Abstract
Translation initiation of hepatitis C virus (HCV) RNA occurs by internal entry of a ribosome into the 5′ nontranslated region in a cap-independent manner. The HCV RNA sequence from about nucleotide 40 up to the N terminus of the coding sequence of the core protein is required for efficient internal initiation of translation, though the precise border of the HCV internal ribosomal entry site (IRES) has yet to be determined. Several cellular proteins have been proposed to direct HCV IRES-dependent translation by binding to the HCV IRES. Here we report on a novel cellular protein that specifically interacts with the 3′ border of the HCV IRES in the core-coding sequence. This protein with an apparent molecular mass of 68 kDa turned out to be heterogeneous nuclear ribonucleoprotein L (hnRNP L). The binding of hnRNP L to the HCV IRES correlates with the translational efficiencies of corresponding mRNAs. This finding suggests that hnRNP L may play an important role in the translation of HCV mRNA through the IRES element.
Internal ribosome binding to mRNAs without 5′-end scanning of the mRNA, which occurs in most cellular mRNAs (18), was first discovered for picornavirus mRNAs (13, 25). Internal ribosome binding requires a cis-acting element on the mRNA termed the internal ribosomal entry site (IRES) (14) and cellular factors that interact specifically with the IRES. Several cellular proteins that direct IRES-dependent translation have been identified (10, 15, 17, 20). Besides picornaviruses, some members of the virus family Flaviviridae also contain IRESs. Hepatitis C virus (HCV) (32, 34), bovine viral diarrhea virus (27), and classical swine fever virus (29) belong to this group.
HCV RNA contains a long 5′ nontranslated region (5′NTR) (341 nucleotides [nt] long in most strains) harboring three to five noninitiating AUG triplets (9, 16). The 5′NTR and part of the core protein-coding region of HCV mRNA contain an IRES element. In other words, HCV RNA stretching from about nt 40 (7, 34) to the N-terminal coding sequence of the core (11, 12, 19, 28) is required for efficient initiation of translation, but the precise borders of this HCV IRES have as yet to be mapped. A complex secondary structure has been proposed for the HCV IRES (4). cis-acting structural elements, such as a helical structure and a pseudoknot-like structure, were suggested to be essential for IRES function (33, 35).
Several cellular proteins were proposed to be involved in HCV IRES-dependent translation. Polypyrimidine tract-binding protein (PTB) has been reported to bind to multiple sites of the HCV IRES, but the role of PTB in translation is still not clear (1, 17). Binding of La antigen to the initiation codon of the HCV IRES was reported to enhance HCV IRES-dependent translation (2). A 25-kDa cellular protein of unknown identity was also shown to bind specifically to the HCV IRES, and its binding affinity was correlated with efficiency of translation initiation (6).
Here we report on a cellular protein that specifically interacts with the 3′ border of the HCV IRES spanning part of the core-coding sequence. This protein, with an apparent molecular mass of 68 kDa, turned out to be heterogeneous nuclear ribonucleoprotein L (hnRNP L). Interestingly, hnRNP L has been shown to interact with PTB in a yeast two-hybrid system and by coprecipitation (8). The binding of hnRNP L to the HCV IRES correlated well with the translational efficiencies of corresponding mRNAs. This result suggests that hnRNP L may play a key role in the translation of HCV mRNA through the IRES element.
MATERIALS AND METHODS
Constructions of plasmids.
The HCV cDNA clones pCV, pKI5, pKIDel1, and pN10 have been previously described by Tsukiyama-Kohara et al. (32). To construct pH(280-E2′) and pH(331-E2′), the pSK(−) vector was treated with KpnI plus T4 polymerase plus PstI and the pCV vector was treated with StuI and PstI or AccI plus Klenow fragment plus PstI, respectively. pH(280-E2′) and pH(331-E2′) were treated with AatII plus PstI and T4 polymerase to construct plasmids pH(280–402) and pH(331–402), respectively. For the construction of pH(18-402)CAT, PCR was performed with the following primers: primer 1, CGGGGTACCGGCGACACTCCACCATAG; primer 2, CGCGGATCCCTGTGGGCGGCGGTTG; primer 3, CGCGGATCCACAACCATGAGCTTGGC; and primer 4, GCTCTAGATTATCACTTATTCAGGCGTAGC.
Primers 1 and 2 and pKI5 were used to amplify nt 18 to 402 of the HCV cDNA (PCR product 1). Primers 3 and 4 and pSK/Bip/CAT, kindly provided by P. Sarnow, were used to amplify the chloramphenicol acetyltransferase (CAT) gene (PCR product 2). The KpnI- and XbaI-digested pSK(−) vector, KpnI- and BamHI-digested PCR product 1, and BamHI- and XbaI-digested PCR product 2 were ligated to generate plasmid pH(18-402)CAT. To amplify corresponding HCV cDNAs, the following primers were used in PCR: primer 5, CGCGGATCCCATGATGCACGGTCTACG; primer 6, CGCGGATCCAGGATTTGTGCT; primer 7, CGCGGATCCGGTTTTTCTTTGAGGTTTAG; and primer 8, CGCGGTACCATGAGCACAAATCCTAAAC.
To construct plasmids pH(18-344)CAT, pH(18-356)CAT, and pH(18-374)CAT, PCR was carried out with primers 1 and 5, primers 1 and 6, and primers 1 and 7, respectively. The PCR products were treated with KpnI and BamHI, and the vector pH(18-402)CAT was treated with KpnI and BamHI to generate plasmids pH(18-344)CAT, pH(18-356)CAT, and pH(18-374)CAT. To construct pH(342-402), PCR was carried out with primers 4 and 8, the amplified PCR product was treated with KpnI and XbaI, and the vector pH(18-402)CAT was treated with KpnI and XbaI. For the construction of dicistronic clones containing a CAT gene with a truncated C terminus followed by HCV IRES and a full-length CAT gene, a fragment of a C-terminally truncated CAT gene was inserted into the unique KpnI site upstream of each monocistronic clone. pSK-hnRNP L and pRSET-hnRNP L were used for the in vitro transcription and purification of hnRNP L, respectively (8).
Purification of hnRNP L.
Escherichia coli BL21(DE3) pLysS was used to produce hnRNP L from plasmid pRSET-hnRNP L. IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 1 mM to the cells at an optical density at 600 nm of 0.25. After further incubating the cells expressing hnRNP L for 5 h at 25°C, the cells were harvested, resuspended in lysis buffer (20 mM Na-phosphate [pH 7.6], 300 mM NaCl, 0.5 mM phenylmethylsulfonyl fluoride, 1 mM β-mercaptoethanol, 10% glycerol) and sonicated. After lysis, the cell extracts were loaded onto a Ni-nitrilotriacetic acid (NTA) agarose column (Qiagen) equilibrated with lysis buffer. The columns were then washed with 5 volumes of lysis buffer containing 70 mM imidazole. The hnRNP L was eluted with 200 mM imidazole. Peak fractions were pooled and loaded onto a poly(U)-Sepharose column. After the resin was washed with lysis buffer containing 0.4 M NaCl, the hnRNP L was eluted with 1.5 M NaCl.
In vitro transcription and in vitro translation.
Plasmid DNAs were purified by the polyethylene glycol precipitation method (31) and then linearized with appropriate restriction enzymes. Linearized DNAs were extracted with phenol-chloroform and ethanol precipitated. The RNAs were transcribed from the linearized DNAs with T7 RNA polymerase (Boehringer Mannheim) for 90 min at 37°C as recommended by the manufacturer. Radioactive RNA probes and biotinylated RNAs were synthesized under similar reaction conditions with [32P]UTP (NEN) or biotin-labeled UTP (Pharmacia Biotech Inc.).
In vitro translations were performed in 12.5-μl reaction mixtures containing 40 nM mRNA in the presence of [35S]methionine (NEN) as described by Rose et al. (30). Cytoplasmic S-10 extract of HeLa S3 cells was prepared as described by Oh et al. (23). Translation reactions were carried out at 30°C for 1 h and analyzed by sodium dodecyl sulfate (SDS)–15% polyacrylamide gel electrophoresis (PAGE). The intensities of the autoradiographic images produced by 35S were enhanced by fluorography with salicylic acid. Gels were dried and exposed to Kodak XAR-5 or Agfa Curix RP1 film for 12 to 18 h.
UV cross-linking.
32P-labeled RNA probes were synthesized by in vitro transcription and isolated by push column chromatography (Stratagene). RNAs (2 × 106 cpm) were incubated with 40 μg of HeLa cell cytoplasmic extract or with 0.3 μg of purified hnRNP L which had been dialyzed into the translation buffer (16 mM HEPES [pH 7.5], 36 mM KCl, 169 mM potassium acetate, 1.2 mM magnesium acetate, 1.6 mM dithiothreitol [DTT], 2.8 mM β-mercaptoethanol). RNA-protein binding was carried out in a 30-μl reaction mixture containing 0.5 mM DTT, 5 mM HEPES (pH 7.6), 75 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 4% glycerol, 20 U of RNasin, and 3 μg of tRNA. After 20 min of incubation at 30°C, the samples were irradiated with UV light on ice for 30 min with a UV-Stratalinker (Stratagene). Unbound RNAs were digested with 5 μl of RNase cocktail (2 μl of RNase A [10 mg/ml], 2 μl of RNase T1 [100 U/ml], 1 μl of RNase V1 [700 U/ml]) at 37°C for 30 min and then analyzed by SDS–12% PAGE.
RNA affinity resin-binding assay: precipitation of proteins with biotinylated RNAs.
Biotinylated RNAs (1 μg) were incubated with 35S-labeled protein translated in vitro or with HeLa cell cytoplasmic extract (40 μg) in KHN buffer (150 mM KCl, 20 mM HEPES [pH 7.5], 0.03% Nonidet P-40, 0.2 mM DTT) for 1 h at 25°C. The mixture was then transferred into 1 ml of KHN buffer containing streptavidin acrylamide beads (Pierce) and incubated at 4°C for 1 h. The beads were collected by centrifugation, washed three times with 1 ml of KHN buffer, resuspended in sample buffer, and boiled for 4 min. The supernatant containing the RNA-bound protein was analyzed by SDS-PAGE.
RNA gel mobility shift assay.
32P-labeled RNAs corresponding to HCV RNAs from nt 228 to 331 and 342 to 402 {[32P]RNA(228–331) and [32P]RNA(342–402), respectively} were used as probes in a gel mobility shift assay. The RNAs (104 cpm) were incubated with purified hnRNP L or bovine serum albumin (BSA). RNA-protein interaction was allowed at 30°C for 20 min in RNA-binding buffer (5 mM HEPES [pH 7.6], 70 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 0.5 mM DTT, 0.7 μg of tRNA). Electrophoresis sample buffer was added, and the samples were loaded onto 5% nondenaturing polyacrylamide gels (pH 8.6) (23). Gels were dried and exposed to X-ray film for autoradiography. For the experiment combining the gel mobility shift assay and immunoblot analysis, cold HCV RNA (nt 18 to 331 or 18 to 402) was incubated with HeLa cell extract (20 μg) in the same binding buffer containing tRNA (10 μg) and then resolved in a 5% nondenaturing gel. Proteins were transferred to a nitrocellulose membrane (Amersham), and immunoblot analysis was performed with monoclonal antibody against human hnRNP L.
Immunoblot analysis.
After RNA-bound proteins were resolved in an SDS–12% polyacrylamide gel or a 5% native gel, the proteins were transferred to nitrocellulose membranes (Amersham). The membranes were incubated overnight at 4°C in blocking solution (20 mM Tris-HCl [pH 7.4]), 150 mM NaCl, 0.5% Tween 20, 5% skim milk) to block nonspecific binding of the antibody. The primary antibody (a monoclonal antibody against hnRNP L, 4D11) was added to the blocking solution, and incubation proceeded for 1 h. The antibody had been generously provided by G. Dreyfuss, University of Pennsylvania School of Medicine, Philadelphia. A horseradish peroxidase-linked anti-mouse immunoglobulin G was used as the secondary antibody. Membrane-bound antibodies were detected by enhanced chemiluminescence (Amersham).
RESULTS
Identification of cellular proteins interacting with the HCV IRES.
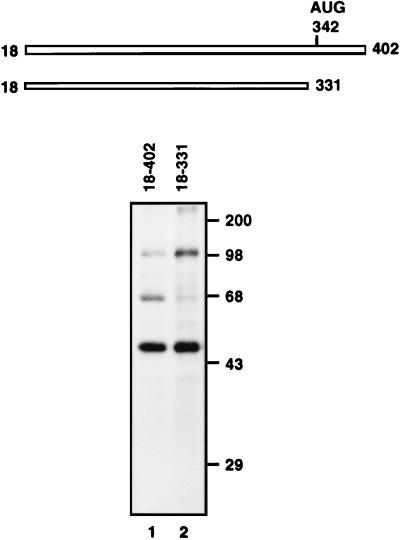
To identify cellular factor(s) interacting with the HCV IRES, UV cross-linking was performed with HeLa cell cytoplasmic extracts as protein sources and the probes [32P]RNA(18–331) and [32P]RNA(18–402). Three proteins with apparent molecular masses of 52, 68, and 100 kDa were found to bind to the HCV probe [32P]RNA(18–402), which encompasses most of the HCV 5′NTR and N-terminal coding sequence of the core protein (Fig. 1, lane 1). On the other hand, only two proteins of 52 and 100 kDa were observed as prominent bands when [32P]RNA(18–331) was used (note that the intensity of the 68-kDa protein band was weakened dramatically upon deletion of nt 332 to 402 [Fig. 1, lane 2]). This indicates that the region of nt 332 to 402 is essential for a strong interaction of the 68-kDa protein with the HCV IRES. The 52-kDa protein may be the La protein, which has been reported to interact specifically with the HCV IRES (2). The identity of the 100-kDa protein remains unknown. We decided to further characterize the 68-kDa protein, since it binds to the region determined to be the 3′ border of the HCV IRES (12, 19, 28).
FIG. 1.
Diagram of probes and detection of cellular proteins interacting with HCV IRES. RNA probes [32P]RNA(18–402) (lane 1) and [32P]RNA(18–331) (lane 2) were cross-linked to HeLa cell cytoplasmic extracts, after which the proteins were analyzed by SDS–12% PAGE. Molecular masses (in thousands) are noted to the right of the gel.
HnRNP L binds to the HCV IRES.
Recently, we have shown that hnRNP L interacts with PTB, a protein that binds to the encephalomyocarditis virus and HCV IRESs (8). We therefore investigated whether the 68-kDa protein, which interacts with the HCV IRES, is identical to hnRNP L, since PTB, which interacts with hnRNP L, was shown to bind to the HCV 5′NTR (1) and since the apparent molecular mass of hnRNP L is also 68 kDa.
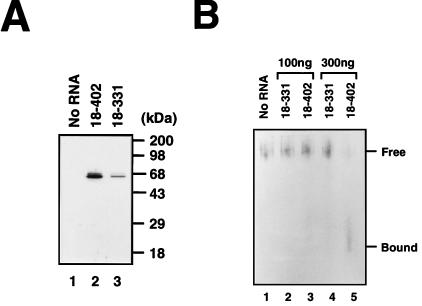
RNA affinity resin-binding assays were carried out with biotinylated HCV RNAs and HeLa cell extract (Fig. 2A). After incubation of HeLa cell extract with the biotinylated RNAs, the RNA-protein complexes were precipitated with streptavidin acrylamide beads. The presence of hnRNP L in the RNA-protein complexes was determined by Western blot analysis with monoclonal anti-hnRNP L antibody (4D11) (Fig. 2A). The RNA containing the complete HCV IRES (nt 18 to 402) bound to hnRNP L much more strongly than HCV RNA lacking the core-coding sequence (nt 18 to 331) (compare lane 2 with lane 3 in Fig. 2A). This result suggests not only that hnRNP L binds to the HCV IRES but also that the RNA segment of HCV spanning nt 332 to 402 is required for the tight binding between the HCV IRES and hnRNP L. The binding pattern of cellular hnRNP L to HCV RNA was similar to that of the 68-kDa protein evident in the UV-cross-linking data (compare Fig. 2A with Fig. 1). Interestingly, at least two hnRNP L-related proteins were recognized by the hnRNP L antibody (Fig. 2A, lanes 2 and 3). This result is probably due to posttranslational modification of hnRNP L, which was suggested by Pinol-Roma et al. (26) based on two-dimensional gel electrophoresis results.
FIG. 2.
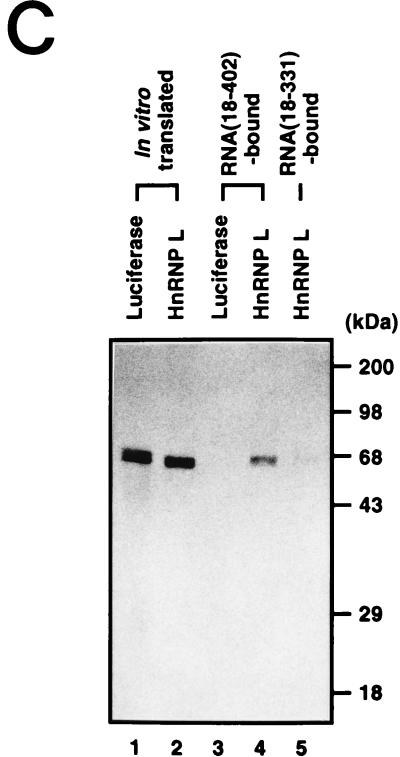
Assignment of the 68-kDa protein band to hnRNP L. (A) Coprecipitation and Western blot analysis. HeLa cell extract was incubated without RNA (lane 1) or with the biotinylated HCV RNA(18–402) (lane 2) and HCV RNA(18–331) (lane 3). The bound proteins were then precipitated with streptavidin acrylamide beads. After being resolved in an SDS–12% polyacrylamide gel, hnRNP L was visualized by Western blot analysis with anti-hnRNP L monoclonal antibody. (B) Combination of RNA mobility shift and Western blot analyses. Aliquots of HeLa cell extract (20 μg) were incubated without RNA (lane 1) or with 100 ng of RNA(18–331) (lane 2), 100 ng of RNA(18–402) (lane 3), 300 ng of RNA(18–331) (lane 4), or 300 ng of RNA(18–402) (lane 5). The samples were electrophoresed in a 5% nondenaturing gel, after which the proteins were blotted onto a nitrocellulose membrane and probed by Western blotting with anti-hnRNP L monoclonal antibody. Free hnRNP L and RNA-bound hnRNP L are marked “Free” and “Bound,” respectively. (C) Coprecipitation of 35S-labeled proteins. The in vitro-translated proteins luciferase and hnRNP L are in lanes 1 and 2, respectively. These 35S-labeled proteins were incubated with biotinylated RNA(18–402) (lanes 3 and 4) and RNA(18–331) (lane 5), after which the RNA-bound proteins were precipitated with streptavidin acrylamide beads. After the samples were washed with binding buffer, the RNA-bound proteins were resolved in an SDS–12% polyacrylamide gel. The protein bands were detected by autoradiography.
To further confirm the interaction between hnRNP L and HCV IRES, a combination experiment with the RNA gel mobility shift assay and Western blot analysis was carried out (Fig. 2B). After incubation of HeLa cell extracts with RNAs corresponding to different parts of HCV IRES, the samples were electrophoresed on a nondenaturing gel. The positions of hnRNP L in the gel were visualized by Western blot analysis with monoclonal anti-hnRNP L antibody (4D11). The free hnRNP L was detected near the well of the gel, because the pI of hnRNP L is 7.4 to 7.7 (5) and the pH of the running buffer was 8.3. RNA-hnRNP L complexes migrated faster due to the highly negatively charged RNA in spite of the larger size of the RNA-protein complex (note that the band labeled “Bound” migrated faster than the band labeled “Free” in lane 5 of Fig. 2B). HCV RNA(18–402), which contains the full length of IRES, formed a much larger amount of RNA-protein complex with hnRNP L than HCV RNA(18–331), which lacks the core-coding sequence (compare lane 4 with lane 5 in Fig. 2B). This indicates that hnRNP L in HeLa cells can bind to the HCV IRES in the presence of other elements of the translational machinery and that the core-coding sequence of HCV plays an important role in the binding of hnRNP L.
The interaction between the HCV IRES and hnRNP L was also confirmed by testing the binding between in vitro-translated hnRNP L and the HCV RNAs (Fig. 2C). 35S-labeled hnRNP L and luciferase were synthesized in a rabbit reticulocyte lysate (RRL) system as shown in Fig. 2C. These proteins were then incubated with the biotinylated HCV RNA(18–402) or RNA(18–331). The RNA-protein complexes were then precipitated with streptavidin acrylamide beads and resolved by SDS-PAGE. 35S-labeled hnRNP L bound strongly to RNA(18–402) (Fig. 2C, lane 4) and weakly to RNA(18–331) (Fig. 2C, lane 5). Under the same conditions, 35S-labeled luciferase bound neither to RNA(18–402) (Fig. 2C, lane 3) nor to RNA(18–331) (data not shown). This result also indicates that hnRNP L interacts with the HCV IRES and that the core-coding sequence is important for that hnRNP L interaction.
Determination of the hnRNP L-binding site within the HCV IRES.
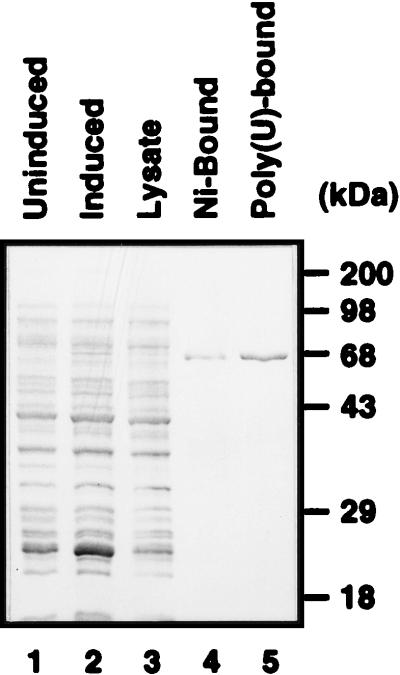
In order to investigate the interaction of hnRNP L with the HCV IRES in more detail, we expressed hnRNP L in E. coli and then purified it using Ni-NTA (Fig. 3, lane 4) and poly(U)-Sepharose (Fig. 3, lane 5) column chromatographies. The identity of the purified hnRNP L was confirmed by Western blot analysis with monoclonal antibody against hnRNP L (data not shown).
FIG. 3.
Purification of hnRNP L. Purified hnRNP L was resolved by SDS–12% PAGE and stained with Coomassie brilliant blue G-250. Protein profiles in uninduced E. coli cells, in induced E. coli cells, in E. coli lysate, in E. coli cells after Ni-NTA column chromatography, and in E. coli cells after poly(U) column chromatography are shown in lanes 1, 2, 3, 4, and 5, respectively.
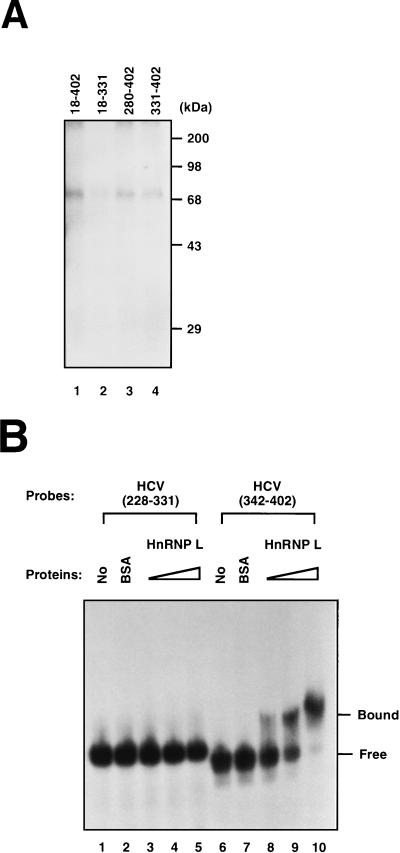
Direct interaction between hnRNP L and the HCV IRES was investigated by a UV cross-linking method with purified hnRNP L and HCV RNAs spanning different regions of the HCV IRES. Purified hnRNP L exhibited a much stronger RNA-binding activity with [32P]RNA(18–402) than with [32P]RNA(18–331) (compare lane 1 with lane 2 in Fig. 4A). Purified hnRNP L also bound to small RNA fragments containing the 3′ region of the HCV IRES {[32P]RNA(280–402) and [32P]RNA(331–402)} (Fig. 4A, lanes 3 and 4), although the intensities of the bands were lower. This reduction in hnRNP L binding is likely due to the elimination of the cooperative binding of hnRNP L to the strong binding site at the core-coding sequence and the weaker binding near the 5′ border of the HCV IRES (data not shown). Protein-protein interaction between hnRNP L molecules, which may support cooperative binding, was detected in a yeast two-hybrid system and by a coprecipitation method (data not shown).
FIG. 4.
Determination of the minimal hnRNP L-binding site in the HCV IRES. (A) UV cross-linking of hnRNP to HCV RNA. [32P]RNA(18–402) (lane 1), [32P]RNA(18–331) (lane 2), [32P]RNA(280–402) (lane 3), and [32P]RNA(331–402) (lane 4) were cross-linked to purified hnRNP L by UV irradiation. The labeled proteins were then analyzed by SDS–12% PAGE. (B) Gel mobility shift assay. RNA gel mobility shift assays were performed with [32P]RNA(228–331) (lanes 1 to 5) and [32P]RNA(342–402) (lanes 6 to 10). Samples in lanes 2 and 7 were incubated with 1 μg of BSA. Samples in lanes 3 and 8, 4 and 9, and 5 and 10 were incubated with 0.15, 0.3, and 0.6 μg of purified hnRNP L, respectively. Samples were analyzed in a 5% nondenaturing polyacrylamide gel. The positions of free RNA probe and protein-bound probe are marked “Free” and “Bound,” respectively.
To test whether the core-coding region is sufficient for the interaction with hnRNP L, the HCV probe [32P]RNA(342–402) was generated and its hnRNP L-binding ability was investigated by the RNA gel mobility shift assay. HnRNP L showed high RNA-binding activity towards [32P]RNA(342–402), which spans the N terminus of the core-coding sequence (Fig. 4B, lanes 8 to 10). However, hnRNP L did not bind to the similarly sized HCV probe [32P]RNA(228–331) (Fig. 4B, lanes 3 to 5). Under the same conditions, BSA bound to neither [32P]RNA(228–331) nor [32P]RNA(342–402) (Fig. 4B, lanes 2 and 6). This result shows that hnRNP L can by itself bind to the 3′ boundary of the HCV IRES corresponding to nt 342 to 402 and that this region is the major hnRNP L-binding site.
Strength of hnRNP L binding to HCV IRES correlates with translation efficiency of an mRNA.
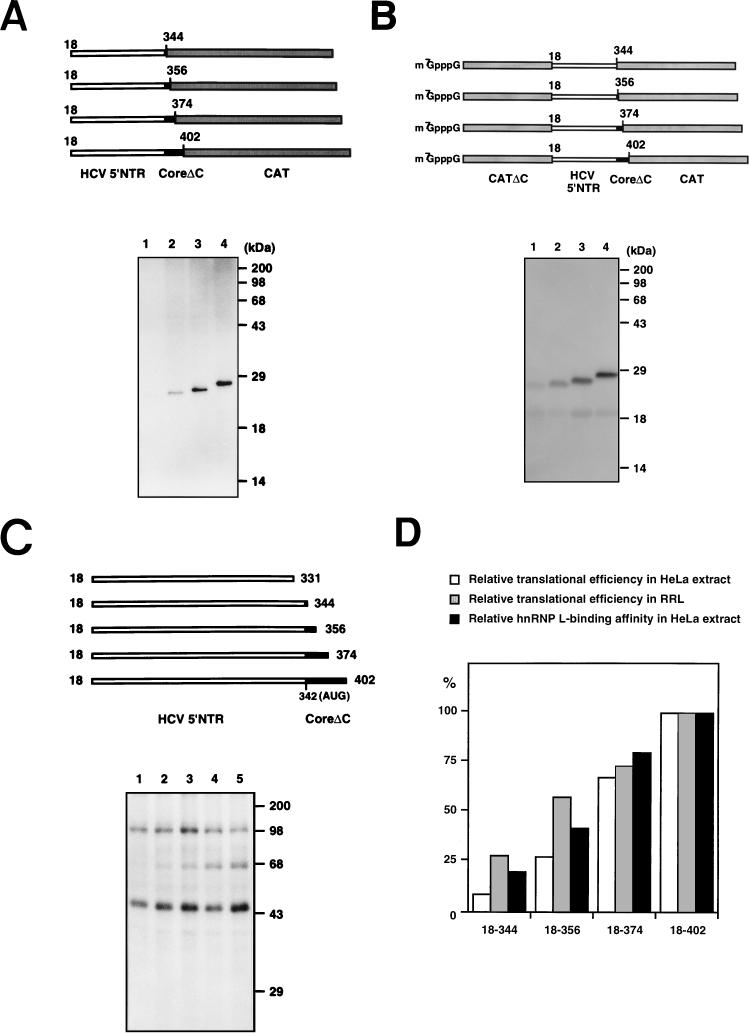
The effect of the core-coding sequence on HCV mRNA translation was examined by serial deletions of the core-coding sequence. The CAT-coding sequence was fused in frame with the truncated core sequence to serve as a reporter gene. Translational efficiencies of the transcripts were examined in HeLa cell extract and in RRL in which newly synthesized proteins were labeled with [35S]methionine. There are equal numbers of methionine residues in each polypeptide. The translational efficiencies of the hybrid genes gradually increased with the expansion of the core sequence both in HeLa extract with a monocistronic configuration (compare lanes 1, 2, 3, and 4 in Fig. 5A) and in RRL with a dicistronic configuration (compare lanes 1, 2, 3, and 4 in Fig. 5B). Binding affinity of hnRNP L to HCV RNA gradually increased along with the length of the core-coding sequence (compare the 68-kDa protein bands in lanes 1 to 5 in Fig. 5C). Translational efficiencies (Fig. 5A and B) and the intensities of the hnRNP L bands (Fig. 5C) of the corresponding mRNAs matched well, as seen in Fig. 5D. These data therefore indicate that hnRNP L, which binds around the 3′ border of the HCV IRES, might enhance HCV mRNA translation.
FIG. 5.
Correlation between translational efficiency and hnRNP L-binding affinity. (A) Translation of monocistronic mRNAs in HeLa cell extract. Uncapped mRNAs were translated in HeLa cell extracts. Translation products were resolved by SDS–15% PAGE. Schematic diagrams of the mRNAs are shown at the top of the panel. CoreΔC, C-terminally truncated core. (B) Translation of dicistronic mRNAs in RRL. Capped dicistronic mRNAs were translated in RRL. Translation products were resolved by SDS–15% PAGE. Schematic diagrams of the mRNAs are shown at the top of the panel. CATΔC, C-terminally truncated CAT gene. (C) UV cross-linking of HCV RNAs. 32P-labeled RNA probes were cross-linked with HeLa cell extracts and then analyzed by SDS–12% PAGE. Schematic diagrams of the RNA probes are shown at the top of the panel. (D) Relative translational efficiencies of the mRNAs and relative affinities of the same set of RNAs for hnRNP L. The intensities of the bands corresponding to the polypeptides shown in panels A and B and the bands corresponding to hnRNP L in panel C were measured with a phosphorimager. The relative translational efficiencies and relative hnRNP L-binding affinities of the HCV RNAs were normalized by considering the activity of HCV RNA(18–402) 100%.
DISCUSSION
Translation of HCV RNA is initiated by internal entry of the ribosome into the HCV IRES. Several cellular proteins that interact specifically with the HCV IRES RNA have been identified (1, 2, 6). PTB, which is required for picornavirus IRES function (17, 22), has been reported to bind to multiple sites on the HCV IRES (1). Ali and Siddiqui (1) observed inhibition of HCV mRNA translation after depletion of PTB and proteins associated with it using an antibody against PTB. They predicted the presence of a PTB-associated protein essential for HCV IRES function, since translation of HCV mRNA could not be restored by addition of purified PTB to the PTB-immunodepleted translation mixture (1). Here we report that hnRNP L binds specifically to the 3′ border region of the HCV IRES (nt 342 to 402) and that binding correlates with the translational efficiency of HCV mRNA. Interestingly, we discovered the hnRNP L-PTB interaction using yeast two-hybrid screening (8). This finding suggests that hnRNP L may be one of the cellular factors facilitating translation of HCV mRNA, possibly in cooperation with PTB.
Core-coding sequence up to nt 33 or more was required for efficient binding of hnRNP L and translation. This result is consistent with the 3′ boundary of the HCV IRES reported by Reynolds et al. (28). Expansion of the core sequence up to nt 61 increased translation and hnRNP L binding further (compare lane 3 with lane 4 in Fig. 5A and B, and lane 4 with lane 5 in Fig. 5C). These findings may indicate that the core sequence segment of nt 34 to 61 contributes at least in part, if not essentially, to the translation of HCV RNA, probably by enhancing the binding of hnRNP L. The requirement of the core-coding sequence for HCV IRES function was also demonstrated in the construction of a hybrid poliovirus containing the HCV IRES element (19). HCV IRES functioning was minimal when only 24 nt of core-coding sequence was included in the hybrid virus. On the other hand, HCV IRES function was dramatically increased when 369 nt of core-coding sequence was included. This indicates that some core-coding sequence is required for optimal IRES function.
Role of hnRNPs in the translation of mRNAs.
hnRNPs are, by definition, nuclear proteins that interact with heterogeneous nuclear RNAs (hnRNAs). Several functions have been suggested for hnRNPs. They mostly relate to RNA functions such as pre-mRNA processing, mRNA translocation from the nucleus to the cytoplasm, and translation (5). The last two functions are attributed to a group of hnRNPs that shuttle between the nucleus and the cytoplasm. Several hnRNPs were reported to play roles in translation. PTB (hnRNP I) was shown to enhance IRES-dependent translation of encephalomyocarditis virus and foot-and-mouth disease virus mRNAs (17, 22). hnRNP E2, which is also known as PCBP2, was required for the efficient translation of poliovirus RNA in HeLa cells (3). On the other hand, hnRNP K and E1 inhibit translation of erythroid 15-lipoxygenase mRNA by binding to the 3′NTR of the mRNA (24). Therefore, it is not surprising to discover other hnRNPs which take part in translation.
In order to participate in HCV IRES-dependent translation, hnRNP L should be present in the cytoplasm at least to some extent. However, hnRNP L was reported to be localized mainly in the nucleus (26). Interestingly, hnRNP L was found in the cytoplasm as well as the nucleus when transcription of cellular mRNA was blocked by poliovirus infection or treatment with actinomycin D (data not shown). These findings suggest that hnRNP L may shuttle between the nucleus and the cytoplasm in a transcription-sensitive manner similar to that of some other hnRNP proteins (for example, hnRNP A1, E, and I [21]). Therefore, hnRNP L may facilitate translation of some mRNAs while it stays in the cytoplasm.
How can hnRNP L facilitate translation of HCV mRNA?
With the limited information of hnRNP L we have, we cannot conclusively infer the molecular mechanism of the translational activation by hnRNP L. However, we can speculate about the role of hnRNP L in translation. First, binding of hnRNP L to HCV mRNA may put the structure of the HCV mRNA into a proper conformation accessible to the translational machinery. The putative conformational change might be induced by interactions among mRNA, PTB, and hnRNP L (note that hnRNP L-PTB, PTB-PTB, PTB-RNA, and hnRNP L-RNA interactions are all possible [8]). Second, hnRNP L that is bound near the initiation codon of HCV mRNA may recruit canonical translational initiation factors or the ribosome through direct interaction. In both cases, hnRNP L might be removed from the mRNA once the translational machinery gets onto the initiation codon. Intriguingly, La protein, which has RNA helicase activity, has been shown to interact with the initiation codon of HCV mRNA and facilitate translation (2). Translational activation of HCV mRNA by La protein might happen by a cleaning up of the prebound initiation factor(s), including hnRNP L, from the mRNA in order to initiate translation after ribosome binding to the mRNA.
ACKNOWLEDGMENTS
We are indebted to G. Dreyfuss, University of Pennsylvania School of Medicine, Philadelphia, for kindly providing us with a monoclonal antibody against hnRNP L, 4D11.
This study was supported in part by grants from the G7 program, the Ministry of Science and Technology, KOESEF BSRI-97-4434, LG Chem., KOESEF, through the SRC for cell differentiation, and the POSTECH/BSRI special fund.
REFERENCES
- 1.Ali N, Siddiqui A. Interaction of polypyrimidine tract-binding protein with the 5′ noncoding region of the hepatitis C virus RNA genome and its functional requirement in internal initiation of translation. J Virol. 1995;69:6367–6375. doi: 10.1128/jvi.69.10.6367-6375.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali N, Siddiqui A. The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc Natl Acad Sci USA. 1997;18:2249–2254. doi: 10.1073/pnas.94.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blyn L B, Towner J S, Semler B L, Ehrenfeld E. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J Virol. 1997;71:6243–6246. doi: 10.1128/jvi.71.8.6243-6246.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown E A, Zhang H, Ping L H, Lemon S M. Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 1992;20:5041–5045. doi: 10.1093/nar/20.19.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dreyfuss G, Matunis M J, Pinol-Roma S, Burd C G. HnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 6.Fukushi S, Kurihara C, Ishiyama N, Hoshino F B, Oya A, Katayama K. The sequence element of the internal ribosome entry site and a 25-kilodalton cellular protein contribute to efficient internal initiation of translation of hepatitis C virus RNA. J Virol. 1997;71:1662–1666. doi: 10.1128/jvi.71.2.1662-1666.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fukushi S, Katayama K, Kurihara C, Ishiyama N, Hoshino F B, Ando T, Oya A. Complete 5′ noncoding region is necessary for the efficient internal initiation of hepatitis C virus RNA. Biochem Biophys Res Commun. 1994;199:425–432. doi: 10.1006/bbrc.1994.1246. [DOI] [PubMed] [Google Scholar]
- 8.Hahm B, Cho O H, Kim J-E, Kim Y K, Kim J H, Oh Y L, Jang S K. Polypyrimidine tract-binding protein interacts with hnRNP L. FEBS Lett. 1998;425:401–406. doi: 10.1016/s0014-5793(98)00269-5. [DOI] [PubMed] [Google Scholar]
- 9.Han J H, Shyamala V, Richman K H, Brauer M J, Irvine B, Urdea M S, Tekamp-Olson P, Kuo G, Choo Q-L, Houghton M. Characterization of the terminal regions of hepatitis C viral RNA: identification of conserved sequences in the 5′ untranslated region and poly(A) tails at 3′ end. Proc Natl Acad Sci USA. 1991;88:1711–1715. doi: 10.1073/pnas.88.5.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hellen C U T, Pestova T V, Litterst M, Wimmer E. The cellular polypeptide p57 (pyrimidine tract-binding protein) binds to multiple sites in the poliovirus 5′ nontranslated region. J Virol. 1994;68:941–950. doi: 10.1128/jvi.68.2.941-950.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Honda M, Brown E A, Lemon S M. Stability of a stem-loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA. 1996;2:955–968. [PMC free article] [PubMed] [Google Scholar]
- 12.Honda M, Ping L H, Rijnbrand R C, Amphlett E, Clarke B, Rowlands D, Lemon S M. Structural requirements for initiation of translation by internal ribosome entry within genome-length hepatitis C virus RNA. Virology. 1996;222:31–42. doi: 10.1006/viro.1996.0395. [DOI] [PubMed] [Google Scholar]
- 13.Jang S K, Kräusslich H G, Nicklin M J H, Duke G M, Palmenberg A C, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jang S K, Davies M V, Kaufman R J, Wimmer E. Initiation of protein synthesis by internal entry of ribosomes into the 5′ nontranslated region of encephalomyocarditis virus RNA in vivo. J Virol. 1989;63:1651–1660. doi: 10.1128/jvi.63.4.1651-1660.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jang S K, Wimmer E. Cap-independent translation of encephalomyocarditis virus RNA: structural elements of the internal ribosomal entry site and involvement of a cellular 57-kD RNA-binding protein. Genes Dev. 1990;4:1560–1572. doi: 10.1101/gad.4.9.1560. [DOI] [PubMed] [Google Scholar]
- 16.Kato N, Hijikata M, Ootsuyama Y, Nakagawa M, Ohkoshi S, Suguyama T, Shimotohno K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A non-B hepatitis. Proc Natl Acad Sci USA. 1990;87:9524–9528. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaminski A, Hunt S L, Patton J G, Jackson R J. Direct evidence that polypyrimidine tract binding protein is essential for internal initiation of translation of encephalomyocarditis virus RNA. RNA. 1995;1:924–938. [PMC free article] [PubMed] [Google Scholar]
- 18.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu H-H, Wimmer E. Poliovirus chimeras replicating under the translational control of genetic elements of hepatitis C virus reveal unusual properties of the internal ribosomal entry site of hepatitis C virus. Proc Natl Acad Sci USA. 1996;93:1412–1417. doi: 10.1073/pnas.93.4.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meerovitch K, Svitkin Y V, Lee H S, Lejbkowicz F, Kenan D J, Chan E K L, Agol V I, Keene J D, Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J Virol. 1993;67:3798–3807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michael W M, Siomi H, Choi M, Pinol-Roma S, Nakielny S, Liu Q, Dreyfuss G. Cold Spring Harbor Symp. Quant Biol. 1995;60:663–668. doi: 10.1101/sqb.1995.060.01.071. [DOI] [PubMed] [Google Scholar]
- 22.Niepmann M. Porcine polypyrimidine tract-binding protein stimulates translation initiation at the internal ribosome entry site of foot-and-mouth-disease virus. FEBS Lett. 1996;388:39–42. doi: 10.1016/0014-5793(96)00509-1. [DOI] [PubMed] [Google Scholar]
- 23.Oh Y L, Hahm B, Kim Y K, Lee H K, Lee J W, Song O-K, Tsukiyama-Kohara K, Kohara M, Nomoto A, Jang S K. Determination of functional domains in polypyrimidine tract-binding protein. Biochem J. 1998;331:169–175. doi: 10.1042/bj3310169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ostareck D H, Ostareck-Lederer A, Wilm M, Thiele B J, Mann M, Hentze M W. mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell. 1997;16:597–606. doi: 10.1016/s0092-8674(00)80241-x. [DOI] [PubMed] [Google Scholar]
- 25.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 26.Pinol-Roma S, Swanson M S, Gall J G, Dreyfuss G. A novel heterogeneous nuclear RNP protein with a unique distribution on nascent transcripts. J Cell Biol. 1989;109:2575–2587. doi: 10.1083/jcb.109.6.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poole T L, Wang C, Popp R A, Potgieter L N, Siddiqui A, Collett M S. Pestivirus translation initiation occurs by internal ribosome entry. Virology. 1995;206:750–754. doi: 10.1016/s0042-6822(95)80003-4. [DOI] [PubMed] [Google Scholar]
- 28.Reynolds J E, Kaminski A, Kettinen H J, Grace K, Clarke B E, Carroll A R, Rowlands D J, Jackson R J. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rijnbrand R, van der Straaten T, van Rijn P A, Spaan W J, Bredenbeek P J. Internal entry of ribosomes is directed by the 5′ noncoding region of classical swine fever virus and is dependent on the presence of an RNA pseudoknot upstream of the initiation codon. J Virol. 1997;71:451–457. doi: 10.1128/jvi.71.1.451-457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rose J K, Trachsel H, Leong K, Baltimore D. Inhibition of translation by poliovirus: inactivation of a specific initiation factor. Proc Natl Acad Sci USA. 1978;75:2732–2736. doi: 10.1073/pnas.75.6.2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Tsukiyama-Kohara K, Lizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang C, Le S Y, Ali N, Siddiqui A. An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5′ noncoding region. RNA. 1995;1:526–537. [PMC free article] [PubMed] [Google Scholar]
- 34.Wang C, Sarnow P, Siddiqui A. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J Virol. 1993;67:3338–3344. doi: 10.1128/jvi.67.6.3338-3344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang C, Sarnow P, Siddiqui A. A conserved helical element is essential for internal initiation of translation of hepatitis C virus RNA. J Virol. 1994;68:7301–7307. doi: 10.1128/jvi.68.11.7301-7307.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]