Abstract
Solid tumors are more than an accumulation of cancer cells. Indeed, cancerous cells create a permissive microenvironment by exploiting non-transformed host cells. Thus, solid tumors rather resemble abnormal organs composed of the cancerous cells itself and the stroma providing the supportive framework. The stroma can be divided into the extracellular matrix consisting of proteoglycans, hyaluronic acid, and fibrous proteins, as well as stromal cells including mesenchymal and immune cells; moreover, it contains various peptide factors and metabolites. Here, we will focus on immune-modulating capacities of the tumor microenvironment.
Keywords: A disintegrin and metalloproteinase (ADAM); Indoleamine-2,3-dioxygenase (IDO); Arginase; Hypoxia; Adenosine; Natural killer group 2 member D (NKG2D) ligands
Introduction
The creation of a permissive microenvironment is frequently observed in cancer [1, 2]. Thus, solid tumors are composed of the transformed cells itself as well as the stroma (from Late Latin: ‘a mattress’) which not only provides a supportive framework, but also helps the transformed cells to escape the immune surveillance. To this end, the cancer cells may comprise only 30 % or less of the cells in the tumor. The stroma consists of (1) the extracellular matrix (ECM) composed of proteoglycans, hyaluronic acid, and fibrous proteins, (2) stromal cells including mesenchymal and immune cells, (3) various peptide factors (e.g., enzymes, chemokines, and cytokines), and (4) metabolites produced by cancerous and stromal cells. Notably, tumors express an array of both cell-surface molecules and soluble factors that influence cells of the immune system. Moreover, immune responses can also be indirectly modulated by either recruiting cells to the tumor site or modifying the function of cells already present in the microenvironment.
The fact that the evolution of tumors for many entities occurs in the presence of inflammation is one of the keys to understand the intrinsic relationship between cancer cells and the stroma [3, 4]. Indeed, this inflammatory state is present in most cases even prior to key tumorigenic genetic alterations and mediates co-evolution of the tumor with numerous cell types [5]. During tumor development, the stroma and the ECM undergo substantial changes as a consequence of interactions between cancerous and stromal cells. To this end, activated fibroblasts constituting the most abundant stromal cells in many tumors secrete numerous cytokines and chemokines that impact directly on cells in the microenvironment and/or attract additional cells to the tumor site; these include IL-6, FOX03, transforming growth factor β (TGFβ), COX-2, vascular endothelial growth factor (VEGF), serum-derived factor 1 (SDF-1), CXCL1/2, and IL-1β [2]. However, these molecules may not only be expressed by fibroblasts but likewise by cancer or immune competent cells. Furthermore, the activity of these factors is also indirectly regulated by the ECM, for example, by protease-mediated release of sequestered cytokines [6].
Tumor-inhibiting versus tumor-promoting microenvironments
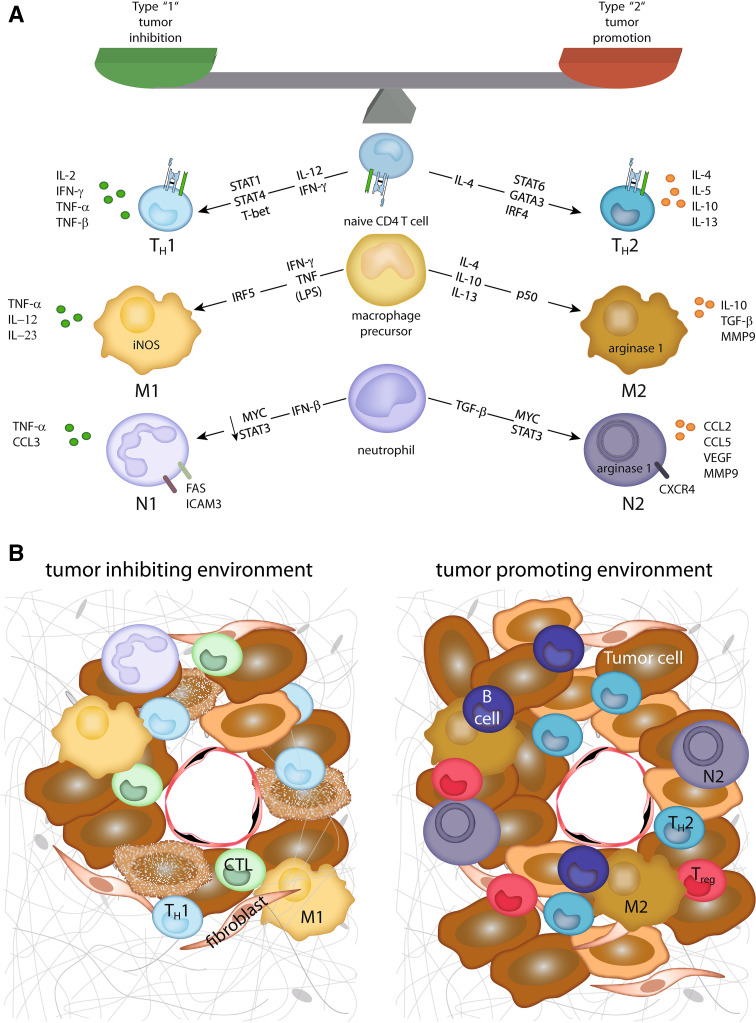
Malignant cells can be detected and destroyed by cells of the immune system, particularly by cells of the adaptive immune system, and there is strong evidence that the immunological recognition impacts on prognosis [7]. Nevertheless, there is even stronger evidence that in many cancer patients, the cancerous cells may escape these immune responses [8, 9]. Many of the immune escape mechanisms are based on the induction of what we presume is an inappropriate response, that is, a response that is not inhibiting but maybe even promoting the tumor (Fig. 1). One of the best explored examples for such inappropriate responses—particularly with respect to adaptive immune responses—is the induction of the two different effector CD4+ T helper cell responses, that is, Th1 and Th2 responses. The Th1 response is fostering cytotoxic responses, for example, by secreting interferon γ (IFNγ) activating the cytolytic activities of macrophages and cytotoxic T lymphocytes (CTL) [10]; Th2 cells are fostering humoral responses, for example, by production of IL4-activating B-cells. Th1 responses are best suited to fight virally infected and transformed cells, whereas Th2 responses will fight extracellular bacteria, parasites, and toxins. Generally, a Th2 response is regarded rather as tumor-promoting as compared to a tumor-inhibiting Th1 response which could potentially lead to tumor clearance by triggering a CTL response against tumor antigens. By now for almost any immune competent cell, there has (at least) a type ‘1’ and type ‘2’ cell been established, for example, besides for T helper cells, for natural killer T (NKT) cells as NKT1 and NKT2 cells, neutrophils as N1 and N2, or macrophages as M1 and M2 (Fig. 1) [11–13]. In all cases, type ‘1’ cells are regarded as tumor-inhibiting and type ‘2’ cells as tumor-promoting. Although it is generally thought that the class of an immune response is tailored to fit the invading pathogen, it has recently been suggested that it is primarily tailored to fit the tissue in which the response occurs [14]. This notion is based on the hypothesis that a microenvironment (including the tumor microenvironment) is not simply a passive recipient of immune protection, but is an active participant in its own defense. Indeed, each cell in the tumor microenvironment—irrespective of the transformation status—holds the capacity to produce immune modulatory signals [2]. Thus, the differentiation and activation status of any immune competent cell that enters the tumor microenvironment will be modulated by signals in the respective microenvironment.
Fig. 1.
Tumor-inhibiting and tumor-promoting microenvironments. a Depicted are the major factors, and transcription factors involved in the differentiation of type 1 or type 2 cell subtypes. In addition, the main effector molecules from these cells contributing to their tumor-inhibiting or tumor-promoting effect, respectively, are displayed. b In a tumor-inhibiting microenvironment, type 1 cells together with cytotoxic T cells (CTL) contribute to tumor cell destruction. In contrast, in a tumor-promoting microenvironment, type 2 cells are accompanied by regulatory T cells (Treg) and B lymphocytes inhibiting a cytotoxic immune reaction and promoting tumor cell growth, angiogenesis, and metastasis
The extracellular matrix
The ECM has a profound impact on the cellular constituents of the tumor microenvironment. To this end, proteolytic enzymes [e.g. matrix metalloproteinases (MMPs) and a disintegrin and metalloproteinase (ADAM)] are key enzymes involved with remodeling of the ECM [15]. While these proteolytic enzymes can degrade numerous components of the ECM, recent studies have implicated MMPs as important mediators of cell–cell communication by virtue of their ability to process multiple non-matrix molecules, such as cytokines and growth factors, to soluble forms that have either enhanced or attenuated activities: Examples include (1) MMPs increase the bioavailability of TGFβ by regulating the release from an inactive extracellular complex [16]; (2) MMPs cleave growth factors and cytokine receptors, for example, the IL-2 receptor (IL-2α) and thereby inhibit proliferation and activation of T cells in the vicinity [17]; additionally, (3) ADAMs cleave NKG2D ligands and cell adhesion molecules such as L-selectin (CD62L) or ICAM-1 [18, 19]. Thus, the presence of proteolytic enzymes influences cancer cell growth, differentiation, metastatic capacity, and resistance to apoptosis as well as the immunologic micromilieu. Notably, within solid tumors, both cancerous and stromal cells express these proteolytic enzymes.
Interestingly, MMP-2 has been identified as a tumor antigen as well: A MMP-2-derived peptide is presented in the context of HLA-A2 on the surface of melanoma cells which can be recognized by cytotoxic CD8+ T cells [20]. More recently, it was demonstrated that MMP-2 also plays a role in polarizing adaptive immune responses. CD4+ T cells recognizing class II restricted MMP2-derived peptides were mainly of Th2 type expressing GATA and secreting TNFα, IL-4, and IL-13 [20]. Notably, the mechanism underlying this Th2 polarization was attributed to both active and inactive MMP-2. While active MMP-2 led to degradation of the type I IFN receptors on dendritic cells (DCs), both inactive and active MMP-2 induced up-regulation of the CD40 ligand on DCs.
Immune suppression by metabolic enzymes: indoleamine-2,3-dioxygenase (IDO), tryptophan 2,3-dioxygenase (TDO), and arginase
Over the past years, it became increasingly obvious that both depletion of essential nutrients and accumulation of immunosuppressive metabolites generate a tumor-permissive microenvironment [1]. Thus, the metabolic changes within the tumor microenvironment help to evade antigen-specific immune responses.
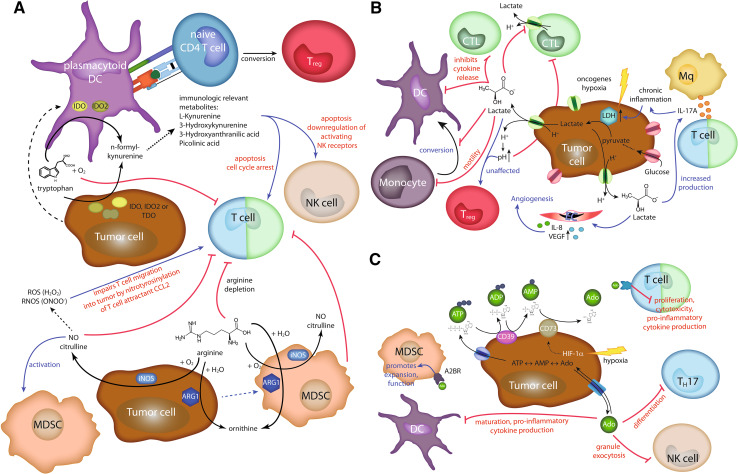
The immune-suppressive capacity of indoleamine-2,3-dioxygenase has been demonstrated in different physiologic and pathologic situations including cancer (Fig. 2) [21]: IDO restrains potentially harmful inflammatory reactions directly by degradation of the essential amino acid tryptophan and indirectly by recruitment of regulatory T cells. In addition, IDO generates immune tryptophan metabolites that have direct toxic effects on CTL and Th1 cells [22]. Hence, IDO may shift T cell polarization toward a Th2 response. Treg-mediated immune suppression is mediated by interaction of B7 expressed by IDO+ DC with CTLA4 on regulator T cells (Tregs) inducing Treg proliferation. Moreover, IDO blocks the IL-6-mediated reprogramming of mature Tregs to cells resembling pro-inflammatory Th17 cells [23]. IDO expression within solid tumors has been reported for both cancerous and stromal cells [21]. In addition, IDO elevation occurs in a subset of plasmacytoid DCs in tumor-draining lymph nodes [22]. IDO may not only be expressed in the tumor microenvironment itself, but also in the tumor-draining lymph node where it may help to create the pre-metastatic niche. Accordingly, IDO expression in tumor cells has been demonstrated to correlate with a decreased serum tryptophan concentration which is associated with an impaired prognosis [24]. Notably, IDO is also involved in both tumor vascularization and IL-6-dependent myeloid-derived suppressor cell (MDSC)-driven immune escape [25, 26].
Fig. 2.
Metabolic enzymes and metabolites in immunosuppression. a Immunosuppression by metabolism of the amino acids tryptophan (upper part) or arginine (lower part). Tumor cells can express the enzymes indoleamine-2,3-dioxygenase (IDO), indoleamine-2,3-dioxygenase-like protein (IDO2), and tryptophan 2,3-dioxygenase (TDO). All these enzymes are able to catalyze the first step in kynurenine pathway of tryptophan metabolism. In addition, plasmacytoid DCs in the draining lymph nodes of cancer patients can express IDO and IDO2. The general depletion of tryptophan impairs T cell proliferation. Furthermore, immune modulatory tryptophan metabolites can induce apoptosis and affect activity of T and NK cells. IDO expressing plasmacytoid DCs can also trigger regulatory T cell (Treg) differentiation. Similarly, arginine depletion by arginase I (ARG1) or inducible nitric oxide synthase (iNOS) impairs T cell activation. iNOS can further suppress immune responses by recruitment and activation of MDSC. NO can be further converted into reactive oxygen species (ROS) and reactive nitrogen and oxygen species (RNOS). The latter can impair T cell migration into the tumor by nitrotyrosinylation of the chemokine CCL2. b Tumor cells primarily rely on glycolysis. One important enzyme involved is LDH converting pyruvate into lactate. Its expression is up-regulated by hypoxia or oncogenes. Lactate is secreted from the cells via monocarboxylate transporters accompanied by H+ transport decreasing the extracellular pH. Lactate affects many different processes like inhibition of cytotoxic T cell responses, causing chronic inflammation via triggering enhanced IL-17A cytokine secretion, or increasing secretion of angiogenic factors by endothelial cells (EC). c Immune suppression by adenosine (Ado). Extracellular ATP is converted by CD39 into ADP and then into AMP. CD73 converts AMP into Ado. Ado can be exported and imported from the tumor cell. HIF-1α enhances CD73 expression. Ado interaction with adenosine A2A receptor (A2AR) on T cells impairs their activity, whereas the binding to adenosine A2B receptor (A2BR) on MDSC promotes their recruitment and function. In addition, Ado inhibits the differentiation of Th17 cells, the activity of NK cells, and the maturation of DCs
Another means to degrade tryptophan, and thereby, resist immune rejection is the expression of tryptophan 2,3-dioxygenase (TDO), a homo-tetrameric heme-containing cytosolic enzyme. While under physiologic conditions, TDO is almost exclusively expressed in the liver, tumors of different tissue origins such as melanoma, bladder cancer, and glioblastoma express TDO [27]. The production of kynurenine by TDO inhibits anti-tumor immune responses. Interestingly, blocking of both TDO and IDO might turn out to be complementary, not redundant: In a series of more than a hundred human tumor cell lines, one-third expressed only TDO and another third only IDO [27]. Similarly, tumor-associated tryptophan hydroxylase-1 (Tph-1), a synthase that catalyzes the conversion of tryptophan to serotonin, is a potent regulator of immunity [28].
Besides manipulating the metabolism of tryptophan, l-arginine depletion by arginase or inducible nitric oxide synthase (iNOS) results in suppression of tumor-specific T cell responses [29]. Arginase expression can be induced by cyclooxygenase-2, thus explaining the correlation of increased concentrations of prostaglandin-2 with the suppression of T cell activation. Notably, aberrant induction of COX-2 and up-regulation of the prostaglandin cascade have additional functions in carcinogenesis which are beyond the scope of this review [30]. In addition to cancerous cells, in the tumor microenvironment macrophages, granulocyte and MDSCs may express arginase and iNOS.
Hypoxia and the ‘Warburg effect’
Hypoxia has an established role in tumor cell stemness and invasiveness, radio- and chemo-resistance, as well as in generating an immune-permissive microenvironment [31]. Many of these hypoxia-induced changes are linked to reduced nitric oxide (NO) signaling [32]. The primary source of NO is the nitric oxide synthase (NOS), which has three isoforms; two are constitutively expressed, and one is inducible. The inducible form iNOS produces NO for prolonged periods of time in a calcium-independent manner. Levels of NO produced by iNOS in the microenvironment of the cell can range from as low as 10 nM to μM amounts for days. Notably, NO has a strong impact on several immune competent cells. For example, tumor-expressed iNOS leads to recruitment and induction of functional MDSC in a spontaneous murine melanoma model [33]. This effect is mediated by modulation of tumor VEGF secretion and up-regulation of STAT3 and ROS in MDSCs.
Notably, T cells are exposed to different oxygen tensions during their development and while migrating between blood and tissue. Under hypoxic conditions, T cells increase the expression of genes that are regulated by HIF [31]. While hypoxia enhances the transcription of hypoxia-responsive element (HRE)-containing genes, for example, VEGF, glycolytic enzymes, it inhibits the accumulation of non-HRE-containing genes, such as IL-2 and IFNγ during TCR-driven activation. Thus, T cell activation under hypoxic conditions in vivo may lead to different patterns of cytokine secretion.
Cancer cells primarily rely on glycolysis rather than on oxidation of pyruvate in mitochondria for energy production prevalent in normal cells, a phenomenon known as ‘Warburg effect.’ It is associated with an increased expression of glycolytic enzymes, for example, lactate dehydrogenase (LDH) and pyruvate kinase, as well as an enhanced glucose uptake [34]. The ‘Warburg effect’ is caused extrinsically by hypoxia and intrinsically by activated oncogenes; notably, both ways may act in synergism. For example, over-expression of myc leads to up-regulation of glycolytic enzymes, and the hypoxia-inducible factor (HIF) collaborates with myc to induce additional genes such as LDH. The ‘Warburg effect’ causes an increased lactate production, and high lactate levels have immune modulatory properties: (1) Lactate inhibits the differentiation of monocytes to DCs, (2) induces IL-23, a tumor-promoting cytokine involved in the generation of Th17 cells [35, 36], and (3) directly inhibits CTLs [5]. Furthermore, both the tumor cells and the activated T cells rely on and thus compete for glucose in the tumor microenvironment [37].
Adenosine
An additional mechanism for cancer-induced immune suppression is the accumulation of adenosine at the tumor site. Adenosine exerts organ- and cyto-protective functions such as stimulation of angiogenesis as well as inhibition of both inflammatory reactions and adaptive immune responses [38]. There are several independent sources of adenosine in the tumor microenvironment, for example, cell death and nucleotide degradation, hypoxia and ATP breakdown, ATP/ADP release and subsequent dephosphorylation, AMP release, and S-adenosylhomocysteine hydrolysis. Indeed, each of these mechanisms individually or in combination provides a continuous supply of adenosine to the tumor microenvironment [39].
It is not so much the mere amount of adenosine but rather its balance with ATP that impacts immune homeostasis: Adenosine suppresses immune responses through the activation of G-protein-coupled receptors [40], whereas ATP acts as a danger signal released by damaged and dying cells [39]. To this end, ATP initiates immune responses through the ligation of P2X and P2Y purinoreceptors. There are four receptors for adenosine: the pertussis toxin sensitive A1 and A3 receptors, which signal via decreased cAMP as well as phosphoinositide 3-kinase (PI3K) and protein kinase (PK) C pathways, and the adenylate cyclase activating A2A and A2B receptors, which signal via increased cAMP. All receptors stimulate the mitogen-activated protein kinase (MAPK) pathway. Under physiological conditions, adenosine acts through the high-affinity A1 and A2A receptors, but under pathological circumstances with high adenosine concentrations, the low-affinity A2B and A3 receptors become relevant. Numerous immune competent cells express adenosine receptors including B, T, NKT and NK cells, macrophages, DCs, neutrophils, as well as mast cells [40]. Human CD4+ and CD8+ T cells express A2A, A2B, and A3 receptors; their expression is up-regulated upon activation. The A2A receptor is dominant for suppression of T cell responses by inhibiting proliferation, cytotoxicity, and secretion of pro-inflammatory cytokines such as IL-2, TNF-alpha, and macrophage inflammatory protein-1a [40]. Notably, mice lacking the A2A receptor have both a significantly delayed growth of lymphoma cells when compared to wild type mice and when specifically immunized they are characterized by an enhanced protection to subsequent tumor challenges. This protection was associated by an increased frequency of tumor-reactive CTLs at the vaccine-site-draining lymph node [41]. Similarly, pharmacological blockade of the A2A receptor substantially increases the efficacy of antitumor T cell-mediated immunity in mice [42].
The downstream signaling of the A2A receptor is not completely understood but involves inhibition of NF-κB activity. Furthermore, adenosine inhibits the adhesion of cytolytic lymphocytes to cancer cells as well as granule exocytosis by natural killer cells [43]. It also prevents the development of Th17 responses within the tumor microenvironment [44]. Adenosine also inhibits the maturation of DCs and their production of pro-inflammatory cytokines. Indeed, DCs differentiating in the presence of adenosine rather suppress than activate anti-tumor immunity. Finally, adenosine promotes expansion and function of MDSCs via ligation of A2B receptors [45]. Consequently, conversion of ATP into adenosine is strictly regulated. The conversion of ATP into AMP is catalyzed by nucleoside triphosphate diphosphohydrolase 1 (CD39). Subsequently, dephosphorylation of AMP catalyzed by 5′-nucleotidase (5′-NT) generates adenosine; several forms of 5′-NT have been described, but only two of them, that is, cytosolic 5′-NT-I and ecto-5′-NT seem to participate in adenosine generation, with ecto-5′-NT (CD73) being most relevant for the tumor microenvironment [46]. Since the metabolism of AMP into adenosine can only be reversed in the cell, CD73 is an important checkpoint for this process. Several factors present in the tumor microenvironment can induce CD73 including hypoxia. Notably, a HRE-element is present within the CD73 promoter. In addition, humoral factors such as type I IFNs, TNF-α, IL-1β, PGE2, TGF-β, and agonists of the Wnt signaling pathway have been demonstrated to up-regulate CD73 expression [47].
Expression of ligands for immune inhibiting receptors
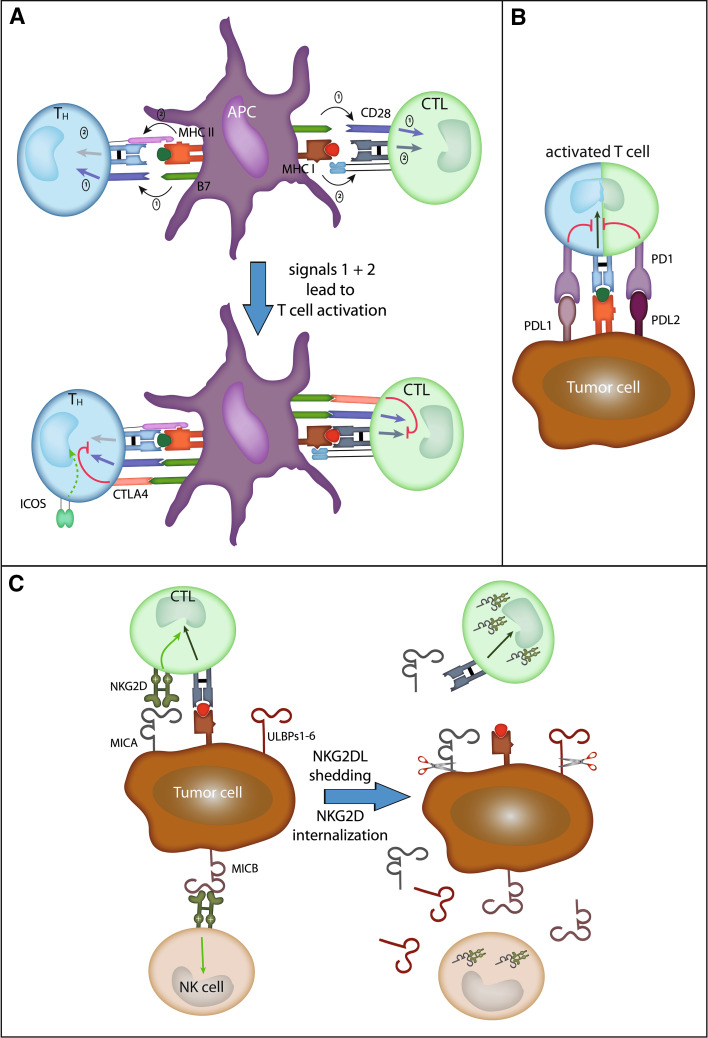
The tumor-specific adaptive immune response to cancer is often ineffective due to the fact that tumors develop multiple resistance mechanisms, for example, expression of ligands for inhibitory immune checkpoints (Fig. 3a) [48]. Consequently, several immune therapeutic approaches for cancer are targeting the immune checkpoint such as the anti-CTLA-4 antibody ipilimumab (Yervoy®) which has been recently registered for the treatment of patients with advanced melanoma [49]. However, it still remains to be established which are the most suitable combination partners for anti-CTLA-4 antibodies and if there are any predictive biomarkers for this therapy [50]. Notably, ipilimumab has been combined or is currently tested with different chemotherapy regimens [51–53], radiotherapy [54], vaccines [55, 56], other immune-modulating agents, and different targeted agents (http://www.clinicaltrials.gov). The identification of immunological markers, such as absolute lymphocyte counts (ALC), CD4+ T cell differentiation, Th17 cells, and induction of ICOS, which correlate with clinical benefit, will be important in the future to select the patients most likely to respond to therapy [50, 54, 55, 57, 58]. However, most potential biomarkers have been identified in retrospective analysis of phase I and phase II clinical studies; thus, they have to be validated in a prospective fashion. Importantly, several lines of evidence suggest that an immune-active tumor microenvironment favors clinical response to ipilimumab [59, 60]. It was demonstrated that a higher baseline expression of a number of immune-related genes, for example, interferon-inducible genes, and Th1 and cytotoxic T cell-associated markers, correlates with clinical response following ipilimumab treatment [59]. In contrast, ipilimumab appears to be equally effective in both the wild type and BRAF-V600E-mutated melanoma patients [61].
Fig. 3.
Ligand-mediated lymphocyte control. a Two signals are needed for naive T cell activation: Firstly, CD80 (B7-1) or CD86 (B7-2) has to trigger CD28 signaling, and secondly, the T cell receptor has to interact with the respective peptide/MHC complex. Shortly after activation, CTLA-4 is up-regulated on the T cell-surface competing with CD28 for ligand binding and in turn delivers an inhibitory signal to the T cell. b PD-1 is an inhibitory receptor on activated T cells. Its ligands PDL1 and PDL2 can be expressed on peripheral tissues during inflammation, but also on tumor cells. c In contrast, NKG2D is an activating receptor on NK cells and delivers a co-stimulatory signal to cytotoxic cells (CTLs). Human ligands induced by viral infection, cytokines, or DNA damage are MICA, MICB, and the ULBP1–6. Shedding of these ligands (sNKG2DL) by tumor cells leads to NKG2D receptor internalization thereby preventing their activating effect
Programmed death protein 1 (PD-1) is a cell-surface membrane protein of the immunoglobulin superfamily which is up-regulated by activated T cells and negatively regulates their function (Fig. 3b) [62]. It was first discovered as a transmembrane protein that is highly expressed in apoptotic T cells. Subsequently, PD-1 was identified as a marker for exhausted T cells. The PD-1 ligands PD-L1 (B7-H1) and PD-L2 (B7-DC) are members of the B7-family. Engagement of PD-1 by its ligands delivers inhibitory signals through activating phosphatases, resulting in dephosphorylation of key elements in the T cell activation pathway, ultimately leading to down-regulating proliferation, survival, and cytokine production [62]. Consequently, blockade of the PD-1 pathway restores cytokine production and proliferation of T cells.
Recent studies demonstrated that the PD-1/PD-L system constitutes one of the immune checkpoint pathways that tumors frequently exploit to suppress the function of tumor-infiltrating T cells. PD-1 is up-regulated on tumor-infiltrating T lymphocytes, and PD1 ligands are expressed in numerous solid tumors. Indeed, there is a strong correlation between the expression of PD-1 ligands in tumors and poor prognosis. Importantly, inhibition of the interaction of PD-1 and its ligands boosts T cell responses in vitro and in vivo [63, 64]. Furthermore, the therapeutic efficacy of PD-1 blockade depends on PD-L1 expression in the individual patient [63]. It has also been demonstrated that reduction of PD-L1 expression on DCs by means of siRNA-lipid nanoparticles results in superior APC function [65].
In addition, other molecules of the B7 family are expressed by tumor cells, for example, B7-H3 and B7-H4 [66]. Available data suggest that these ligands take part in inhibiting immune responses; however, the respective receptors have not been characterized yet [67]. Albeit numerous studies have addressed the question whether the expression of B7-H3 and B7-H4 correlates with prognosis, and the obtained results are not conclusive.
Expression and shedding of NKG2D ligands
The stimulatory natural killer group 2 member D (NKG2D) lymphocyte receptor and its ligands are important mediators of tumor immunity but also are instrumental in promoting tumor immune evasion and immune suppression. In humans, the ligands for NKG2D are the UL16-binding protein (ULBP) 1–6, and the stress-inducible MHC related protein A (MICA) and B (MICB). NKG2D ligands are absent from the surface of most normal cells but are induced by generic responses to cellular stress in diseased cells; for example, their expression can be induced by DNA damage [68]. Indeed, most cancer cell lines and tumor cells in biopsies are positive for NKG2D ligands with MICA being the most highly expressed one. In accordance with this notion, we have previously demonstrated that melanocytic lesions are negative for expression of MICA, whereas primary melanomas were positive in 31 of 40 primary lesions examined [69].
Numerous studies have shown that ligand expression leads to recognition and killing of tumor cells by NK cells [70]. Moreover, activated T cells express NKG2D, and NKG2D signaling seems to be co-stimulatory to TCR engagement. As a consequence, NKG2D ligand expression by cancer cells should lead to improved immune recognition by NK and T cells. However, a sustained surface expression of NKG2D ligands has been proposed as a possible mechanism of suppression of NK cell function (Fig. 3c). Indeed, in patients with tumors expressing NKG2D ligands, NKG2D expression by tumor-infiltrating and circulating cytotoxic cells is low; moreover, its function is often compromised. This can be ascribed to trans-acting effects of soluble NKG2D ligands shed from tumor cells by proteolytic enzymes [71]. Notably, hypoxia increases the shedding of NKG2D ligands through impaired NO signaling [72]. Recently, a mechanistic link between hypoxia-induced accumulation of the α-subunit of HIF-1, increased expression of ADAM10, and decreased surface MICA levels leading to tumor cell resistance to lysis mediated by innate immune effectors, had been demonstrated [73]. Nitric oxide mimetic agents interfered with the hypoxia-induced accumulation of HIF-1α and with the hypoxia-induced up-regulation of ADAM10 expression required for decreased surface MICA expression and resistance to lysis. Moreover, enhanced expression of ADAMs has been described in various inflammatory diseases, suggesting that the inflammatory microenvironment of tumors also contributes to the shedding of NKG2D ligands [74].
Binding of soluble NKG2D ligands leads to internalization of NKG2D and its subsequent degradation. Indeed, soluble NKG2D ligands have been detected in cancer patient sera from both hematological and solid cancers, and also, MICB and ULBP-2 have been found at elevated levels in cancer patient sera. For example, it was demonstrated that soluble ULBP-2 is a marker for poor prognosis in melanoma patients pointing to a functional impact of soluble NKG2D ligands in sera from cancer patients [75]. In the latter study, however, MICA serum levels did not correlate with the clinical course of disease. Interestingly, NKG2D ligands not only differ in their expression pattern, but also in their membrane association. NKG2D ligands may be membrane anchored or glycosylphosphatidylinositol-linked. Thus, some NKG2D ligands may be easier shed. Moreover, substantial polymorphic variation in the sequences that associate the NKG2D ligand MICA with the membrane has been reported, suggesting that polymorphisms in NKG2D ligands cause a relevant person-to-person variation to which end they are shed. In addition, it has been recently reported that melanoma cells may interfere with NK cell function by down-regulating the surface expression of activating receptors, including NKG2D; this inhibitory effect is primarily mediated by IDO and PGE2. Another angle added to the tale stems from recent data, demonstrating that cancer cells may also express NKG2D and that signaling upon ligand binding stimulates tumor growth [76]. These observations help to explain the selection of tumor cells that sustain NKG2D ligands expression.
Tumor-derived membrane vesicles
Once regarded as cellular ‘debris,’ extracellular vesicles are now realized as an important means of cell-to-cell communication and receive particular attention in cancer research [77]. Indeed, tumors are characterized by constitutive secretion of various forms of membrane vesicles. These comprise ‘exosomes,’ ‘microvesicles,’ and ‘membrane particles.’
Taylor et al. [78] initially described the release of nano-sized membranous vesicles by viable cells over three decades ago for cancer. This notion has been confirmed in different cancers and has been extended to non-transformed cells. The biogenesis of exosomes begins with endosomes fusing to form multivesicular bodies (MVBs). Through the inward budding of the MVB membrane, intraluminal vesicles are formed, which, in the process of invagination, enclose various endoplasmic components. Finally, exosomes are released by MVB fusion with the cell membrane in an ATP-dependent process into extracellular space vesicles with a double membrane [79]. The name ‘exosomes’ originate from the description of neoplastic cell line-derived exfoliated vesicles, which mirrored the 5′-nucleotidase activity of the parent cells, that is, ‘exfoliated membrane vesicles with 5′-nucleotidase activity’ [80].
Unfortunately, the nomenclature for the different types of membranous vesicles secreted by cells is not always used with the required care [81]. Indeed, the terms ‘exosomes,’ ‘microvesicles,’ and ‘membrane particles’ are often used interchangeably. However, ‘exosomes’ are generated by exocytic fusion of MVBs, have a diameter of 30–100 nm, a density of 1.13–1.19 g/mL in a sucrose gradient (in which they can be sedimented at 100,000×g), whereas ‘microvesicles’ are shed from the plasma membrane, have a relatively larger size (100–1,000 nm) than exosomes, and can be sedimented at 10,000×g; ‘membrane particles’ refer to vesicles that also originate from plasma membrane, but have a small size similar to exosomes [82].
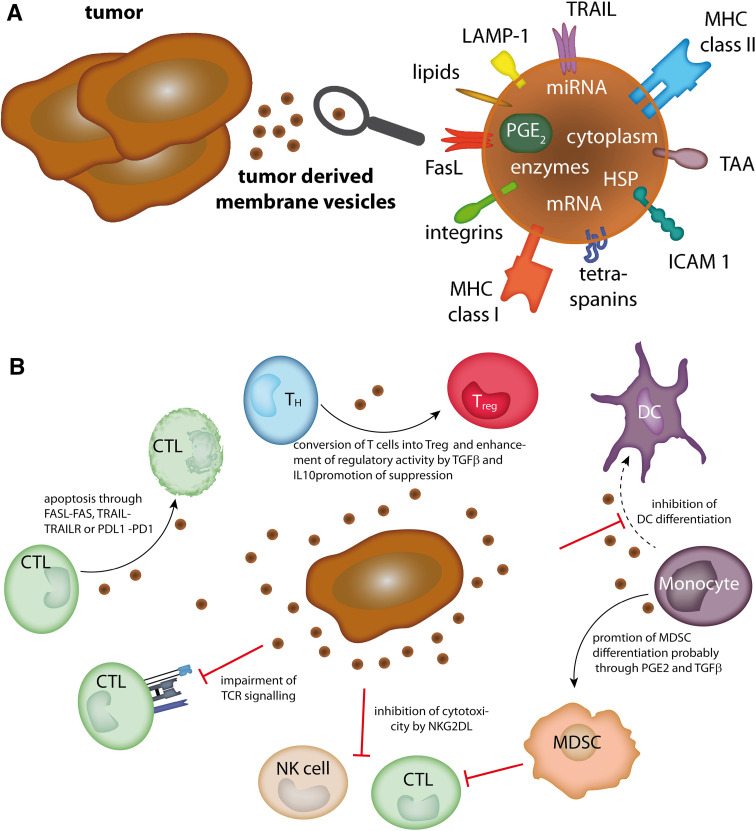
Tumor-derived membrane vesicles contain cytosolic and membrane proteins as well as functional RNA molecules such as mRNA and microRNAs derived from the parental cells (Fig. 4a), though ‘exosomes,’ ‘microvesicles,’ and ‘membrane particles’ may vary with respect to their content. For example, due to their biogenesis, exosomes contain proteins involved in MVB formation, for example, Alix, Tsg101, and clathrin. Moreover, the protein content of exosomes is generally enriched for certain molecules, including targeting/adhesion, membrane trafficking molecules, chaperones (e.g., Hsp70 and Hsp90), signal transduction proteins, and cytoplasmic enzymes [83]. Already, in the 1980s, it was suggested that tumor-derived membrane vesicles have immunosuppressive properties. Indeed, tumor-derived exosomes often contain immunosuppressive proteins such as FasL and TRAIL [83]. Moreover, exosomes can directly inhibit adaptive cytotoxic cells as well as promote the induction of Tregs and MDSCs [84]; these effects have been attributed to the presence of NKG2D ligands, death receptor ligands, and membrane-bound TGFβ on these vesicles (Fig. 4b).
Fig. 4.
Tumor-derived membrane vesicles. a Molecules present on or in tumor-derived membrane vesicles. b Immune-modulating effects of tumor-derived membrane vesicles (see text for details). CTL cytotoxic lymphocyte, DC dendritic cell, FasL Fas ligand, HSP heat-shock protein, ICAM 1 intercellular adhesion molecule 1, LAMP-1 lysosome-associated membrane protein 1, MDSC myeloid-derived suppressor cell, MHC major histocompatibility complex, miRNA microRNA, mRNA messenger RNA, PD1 programmed cell death protein 1, PDL1 programmed cell death 1 ligand 1, PG prostaglandin, TAA tumor-associated antigen, TCR T cell receptor, T H T helper cell, T reg regulatory T cell, TRAIL tumor necrosis factor-related apoptosis-inducing ligand
Conclusions
Feed-back mechanisms and counter-regulatory responses are essential for the homeostasis of the immune system. Without such a control of the intensity and extent of immune responses, severe damage to the host as exemplified by autoimmune diseases would be more prevalent. However, these counter-regulatory mechanisms bear the danger that the ability of the host to mount an effective immune response against the tumor is impaired. Unfortunately, as discussed here, tumor cells exploit numerous of these mechanisms in order to create a tumor-promoting immune-suppressive microenvironment. Future immune-modulating therapeutic approaches have to take this notion into account; notably, one of the most effective immune-modulating therapies, that is, the blockade of the PD1/PDL-1 interaction, is based on the inhibition of immune inhibitory feed-back mechanisms [63].
Conflict of interest
Jürgen C. Becker is a paid Advisor for and Member of the Speakers Bureau of Bristol-Myers Squibb, Glaxo-Smith-Kline, Leo Pharma, and Novartis. All other authors do not have any conflict of interest.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Pietras K, Östman A. Hallmarks of cancer: interactions with the tumor stroma. Exp Cell Res. 2010;316:1324–1331. doi: 10.1016/j.yexcr.2010.02.045. [DOI] [PubMed] [Google Scholar]
- 3.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Visser KE, Korets LV, Coussens LM. De novo carcinogenesis promoted by chronic inflammation is B lymphocyte dependent. Cancer Cell. 2005;7:411–423. doi: 10.1016/j.ccr.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 5.Colotta F, Allavena P, Sica A, et al. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis. 2009;30:1073–1081. doi: 10.1093/carcin/bgp127. [DOI] [PubMed] [Google Scholar]
- 6.Egeblad M, Nakasone ES, Werb Z. Tumors as organs: complex tissues that interface with the entire organism. Dev Cell. 2010;18:884–901. doi: 10.1016/j.devcel.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenberg SA. Raising the bar: the curative potential of human cancer immunotherapy. Sci Transl Med. 2012;4:127ps8. doi: 10.1126/scitranslmed.3003634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kerkar SP, Restifo NP. Cellular constituents of immune escape within the tumor microenvironment. Cancer Res. 2012;72:3125–3130. doi: 10.1158/0008-5472.CAN-11-4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bronte V, Mocellin S. Suppressive influences in the immune response to cancer. J Immunother. 2009;32:1–11. doi: 10.1097/CJI.0b013e3181837276. [DOI] [PubMed] [Google Scholar]
- 10.Andersen MH, Schrama D, thor Straten P, Becker JC. PS_JID_5700001.indd. J Invest Dermatol. 2006;126:32–41. doi: 10.1038/sj.jid.5700001. [DOI] [PubMed] [Google Scholar]
- 11.Hajishengallis G, Chavakis T. Endogenous modulators of inflammatory cell recruitment. Trends Immunol. 2013;34:1–6. doi: 10.1016/j.it.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brennan PJ, Brigl M, Brenner MB. Invariant natural killer T cells: an innate activation scheme linked to diverse effector functions. Nat Rev Immunol. 2013;13:101–117. doi: 10.1038/nri3369. [DOI] [PubMed] [Google Scholar]
- 13.Mantovani A. The yin-yang of tumor-associated neutrophils. Cancer Cell. 2009;16:173–174. doi: 10.1016/j.ccr.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 14.Matzinger P, Kamala T. Tissue-based class control: the other side of tolerance. Nat Rev Immunol. 2011;11:221–230. doi: 10.1038/nri2940. [DOI] [PubMed] [Google Scholar]
- 15.Hofmann UB, Houben R, Bröcker E-B, Becker JC. Role of matrix metalloproteinases in melanoma cell invasion. Biochimie. 2005;87:307–314. doi: 10.1016/j.biochi.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 16.Yu Q, Stamenkovic I. Cell surface-localized matrix metalloproteinase-9 proteolytically activates TGF-beta and promotes tumor invasion and angiogenesis. Gen Dev. 2000;14:163–176. [PMC free article] [PubMed] [Google Scholar]
- 17.Sheu BC, Hsu SM, Ho HN, et al. A novel role of metalloproteinase in cancer-mediated immunosuppression. Cancer Res. 2001;61:237–242. [PubMed] [Google Scholar]
- 18.Boutet P, Agüera-González S, Atkinson S, et al. Cutting edge: the metalloproteinase ADAM17/TNF-alpha-converting enzyme regulates proteolytic shedding of the MHC class I-related chain B protein. J Immunol. 2009;182:49–53. doi: 10.4049/jimmunol.182.1.49. [DOI] [PubMed] [Google Scholar]
- 19.Scheller J, Chalaris A, Garbers C, Rose-John S. ADAM17: a molecular switch to control inflammation and tissue regeneration. Trends Immunol. 2011;32:380–387. doi: 10.1016/j.it.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 20.Godefroy E, Manches O, Dréno B, et al. Matrix metalloproteinase-2 conditions human dendritic cells to prime inflammatory TH2 cells via an IL-12- and OX40L-dependent pathway. Cancer Cell. 2011;19:333–346. doi: 10.1016/j.ccr.2011.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Godin-Ethier J, Hanafi LA, Piccirillo CA, Lapointe R. Indoleamine 2,3-dioxygenase expression in human cancers: clinical and immunologic perspectives. Clin Cancer Res. 2011;17:6985–6991. doi: 10.1158/1078-0432.CCR-11-1331. [DOI] [PubMed] [Google Scholar]
- 22.Platten M, Wick W, Van den Eynde BJ. Tryptophan catabolism in cancer: beyond IDO and tryptophan depletion. Cancer Res. 2012;72:5435–5440. doi: 10.1158/0008-5472.CAN-12-0569. [DOI] [PubMed] [Google Scholar]
- 23.Zou W, Restifo NP. TH17 cells in tumour immunity and immunotherapy. Nat Rev Immunol. 2010;10:248–256. doi: 10.1038/nri2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinlich G, Murr C, Richardsen L, et al. Decreased serum tryptophan concentration predicts poor prognosis in malignant melanoma patients. Dermatology. 2007;214:8–14. doi: 10.1159/000096906. [DOI] [PubMed] [Google Scholar]
- 25.Smith C, Chang MY, Parker KH, et al. IDO is a nodal pathogenic driver of lung cancer and metastasis development. Cancer Discov. 2012;2:722–735. doi: 10.1158/2159-8290.CD-12-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersen MH. The specific targeting of immune regulation: T-cell responses against indoleamine 2,3-dioxygenase. Cancer Immunol Immunother. 2012;61:1289–1297. doi: 10.1007/s00262-012-1234-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pilotte L, Larrieu P, Stroobant V, et al. Reversal of tumoral immune resistance by inhibition of tryptophan 2,3-dioxygenase. Proc Nat Acad Sci. 2012;109:2497–2502. doi: 10.1073/pnas.1113873109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nowak EC, de Vries VC, Wasiuk A, et al. Tryptophan hydroxylase-1 regulates immune tolerance and inflammation. J Exp Med. 2012;209:2127–2135. doi: 10.1084/jem.20120408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bronte V, Zanovello P. Regulation of immune responses by l-arginine metabolism. Nat Rev Immunol. 2005;5:641–654. doi: 10.1038/nri1668. [DOI] [PubMed] [Google Scholar]
- 30.Rundhaug JE, Simper MS, Surh I, Fischer SM. The role of the EP receptors for prostaglandin E2 in skin and skin cancer. Cancer Metastasis Rev. 2011;30:465–480. doi: 10.1007/s10555-011-9317-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chouaib S, Messai Y, Couve S, et al. Hypoxia promotes tumor growth in linking angiogenesis to immune escape. Front Immunol. 2012;3:21. doi: 10.3389/fimmu.2012.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wink DA, Hines HB, Cheng RYS, et al. Nitric oxide and redox mechanisms in the immune response. J Leuk Biol. 2011;89:873–891. doi: 10.1189/jlb.1010550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jayaraman P, Parikh F, Lopez-Rivera E, et al. Tumor-expressed inducible nitric oxide synthase controls induction of functional myeloid-derived suppressor cells through modulation of vascular endothelial growth factor release. J Immunol. 2012;188:5365–5376. doi: 10.4049/jimmunol.1103553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singer K, Gottfried E, Kreutz M, Mackensen A. Suppression of T-cell responses by tumor metabolites. Cancer Immunol Immunother. 2011;60:425–431. doi: 10.1007/s00262-010-0967-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirschhaeuser F, Sattler UGA, Mueller-Klieser W. Lactate: a metabolic key player in cancer. Cancer Res. 2011;71:6921–6925. doi: 10.1158/0008-5472.CAN-11-1457. [DOI] [PubMed] [Google Scholar]
- 36.Shime H, Yabu M, Akazawa T, et al. Tumor-secreted lactic acid promotes IL-23/IL-17 proinflammatory pathway. J Immunol. 2008;180:7175–7183. doi: 10.4049/jimmunol.180.11.7175. [DOI] [PubMed] [Google Scholar]
- 37.Cham CM, Gajewski TF. Glucose availability regulates IFN-gamma production and p70S6 kinase activation in CD8+ effector T cells. J Immunol. 2005;174:4670–4677. doi: 10.4049/jimmunol.174.8.4670. [DOI] [PubMed] [Google Scholar]
- 38.Mandapathil M, Szczepanski MJ, Szajnik M, et al. Adenosine and prostaglandin E2 cooperate in the suppression of immune responses mediated by adaptive regulatory T cells. J Biol Chem. 2010;285:27571–27580. doi: 10.1074/jbc.M110.127100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stagg J, Smyth MJ. Extracellular adenosine triphosphate and adenosine in cancer. Oncogene. 2010;29:5346–5358. doi: 10.1038/onc.2010.292. [DOI] [PubMed] [Google Scholar]
- 40.Haskó G, Linden J, Cronstein B, Pacher P. Adenosine receptors: therapeutic aspects for inflammatory and immune diseases. Nat Rev Drug Discov. 2008;7:759–770. doi: 10.1038/nrd2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waickman AT, Alme A, Senaldi L, et al. Enhancement of tumor immunotherapy by deletion of the A2A adenosine receptor. Cancer Immunol Immunother. 2012;61:917–926. doi: 10.1007/s00262-011-1155-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su Y, Jackson EK, Gorelik E. Receptor desensitization and blockade of the suppressive effects of prostaglandin E(2) and adenosine on the cytotoxic activity of human melanoma-infiltrating T lymphocytes. Cancer Immunol Immunother. 2011;60:111–122. doi: 10.1007/s00262-010-0924-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Häusler SFM, Montalbán del Barrio I, Strohschein J, et al. Ectonucleotidases CD39 and CD73 on OvCA cells are potent adenosine-generating enzymes responsible for adenosine receptor 2A-dependent suppression of T cell function and NK cell cytotoxicity. Cancer Immunol Immunother. 2011;60:1405–1418. doi: 10.1007/s00262-011-1040-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kryczek I, Wu K, Zhao E, et al. IL-17+ regulatory T Cells in the microenvironments of chronic inflammation and cancer. J Immunol. 2011;186:4388–4395. doi: 10.4049/jimmunol.1003251. [DOI] [PubMed] [Google Scholar]
- 45.Ryzhov S, Novitskiy SV, Goldstein AE, et al. Adenosinergic regulation of the expansion and immunosuppressive activity of CD11b+ Gr1+ Cells. J Immunol. 2011;187:6120–6129. doi: 10.4049/jimmunol.1101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beavis PA, Stagg J, Darcy PK, Smyth MJ. CD73: a potent suppressor of antitumor immune responses. Trends Immunol. 2012;33:231–237. doi: 10.1016/j.it.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 47.Jin D, Fan J, Wang L, et al. CD73 on tumor cells impairs antitumor T-cell responses: a novel mechanism of tumor-induced immune suppression. Cancer Res. 2010;70:2245–2255. doi: 10.1158/0008-5472.CAN-09-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garber K. Beyond ipilimumab: new approaches target the immunological synapse. J Nat Cancer Inst. 2011;103:1079–1082. doi: 10.1093/jnci/djr281. [DOI] [PubMed] [Google Scholar]
- 49.Cameron F, Whiteside G, Perry C. Ipilimumab: first global approval. Drugs. 2011;71:1093–1104. doi: 10.2165/11594010-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 50.Callahan MK, Wolchok JD, Allison JP. Anti-CTLA-4 antibody therapy: immune monitoring during clinical development of a novel immunotherapy. Semin Oncol. 2010;37:473–484. doi: 10.1053/j.seminoncol.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robert C, Thomas L, Bondarenko I, et al. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N Engl J Med. 2011;364:2517–2526. doi: 10.1056/NEJMoa1104621. [DOI] [PubMed] [Google Scholar]
- 52.Reck M, Bondarenko I, Luft A, et al. Ipilimumab in combination with paclitaxel and carboplatin as first-line therapy in extensive-disease-small-cell lung cancer: results from a randomized, double-blind, multicenter phase 2 trial. Ann Oncol. 2013;24:75–83. doi: 10.1093/annonc/mds213. [DOI] [PubMed] [Google Scholar]
- 53.Di Giacomo AM, Ascierto PA, Pilla L, et al. Ipilimumab and fotemustine in patients with advanced melanoma (NIBIT-M1): an open-label, single-arm phase 2 trial. Lancet Oncol. 2012;13:879–886. doi: 10.1016/S1470-2045(12)70324-8. [DOI] [PubMed] [Google Scholar]
- 54.Postow MA, Callahan MK, Barker CA, et al. Immunologic correlates of the abscopal effect in a patient with melanoma. N Engl J Med. 2012;366:925–931. doi: 10.1056/NEJMoa1112824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Santegoets SJAM, Stam AGM, Lougheed SM, et al. T cell profiling reveals high CD4(+)CTLA-4 (+) T cell frequency as dominant predictor for survival after prostate GVAX/ipilimumab treatment. Cancer Immunol Immunother. 2012 doi: 10.1007/s00262-012-1330-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hodis R, Watson IR, Kryukov GV, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Downey SG, Klapper JA, Smith FO, et al. Prognostic factors related to clinical response in patients with metastatic melanoma treated by CTL-associated antigen-4 blockade. Clin Cancer Res. 2007;13:6681–6688. doi: 10.1158/1078-0432.CCR-07-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liakou CI, Kamat A, Tang DN, et al. CTLA-4 blockade increases IFNgamma-producing CD4+ ICOShi cells to shift the ratio of effector to regulatory T cells in cancer patients. Proc Nat Acad Sci. 2008;105:14987–14992. doi: 10.1073/pnas.0806075105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ji R-R, Chasalow SD, Wang L, et al. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunol Immunother. 2012;61:1019–1031. doi: 10.1007/s00262-011-1172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hamid O, Schmidt H, Nissan A, et al. A prospective phase II trial exploring the association between tumor microenvironment biomarkers and clinical activity of ipilimumab in advanced melanoma. J Transl Med. 2011;9:204. doi: 10.1186/1479-5876-9-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shahabi V, Whitney G, Hamid O, et al. Assessment of association between BRAF-V600E mutation status in melanomas and clinical response to ipilimumab. Cancer Immunol Immunother. 2012;61:733–737. doi: 10.1007/s00262-012-1227-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.la Fuente de H, Cibrián D, Sánchez-Madrid F. Immunoregulatory molecules are master regulators of inflammation during the immune response. FEBS Lett. 2012;586:2897–2905. doi: 10.1016/j.febslet.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brahmer JR, Tykodi SS, Chow LQM, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hobo W, Novobrantseva TI, Fredrix H, et al. Improving dendritic cell vaccine immunogenicity by silencing PD-1 ligands using siRNA-lipid nanoparticles combined with antigen mRNA electroporation. Cancer Immunol Immunother. 2012 doi: 10.1007/s00262-012-1334-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seliger B, Quandt D. The expression, function, and clinical relevance of B7 family members in cancer. Cancer Immunol Immunother. 2012;61:1327–1341. doi: 10.1007/s00262-012-1293-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yi KH, Chen L. Fine tuning the immune response through B7-H3 and B7-H4. Immunol Rev. 2009;229:145–151. doi: 10.1111/j.1600-065X.2009.00768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Textor S, Fiegler N, Arnold A, et al. Human NK cells are alerted to induction of p53 in cancer cells by upregulation of the NKG2D ligands ULBP1 and ULBP2. Cancer Res. 2011;71:5998–6009. doi: 10.1158/0008-5472.CAN-10-3211. [DOI] [PubMed] [Google Scholar]
- 69.Vetter CS, Groh V, thor Straten P, et al. Expression of stress-induced MHC class I related chain molecules on human melanoma. J Invest Dermatol. 2002;118:600–605. doi: 10.1046/j.1523-1747.2002.01700.x. [DOI] [PubMed] [Google Scholar]
- 70.Champsaur M, Lanier LL. Effect of NKG2D ligand expression on host immune responses. Immunol Rev. 2010;235:267–285. doi: 10.1111/j.0105-2896.2010.00893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Waldhauer I, Goehlsdorf D, Gieseke F, et al. Tumor-associated MICA is shed by ADAM proteases. Cancer Res. 2008;68:6368–6376. doi: 10.1158/0008-5472.CAN-07-6768. [DOI] [PubMed] [Google Scholar]
- 72.Siemens DR, Hu N, Sheikhi AK, et al. Hypoxia increases tumor cell shedding of MHC class I chain-related molecule: role of nitric oxide. Cancer Res. 2008;68:4746–4753. doi: 10.1158/0008-5472.CAN-08-0054. [DOI] [PubMed] [Google Scholar]
- 73.Barsoum IB, Hamilton TK, Li X, et al. Hypoxia induces escape from innate immunity in cancer cells via increased expression of ADAM10: role of nitric oxide. Cancer Res. 2011;71:7433–7441. doi: 10.1158/0008-5472.CAN-11-2104. [DOI] [PubMed] [Google Scholar]
- 74.Pruessmeyer J, Ludwig A. The good, the bad and the ugly substrates for ADAM10 and ADAM17 in brain pathology, inflammation and cancer. Semin Cell Dev Biol. 2009;20:164–174. doi: 10.1016/j.semcdb.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 75.Paschen A, Sucker A, Hill B, et al. Differential clinical significance of individual NKG2D ligands in melanoma: soluble ULBP2 as an indicator of poor prognosis superior to S100B. Clin Cancer Res. 2009;15:5208–5215. doi: 10.1158/1078-0432.CCR-09-0886. [DOI] [PubMed] [Google Scholar]
- 76.Benitez AC, Dai Z, Mann HH, et al. Expression, signaling proficiency, and stimulatory function of the NKG2D lymphocyte receptor in human cancer cells. Proc Nat Acad Sci. 2011;108:4081–4086. doi: 10.1073/pnas.1018603108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rak J. Extracellular vesicles—biomarkers and effectors of the cellular interactome in cancer. Front Pharmacol. 2013;4:21. doi: 10.3389/fphar.2013.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Taylor DD, Doellgast GJ. Quantitation of peroxidase-antibody binding to membrane fragments using column chromatography. Anal Biochem. 1979;98:53–59. doi: 10.1016/0003-2697(79)90704-8. [DOI] [PubMed] [Google Scholar]
- 79.Taylor DD, Gercel-Taylor C. Exosomes/microvesicles: mediators of cancer-associated immunosuppressive microenvironments. Semin Immunopathol. 2011;33:441–454. doi: 10.1007/s00281-010-0234-8. [DOI] [PubMed] [Google Scholar]
- 80.Filipazzi P, Bürdek M, Villa A, et al. Recent advances on the role of tumor exosomes in immunosuppression and disease progression. Sem Cancer Biol. 2012;22:342–349. doi: 10.1016/j.semcancer.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 81.Huber V, Filipazzi P, Iero M, et al. More insights into the immunosuppressive potential of tumor exosomes. J Transl Med. 2008;6:63. doi: 10.1186/1479-5876-6-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.van der Heyde HC, Gramaglia I, Combes V, et al. Flow cytometric analysis of microparticles. Methods Mol Biol. 2011;699:337–354. doi: 10.1007/978-1-61737-950-5_16. [DOI] [PubMed] [Google Scholar]
- 83.Whiteside TL. Immune modulation of T-cell and NK (natural killer) cell activities by TEXs (tumour-derived exosomes) Biochem Soc Trans. 2013;41:245–251. doi: 10.1042/BST20120265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xiang X, Poliakov A, Liu C, et al. Induction of myeloid-derived suppressor cells by tumor exosomes. Int J Cancer. 2009;124:2621–2633. doi: 10.1002/ijc.24249. [DOI] [PMC free article] [PubMed] [Google Scholar]