTA GAATTCGGCAGC GT-3′), and 4X12-5 (5′-AACGGCATAAAG CTTGACGATTAC ATTGCTAGGACA TCTTTGCCCACC TGCAGGTTCACC CA-3′). Oligonucleotide 4X12-2 was 5′-32P-end labeled by using T4 polynucleotide kinase and γ-32P. The labeled oligonucleotide was mixed with an excess of various combinations of partially complementary oligonucleotides as follows: no oligonucleotide for single-stranded DNA; 4X12-1 for a pseudo-Y-forked substrate; 4X12-1 plus 4X12-3 for a three-way junction; 4X12-1 plus 4X12-3 plus 4X12-4 for a four-way cruciform structure; and 4X12-5 for a double-stranded linear structure. Reaction mixtures containing branched structures and 20 U of T4 Endo VII were incubated at 37°C in 50 mM Tris (pH 8)–10 mM MgCl2–10 mM 2-mercaptoethanol (BME)–100 μg of bovine serum albumin per ml for 20 min. Reaction mixtures containing branched structures and 20 ng (0.04 U) of either UL12, mutant D340E, or mutant G336A/S338A were incubated at 37°C in 50 mM Tris (pH 9)–2 mM MgCl2–5 mM BME for 15 min. Reactions were stopped by the addition of 50 mM EDTA, 10% glycerol, 0.1% bromphenol blue, and 0.1% xylene cyanol, and the products were subjected to electrophoresis through a nondenaturing 8% polyacrylamide gel. Following electrophoresis, gels were dried and exposed to film.
Isolation of DNA.
Well DNA was isolated as described in reference 45 with some modifications. Vero cells in 60-mm-diameter plates were infected at a multiplicity of infection of 5 PFU/cell and harvested 18 h postinfection. Infected cells were suspended in 1% low-melting point (LMP) agarose in phosphate-buffered saline. Agarose plugs were incubated overnight at 37°C in 0.4 M EDTA (pH 9.5)–1% sodium dodecyl sulfate (SDS)–1 mg of proteinase K per ml, followed by five washes with TE (10 mM Tris [pH 8], 1 mM EDTA) at 50°C. Following PFGE, plugs were recovered, incubated in PacI digestion buffer, and treated with PacI to digest cellular DNA (the KOS genome contains no PacI sites). Plugs were then subjected to another round of PFGE, recovered, and stored in TE at 4°C. To isolate well DNA from HCMV-infected cells, confluent monolayers of 106 HFF cells were infected with HCMV at a multiplicity of infection of >5 PFU/cell and harvested 72 h postinfection. Infected cells were treated as described above except that no PacI digestion was performed before the second round of PFGE.
DNA released from the well of a pulsed-field gel by alkaline nuclease was isolated as follows. Well DNA was incubated with buffer or treated with UL12, subjected to PFGE as described above except that LMP agarose was used, and stained with ethidium bromide. Untreated well DNA and released DNA in the treated lane were excised from the LMP agarose. For BamHI digestion, agarose plugs were treated with BamHI and then with GELase (Epicenter Technologies, Madison, Wis.). DNA was precipitated in ethanol, resuspended in 20 μl of TE, and subjected to electrophoresis in a 0.8% agarose gel. The gel was Southern blotted and probed with 32P-labeled KOS genomic DNA labeled with random hexamers as recommended by the manufacturer (Boehringer GmbH, Mannheim, Germany).
Virion DNA was isolated from KOS virions, which were prepared as described previously (70). Virions were resuspended in 1% LMP agarose and cast into plugs for PFGE. Plugs were lysed as described above for infected cells.
Enzymatic treatment of well DNA.
Digestion of well DNA with various enzymes was typically performed as follows. Plugs were cut into 3- by 20-mm blocks, each containing the equivalent of 8 × 104 infected cells. Plugs were incubated overnight at 4°C in the optimum buffer for each enzyme. The buffer was then replaced with 300 μl of fresh buffer and enzyme, and the mixture was incubated on ice for 1 h to permit diffusion and then incubated at 37°C for 2 h. Plugs were washed once with 0.5 M EDTA and four times with TE and then were either incubated with buffer for the next reaction or subjected to PFGE as described previously (45). Southern blots were hybridized to 32P-labeled KOS genomic DNA or a fragment of the HCMV genome. For the time course experiment, reactions were stopped by adding 0.5 M EDTA to the plugs and placing them on ice for 1 h and then the plugs were incubated in TE–1% SDS–1 mg of proteinase K per ml at 37°C overnight. Following SDS-proteinase K treatment, plugs were washed with TE.
UL12 reactions were typically performed with an agarose plug containing well DNA isolated from 8 × 104 infected cells, with 1.2 μg (2.4 U) of UL12 in AN buffer (50 mM Tris-HCl [pH 9], 5 mM MgCl2, 5 mM BME, 100 μg of bovine serum albumin per ml). Staphylococcal nuclease digestion was performed in the buffer described in reference 65. DNase I digestion was performed in 0.1 M sodium acetate (pH 5.0)–5 mM MgCl2. Exonuclease V (exo V) reactions were performed in 67 mM glycine-NaOH (pH 9.4)–30 mM MgCl2–8.3 mM BME–0.5 mM ATP. S1 nuclease reactions were performed in 30 mM NaAc (pH 4.6)–1 mM ZnAc–5% glycerol. For the S1 nuclease control experiments, 1 μg of pUC119 was linearized with BamHI, embedded in plugs of LMP agarose, and treated as described for well DNA. After the reaction was stopped, DNA in the agarose plugs was subjected to electrophoresis on a 0.8% agarose gel, stained with ethidium bromide, and photographed.
RESULTS
UL12 is not a simple resolvase.
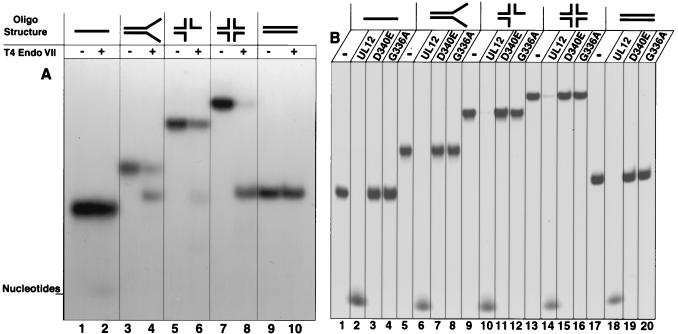
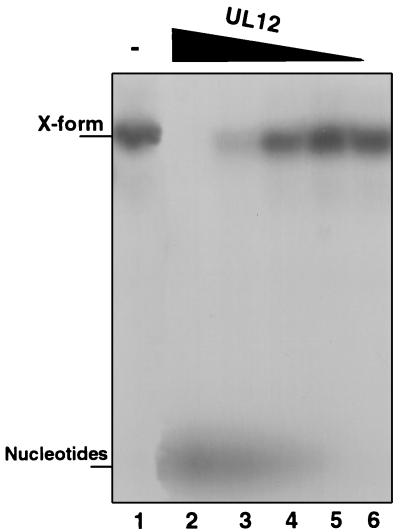
We have previously proposed that UL12 may act as a cruciform resolvase, in a manner analogous to the activities of T4 Endo VII or Escherichia coli RuvC (45). This hypothesis was tested by constructing artificial branched structures with oligonucleotides as described previously (4). As a control, these substrates were first incubated with purified T4 Endo VII, which exhibits a well-characterized resolvase activity (49). Figure 1A shows that T4 Endo VII specifically cleaves cruciform structures into a duplex linear form (lanes 7 and 8). T4 Endo VII also exhibits resolving activity on other branched substrates (Fig. 1A, lanes 3 to 6). The activity of alkaline nuclease was then compared to that of T4 Endo VII by incubating the same structures with alkaline nuclease purified from recombinant baculovirus-infected insect cells as described previously (21). In contrast to T4 Endo VII, UL12 simply degrades branched structures (Fig. 1B, lanes 2, 6, 10, 14, and 18). This result suggests either that UL12 is not a resolvase or that the exonuclease activity of UL12 masks any cruciform-resolving activity. In order to examine whether the endonuclease activity of UL12 could act as a resolvase in the absence of exonuclease activity, we incubated the branched structures with two previously described mutant UL12 proteins which exhibit no exonuclease activity (21). We have previously shown that the mutant D340E protein preparation retains some residual endonuclease activity, while the mutant G336A/S338A protein preparation exhibits no endonuclease activity (21). When these mutant proteins were incubated with the artificial structures, no resolvase activity was observed (Fig. 1B, lanes 3, 4, 7, 8, 11, 12, 15, 16, 19, and 20). Furthermore, these results show that the exonucleolytic degradation exhibited by the wild-type protein was not due to a contaminating nuclease. Finally, in order to determine whether wild-type UL12 could exhibit some resolvase activity under conditions that minimize exonuclease activity, branched structures were incubated with various amounts of UL12 (0.012 to 1.2 U) and at pH 8 with Mn2+, which has previously been reported to enhance endonuclease activity at the expense of exonuclease activity (25). Figure 2 shows that no evidence for an internal cleavage event was seen even under limiting exonuclease conditions. Although some structure-specific endonucleases exhibit exonuclease activity (42), the exonuclease activity is typically less active and more difficult to detect than that of the endonuclease. This is not the case for UL12. The simplest explanation for the result described here is that alkaline nuclease does not act as a simple resolvase.
FIG. 1.
Alkaline nuclease activity on preformed branched substrates. Branched structures were constructed from oligonucleotides as described under Materials and Methods. (A) Branched structures were treated with T4 Endo VII. Lane assignments for the structures used are as follows: lanes 1 and 2, single-stranded linear DNA; lanes 3 and 4, forked DNA (pseudo-Y shaped) in which the upper single-stranded end is 3′; lanes 5 and 6, three-way junction DNA in which the rightmost single-stranded end is 3′ and the lower single-stranded end is 5′; lanes 7 and 8, cruciform DNA (X-shaped form); lanes 9 and 10, double-stranded linear DNA. (B) Branched structures were treated with either wild-type alkaline nuclease (lanes 2, 6, 10, 14, and 18), mutant D340E (lanes 3, 7, 11, 15, and 19), or mutant G336A/S338A (lanes 4, 8, 12, 16, and 20). For lanes labeled with a minus sign, structures were not treated.
FIG. 2.
Alkaline nuclease activity on cruciform substrates in the presence of MnCl2. Cruciform (X-shaped) structures were treated with alkaline nuclease in a pH 8 reaction buffer containing 5 mM MnCl2. Lane 1, no enzyme; lane 2, 1.2 U of UL12; lane 3, 0.12 U of UL12; lane 4, 0.012 U of UL12; lane 5, 0.0012 U of UL12; lane 6, 0.00012 U of UL12. Reaction products were subjected to nondenaturing 8% polyacrylamide gel electrophoresis.
Well DNA can be released into a pulsed-field gel by UL12.
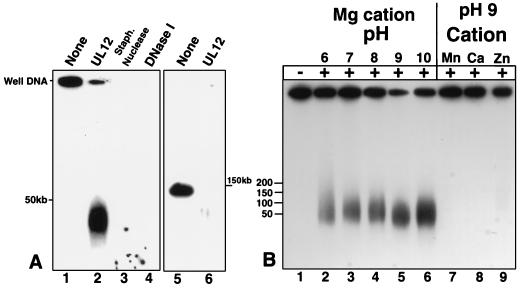
Genetic evidence has suggested that alkaline nuclease plays a role in the correct processing of DNA replication intermediates. Replication intermediates were isolated as well DNA as described previously (45) and treated with UL12 (Fig. 3A, lane 2). After UL12 digestion for 2 h under optimal conditions (pH 9 with Mg2+ as the cation), a significant amount of well DNA was released as a heterogeneous smear ranging in size from 10 to 50 kb. In contrast, lanes 3 and 4 show that both staphylococcal nuclease and DNase I can degrade well DNA to completion. Alkaline nuclease is known to degrade both single- and double-stranded DNA and even closed circular supercoiled plasmids (due to endonuclease activity that nicks this substrate) (7, 21, 25, 27, 37, 69). At pH 9 in the presence of magnesium, alkaline nuclease degrades equivalent amounts of agarose-embedded salmon sperm DNA (data not shown) and virion DNA (Fig. 3A, lane 6) to completion. Well DNA therefore appears unique in its partial resistance to UL12 activity. The ability of UL12 to release well DNA was not dependent on the concentration of UL12, the concentration of DNA in agarose, or the time postinfection at which well DNA was harvested from infected cells (data not shown). Lanes 2 to 6 of Fig. 3B show that in the presence of magnesium, UL12 releases well DNA into the same heterogeneous smear over a pH range of 6 to 10, despite the fact that alkaline nuclease is more than 100-fold more active at high pH than at low pH (7, 26, 69). The released product in Fig. 3B migrates at a slightly different molecular weight than that in Fig. 3A; despite this variation between experiments, no differences in migration were observed within any experiment, and the observed differences may be due to temperature variation during electrophoresis. Manganese has previously been reported to enhance UL12 endonucleolytic activity and to repress exonucleolytic activity (25). When manganese is substituted for magnesium at pH 9, no releasing activity is detected (Fig. 3B, lane 7). In addition, neither calcium nor zinc can substitute for magnesium (lanes 8 and 9). These results suggest that the exonucleolytic activity of UL12, rather than the endonucleolytic activity, is responsible for the release of well DNA into a pulsed-field gel.
FIG. 3.
Alkaline nuclease activity on well DNA. Well DNA was prepared from KOS-infected cells as described under Materials and Methods. (A) Well DNA was either untreated (lane 1) or treated with UL12 (lane 2), staphylococcal nuclease (lane 3), or DNase I (lane 4) as described under Materials and Methods. Virion DNA was untreated (lane 5) or treated with UL12 (lane 6). (B) Well DNA was treated with UL12 in AN buffer, either unmodified (lane 5) or with the following modifications in pH and/or components: 50 mM N,N-methylenebisacrylamide (BIS)–Tris, pH 6 (lane 2); 50 mM BIS-Tris, pH 7 (lane 3); 50 mM Tris, pH 8 (lane 4); 50 mM 3-(cyclohexylamino)-1-propanesulfonic acid (CAPS), pH 10 (lane 6); 5 mM MnCl2 (lane 7); 5 mM CaCl2 (lane 8); 5 mM ZnCl2 (lane 9). Samples were subjected to PFGE in 1.3% agarose gels. Southern blots of the gels were probed with 32P-labeled KOS genomic DNA. The molecular weight ladder was the Bio-Rad lambda ladder pulsed-field gel marker (1 Mb to 50 kb). Lane 1, no treatment.
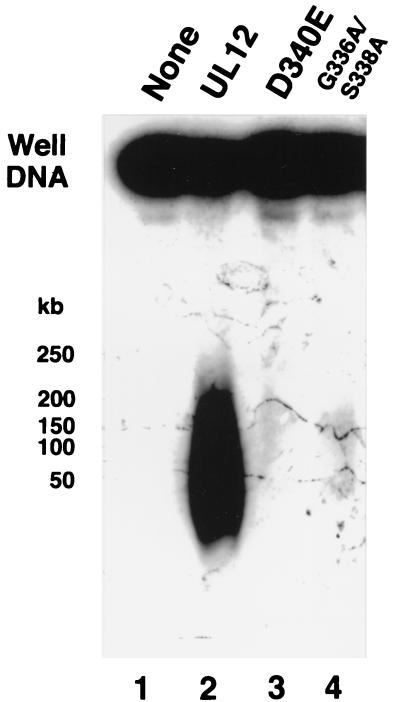
The releasing activity was further analyzed with the two mutant UL12 proteins described above, which exhibit no exonuclease activity. Well DNA was incubated with wild-type UL12, mutant D340E, or mutant G336A/S338A. For this experiment, electrophoresis conditions which would be expected to broaden the released smear were chosen and the blot was overexposed in order to reveal any specific bands that may be present. Figure 4 shows that neither mutant protein can release well DNA into a pulsed-field gel. The releasing reaction therefore requires the exonuclease activity of UL12.
FIG. 4.
Activities of mutant forms of alkaline nuclease on well DNA. Well DNA was prepared from KOS-infected cells. Plugs were incubated at 4°C overnight in AN buffer. The buffer was replaced with fresh buffer, and then the plugs were treated with UL12, mutant D340E, or mutant G336A/S338A. Plugs were subjected to PFGE in a 1% agarose gel. A Southern blot of the gel was probed with KOS genomic DNA. The blot was overexposed to show any faint bands or activity.
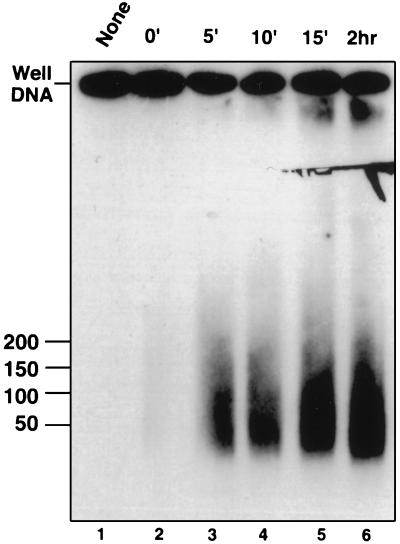
A time course experiment was performed to look for intermediates that might form during the UL12 releasing reaction. Lane 2 of Fig. 5 shows that in plugs incubated at 4°C, no well DNA was released despite the presence of UL12. After 5 min at 37°C, the heterogeneous smear is released into the well (lane 3). No intermediates between well DNA and the final smear were observed even at the 5-min time point. The product after 2 h of digestion (lane 6) migrates in the same molecular weight range as that produced after 5 min (lane 3). One explanation for this result is that UL12 is highly processive and acts upon one molecule to completion before diffusing through the agarose to another. Under this scenario, any molecule released into the well will have been processed to completion; this may explain the absence of reaction intermediates seen in Fig. 5. In support of this hypothesis, the Epstein-Barr virus DNase has been reported to act processively on double-stranded DNA (43).
FIG. 5.
Time course of alkaline nuclease activity on well DNA. Agarose plugs containing well DNA were incubated overnight at 4°C in AN buffer. The buffer was replaced with fresh buffer, and UL12 was added to samples 2 to 6. Following 1 h on ice, samples were subjected to 37°C (time zero). At various times, reactions were stopped as described under Materials and Methods. Plugs were subjected to PFGE in a 1% agarose gel. A Southern blot of the gel was probed with KOS genomic DNA. Reaction times: 2 h (lane 1), 0 min (lane 2), 5 min (lane 3), 10 min (lane 4), 15 min (lane 5), and 2 h (lane 6).
Released DNA is not sequence specific.
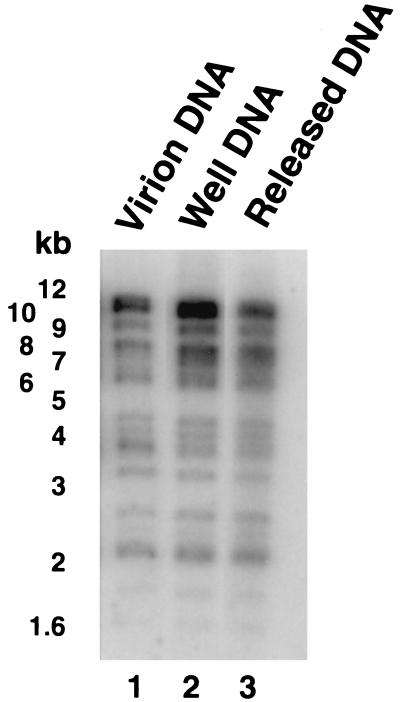
One explanation for the “resistance” of released DNA to further UL12 digestion is that specific regions of replicating HSV-1 DNA may block UL12 activity. For example, the nuclease activity of the E. coli RecBCD enzyme is attenuated at specific chi sequences (40). If such sequences exist for HSV-1, then released DNA may represent only a subset of HSV-1 genomic sequences. To test this hypothesis, released DNA was excised from a LMP agarose gel following PFGE and digested with BamHI (42 recognition sites per genome). BamHI treatment produces a well-characterized pattern of restriction fragments (44, 56). Figure 6 shows the restriction pattern following BamHI digestion of virion DNA, well DNA, and UL12-released DNA. No major differences between well DNA and released DNA were observed in the pattern of bands. An enhanced band migrating below the 4-kb marker in BamHI-digested virion DNA (lane 1) probably represents the unique short terminus. Released DNA does not appear to represent a specific subset of sequences from the viral genome, which suggests that no specific sequences are protected from UL12 activity. This result is consistent with the previous finding that virion DNA is completely degraded by UL12 (Fig. 3A, lane 6).
FIG. 6.
BamHI digestion of released DNA. Well DNA was treated with UL12 or was left untreated and subjected to PFGE in a 1.3% LMP agarose gel. Released DNA and untreated well DNA were excised from the gel, digested with BamHI, treated with GELase, and ethanol precipitated. DNA was resuspended in TE and subjected to electrophoresis on a 0.8% agarose gel. Virion DNA was digested with BamHI as a control. A Southern blot of the gel was probed with KOS genomic DNA. The molecular weight ladder was a GIBCO 1-kb DNA ladder.
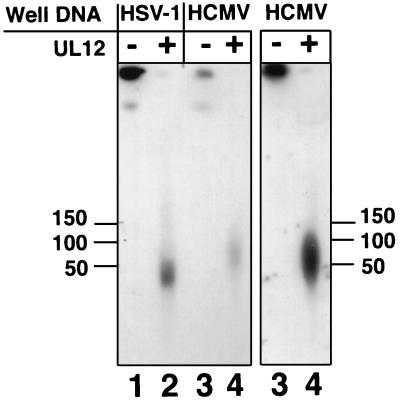
We additionally examined the specificity of UL12 activity by incubating it with well DNA from cells infected with the distantly related herpesvirus HCMV. The HCMV genome is 230 kb, in contrast to the 152-kb genome of HSV-1, and limited homology exists between the two virus genomes (28, 47). The replication fork machinery, however, is conserved, and replicating DNA from cells infected with HCMV exhibits properties similar to those of DNA from cells infected with HSV-1 (48, 52, 60). Well DNA was harvested from HCMV-infected cells and treated with UL12. Lane 4 of Fig. 7 shows that UL12 releases HCMV well DNA into the pulsed-field gel, although the released DNA migrates at a higher molecular weight than released DNA from HSV-1 well DNA (compare lanes 2 and 4). These data imply that the ability of UL12 to release well DNA into molecules which are resistant to further UL12 digestion is dependent upon the structure of replicating DNA, rather than upon the DNA sequence.
FIG. 7.
Alkaline nuclease activity on HCMV well DNA. Well DNA from HSV-1-infected cells or HCMV-infected cells was subjected to treatment with UL12. Following treatment, plugs were subjected to PFGE in a 1.3% agarose gel. A Southern blot of the gel was probed first with KOS genomic DNA (left panel) and then was stripped and reprobed with an HCMV-specific probe (right panel).
Well DNA exists in a structure which is resistant to exonuclease activity.
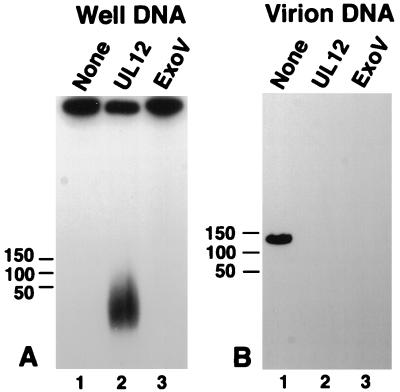
Well DNA appears to contain a structural modification that prevents its complete digestion by UL12. To probe the structure of replicating DNA, we tested its susceptibility to M. luteus exo V, a double-stranded end-specific exonuclease which digests linear but not circular molecules (51, 71). Umene and Nishimoto have previously used this enzyme to examine early circularization of the HSV-1 genome (73). Lane 3 of Fig. 8A shows that well DNA appears to be resistant to exo V digestion. In contrast, virion DNA was completely degraded under the same conditions (Fig. 8B). This result suggests that well DNA may not contain accessible double-stranded ends. Although well DNA exhibits unique long termini, these may be relatively infrequent (41, 45, 61, 83); the bulk of the DNA in replication intermediates may exhibit no termini or other free ends. Alternatively, it is possible that well DNA contains double-stranded ends but that an altered secondary structure at or near these ends confers resistance to exo V digestion.
FIG. 8.
Exo V activity on well and virion DNA. (A) Well DNA from KOS-infected cells was subjected to treatment with either UL12 or exo V, a double-stranded end-specific exonuclease. (B) KOS virion DNA suspended in a plug of agarose was subjected to treatment with either UL12 or exo V. Following treatment, plugs were subjected to PFGE in 1.3% agarose gels. Southern blots of the gels were probed with KOS genomic DNA.
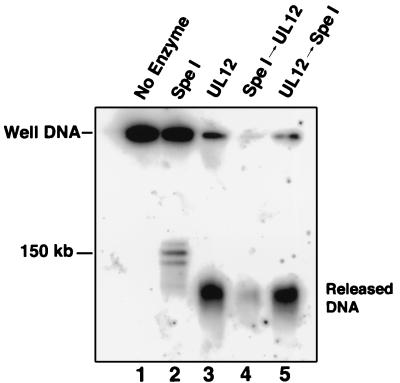
We hypothesized that a lack of double-stranded ends in well DNA may be responsible for the resistance of this DNA to complete UL12 digestion. To test this model, we pretreated well DNA with the restriction enzyme SpeI and then digested it with UL12 (Fig. 9). Control lane 2 shows that SpeI releases three bands (due to HSV-1 genomic inversion [45]), and control lane 5 shows that SpeI treatment following UL12 digestion has little effect. Lane 4 of Fig. 9 shows that following SpeI treatment, well DNA is much more susceptible to digestion by UL12. Therefore, UL12 is capable of digesting well DNA provided that double-strand breaks are available. This experiment further supports the notion that resistance to UL12 digestion is not sequence specific.
FIG. 9.
Alkaline nuclease activity on well DNA following the introduction of double-strand breaks. Well DNA was incubated with SpeI, UL12, or both enzymes in succession (arrows) as shown. All samples were subjected to a series of three incubations: SpeI buffer, AN buffer, and SpeI buffer. Samples were washed with TE between treatments. DNA was subjected to PFGE in a 1.3% agarose gel. A Southern blot of the gel was probed with KOS genomic DNA.
Well DNA contains regions of single-strandedness.
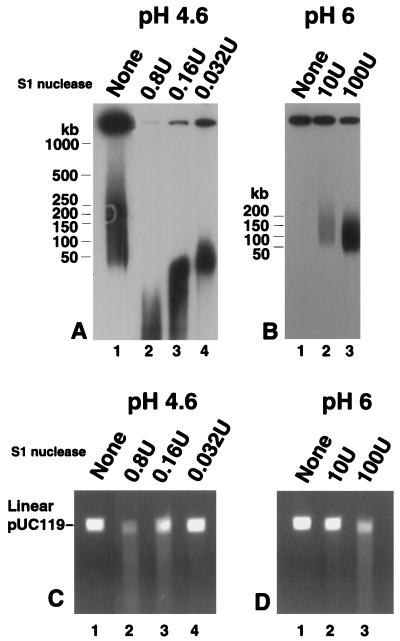
DNA containing single-stranded regions, such as nicks or gaps, would be expected to act as a substrate for UL12 but not exo V. A number of reports have suggested that nicks and gaps are present in both virion and replicating DNA (3, 17, 24, 31, 36, 57, 66, 80, 81). We examined whether well DNA contains single-stranded regions by treating it with S1 nuclease, a single-strand-specific endonuclease which cuts opposite nicks and gaps and at other regions of single-strandedness. Figure 10A shows that well DNA is, in fact, susceptible to S1 nuclease digestion. Under optimal S1 nuclease conditions (pH 4.6), as little as 0.032 U was sufficient to release the bulk of well DNA into a pulsed-field gel (Fig. 10A, lane 4). Increasing concentrations produced progressively smaller fragments (Fig. 10A, lanes 2 and 3). Unexpectedly, incubation of well DNA in S1 nuclease buffer alone resulted in some DNA breakage (Fig. 10A, lane 1). We therefore determined conditions under which well DNA would remain at the gel origin but S1 nuclease would retain activity. At pH 6, well DNA appears to remain intact in the absence of enzyme (Fig. 10B, lane 1). S1 nuclease does remain active under these conditions, although 100- to 1,000-fold-larger amounts of enzyme are necessary (Fig. 10B, lanes 2 and 3). S1 nuclease releases a heterogeneous smear of DNA into a pulsed-field gel. Next, we asked whether S1 nuclease behaves in a single-strand-specific manner under the reaction conditions used in this experiment. S1 nuclease activity on a linearized double-stranded plasmid was examined. Linear pUC119 was embedded in agarose, incubated in S1 nuclease buffer at either pH 4.6 or pH 6, treated with S1 nuclease, and subjected to gel electrophoresis. Figure 10C shows that at pH 4.6, high concentrations of S1 nuclease do in fact degrade double-stranded DNA (Fig. 10C, lane 2) but that lower concentrations have little effect (Fig. 10C, lanes 3 and 4). Similarly, at pH 6, 10 U of S1 nuclease does not digest the plasmid (Fig. 10D, lane 2), while this amount is sufficient to release well DNA into the pulsed-field gel (Fig. 10B, lane 2). Thus, the releasing activity observed in Fig. 10A, lanes 3 and 4, and Fig. 10B, lane 2, is probably due to digestion of single-stranded, not double-stranded, DNA. Furthermore, the absence of specific bands suggests that single-stranded regions are located randomly throughout the genome, consistent with the findings of Wilkie et al. (81).
FIG. 10.
S1 nuclease activity on well DNA. Well DNA isolated from KOS-infected cells was subjected to treatment with S1 nuclease as follows. (A) Well DNA was incubated in S1 nuclease buffer containing 30 mM NaAc, pH 4.6, and then subjected to S1 nuclease treatment as indicated above the lanes. (B) Well DNA was incubated with S1 nuclease buffer containing 50 mM BIS–Tris, pH 6, and then subjected to S1 nuclease treatment as indicated above the lanes. Following treatment, plugs were subjected to PFGE in 1.3% agarose gels. Southern blots of the gels were probed with KOS genomic DNA. The molecular weight ladder was a Bio-Rad lambda ladder. Plasmid pUC119 was linearized with BamHI, embedded in plugs of 1% LMP agarose, and treated with S1 nuclease as for panel A (C) or as for panel B (D). Following treatment, plugs were subjected to electrophoresis in a 0.8% agarose gel. The gel was stained with ethidium bromide and photographed.
DISCUSSION
Several observations regarding alkaline nuclease activity and the structure of replicating DNA are made in this report. (i) Purified HSV-1 UL12 exhibits exonucleolytic activity on preformed cruciforms and no detectable resolvase activity. (ii) Alkaline nuclease can release replicating HSV-1 DNA into a pulsed-field gel in an exonucleolytic reaction which does not appear to exhibit any sequence specificity. (iii) The DNA released by UL12 digestion migrates as a smear of smaller-than-unit-length DNA which is resistant to further digestion with UL12. (iv) Well DNA is resistant to digestion with exo V, which acts at double-stranded termini, suggesting that replicating DNA does not contain accessible double-stranded ends. (v) Well DNA is susceptible to digestion with S1 nuclease, confirming previous reports that replicating DNA contains nicks and gaps.
Previous studies with a UL12-null mutant virus led to the proposal that alkaline nuclease acts to resolve recombination intermediates generated as a result of DNA replication (45). An artificial cruciform was treated with purified UL12, and rather than being resolved into linear forms as would be expected from a resolvase, the substrate was degraded exonucleolytically. Although it is possible that in vivo other viral or cellular proteins may modulate the exonucleolytic activity of UL12 resulting in an alteration of its activity, the simplest explanation for these results is that alkaline nuclease does not function as a simple cruciform resolvase. This explanation is consistent with our recent finding that the exonucleolytic activity of UL12 is required in vivo (21). Both biochemical and genetic evidence indicates, therefore, that exonuclease activity is important for UL12 function.
Although UL12 digested virion DNA and salmon sperm DNA to completion, it did not completely degrade well DNA. UL12 instead released well DNA into the gel, generating a wide band migrating as smaller-than-unit-length molecules which were resistant to further digestion. This unexpected result has two significant implications: (i) UL12 removes the structural features that prevent well DNA from migrating into a pulsed-field gel, and (ii) well DNA contains structures that prevent UL12 from completely degrading it. UL12 releases a similar heterogeneous smear from well DNA isolated from UL12− and UL6− mutant infected cells (data not shown), suggesting that neither the cleavage and packaging machinery nor UL12 is responsible for the generation of these structural features. The structures that confer UL12 resistance are probably a result of the process of DNA replication itself.
The releasing activity was studied further in an attempt to understand how the exonucleolytic activity of UL12 may function in vivo in the maturation of viral genomes prior to packaging. No intermediates between well DNA and the released product were observed at early times in the reaction, consistent with findings that UL12 acts processively (43, 69). In addition, UL12 does not appear to act in a sequence-specific manner. Released DNA migrates as a heterogeneous smear; no specific region of the genome is preferentially degraded or protected; and UL12 activity on well DNA from HCMV-infected cells is similar to that on well DNA from HSV-1-infected cells despite the large disparity in genome sequence. This finding is consistent with recent work demonstrating that the HCMV UL12 homolog can functionally substitute for UL12 in vivo (19). UL12 probably acts on a particular structure or conformation of replicating DNA which exists in both HSV-1- and HCMV-infected cells rather than on any particular sequence. Finally, the finding that released DNA is smaller than unit length may indicate that the releasing reaction observed in vitro does not completely mimic UL12 activity in vivo, where the activity of UL12 can be modulated or attenuated by interactions with other proteins (43, 72).
This report also demonstrates that alkaline nuclease does not completely degrade well DNA. To our knowledge, well DNA is the first published DNA substrate that is not digested to completion by this enzyme. As UL12 exhibits no sequence specificity, well DNA must contain a structural modification that prevents UL12 progression. It is possible that some stretches of well DNA contain unusual bases, such as covalent DNA modifications, which block UL12 progression. This model is not consistent, however, with the observation that the introduction of double-stranded breaks renders the bulk of well DNA sensitive to UL12 digestion (Fig. 8). No stretches of DNA are inherently resistant to UL12 activity. Another possibility is that one or more proteins are covalently attached to well DNA and amino acid residues that survive proteinase K treatment prevent exonucleolytic progression. It has previously been reported that up to four proteins may be tightly (although not covalently) bound near the ends of virion DNA (30, 82), and it is possible that replicating DNA may also exist in a protein-bound form. Alternatively, unusual secondary structures may form during DNA replication at random positions throughout the genome and these structures may prevent UL12 progression. Whether well DNA is protected by covalently attached amino acid residues or by specific secondary structures, the introduction of double-strand breaks may provide UL12 with loading sites from which it can digest most of the DNA.
We probed the structure of replicating DNA by using a series of endo- and exonucleases with defined biochemical activities. The apparent resistance of well DNA to exo V activity suggests that replicating DNA contains few if any accessible double-stranded ends. It has been known for some time that HSV-1 DNA becomes circular soon after infection (20, 32, 55), and Zhang et al. (83) have proposed that DNA replication may proceed by a double-rolling-circle mechanism similar to that used by the 2μm plasmid of Saccharomyces cerevisiae (6). If the genome is maintained in a circular form during replication, no double-stranded ends would be present in replicating DNA. Alternatively, as mentioned above, the replicating genome may contain ends that are structurally modified in some way which prevents exonucleolytic progression. In contrast, replicating DNA is susceptible to a releasing reaction by UL12 and S1 nuclease, suggesting that replicating DNA contains nicks, gaps, or other internal structures that are inaccessible to exo V.
Although no sequence homology between UL12 and other known exo- or endonucleases can be detected, alkaline nuclease is well conserved throughout the herpesvirus family, suggesting that it plays an important role in the life cycle of herpesviruses. The growth properties of null and other mutants of alkaline nuclease confirm that it is essential for efficient production of progeny virus (11, 19, 46, 53, 79). The major defect in UL12-null mutant infections is manifested at the point of capsid egress from the nucleus; in the absence of UL12, few if any DNA-containing capsids appear in the cytoplasm (63). We believe that the defect in egress is due to a defect in genome maturation, which in turn affects the stability of DNA-containing capsids. Specifically, the DNA which accumulates in cells infected with UL12-null mutants is even more complex than that in cells infected with wild-type viruses (45). Furthermore, when well DNA is isolated from cells infected with wild-type and AN-1 mutant viruses and the agarose is digested, AN-1 well DNA, as observed by electron microscopy, appears to be much shorter (approximately 6 kb) than that isolated from wild-type virus-infected cells (approximately 66 kb) (21a). This result suggests that while wild-type well DNA is fragile and shears upon isolation from agarose, AN-1 well DNA is up to 10-fold more fragile. We propose that in both wild-type and AN-1 virus infections, the complexity and fragility of viral replicating DNA may be caused by the existence of aberrant structures which both prevent pulsed-field gel migration and are fragile to manipulation. These structural features are apparently present with an increased frequency in the absence of UL12.
Although the nature of these putative aberrant structures remains unclear, they may be remnants of recombination or replication. Two mechanisms could explain their enhanced frequency in replicating DNA from cells infected with the AN-1 mutant. First, UL12 may normally act to resolve or process aberrant structures following replication. Although UL12 may not act as a cruciform resolvase, its exonucleolytic activity may function to remove such structures as a form of DNA repair. A role for exonucleases in secondary structure repair has been proposed for other systems (16). On the other hand, UL12 may act during replication to prevent the formation of aberrant structures. By analogy with bacteriophage T4, late replication may be initiated by strand invasion of 3′ ends, which can then be extended to form productive replication forks. Sheaffer et al. have proposed that alkaline nuclease may act at the 5′ ends of lagging strands to prevent 5′-end strand invasion, which might lead to junctions that cannot promote further replication (64). Precedent for such a mechanism appears in the E. coli system: only 3′ invading strands are productive for RecA/single-stranded binding protein-mediated joint formation (14, 18, 38), and the 5′ single-strand exonuclease RecJ appears to enhance strand transfer (9) by removing 5′ invading strands (18). Although DNA replication and recombination appear to proceed at wild-type levels in the absence of alkaline nuclease activity, it is possible that redundant mechanisms exist to initiate these processes. In the absence of alkaline nuclease, the presence of unresolvable 5′ junctions may result in structures which cannot be stably packaged into capsids. Regardless of the mechanism, our results are consistent with a model in which UL12 acts to remove potentially harmful structures from HSV-1 replicating DNA. Further characterization of these structures will clearly be important to further understand not only the mechanism of UL12 activity but also the role of recombination in herpesvirus DNA replication.
ACKNOWLEDGMENTS
We thank Börries Kemper for providing T4 Endo VII. We thank Elizabeth Mostello, Richard Zeff, and John Shanley for providing HCMV-infected HFF cells. We thank members of our laboratory for helpful discussions of the work and the manuscript.
This investigation was supported by Public Health Service grants AI21747 and AI37549 and by a Faculty Research Grant from the University of Connecticut Health Center Research Advisory Committee.
REFERENCES
- 1.Bataille D, Epstein A. Herpes simplex virus replicative concatemers contain L components in inverted orientation. Virology. 1994;203:384–388. doi: 10.1006/viro.1994.1498. [DOI] [PubMed] [Google Scholar]
- 2.Bataille D, Epstein A L. Herpes simplex virus type 1 replication and recombination. Biochimie. 1995;77:787–795. doi: 10.1016/0300-9084(96)88197-1. [DOI] [PubMed] [Google Scholar]
- 3.Ben-Porat T, Stehn B, Kaplan A S. Fate of parental herpesvirus DNA. Virology. 1976;71:412–422. doi: 10.1016/0042-6822(76)90369-x. [DOI] [PubMed] [Google Scholar]
- 4.Benson F E, West S C. Substrate specificity of the Escherichia coli RuvC protein. J Biol Chem. 1994;269:5195–5201. [PubMed] [Google Scholar]
- 5.Boehmer P E, Lehman I R. Herpes simplex virus DNA replication. Annu Rev Biochem. 1997;66:347–384. doi: 10.1146/annurev.biochem.66.1.347. [DOI] [PubMed] [Google Scholar]
- 6.Broach J R, Volkert F C. Circular DNA plasmids of yeasts. In: Broach J R, Pringle J R, Jones E W, editors. The molecular and cellular biology of the yeast saccharomyces: genome dynamics, protein synthesis, and energetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 297–331. [Google Scholar]
- 7.Bronstein J C, Weber P C. Purification and characterization of herpes simplex virus type 1 alkaline exonuclease expressed in Escherichia coli. J Virol. 1996;70:2008–2013. doi: 10.1128/jvi.70.3.2008-2013.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carmichael E P, Kosovsky M J, Weller S K. Isolation and characterization of herpes simplex virus type 1 host range mutants defective in viral DNA synthesis. J Virol. 1988;62:91–99. doi: 10.1128/jvi.62.1.91-99.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corrette-Bennett S E, Lovett S T. Enhancement of RecA strand-transfer activity by the RecJ exonuclease of Escherichia coli. J Biol Chem. 1995;270:6881–6885. doi: 10.1074/jbc.270.12.6881. [DOI] [PubMed] [Google Scholar]
- 10.Deshmane S L, Raengsakulrach B, Berson J F, Fraser N W. The replicating intermediates of herpes simplex virus type 1 DNA are relatively short. J Neurovirol. 1995;1:165–176. doi: 10.3109/13550289509113963. [DOI] [PubMed] [Google Scholar]
- 11.deWind N, Peeters B P H, Zijderveld A, Gielkens A L J, Berns A J M, Kimman T G. Mutagenesis and characterization of a 41-kilobase-pair region of the pseudorabies virus genome: transcription map, search for virulence genes, and comparison with homologs of herpes simplex virus type 1. Virology. 1994;200:784–790. doi: 10.1006/viro.1994.1242. [DOI] [PubMed] [Google Scholar]
- 12.Dutch R E, Bianchi V, Lehman I R. Herpes simplex virus type 1 DNA replication is specifically required for high-frequency homologous recombination between repeated sequences. J Virol. 1995;69:3084–3089. doi: 10.1128/jvi.69.5.3084-3089.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dutch R E, Bruckner R C, Mocarski E S, Lehman I R. Herpes simplex virus type 1 recombination: role of DNA replication and viral a sequences. J Virol. 1992;66:277–285. doi: 10.1128/jvi.66.1.277-285.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dutreix M, Rao B J, Radding C M. The effects on strand exchange of 5′ versus 3′ ends of single-stranded DNA in RecA nucleoprotein filaments. J Mol Biol. 1991;219:645–654. doi: 10.1016/0022-2836(91)90661-o. [DOI] [PubMed] [Google Scholar]
- 15.Flemming M, Deumling B, Kemper B. Function of gene 49 of bacteriophage T4 III. Isolation of Holliday structures from very fast-sedimenting DNA. Virology. 1993;196:910–913. doi: 10.1006/viro.1993.1557. [DOI] [PubMed] [Google Scholar]
- 16.Fraser M J. Endo-exonucleases: enzymes involved in DNA repair and cell death? Bioessays. 1994;16:761–766. doi: 10.1002/bies.950161011. [DOI] [PubMed] [Google Scholar]
- 17.Frenkel N, Roizman B. Separation of the herpesvirus deoxyribonucleic acid duplex into unique fragments and intact strand on sedimentation in alkaline gradients. J Virol. 1972;10:565–572. doi: 10.1128/jvi.10.4.565-572.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedman-Ohana R, Cohen A. Heteroduplex joint formation in Escherichia coli recombination is initiated by pairing of a 3′-ending strand. Proc Natl Acad Sci USA. 1998;95:6909–6914. doi: 10.1073/pnas.95.12.6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao, M., B. J. Robertson, P. J. McCann III, D. R. O’Boyle II, S. K. Weller, W. W. Newcomb, J. C. Brown, and S. P. Weinheimer. Functional conservation of the alkaline nuclease of herpes simplex type-1 and human cytomegalovirus. Virology, in press. [DOI] [PubMed]
- 20.Garber D A, Beverley S M, Coen D M. Demonstration of circularization of herpes simplex virus DNA following infection using pulsed field gel electrophoresis. Virology. 1993;197:459–462. doi: 10.1006/viro.1993.1612. [DOI] [PubMed] [Google Scholar]
- 21.Goldstein J N, Weller S K. The exonuclease activity of HSV-1 UL12 is required for in vivo function. Virology. 1998;244:442–457. doi: 10.1006/viro.1998.9129. [DOI] [PubMed] [Google Scholar]
- 21a.Goldstein, J. N., and S. K. Weller. Unpublished data.
- 22.Hammarsten O, Yao X, Elias P. Inhibition of topoisomerase II by ICRF-193 prevents efficient replication of herpes simplex virus type 1. J Virol. 1996;70:4523–4529. doi: 10.1128/jvi.70.7.4523-4529.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henderson J O, Ball-Goodrich L J, Parris D S. Structure-function analysis of the herpes simplex virus type 1 UL12 gene: correlation of deoxyribonuclease activity in vitro with replication function. Virology. 1998;243:247–259. doi: 10.1006/viro.1998.9054. [DOI] [PubMed] [Google Scholar]
- 24.Hirsch I, Roubal J, Vonka V. Replicating DNA of herpes simplex virus type 1. Intervirology. 1976;7:155–175. doi: 10.1159/000149949. [DOI] [PubMed] [Google Scholar]
- 25.Hoffmann P J. Mechanism of degradation of duplex DNA by the DNase induced by herpes simplex virus. J Virol. 1981;38:1005–1014. doi: 10.1128/jvi.38.3.1005-1014.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffmann P J, Cheng Y-C. The deoxyribonuclease induced after infection of KB cells by herpes simplex virus type 1 or type 2. J Biol Chem. 1978;253:3557–3562. [PubMed] [Google Scholar]
- 27.Hoffmann P J, Cheng Y-C. DNase induced after infection of KB cells by herpes simplex virus type 1 or type 2. J Virol. 1979;32:449–457. doi: 10.1128/jvi.32.2.449-457.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang E-S, Pagano J S. Human cytomegalovirus. II. Lack of relatedness to DNA of herpes simplex I and II, Epstein-Barr virus, and nonhuman strains of cytomegalovirus. J Virol. 1974;13:642–645. doi: 10.1128/jvi.13.3.642-645.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huberman J A. Visualization of replicating mammalian and T4 bacteriophage DNA. Cold Spring Harbor Symp Quant Biol. 1968;33:509–524. doi: 10.1101/sqb.1968.033.01.059. [DOI] [PubMed] [Google Scholar]
- 30.Hyman R W. Identification of proteins tightly bound to herpes simplex virus DNA. Virology. 1980;105:254–255. doi: 10.1016/0042-6822(80)90174-9. [DOI] [PubMed] [Google Scholar]
- 31.Hyman R W, Oakes J E, Kudler L. In vitro repair of the preexisting nicks and gaps in herpes simplex virus DNA. Virology. 1977;76:286–294. doi: 10.1016/0042-6822(77)90303-8. [DOI] [PubMed] [Google Scholar]
- 32.Jacob R J, Morse L S, Roizman B. Anatomy of herpes simplex virus DNA. XII. Accumulation of head-to-tail concatemers in nuclei of infected cells and their role in the generation of the four isomeric arrangements of viral DNA. J Virol. 1979;29:448–457. doi: 10.1128/jvi.29.2.448-457.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacob R J, Roizman B. Anatomy of herpes simplex virus DNA. VIII. Properties of the replicating DNA. J Virol. 1977;23:394–411. doi: 10.1128/jvi.23.2.394-411.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jean J-H, Blankenship M L, Ben-Porat T. Replication of herpesvirus DNA. I. Electron microscopic analysis of replicative structures. Virology. 1977;79:281–291. doi: 10.1016/0042-6822(77)90355-5. [DOI] [PubMed] [Google Scholar]
- 35.Kemper B, Brown D T. Function of gene 49 of bacteriophage T4. II. Analysis of intracellular development and the structure of very fast sedimenting DNA. J Virol. 1976;18:1000–1015. doi: 10.1128/jvi.18.3.1000-1015.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kieff E D, Bachenheimer S L, Roizman B. Size, composition, and structure of the deoxyribonucleic acid of herpes simplex virus subtypes 1 and 2. J Virol. 1971;8:125–132. doi: 10.1128/jvi.8.2.125-132.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knopf C W, Weisshart K. Comparison of exonucleolytic activities of herpes simplex virus type-1 DNA polymerase and DNase. Eur J Biochem. 1990;191:263–273. doi: 10.1111/j.1432-1033.1990.tb19119.x. [DOI] [PubMed] [Google Scholar]
- 38.Konforti B B, Davis R W. 3′ homologous free ends are required for stable joint molecule formation by the RecA and single-stranded binding proteins of Escherichia coli. Proc Natl Acad Sci USA. 1987;84:690–694. doi: 10.1073/pnas.84.3.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kosak H G, Kemper B W. Large-scale preparation of T4 endonuclease VII from over-expressing bacteria. Eur J Biochem. 1990;194:779–784. doi: 10.1111/j.1432-1033.1990.tb19469.x. [DOI] [PubMed] [Google Scholar]
- 40.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lamberti C, Weller S K. The herpes simplex virus type 1 UL6 protein is essential for cleavage and packaging but not for genomic inversion. Virology. 1996;226:403–407. doi: 10.1006/viro.1996.0668. [DOI] [PubMed] [Google Scholar]
- 42.Lieber M R. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination, and repair. Bioessays. 1997;19:233–240. doi: 10.1002/bies.950190309. [DOI] [PubMed] [Google Scholar]
- 43.Lin S-F, Hsu T-Y, Liu M-Y, Lin L-S, Yang H-L, Chen J-Y, Yang C-S. Characterization of Epstein-Barr virus DNase and its interaction with the major DNA binding protein. Virology. 1995;208:712–722. doi: 10.1006/viro.1995.1203. [DOI] [PubMed] [Google Scholar]
- 44.Locker H, Frenkel N. BamI, KpnI, and SalI restriction enzyme maps of the DNAs of herpes simplex virus strains Justin and F: occurrence of heterogeneities in defined regions of the viral DNA. J Virol. 1979;32:429–441. doi: 10.1128/jvi.32.2.429-441.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martinez R, Sarisky R T, Weber P C, Weller S K. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J Virol. 1996;70:2075–2085. doi: 10.1128/jvi.70.4.2075-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martinez R, Shao L, Bronstein J C, Weber P C, Weller S K. The product of a 1.9-kb mRNA which overlaps the HSV-1 alkaline nuclease gene (UL12) cannot relieve the growth defects of a null mutant. Virology. 1996;215:152–164. doi: 10.1006/viro.1996.0018. [DOI] [PubMed] [Google Scholar]
- 47.McGeoch D J, Cook S, Dolan A, Jamieson F E, Telford E A R. Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J Mol Biol. 1995;247:443–458. doi: 10.1006/jmbi.1995.0152. [DOI] [PubMed] [Google Scholar]
- 48.McVoy M A, Adler S P. Human cytomegalovirus DNA replicates after early circularization by concatemer formation, and inversion occurs within the concatemer. J Virol. 1994;68:1040–1051. doi: 10.1128/jvi.68.2.1040-1051.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mizuuchi K, Kemper B, Hays J, Weisberg R A. T4 endonuclease VII cleaves Holliday structures. Cell. 1982;29:357–365. doi: 10.1016/0092-8674(82)90152-0. [DOI] [PubMed] [Google Scholar]
- 50.Mosig G. Homologous recombination. In: Karam J D, editor. Molecular biology of bacteriophage T4. Washington, D.C: ASM Press; 1994. pp. 54–82. [Google Scholar]
- 51.Mukai T, Matsubara K, Takagi Y. Isolation of circular DNA molecules from whole cellular DNA by use of ATP-dependent deoxyribonuclease. Proc Natl Acad Sci USA. 1973;70:2884–2887. doi: 10.1073/pnas.70.10.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pari G S, Anders D G. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J Virol. 1993;67:6979–6988. doi: 10.1128/jvi.67.12.6979-6988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patel A H, Subak-Sharpe J H, Stowe N D, Marsden H S, Maclean J B, Dargan D J. Suppression of amber nonsense mutations of herpes simplex virus type 1 in a tissue culture system. J Gen Virol. 1996;77:199–209. doi: 10.1099/0022-1317-77-2-199. [DOI] [PubMed] [Google Scholar]
- 54.Pfüller R, Hammerschmidt W. Plasmid-like replicative intermediates of the Epstein-Barr virus lytic origin of DNA replication. J Virol. 1996;70:3423–3431. doi: 10.1128/jvi.70.6.3423-3431.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poffenberger K L, Roizman B. A noninverting genome of a viable herpes simplex virus 1: presence of head-to-tail linkages in packaged genomes and requirements for circularization after infection. J Virol. 1985;53:587–595. doi: 10.1128/jvi.53.2.587-595.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Post L E, Conley A J, Mocarski E S, Roizman B. Cloning of reiterated and nonreiterated herpes simplex virus 1 sequences as Bam HI fragments. Proc Natl Acad Sci USA. 1980;77:4201–4205. doi: 10.1073/pnas.77.7.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Puvion-Dutilleul F, Pichard E, Laithier M, Puvion E. Cytochemical study of the localization and organization of parental herpes simplex virus type 1 DNA during initial infection of the cell. Eur J Cell Biol. 1989;50:187–200. [PubMed] [Google Scholar]
- 58.Roizman B. The structure and isomerization of herpes simplex virus genomes. Cell. 1979;16:481–494. doi: 10.1016/0092-8674(79)90023-0. [DOI] [PubMed] [Google Scholar]
- 59.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fundamental virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1043–1107. [Google Scholar]
- 60.Sarisky R T, Hayward G S. Evidence that the UL84 gene product of human cytomegalovirus is essential for promoting oriLyt-dependent DNA replication and formation of replication compartments in cotransfection assays. J Virol. 1996;70:7398–7413. doi: 10.1128/jvi.70.11.7398-7413.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Severini A, Morgan A R, Tovell D R, Tyrrell D L. Study of the structure of replicative intermediates of HSV-1 DNA by pulsed-field gel electrophoresis. Virology. 1994;200:428–435. doi: 10.1006/viro.1994.1206. [DOI] [PubMed] [Google Scholar]
- 62.Severini A, Scraba D G, Tyrrell D L J. Branched structures in the intracellular DNA of herpes simplex virus type 1. J Virol. 1996;70:3169–3175. doi: 10.1128/jvi.70.5.3169-3175.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shao L, Rapp L M, Weller S K. Herpes simplex virus 1 alkaline nuclease is required for efficient egress of capsids from the nucleus. Virology. 1993;196:146–162. doi: 10.1006/viro.1993.1463. [DOI] [PubMed] [Google Scholar]
- 64.Sheaffer A K, Weinheimer S P, Tenney D J. The human cytomegalovirus UL98 gene encodes the conserved herpesvirus alkaline nuclease. J Gen Virol. 1997;78:2953–2961. doi: 10.1099/0022-1317-78-11-2953. [DOI] [PubMed] [Google Scholar]
- 65.Sherman G, Bachenheimer S L. Characterization of intranuclear capsids made by ts morphogenic mutants of HSV-1. Virology. 1988;163:471–480. doi: 10.1016/0042-6822(88)90288-7. [DOI] [PubMed] [Google Scholar]
- 66.Shlomai J, Friedmann A, Becker Y. Replicative intermediates of herpes simplex virus DNA. Virology. 1976;69:647–659. doi: 10.1016/0042-6822(76)90493-1. [DOI] [PubMed] [Google Scholar]
- 67.Skaliter R, Lehman I R. Rolling circle DNA replication in vitro by a complex of herpes simplex virus type 1-encoded enzymes. Proc Natl Acad Sci USA. 1994;91:10665–10669. doi: 10.1073/pnas.91.22.10665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Skaliter R, Makhov A M, Griffith J D, Lehman I R. Rolling circle DNA replication by extracts of herpes simplex virus type 1-infected cells. J Virol. 1996;70:1132–1136. doi: 10.1128/jvi.70.2.1132-1136.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Strobel-Fidler M, Francke B. Alkaline deoxyribonuclease induced by herpes simplex virus type 1: composition and properties of the purified enzyme. Virology. 1980;103:493–501. doi: 10.1016/0042-6822(80)90206-8. [DOI] [PubMed] [Google Scholar]
- 70.Szilagyi J F, Cunningham C. Identification and characterization of a novel non-infectious herpes simplex virus-related particle. J Gen Virol. 1991;72:661–668. doi: 10.1099/0022-1317-72-3-661. [DOI] [PubMed] [Google Scholar]
- 71.Takagi Y, Matsubara K, Anai M. A deoxyribonuclease which requires nucleoside triphosphate from micrococcus lysodeikticus. IV. The mode of DNA hydrolysis. Biochim Biophys Acta. 1972;269:347–353. doi: 10.1016/0005-2787(72)90121-9. [DOI] [PubMed] [Google Scholar]
- 72.Thomas M S, Gao M, Knipe D M, Powell K L. Association between the herpes simplex virus major DNA-binding protein and alkaline nuclease. J Virol. 1992;66:1152–1161. doi: 10.1128/jvi.66.2.1152-1161.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Umene K, Nishimoto T. Replication of herpes simplex virus type 1 DNA is inhibited in a temperature-sensitive mutant of BHK-21 cells lacking RCC1 (regulator of chromosome condensation) and virus DNA remains linear. J Gen Virol. 1996;770:2261–2270. doi: 10.1099/0022-1317-77-9-2261. [DOI] [PubMed] [Google Scholar]
- 74.Vlazny D, Kwong A, Frenkel N. Site-specific cleavage/packaging of herpes simplex virus DNA and the selective maturation of nucleocapsids containing full-length viral DNA. Proc Natl Acad Sci USA. 1982;79:1423–1427. doi: 10.1073/pnas.79.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wagner L, Lai E. Separation of large DNA molecules with high voltage pulsed field gel electrophoresis. Electrophoresis. 1994;15:1078–1083. doi: 10.1002/elps.11501501161. [DOI] [PubMed] [Google Scholar]
- 76.Weber P C, Challberg M D, Nelson N J, Levine J, Glorioso J C. Inversion events in the HSV-1 genome are directly mediated by the viral DNA replication machinery and lack sequence specificity. Cell. 1988;54:369–381. doi: 10.1016/0092-8674(88)90200-0. [DOI] [PubMed] [Google Scholar]
- 77.Weller S K. Herpes simplex virus DNA replication and genome maturation. In: Cooper G M, Temin R G, Sugden B, editors. The DNA provirus: Howard Temin’s scientific legacy. Washington, D.C: ASM Press; 1995. pp. 189–213. [Google Scholar]
- 78.Weller S K, Lee K J, Sabourin D J, Schaffer P A. Genetic analysis of temperature-sensitive mutants which define the gene for the major herpes simplex virus type 1 DNA-binding protein. J Virol. 1983;45:354–366. doi: 10.1128/jvi.45.1.354-366.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weller S K, Seghatoleslami R M, Shao L, Rowse D, Carmichael E P. The herpes simplex virus type 1 alkaline nuclease is not essential for viral DNA synthesis: isolation and characterization of a lacZ insertion mutant. J Gen Virol. 1990;71:2941–2952. doi: 10.1099/0022-1317-71-12-2941. [DOI] [PubMed] [Google Scholar]
- 80.Wilkie N M. The synthesis and substructure of herpesvirus DNA: the distribution of alkali-labile single strand interruptions in HSV-1 DNA. J Gen Virol. 1973;21:453–467. doi: 10.1099/0022-1317-21-3-453. [DOI] [PubMed] [Google Scholar]
- 81.Wilkie N M, Clements J B, Macnab J C M, Subak-Sharpe J H. The structure and biological properties of herpes simplex virus DNA. Cold Spring Harbor Symp Quant Biol. 1974;39:657–666. doi: 10.1101/sqb.1974.039.01.079. [DOI] [PubMed] [Google Scholar]
- 82.Wu M, Hyman R W, Davidson N. Electron microscopic mapping of proteins bound to herpes simplex virus DNA. Nucleic Acids Res. 1979;6:3427–3441. doi: 10.1093/nar/6.11.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang X, Efstathiou S, Simmons A. Identification of novel herpes simplex virus replicative intermediates by field inversion gel electrophoresis: implications for viral DNA amplification strategies. Virology. 1994;202:530–539. doi: 10.1006/viro.1994.1375. [DOI] [PubMed] [Google Scholar]