Abstract
Many cancer-testis antigen genes have been identified; however, few human leukocyte antigen (HLA)-A24-restricted cytotoxic T cell (CTL) epitope peptides are available for clinical immunotherapy. To solve this problem, novel tools increasing the efficacy and accuracy of CTL epitope detection are needed. In the present study, we utilized a highly active dendritic cell (DC)-culture method and an in silico HLA-A24 peptide-docking simulation assay to identify novel CTL epitopes from MAGE-A6 and MAGE-A12 antigens. The highly active DCs, called α-type-1 DCs, were prepared using a combination of maturation reagents to produce a large amount of interleukin-12. Meanwhile, our HLA-A24 peptide-docking simulation assay was previously demonstrated to have an obvious advantage of accuracy over the conventional prediction tool, bioinformatics and molecular analysis section. For CTL induction assays, peripheral blood mononuclear cells derived from six cases of HLA-A24+ melanoma were used. Through CTL induction against melanoma cell lines and peptide-docking simulation assays, two peptides (IFGDPKKLL from MAGE-A6 and IFSKASEYL from MAGE-A12) were identified as novel CTL epitope candidates. Finally, we verified that the combination of the highly active DC-culture method and HLA-A24 peptide-docking simulation assay might be tools for predicting CTL epitopes against cancer antigens.
Electronic supplementary material
The online version of this article (doi:10.1007/s00262-012-1298-1) contains supplementary material, which is available to authorized users.
Keywords: MAGE antigen, In silico docking, MHC stabilization, HLA-A24 peptide
Introduction
Malignant melanoma is the best known cancer for which multiple tumor-specific antigens have been identified and used for vaccination [1, 2]. As human leukocyte antigen (HLA)-A2 is the dominant type in Caucasians, HLA-A2-restricted peptide-based cancer immunotherapy has mainly been performed. However, in Asia, HLA-A24 is more common, and clinical immunotherapeutic trials using specific HLA-A24-restricted peptides such as CEA, P53, melanoma-associated antigen (MAGE)-A3 and VEGFR have been performed [3–6].
With regard to cancer-testis antigens, a substantial amount of research mainly regarding MAGE, GAGE, BAGE, etc. has been accumulated, and a family of MAGE antigens and NY-ESO1 are known to be good peptides for cancer vaccination in clinical trials [7–10]. In the current study, our DNA microarray demonstrated MAGE-A6 and MAGE-A12 as specific common antigens in melanomas. Few studies about MAGE-A6 and MAGE-A12 have been conducted because the expression of these antigens is not so common in solid tumors compared with other members of the MAGE family [11–13]. However, quantitative PCR analysis revealed that MAGE-A6 and MAGE-A12 were more frequently expressed in tumors such as melanomas and gliomas than were other MAGE antigens. Taking this observation into account, novel epitopes derived from MAGE-A6 and MAGE-A12 could be good candidates for peptide-based cancer vaccines.
Unfortunately, few HLA-A24 epitope peptides are available for clinical immunotherapy. Therefore, new tools that enhance the efficacy and accuracy of cytotoxic T cell (CTL) epitope detection are needed. In the present study, we utilized a highly active dendritic cell (DC)-culture method and an in silico HLA-A24 peptide-docking simulation assay to identify novel epitopes from MAGE-A6 and MAGE-A12 antigens. A highly active α-type-1 DC culture was reported by Kalinski et al. [14, 15] in which mature DCs stimulated with a combination of maturation reagents were demonstrated to produce a greater amount of interleukin (IL)-12 than conventional DC cultures. Meanwhile, a computer-assisted HLA-A24 peptide-docking simulation program, developed in our laboratory [17], showed an obvious advantage of accuracy over the conventional prediction tool.
Finally, we verified that the combination of these methods might be a good tool for detecting CTL epitopes against cancer antigens.
Materials and methods
Reagents and cell lines
Recombinant human (rh) granulocyte macrophage colony-stimulating factor (GM-CSF), rh-IL-1β, rhIL-2, rhIL-4, rhIL-7, and tumor necrosis factor (TNF)-α were purchased from Pepro Tech Inc. (Rocky Hill, NJ).
Interferon (IFN)-α and IFN-γ were purchased from Sumitomo and Shionogi Pharmaceutical Co, Ltd, Japan, respectively. Poly I/C was obtained from Amersham Biosciences Corp. (Piscateway, NJ). TISI and T2-A24 cells were used for the IFN-γ production assay and in vitro CTL stimulation, respectively, as described previously. Melanoma cell lines and human melanocytes [16] were maintained at 37 °C in a humidified atmosphere of 5 % CO2 in RPMI1640 medium (SIGMA, St. Louis, MO) supplemented with 10 % fetal bovine serum, penicillin, and streptomycin. Human melanoma cell lines and melanocyte-derived mRNAs were used for the gene chip microarray analysis. Mouse monoclonal antibodies (MoAbs) to human HLA-ABC (class I), HLA-DR, DP, DQ (class II), CD4, CD8, and CD56 were all purchased from Pharmingen (San Diego, CA, USA). Mouse MoAbs to human HLA-A24 were obtained from One Lambda, Inc. (Canoga Park, CA, USA). For immunostaining, rabbit anti-human MAGE-A6 and MAGE-A12 polyclonal antibodies were purchased from Abcam Co. Ltd (Cambridge, UK).
DNA microarray analysis
Total RNA from melanoma cell lines was extracted using the NucleoSpin RNAII kit (TakaRa Bio Inc., Shiga, Japan) according to the manufacturer’s instructions. Ten micrograms of total RNA, qualified using the Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA), was amplified to 100 μg of cRNA and hybridized to the high-density oligonucleotide array (GeneChip Human Genome U133 plus 2.0 array; Affymetrix, Santa Clara, CA, USA). The intensity for each feature of the array was calculated using GeneSpringGX ver11 (Agilent) software. For calculating the change of average intensity difference, normalization for all probe sets was performed.
Quantitative PCR analysis
The real-time PCR analysis of MAGE-A3, MAGE-A6, and MAGE-A12 antigens in human melanoma cell lines using the 7500 Real-Time PCR System (Applied Biosystems, Foster, CA, USA) was done as described previously. Briefly, all PCR primers and TaqMan probes were purchased from Applied Biosystems. Complementary DNA was synthesized from 100 ng of the total RNA, and quantitative PCRs were carried out using a TaqMan RNA-to-Ct 1-Step kit (Applied Biosystems). As to the expression of MAGE-A3, MAGE-A6, and MAGE-A12 antigens in melanoma tumor tissue, the regular non-quantitative RT-PCR method was utilized as described before [17].
Synthetic peptides
MAGE-A6 and MAGE-A12 peptide sequences with the HLA-A24-binding motif were searched for at the web site of the bioinformatics and molecular analysis section (BIMAS), HLA-peptide binding predictions. The top six peptides (Table 1) were selected and synthesized using the method reported by Knorr et al. [18]. MAGE-A6-3 and MAGE-A12-3 showed the same sequence, and thus MAGE-A12-3 was deleted from the list. The other synthetic peptides used in the present study are CMVpp65341–349 (QYDPVAALF), and gp100152–160 (VWKTWGQYW).
Table 1.
MAGE-A6 and MAGE-A12 peptide sequences with HLA-A24-binding motif
| Peptide name | Amino acid sequence | Position | Amino acid length | BIMAS score | MHC stabilization assay |
|---|---|---|---|---|---|
| MAGE-A6-1 | QYFVQENYL | 248–256 | 9 | 240 | 2.26 |
| MAGE-A6-2 | SYVKVLHHM | 282–290 | 9 | 53 | 2.71 |
| MAGE-A6-3 | EFLWGPRAL | 270–278 | 9 | 30 | 1.15 |
| MAGE-A6-4 | IFGDPKKLL | 238–246 | 9 | 24 | 2.14 |
| MAGE-A6-5 | TFPDLESEF | 97–105 | 9 | 24 | 1.33 |
| MAGE-A6-6 | IFSKASDSL | 150–158 | 9 | 20 | 1.47 |
| MAGE-A12-1 | SYVKVLHHL | 282–290 | 9 | 420 | 1.19 |
| MAGE-A12-2 | EYLQLVFGI | 156–164 | 9 | 90 | 2.49 |
| MAGE-A12-4 | TFPDLETSF | 97–105 | 9 | 22 | 1.96 |
| MAGE-A12-5 | IFSKASEYL | 150–158 | 9 | 20 | 2.79 |
| MAGE-A12-6 | VFAHPRKLL | 238–246 | 9 | 20 | 1.93 |
| CMVpp65 | QYDPVAALF | 341–349 | 9 | 168 | 3.04 |
| gp100 | VWKTWGQYW | 152–160 | 9 | 0.1 | 0.75 |
Alpha-type-1 DC culture
Highly active α-type-1 DCs were prepared using adherent monocyte-enriched cell cultures. The cytokines added on day 5 were GM-CSF (40 ng/ml) and IL-4 (40 ng/ml) in combination with TNF-α (50 ng/ml), IL-1β (25 ng/ml), IFN-α (3000 U/ml), IFN-γ (1,000 U/ml), and polyI/C (20 μg/ml).
MHC stabilization assay
The protocol of the MHC stabilization assay was described previously [16]. Briefly, 2 × 105 T2-A24 cells suspended in 200 μl of Iscove’s modified Dulbecco’s medium (IMDM) (Gibco, Paisley, UK) containing 0.1 % fetal bovine serum (FBS) were incubated with each peptide at 10 μM at 26 °C for 16 h and at 37 °C for 3 h. The cells were stained with mouse anti-HLA-A24 MoAb and FITC-labeled anti-mouse IgG, and analyzed on a flow cytometer. The mean fluorescence intensity (MFI) increase was calculated as follows: MFI increase = (MFI with the given peptide—MFI without peptide)/(MFI without peptide).
CTL induction cultures
Cultures of PBMCs from six HLA-A*2402+ malignant melanoma patients were used for in vitro CTL induction (The clinical research using PBMCs and tumor tissues from melanoma patients was approved by the Institutional Review Board of Shizuoka Cancer Center, Shizuoka, Japan. All patients gave written informed consent). The melanoma patients consisted of four males and two females, and mean age was 58 ± 17. All patients were metastatic, clinical stage IV. CTL induction cultures were described previously [17]. PBMCs from MEL-001 were used for CTL induction assays against melanoma cell lines. MEL-003-derived CTLs given two rounds of DC stimulation using MAGE-A6-4 were utilized in the Abs-blocking experiment.
IFN-γ production assay
TISI cells were incubated overnight with the HLA-A24 peptide at 20 μg/ml suspended in PBS (+) containing 1 % HSA and used as target cells. When melanoma cell lines were used as target cells, they were stimulated with human IFN-γ at 50 ng/ml overnight and then subjected to the co-culture with CTLs. Melanoma cell lines used as target cells are as follows: TDMM1 (HLA-A24+, MAGE A6+, MAGE A12+), A375 (HLA-A2+, MAGE A6+, MAGE A12+), and MMG-1 (HLA-A24+, MAGE A6− MAGE A12+). Cultured CTLs (1 × 105) and HLA-A24 peptide-pulsed TISI cells (1 × 105) were co-incubated in a round-bottomed 96-well microculture plate for 24 h. Finally, supernatants were collected, and IFN-γ levels were measured using an ELISA kit specific for human IFN-γ (Biosource, Camallilo, CA, USA).
Effect of various antibodies on IFN-γ production by MAGE-A6-4 peptide-specific CTLs
Blocking of the MEL-003-derived MAGE-A6-4 peptide-specific CTL activity against peptide-pulsed TISI cells was achieved by incubating target cells with anti-HLA-A24 and anti-HLA-class II antibodies or effector cells with anti-CD4, anti-CD8, and anti-CD56 antibodies at a dose of 10 μg/ml, respectively, for 1 h at 37 °C before the start of co-culture. After washing, Ab-treated CTLs and target cells were co-cultured for 24 h at 37 °C, and IFN-γ levels in the supernatant were measured.
Immunohistochemistry (IHC)
The tissue specimen from MEL-T5 tumor was used for the IHC analysis. For immunostaining, anti-human MAGE-A6 and MAGE-A12 polyclonal antibodies as primary antibody and goat anti-rabbit IgG antibody as secondary antibody were used. Horseradish peroxidase (HRP) and hydrogen peroxide were utilized for color development according to the manufacturer’s instructions (Vectastatin kit, Vector Lab., CA, USA).
In silico docking simulation assay for epitope peptide binding to HLA-A24 protein
Briefly, to build the initial 3D structure of any given A24-restricted epitope candidate peptide, we used homology-modeling software, MODELLER (version 9v5), with the structure of the hTERT epitope as a template from the Protein Data Bank (PDB). Then, we predicted the affinity between HLA-A24 and synthesized MAGE-A6 and MAGE-A12 peptides by a receptor-ligand docking simulation based on a Lamarckian genetic algorithm, AutoDock (version 4.0). In AutoDock, the affinity between a receptor and ligand is calculated as the Gibbs free energy of binding (ΔG). For each of the epitope candidate peptides, after Kollman charges are added, the docking simulation for HLA-A24 was run 50 times, in each of which one million energy evaluations were performed based on the genetic algorithm. Finally, we obtained the mean of the top three values (the three lowest ΔGs) among the 50 runs as the affinity value for peptide binding.
Statistical analysis
Statistical difference was analyzed using Student’s t test. Values of P < 0.05 were considered statistically significant.
Results
Gene chip microarray analysis
MAGE-A2, MAGE-A3, MAGE-A6, and MAGE-A12 antigen genes were all detected within the top 30 genes specifically expressed in melanoma cell lines, not in melanocytes (Supplementary Fig. 1). The magnitude of gene expression was normalized by the gene expression relative to melanocytes (intensity of melanocytes rated as 1). The MAGE-A6 and MAGE-A12 genes were identified in eight of 10 melanoma cell lines, and the intensity of their expression was more than 100-fold higher than that in melanocytes.
Quantitative PCR analysis of MAGE-A6 and MAGE-A12 genes in melanoma tissue
MAGE-A6 and MAGE-A12 mRNA were frequently identified in melanoma cell lines, but not in melanocytes as shown in Fig. 1a. These antigen genes were also verified frequently like MAGE-A2 and MAGE-A3 antigens in the tumor tissue from advanced metastatic melanoma patients using non-quantitative PCR (Fig. 1b).
Fig. 1.

MAGE-A6 and MAGE-A12 gene expression in melanoma cell lines and tumor tissues. a Quantitative PCR analysis of MAGE-A6 and MAGE-A12 gene expression in melanoma cell lines. The level of target mRNA expression was normalized to the expression of the beta-actin gene. b MAGE antigen gene expression in melanoma tumor tissues. The G-361 cell line was used as a positive control for PCR experiments. The sizes of PCR gene products are shown at the right. DNA sequences of MAGE-specific primers were described previously [16]. c MAGE-A6 and MAGE-A12 protein expression in melanoma tumor tissue (MEL-T5). H-E; hematoxylin–eosin, magnification; × 100
IHC analysis
The MEL-T5 tumor was shown to be positive for both MAGE-A6 and MAGE-A12 protein expression. The staining pattern was homogeneous in the tumor area (Fig. 1c).
MHC stabilization assay
Stabilization assays demonstrated that the MAGE-A6-1, MAGE-A6-2, MAGE-A6-4, MAGE-A12-2 and MAGE-A12-5 A24 peptides had moderate affinity; however, the others had low affinity, which was not similar to the BIMAS prediction. Meanwhile, CMVpp65 HLA-A24 with a high BIMAS score also showed a high affinity score in the MHC stabilization assay (Table 1).
In silico docking simulation assay for MAGE-A6 and MAGE-A12 HLA-A24 peptides
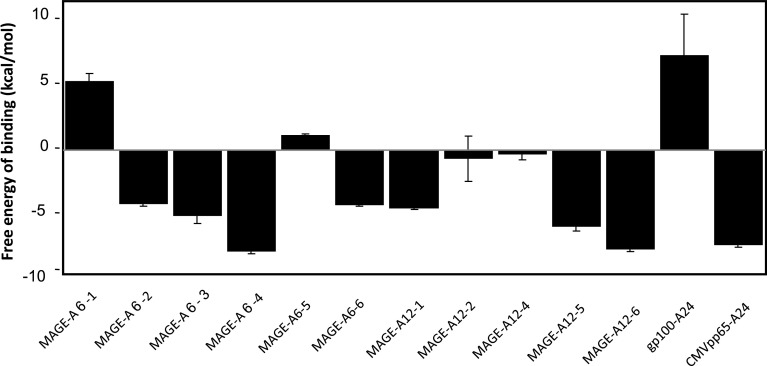
Our docking simulation revealed that four peptides, MAGE-A6-4, MAGE-A12-5, MAGE-A12-6, and CMVpp65, showed high affinity with a ΔG of more than 5 (Fig. 2). MAGE-A6-4 showed the highest affinity among the docking-positive peptides. MAGE-A6-1, MAGE-A6-5, MAGE-A12-2, and MAGE-A12-4 were docking-negative like gp100 A24.
Fig. 2.
Characterization of MAGE-A6 and MAGE-A12 HLA-A24-restricted peptides using the peptide-HLA-docking simulation assay. The affinity between a receptor and ligand is calculated as the Gibbs free energy of binding (ΔG). For every epitope candidate peptide, the docking simulation for HLA-A24 was run 50 times, each time with one million energy evaluations based on the genetic algorithm. Finally, the mean of the top three values (the three lowest ΔGs) among the 50 runs was taken as the affinity value for peptide binding
Alpha-type-1 DC culture and CTL induction assay
In order to confirm the α-type-1 DC’s strong activity for CTL induction, we performed melanoma peptide-specific CTL induction cultures based on various types of activated mature DCs from MEL-001 in an early experiment, where α-type-1 DCs stimulated by TNF-α, IL-1β, IFN-α, IFN-γ, and poly I/C demonstrated greater CTL-inducing activity than any other type of DC (data not shown).
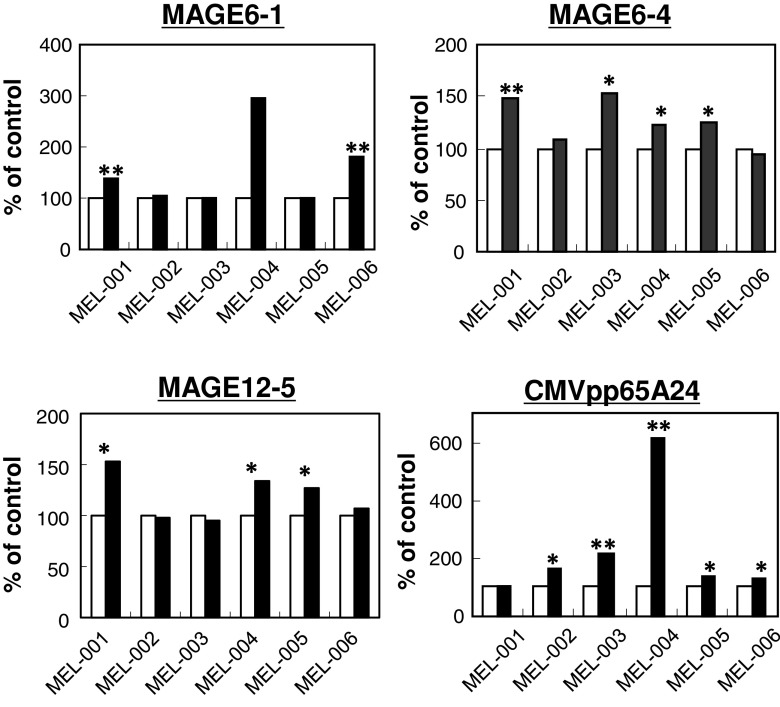
Using α-type-1 DC-based CTL induction cultures for MAGE-A6 and MAGE-A12 A24-binding peptides, MAGE-A6-1, MAGE-A6-4, and MAGE-A12-5 were found to be moderate-to-strong CTL-inducing peptides. MAGE-A6-1 induced a positive CTL reaction in two cases, and the other two peptides induced a response in four and three of six HLA-A24+ melanomas, respectively (Fig. 3, Table 2). Conversely, the positive response rate against the CMVpp65 peptide as a positive control was very high, 83 % (five of six cases).
Fig. 3.
CTL-inducing activity of MAGE-A6 and MAGE-A12 HLA-A24-binding peptides. HLA-A*2402+ PBMCs from six melanoma patients were used for CTL induction. Each column shows the mean IFN-γ level in supernatant from co-cultures of CTLs and peptide-pulsed TISI cells in triplicate. Open column, cultures without peptide; closed column, cultures with peptide. * P < 0.05, ** P < 0.01, statistically significant
Table 2.
CTL-inducing activity of MAGE-A6- and MAGE-A12 A24-binding peptides in six melanoma patients
| Peptide | Positive cases (%) | Peptide | Positive cases (%) |
|---|---|---|---|
| 6-1 | 2/6 (33) | 12-1 | 1/6 (0) |
| 6-2 | 1/6 (17) | 12-2 | 1/6 (17) |
| 6-3 | 0/6 (0) | 12-4 | 0/6 (0) |
| 6-4 | 4/6 (67) | 12-5 | 3/6 (50) |
| 6-5 | 0/6 (0) | 12-6 | 0/6 (0) |
| 6-6 | 1/6 (17) | CMVpp65 A24 | 5/6 (83) |
Antibody-mediated inhibitory effect on IFN-γ production by MAGE-A6-4-specific CTLs
Anti-HLA-class I and anti-CD8 antibodies exhibited a significant inhibitory effect on IFN-γ production by MAGE-A6-4 peptide-specific CTLs. Meanwhile, anti-HLA-class II, anti-CD4, and anti-CD56 antibodies had no inhibitory effect on IFN-γ production by CTLs (Fig. 4).
Fig. 4.
Antibody-mediated inhibition of IFN-γ production by MAGE-A6-4 peptide-specific CTLs. CTLs given two rounds of α-type-1 DC stimulation derived from MEL-003 PMBCs were used for various antibody-mediated inhibition experiments using MAGE-A6-4 peptide-pulsed TISI cells as target cells. +a; TISI cells without peptide; cont., isotype control antibody. * P < 0.05, statistically significant
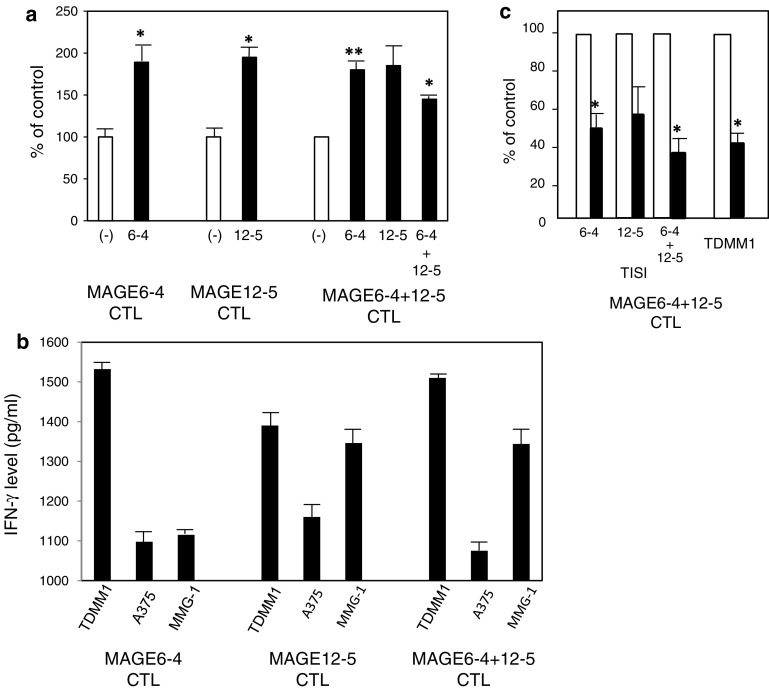
MAGE-A6-4 and MAGE-A12-5 peptide-specific CTL activity against melanoma cell lines
MEL001 PBMC-derived CTLs stimulated with MAGE-A6 or MAGE-A12 alone responded to TISI cells treated with each peptide. CTLs stimulated simultaneously with both peptides responded moderately to TISI cells treated with both peptides (Fig. 5a). Both MAGE-A6-4 and MAGE-A12-5 peptide-specific CTLs also demonstrated specific IFN-γ production against TDMM1 cells (HLA-A24+, MAGE A6+, MAGE A12+), not A375 cells (HLA-A2+, MAGE A6+, MAGE A12+), which was shown to be HLA-class I-restricted (Fig. 5b, c). IFN-γ production against MMG-1 (HLA-A24+, MAGE A6− MAGE A12+) was moderate, but there was a tendency for higher production in MAGE-A12-5 peptide-specific CTLs.
Fig. 5.
MAGE-A6 and/or MAGE-A12 A24 peptide-specific CTL (derived from MEL-001) activity against TISI cells and melanoma cell lines. a CTL activity against TISI cells treated with MAGE-A6-4, MAGE-A12-5 or both. (−), TISI cells without peptide. b CTL activity against melanoma cell lines including TDMM1 (HLA-A24+, MAGE A6+, MAGE A12+), A375 (HLA-A2+, MAGE A6+, MAGE A12+) and MMG-1 (HLA-A24+, MAGE A6− MAGE A12+). c Impact of anti-HLA-A24 MoAb on both MAGE-A6-4 and MAGE-A12-5 peptide-specific CTL activity against TISI cells and a melanoma cell line. Open column, isotype control antibody; closed column, anti-HLA-A24 antibody. * P < 0.05, ** P < 0.01, statistically significant
Discussion
Few studies about MAGE-A6 and MAGE-A12 have been conducted because these antigens are not as common in solid tumors as other members of the MAGE family. However, our quantitative PCR analysis revealed that MAGE-A6 and MAGE-A12 were more frequently expressed in tumors such as melanomas and gliomas, etc. than other MAGE antigens. These results suggested MAGE-A6 and MAGE-A12 to be potential cancer target antigens and that the investigation of specific CTL epitope peptides should contribute to the development of a novel HLA-A24 peptide-based immunotherapy.
Many MAGE antigen-derived HLA-A24-binding CTL epitopes have been reported, and several HLA-A24 binders have actually been utilized in clinical trials against solid cancers [19–21]. Notably, MAGE-A4- and MAGE-A10-derived CTL epitopes, novel peptides among MAGE antigen-derived HLA-A24 binders, were demonstrated to be a good CTL inducer by Jia et al. and other researchers [22, 23]. Interestingly, Graff-Dubois et al. [24] reported that HLA-A*0201-restricted epitopes were shared by MAGE-A1, MAGE-A2, MAGE-A3, MAGE-A4, MAGE-A6, MAGE-A10, and MAGE-A12 tumor antigens and induced CTL by the epitope peptides recognized human HLA-A0201+MAGE-A+ tumor cells of various histological origins. However, our MAGE-A6 and MAGE-A12 A24 peptide sequences were different from theirs. Importantly, the MAGE-A6-4 and MAGE-A12-5 peptides identified as CTL epitopes in the present study were demonstrated to have CTL-inducing activity against a HLA-A24+ MAGE-A6+ MAGE-A12+ melanoma cell line as well as a HLA-A24+ human B-lymphoblastoid cell line, TISI cells.
Here, based on a highly active DC culture method and docking simulation assay, specific MAGE-A6 and MAGE-A12 HLA-A24 epitopes were identified, making it the first MAGE cancer antigen-related study. The highly active DCs, called α-type-1, were obtained from 7-day cultures of monocyte-enriched cells stimulated with a cocktail of reagents (GM-CSF, IL-4, TNF-α, IL-1β, IFN-α, IFN-γ, and poly I/C). The culture of α-type-1 DCs was described by Kalinski et al. [14], in which α-type-1 DCs showed remarkably higher IL-12 production and greater CTL-inducing activity than the standard DC culture. In our experiments using melanoma-associated HLA-A24 peptides, α-type-1 DCs also demonstrated strong CTL-inducing activity even in serum-free cultures. The current results regarding MAGE-A6 and MAGE-A12 CTL epitope detection suggest that α-type-1 DCs may strengthen the weak CTL-inducing activity of HLA-A24 peptides.
In the present study, using a docking simulation assay, the peptides including MAGE-A6-4, MAGE-A12-5, MAGE-A12-6, and CMVpp65 showed high affinity of docking with a ∆G of more than 5 as shown in Fig. 2. Meanwhile, CTL induction assays revealed that MAGE-A6-1, MAGE-A6-4, and MAGE-A12-5 were positive CTL inducers. MAGE-A6-4 and MAGE-A12-5 were efficiently recognized as positive in both assays. Considering that positive MAGE-A6 and MAGE-A12 peptides with CTL-inducing activity and docking affinity showed moderate affinity in MHC stabilization and BIMAS, conventional approaches do not necessarily indicate genuine HLA-binding epitopes. Therefore, docking simulation assays might serve as a novel prediction tool supporting conventional methods.
Unfortunately, some discrepancies in affinity between CTL induction or MHC stabilization and docking assays are sometimes found. The reason for the discrepancy has yet to be elucidated; however, the rigid pocket simulated for the HLA protein may have affected the docking conformation and affinity. Docking simulation, which allows a pocket to be flexible, is a challenge for future computer technology. In summary, two HLA-A24 peptides having strong CTL-inducing activity were successfully recognized as positive binders in peptide-docking simulation assays, which indicates our docking assay is gradually improving in accuracy compared with previous studies regarding CEA epitopes.
Finally, in the present study, we demonstrated that the combination of a highly active DC culture and in silico peptide-docking assay might improve the detection of CTL epitopes against cancer antigens. Using this new tool, the detection of more novel CTL epitopes regarding cancer-testis antigens can be expected in the near future.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Expanded view of the subset of genes whose expression was specifically increased in melanoma cell lines, not in melanocytes, from a high density oligonucleotide assay. Each row corresponds to a single gene, and the columns correspond to gene expression levels in different samples. The color saturation reflects the fold increase of expression intensity compared to that of melanocytes ranging from blue to red. The top 30 genes specifically expressed in melanoma (positive in more than 6 of 10 melanoma cell lines), not in melanocytes, are shown. The MAGE-A1, -A2, -A3 and -A12 genes were all included in the top 30. Supplementary material 1 (PPT 373 kb)
Acknowledgments
We thank Dr. Mochizuki for supplying several synthetic peptides and technical assistance. This work was supported by a grant from the Cooperation of Innovative Technology and Advanced Research in Evolutional Area (CITY AREA) program from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Conflict of interest
The authors declare that they have no conflict of interest.
Abbreviation
- HLA
Human leukocyte antigen
- DC
Dendritic cell
- CTL
Cytotoxic T cell
- BIMAS
Bioinformatics and molecular analysis section
- PBMC
Peripheral blood mononuclear cell
- CMV
Cytomegalovirus
References
- 1.Rosenberg SA, Yang JC, Restifo NP. Cancer immunotherapy: moving beyond current vaccine. Nat Med. 2004;10:909–915. doi: 10.1038/nm1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nestle FO, Alijagic S, Gilliet M, Sun Y, Grabbe S, Dummer R, Burg G, Schadendorf D. Vaccination of melanoma patients with peptide- or tumor lysate-pulsed dendritic cells. Nat Med. 1998;4:328–332. doi: 10.1038/nm0398-328. [DOI] [PubMed] [Google Scholar]
- 3.Matsuda K, Tsunoda T, Tanaka H, Umano Y, Tanimura H, Nukaya I, Takesako K, Yamaue H. Enhancement of cytotoxic T-lymphocyte responses in patients with gastrointestinal malignancies following vaccination with CEA peptide-pulsed dendritic cells. Cancer Immunol Immunother. 2004;53:609–616. doi: 10.1007/s00262-003-0491-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eura M, Chikamatsu K, Katsura F, Obata A, Sobao Y, Takiguchi M, Song Y, Appella E, Whiteside TL, Deleo AB. A wild-type sequence p53 peptide presented by HLA-A24 induces cytotoxic T lymphocytes that recognize squamous cell carcinoma of the head and neck. Clin Cancer Res. 2000;6:979–986. [PubMed] [Google Scholar]
- 5.Ueda Y, Shimizu K, Itoh T, Fuji N, Naito K, Shiozaki A, Yamamoto Y, Shimizu T, Iwamoto A, Tamai H, Yamagishi H. Induction of peptide-specific immune response in patients with primary malignant melanoma of the esophagus after immunotherapy using dendritic cells pulsed with MAGE peptides. Jpn J Clin Oncol. 2007;37:140–145. doi: 10.1093/jjco/hyl136. [DOI] [PubMed] [Google Scholar]
- 6.Wada S, Tssunoda T, Baba T, Primus FJ, Kuwano H, Shibuya M, Tahara H. Rationale for antiangiogenic cancer therapy with vaccination using epitope peptides derived from human vascular endothelial growth factor receptor 2. Cancer Res. 2005;65:4936–4946. doi: 10.1158/0008-5472.CAN-04-3759. [DOI] [PubMed] [Google Scholar]
- 7.Banchereau J, Palucka AK, Dhodapkar M, Burkeholder S, Taquet N, Rolland A, Taquet S, Coquery S, Wittkowski KM, Bhardwaj N, Pineiro L, Steinman R, Fay J. Immune and clinical responses in patients with metastatic melanoma to CD34(+) progenitor-derived dendritic cell vaccine. Cancer Res. 2001;61(17):6451–6458. [PubMed] [Google Scholar]
- 8.Akiyama Y, Tanosaki R, Inoue N, Shimada M, Hotate Y, Yamamoto A, Yamazaki N, Kawashima I, Nukaya I, Takesako K, Maruyama K, Takaue Y, Yamaguchi K. Clinical response in Japanese metastatic melanoma patients treated with peptide cocktail-pulsed dendritic cells. J Transl Med. 2005;3:4. doi: 10.1186/1479-5876-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kakimi K, Isobe M, Uenaka A, Wada H, Sato E, Doki Y, Nakajima J, Seto Y, Yamatsuji T, Naomoto Y, Shiraishi K, Takigawa N, Kiura K, Tsuji K, Iwatsuki K, Oka M, Pan L, Hoffman EW, Old LJ, Nakayama E (2011) A phase I study of vaccination with NY-ESO-1f peptide mixed with Picibanil OK-432 and Montanide ISA-51 in patients with cancers expressing the NY-ESO-1 antigen. Feb3. doi:10.1002/ijc.25955 [DOI] [PubMed]
- 10.Tyagi P, Mirakhur B. MAGRIT: the largest-ever phase III lung cancer trial aims to establish a novel tumor-specific approach to therapy. Clin Lung Cancer. 2009;10(5):371–374. doi: 10.3816/CLC.2009.n.052. [DOI] [PubMed] [Google Scholar]
- 11.Tsai JR, Chong IW, Chen YH, Yang MJ, Sheu CC, Chang HC, Hwang JJ, Hung JY, Lin SR. Differential expression profile of MAGE family in non-small-cell lung cancer. Lung Cancer. 2007;56(2):185–192. doi: 10.1016/j.lungcan.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 12.Chung FY, Cheng TL, Chang HJ, Chiu HH, Huang MY, Chang MS, Chen CC, Yang MJ, Wang JY, Lin SR. Differential gene expression profile of MAGE family in taiwanese patients with colorectal cancer. J Surg Oncol. 2010;102(2):148–153. doi: 10.1002/jso.21580. [DOI] [PubMed] [Google Scholar]
- 13.Karn T, Pusztai L, Ruckhaberle E, Liedtke C, Muller V, Schmidt M, Metzler D, Wang J, Coombes KR, Gatje R, Hanker L, Solbach C, Ahr A, Holtrich U, Rody A, Kaufmann M. Melanoma antigen family A identified by the biomodality index defines a subset of triple negative breast cancers as candidates for immune response augmentation. Eur J Cancer. 2011;48(1):12–23. doi: 10.1016/j.ejca.2011.06.025. [DOI] [PubMed] [Google Scholar]
- 14.Mailliard RB, Wankowicz-Kakinska A, Cai Q, Wesa A, Hilkens CM, Kapsenberg ML, Kirkwood JM, Storkus WJ, Kalinski P. α-type-1 polarized dendritic cells: a novel immunization tool with optimized CTL-inducing activity. Cancer Res. 2004;64(17):5934–5937. doi: 10.1158/0008-5472.CAN-04-1261. [DOI] [PubMed] [Google Scholar]
- 15.Fujita M, Zhu X, Ueda R, Sasaki K, Kohanbash G, Kastenhuber ER, McDonald HA, Gibson GA, Watkins SC, Muthuswamy R, Kalinski P, Okada H. Effective immunotherapy against murine gliomas using rtpe 1 polarizing dendritic cells-significant roles of CXCL10. Cancer Res. 2009;69(4):1587–1595. doi: 10.1158/0008-5472.CAN-08-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamura Y, Tai S, Oshita C, Iizuka A, Ashizawa T, Saito S, Yamaguchi S, Kondo H, Yamaguchi K, Akiyama Y. Analysis of HLA-A24-restricted peptides of carcinoembryonic antigen using a novel structure-based peptide-HLA docking algorithm. Cancer Sci. 2011;102(4):690–696. doi: 10.1111/j.1349-7006.2011.01866.x. [DOI] [PubMed] [Google Scholar]
- 17.Akiyama Y, Maruyama K, Nara N, Mochizuki T, Yamamoto A, Yamazaki N, Kawashima I, Nukaya I, Takesako K, Yamaguchi K. Cytotoxic T cell induction against human malignant melanoma cells using HLA-A24-restricted melanoma peptide cocktail. Anticancer Res. 2004;24:571–577. [PubMed] [Google Scholar]
- 18.Knorr R, Trzeciak A, Bannwarth W, Gillessen D. New coupling reagents in peptide chemistry. Tetrahedron Lett. 1989;30:1927–1930. doi: 10.1016/S0040-4039(00)99616-3. [DOI] [Google Scholar]
- 19.Fujie T, Tahara K, Tanaka F, Mori M, Takesako K, Akiyoshi T. A MAGE-1-encoded HLA-A24-binding synthetic peptide induces specific anti-tumor cytotoxic T lymphocytes. Int J Cancer. 1999;80(2):169–172. doi: 10.1002/(SICI)1097-0215(19990118)80:2<169::AID-IJC1>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 20.Tahara K, Takesako K, Sette A, Celis E, Kitano S, Akiyoshi T. Identification of a MAGE-2-encoded human leukocyte antigen-A24-binding synthetic peptide that induces specific antitumor cytotoxic T lymphocytes. Clin Cancer Res. 1999;5(8):2236–2241. [PubMed] [Google Scholar]
- 21.Tanaka F, Fujie T, Tahara K, Mori M, Takesako K, Sette A, Celis E, Akiyoshi T. Induction of antitumor cytotoxic T lymphocytes with a MAGE-3-encoded synthetic peptide presented by human leukocyte antigen-A24. Cancer Res. 1997;57:4465–4468. [PubMed] [Google Scholar]
- 22.Jia ZC, Ni B, Huang ZM, Tian Y, Tang J, Wang JX, Fu XL, Wu YZ (2010) Identification of two novel HLA-A*0201-restricted CTL epitopes derived from MAGE-A4. Clin Dev Immunol Epub 2011 Feb14 [DOI] [PMC free article] [PubMed]
- 23.Jia ZC, Tian Y, Huang ZM, Wang JX, Fu XL, Ni B, Wu YZ. Identification of a new MAGE-A10 antigenic peptide presented by HLA-A*0201 on tumor cells. Cancer Biol Ther. 2011;11(4):395–400. doi: 10.4161/cbt.11.4.14100. [DOI] [PubMed] [Google Scholar]
- 24.Graff-Dubois S, Faure O, Gross DA, Alves P, Scardino A, Chouaib S, Lemonnier FA, Kosmatopoulos K. Generation of CTL recognizing an HLA-A*0201-restricted epitope shared by MAGE-A1, -A2, -A3, -A4, -A6, -A10, and -A12 tumor antigens: implication in a broad-spectrum tumor immunotherapy. J Immunol. 2002;169(1):575–580. doi: 10.4049/jimmunol.169.1.575. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded view of the subset of genes whose expression was specifically increased in melanoma cell lines, not in melanocytes, from a high density oligonucleotide assay. Each row corresponds to a single gene, and the columns correspond to gene expression levels in different samples. The color saturation reflects the fold increase of expression intensity compared to that of melanocytes ranging from blue to red. The top 30 genes specifically expressed in melanoma (positive in more than 6 of 10 melanoma cell lines), not in melanocytes, are shown. The MAGE-A1, -A2, -A3 and -A12 genes were all included in the top 30. Supplementary material 1 (PPT 373 kb)