Abstract
Maml1 is emerging as a coactivator of many signaling pathways, including the Notch and Wnt pathways. Targeting Maml1 in melanoma cells efficiently knocks down the downstream transcriptional repressors Hey1 and Hes1, resulting in melanoma cell senescence, cellular differentiation, and increased melanin production. Significantly, higher IFNβ and chemokine gene transcripts have been observed, together with increased STAT1 and decreased STAT3 and NF-κB signaling activities. Although decreased cell proliferation contributes to slower tumor growth in vivo, the depletion of NK and CD8+ T cells in an shMaml1-B16 tumor carrier mouse leads to more rapid tumor growth than that observed in control shC002-B16 tumors. This result demonstrates that the knockdown of Maml1 transcription and function contributes to increased immune surveillance. The knockdown of Maml1 transcription in the human melanoma cell line M537 also results in senescence, IFNβ upregulation, increased chemokine gene expression, and greater NK and CD8+ T cell migration in a transwell system. This study demonstrated that targeting Maml1-induced tumor cell senescence and differentiation may alter the tumor microenvironment and cytokine and chemokine profiles and may also promote innate and adaptive immune cell infiltration and function.
Keywords: shMaml1, Cellular senescence, Innate and adaptive immune cells, IFNβ and chemokines
Introduction
The developmental proteins such as Notch, Wnt, and Hedgehog are key regulators of cell fate, proliferation, and differentiation. Their signaling pathways are related and are frequently activated in neoplasms from cancer stem cells that are capable of self-renewal, which often leads to the development of tumor resistance to therapy, tumor recurrence, and metastasis [1–4]. The Notch receptor proteins are highly expressed in melanoma cells and are constitutively cleaved to release the intracellular domain (NICD). Mastermind-like protein-1 (Maml1) directly binds to the most conserved ankyrin repeat domain of the four Notch receptors (NICD1–4), and Maml1 also interacts with CSL to form a stable DNA-binding ternary complex, thereby leading to the transcriptional activation of Hes1 and Hey1 [4–8]. A recent study discovered that Maml1 also participates in the Wnt/β-catenin signaling pathway independently of the Notch signaling pathway. Maml1 is recruited by β-catenin to the cyclin D1 and c-Myc promoters to activate tumor cell survival [9]. In neural precursor cells, Maml1, β-catenin, and NICD form a complex within the promoter region of the Hes1 gene, inducing its expression and simultaneously promoting cellular proliferation and inhibiting differentiation [10]. These studies support a new role for Maml1 as an essential component of Notch and Wnt/β-catenin signaling-mediated tumorigenesis and suggest that Maml1 could be an alternative target for both the Notch and Wnt/β-catenin signaling pathways.
Hes1 encodes a basic helix-loop-helix transcription factor that functions as a classical effector of the Notch signaling pathway [4–8]. However, recent reports indicated that, in stem-like cells, Hedgehog elicited a strong Hes1 response independent of canonical Notch signaling [11–14]. Elsewhere, Schreck’s group found that targeting the Notch pathway using a g-secretase inhibitor increases Hedgehog activity, and the inhibition of both Notch and Hedgehog dramatically decreases the growth of glioblastoma and melanoma cell lines and low-passage neurospheres derived from primary human tumors [15]. The Hes1 signaling pathway is activated in several types of tumor cells. Activation of this signaling pathway allows these tumor cells to evade differentiation; therefore, irreversible senescence contributes to tumorigenesis [16, 17]. Maml1 is an essential coactivator that facilitates Notch-induced Hes1 and Hey1 transcription [7, 18]. Recent studies have indicated that Maml1 directly regulates NF-κB signaling and cell survival [19]. All of these data support our hypothesis that targeting Maml1 may regulate the Notch, Wnt, and Hedgehog signaling pathways simultaneously and may serve as a novel anticancer strategy by inhibiting cell proliferation and promoting cell differentiation and irreversible senescence [19–21].
Previously, we used siMaml1 to inhibit the transcriptional activation of Hes1/Hey1 in melanoma cells, leading to the upregulation of multiple cytokines, including CCL2 [7]. However, researchers found that due to the low siRNA concentrations caused by cell division and intracellular siRNA turnover, the duration of siRNA silencing is short [22]. To analyze the effects of Maml1 on cell differentiation and senescence, the Maml1 gene must be silenced for a prolonged period of time. To accomplish this goal, we transfected several short-hairpin Maml1 (shMaml1) lentiviral expression plasmids to achieve sustained gene silencing, and we established a stable shMaml1-transfected B16 melanoma cell line (shMaml1-B16). We found that there were only a fraction of shMaml1-B16 cells undergoing apoptosis similar to the control plasmid shC002-B16-transfected cells or the parent B16 cells. However, we observed more shMaml1-B16 cells with increased senescence-associated-β-galactosidase (SA-β-Gal) staining and increased cell differentiation based on increased melanin production. These cells secrete several immune-stimulating cytokines and chemokines in vitro. In syngeneic mice, shMaml1-B16 cells formed tumors more slowly than control cells. Flow cytometry showed increased infiltration of immune cells, such as DC, NK, and CD8+ T cells, in these tumors.
Materials and methods
Establishment of shMaml1-B16 and shC002-B16 cells
MISSION shRNA clones (clone ID sh138: TRCN0000115138 targeting NM_175334.2-1187s1c1; clone ID sh139: TRCN0000115139 targeting NM_175334.2-1019s1c1; clone ID sh140: TRCN0000115140 targeting NM_175334.2-2544s1c1; clone ID shmix: mixture of TRCN0000115138, TRCN0000115139, and TRCN0000115140; human clone ID sh353: TRCN0000003353 targeting NM_014757.x-585s1c1; and clone ID sh356: TRCN0000003356 targeting NM_014757.x-1907s1c1) are sequence-verified, lentiviral plasmids (pLKO.1-puro) purchased from Sigma-Aldrich. B16F10 cells were transfected, and stable shMaml1-B16 cells were selected under optimized conditions using puromycin resistance (1 μg/ml) according the manufacturer’s protocol. Briefly, the siGENOME SMARTpool (10 μl) was diluted in 90 μl of serum-free DMEM containing transfection reagents, incubated at room temperature for 15 min, and added to a 2 × 105 cells/ml cell culture at a range of concentrations. Transfections were conducted twice on consecutive days, and the cells were allowed to continue to grow for an additional 2 days at 37 °C in the presence of 5 % CO2 before the puromycin selection. The nontarget, control lentiviral plasmid shC002 validated the specific knockdown of Maml1 by MISSION shMaml1.
Detection of SA-β-gal activity using confocal microscopy
SA-β-gal activity was measured at pH 6.0 for B16 cells and pH 5.5 for mouse tissue, as described previously. Briefly, the adherent cells were fixed with 0.5 % glutaraldehyde in PBS for 15 min, washed with PBS containing 1 mM MgCl2, and stained for 5–6 h in PBS containing 1 mM MgCl2, 1 mg/ml X-Gal (Roche, Indianapolis, IN, USA), 5 mM potassium ferricyanide, and 5 mM potassium ferrocyanide (Sigma-Aldrich, St. Louis, MO, USA). The images were photographed at several magnifications using a Nikon camera.
Analysis of gene expression using RT-qPCR
The cells and tumors were homogenized in 1 ml Trizol (Invitrogen, Grand Island, NY, USA), and total RNA was precipitated from the aqueous phase using isopropyl alcohol after performing a chloroform separation. After DNase I treatment, cDNA was synthesized from 1 μg of total RNA using MLV-Reverse Transcriptase according to the manufacturer’s directions. Real-time PCR (RT-qPCR) was performed using an ABI PRISM® 7300 Sequence Detection System with a Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA). Changes in gene expression were calculated using comparative Ct values with β-actin as an endogenous gene control. The ΔCT was calculated, and the relative expression of each cytokine and chemokine was compared. Prevalidated primers and probes specific for Maml1 (Mm00614627_m1), Hey1 (Mm00468865_m1), Hes1 (Mm01342805_m1), CCL2 (Mm00441242_m1), CCL5 (Mm01302427_m1), CXCL9 (Mm00434946_m1), CXCL10 (Mm00445235_m1), CXCL11 (Mm00444662_m1), CCL22 (Mm00436439_m1), and β-actin (Mm00525061_m1) were purchased from Applied Biosystems.
Immunoblotting and signaling inhibitors
Preparation of cytosolic extracts was carried out using a Nuclear Extract Kit (Active Motif, CA, USA) according to the manufacturer’s recommendations. The concentrations of proteins in the nuclear and cytosolic fractions were measured using a microplate version of the Lowry assay (Micro BCA Protein Assay Reagent Kit, Pierce, IL, USA) with bovine serum albumin (BSA) standards. The lysates were separated using SDS-PAGE on a 10 % gel and then transferred to a nitrocellulose membrane. The membrane was blocked with blocking buffer (5 % nonfat dry milk in TBST (20 mM Tris–HCl, pH 7.5, and 0.5 % Tween 20)) for 1 h at room temperature and then incubated overnight with one of the following primary antibodies diluted in the blocking buffer: anti-phospho-STAT1, anti-phospho-STAT3, anti-phospho-IκBα (1:500; Cell Signaling Technologies, Beverly, MA, USA), rabbit anti-STAT1, anti-STAT3, anti-NFκB p65, or anti-β-actin (1:500; Santa Cruz Biotechnologies, Santa Cruz, CA, USA). An HRP-conjugated, anti-rabbit secondary antibody (Amersham, Arlington Heights, IL, USA) was used at a 1:10,000 dilution for 1 h at room temperature. The blots were visualized using an Amersham enhanced chemiluminescence (ECL) kit. A highly selective inhibitor of IκB kinase (BMS-345541), the STAT1 inhibitor fludarabine (Sigma-Aldrich, St. Louis, MO, USA), and the cell-permeable peptide STAT3 inhibitor LLL12 (BioVision, Mountain view, CA, USA) were applied to the cell cultures when necessary.
Tumor inoculation and tumor-infiltrating cell treatment and analysis
All mice were bred and maintained in a pathogen-free facility at the Medical Center. The mice were handled in accordance with the animal experimental guidelines set by the Institute of Animal Care and Use Committee (IACUC) for all experiments. C57BL/6 wild-type mice (6–7 weeks old) were purchased from the Laboratory Animal Center of Southern Medical University (Guangzhou, China). The mice were subcutaneously injected with shMaml1-B16, shC002-B16, and B16 cells (1 × 106) on the leg. Tumor size was measured on three orthogonal axes (a, b, and c) every 3–4 days, and tumor volumes were calculated as abc/2. The tumors were fixed in 10 % neutral formalin and embedded, and serial sections were histologically stained with H&E and were subjected to immunohistochemistry to visualize CD11b myeloid, F4/80 macrophage, and CD8+ T cells. CD8+ T cells or NK cells were depleted by injecting 100 μg/kg of anti-CD8 (clone 2.43.1) or anti-NK1.1 (clone PK136, BD Pharmingen, CA) antibodies, respectively, administered on days 14 and 21. Depletion was confirmed by flow cytometry analysis of peripheral blood using BD Pharmingen fluorescent-labeled anti-CD8α-FITC and anti-NK-PerCP antibodies [23].
Statistical analysis
One-way ANOVA and Dunnett’s posttest were performed using GraphPad InStat version 3.05 to compare the average volumes of tumors in different experimental groups. Two-tailed Student’s t-tests were performed to compare the means of the different treatment groups. p < 0.05 was accepted as significant.
Results
ShMaml1-B16 cells demonstrated greater cell differentiation and senescence
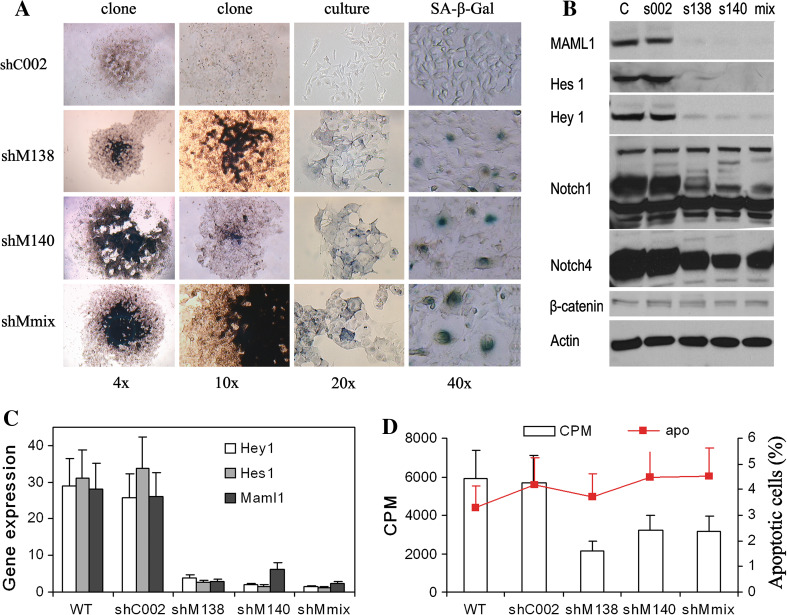
Stable clones of the shMaml1-B16 cell line (shM138, shM140, shMmix) were established. Compared to the nontarget plasmid shC002-B16, the successful targeting of Maml1-promoted cell differentiation was demonstrated by increased melanin production. ShMaml1-B16 cells exhibited a large, flat morphology indicative of senescence, which was confirmed by enhanced SA-β-gal staining (Fig. 1a). A protein analysis of the B16 cells demonstrated the constitutive cleavage and high levels of expression of Notch 1 and Notch 4 but showed only low levels of expression of the Wnt signaling protein β-catenin, suggesting a dominant role for Notch signaling in B16 melanoma cells (we could not detect Hedgehog signaling protein, data not shown) (Fig. 1b). Total RNA was isolated from stable shMaml1-B16, shC002-B16, and parental B16 tumor cells, and RT-qPCR showed that the transcriptional repressors Hey1 and Hes1 abolished downstream Notch signaling (Fig. 1c). ShMaml1-B16 cells proliferated less than shC002-B16 or parental B16 tumor cells, but the difference in apoptotic rates among all cell lines was not significant in vitro (Fig. 1d).
Fig. 1.
Establishment and characterization of a stable shMaml1-B16 cell line. a ShMaml1-B16 cells exhibited more intracellular melanin production and SA-β-gal staining than control cells. b Western blot of Notch and Wnt signaling proteins in B16 cells. c Successful knockdown of the transcriptional repressors Hey1 and Hes1, as well as Maml1, in shMaml1-B16 cells. d A lower proliferation rate but similar apoptosis rates are shown in shMaml1-B16 cells relative to shC002-B16 and parental WT B16 cells
Senescent shMaml1-B16 cells exhibited upregulated IFNβ and chemokine gene transcripts with increased STAT1 activity
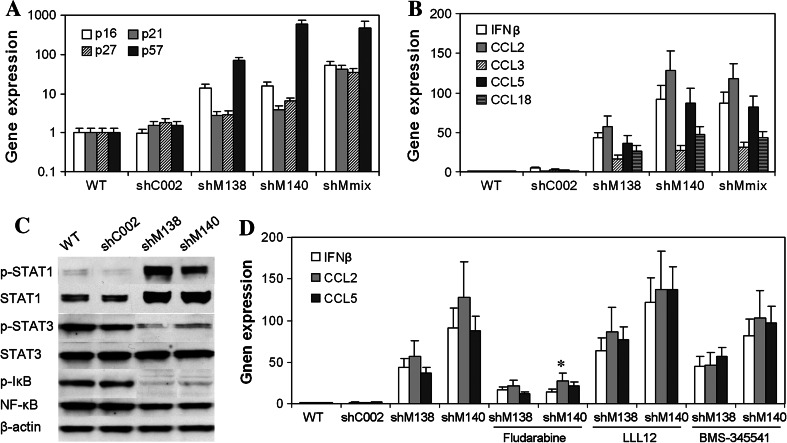
Compared to shC002-B16 or parent B16 tumor cells, shMaml1-B16 cells showed greater expression of senescence-related cell cycle regulatory genes, including the cyclin-dependent kinase inhibitors p16 INK4A, p21 Cip1/Waf1, p27 Kip1, and p57 Kip2 (Fig. 2a). The expression levels of the IFNβ, CCL2, CCL5, and CCL18 transcripts were also increased in shMaml1-B16 cells relative to the shC002-B16 or parent B16 tumor cells (Fig. 2b).
Fig. 2.
Changes in cytokine and chemokine expression in senescent shMaml1-B16 cells. a Upregulation of the cyclin-dependent kinase inhibitors p16 INK4A, p21 Cip1/Waf1, p27 Kip1, and p57 Kip2 in shMaml1-B16 cells compared to shC002-B16 or parent B16 tumor cells. b Upregulation of IFNβ, CCL2, CCL5, and CCL18 transcripts in shMaml1-B16 cells. c Levels of total and phosphorylated STAT1 were increased in shMaml1-B16 cells, but the phosphorylation of STAT3 and IκBα was inhibited. d Inhibition of STAT1 with fludarabine blocked the expression of CCL2, CCL5, and IFNβ in shMaml1-B16 cells, *p < 0.01
STATs and NF-κB have been reported to play essential roles in the synergistic transcriptional activation of multiple chemokines [23–25]. We analyzed the expression and activation of these signaling pathways in shMaml1-B16, shC002-B16, and parental B16 cells. Western blot analysis showed an increase in both the total and phosphorylated levels of STAT1 in the shMaml1-B16 cells, but the phosphorylation of STAT3 and IκBα was inhibited (Fig. 2c). In the same cell line, inhibition of STAT1 with fludarabine blocked the expression of CCL2, CCL5, and IFNβ; however, inhibition of STAT3 using LLL12 or inhibition of IκBα kinase using BMS-345541 did not affect the expression of CCL2, CCL5, and IFNβ (Fig. 2d).
Senescent shMaml1-B16 cells showed slower growth and increased immune cell infiltration in vivo
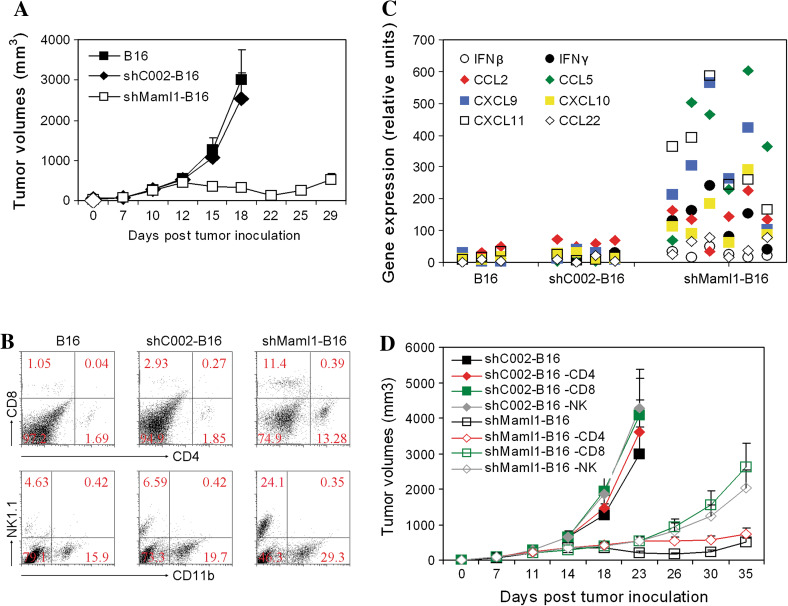
C57BL/6 mice were inoculated with ShMaml1-B16, shC002-B16, or parental B16 tumor cells (1 × 106). Tumor growth was observed, and the mean tumor volumes were calculated. The ShMaml1-B16 tumors were smaller than both the shC002-B16 and parental B16 tumors at day 18 (p = 0.015, p = 0.013) (Fig. 3a). RT-qPCR of tumor lysates showed a higher expression of IFNγ, IFNβ, CCL2, CCL5, CXCL9, CXCL10, and CXCL11 in the shMaml1-B16 tumors, thereby demonstrating that these cells are in an immunostimulatory microenvironment (Fig. 3b). Flow cytometry showed a higher frequency of CD4+ T, CD8+ T, and NK cells in the shMaml1-B16 tumors compared to the frequency in the shC002-B16 or parental B16 tumor cells (Fig. 3c). Depletion of NK (p = 0.026) and CD8+ T (p = 0.021) cells in the shMaml1-B16 tumor carrier mice led to a tumor growth rate that resembled the shC002-B16 tumor growth rate. These data demonstrate that a knockdown of Maml1 transcription and function contributes to slowed tumor growth not only by lowering the relative tumor cell proliferation rate but also through an increased infiltration of innate and adaptive immune cells (Fig. 3d). CD4 depletion (p = 0.144) did not change the growth rate of the shMaml1-B16 tumors. Depletion of NK (p = 0.175) or CD8+ T (p = 0.217) cells in the control shC002-B16 tumor carriers did not significantly change the tumor growth rate.
Fig. 3.
Inhibition of senescent shMaml1-B16 tumor growth and increased immune cell infiltration. a Growth curves of shMaml1-B16, shC002-B16, or parental B16 tumor cells in syngeneic C57BL/6 mice. b RT-qPCR of immune cytokine and chemokine expression in shMaml1-B16 tumor lysates compared to shC002-B16 or parental WT B16 tumor cells. c Flow cytometry of tumor-infiltrating cells in shMaml1-B16 tumors. d Depletion of NK and CD8+ T cells accelerated shMaml1-B16 tumor growth
Induction of cellular senescence in human melanoma cells transfected with shMaml1
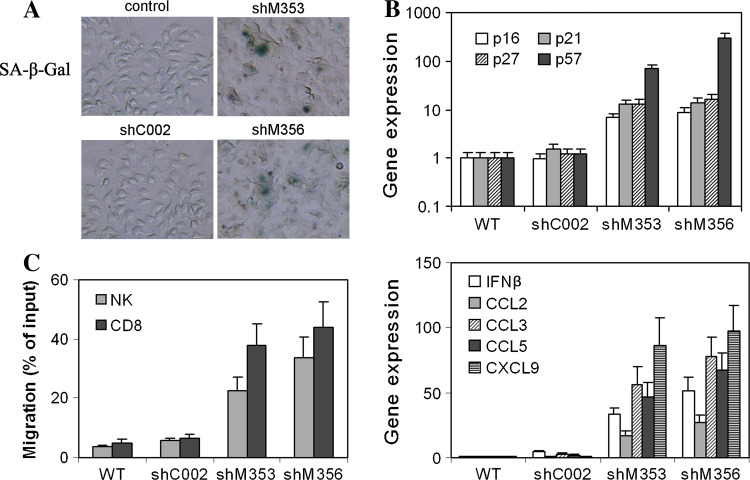
Stable clones of the shMaml1-M537 cell line (shM353, shM356) were established. Compared to the nontarget plasmid shC002-M537, the successful targeting of Maml1-promoted cell senescence was indicated by the large, flat morphology of the cells and enhanced SA-β-gal staining (Fig. 4a). Total RNA was isolated from stable shMaml1-M537, shC002-M537, and parental M537 tumor cells, and RT-qPCR confirmed that the Hey1/Hes1 and Maml1 transcriptional repressors were knocked down (data not shown). The cyclin-dependent kinase inhibitors p16 INK4A, p21 Cip1/Waf1, and p57 Kip2 were upregulated compared to the shC002-M537 or parent M537 tumor cells (Fig 4b, upper panel). Additionally, the expression of the IFNβ, CCL2, CCL3, CCL5, and CXCL9 transcripts in the shMaml1-M537 cells was increased compared to the expression of these transcripts in the shC002-M537 or parental M537 tumor cells (Fig 4b, lower panel). Finally, in a transwell system, the shMaml1-M537 cells attracted more NK (p < 0.05) and CD8+ T (p < 0.01) cells compared to the shC002-M537 or parental M537 tumor cells (Fig 4c).
Fig. 4.
Senescent shMaml1-M537 cells expressed multiple cytokines and are more chemoattractive to NK and CD8+ T cells. a ShMaml1-M537 exhibited more SA-β-gal staining cells than control cells. b Upregulation of the cyclin-dependent kinase inhibitors p16 INK4A, p21 Cip1/Waf1, p27 Kip1, and p57 Kip2 in shMaml1-M537 cells compared to shC002-M537 or parent M537 tumor cells (upper panel). Upregulation of IFNβ, CCL2, CCL5, and CXCL9 transcripts in shMaml1-M537 cells (lower panel). c Migration assay of NK and CD8+ T cells in a transwell system that contains shC002-M537, shC002-M537, or parental M537 cells
Discussion
In this study, we show that a knockdown of Maml1 in melanoma cells results in tumor cell senescence and differentiation, leading to an upregulation of multiple immune cytokines and chemokines in the tumor microenvironment. This investigation further demonstrated that disrupting Maml1–Hes1-induced transcription repression improved immune surveillance and eventually contributed to the elimination of senescent tumor cells [23]. Wnt signaling results in the repression of p21 Cip1/Waf1, whereas Notch signaling represses p27 Kip1 and p57 Kip2 in part through its target gene Hes1 [26, 27]. We knocked down Maml1, a coactivator known to participate in both Notch- and Wnt-activated Hes1 transcription, in B16 melanoma cells. The knockdown effectively repressed the cyclin-dependent kinase inhibitors p16 INK4A, p21 Cip1/Waf1, p27 Kip1, and p57 Kip2, resulting in decreased cell proliferation and senescence [14, 17, 27]. Maml1 is emerging as a coactivator for many other pathways, but this report is the first to show that the inhibition of Maml1–Hes1 signaling induces cellular senescence and immune surveillance.
Maml1 was shown to cause the phosphorylation of IκBα and affect IκBα stability. Maml1 also acts as a coactivator of the NF-κB subunit p65 in NF-κB-dependent transcription. These findings indicate that Maml1 is a novel modulator of NF-κB signaling and has a role in the regulation of cell survival [28, 29]. The activation or suppression of Notch1 and Hes1 has been correlated with the phosphorylation and activation of STAT3, and targeting the STAT3/p63/Notch axis induces several types of tumor cell differentiation [30–32]. Additionally, interactions between activated STAT3 and constitutively active NF-κB have been studied in tumors by Yu’s group, and STAT3 was shown to be a target for enhancing anti-tumor immunity [33–36]. Here, we show that targeting the Notch/Wnt coactivator Maml1 results in melanoma cell differentiation and senescence that not only inhibits the activation of both STAT3 and NF-κB but also stimulates the activity of STAT1. Consequently, the expression of immune cytokines and chemokines is significantly increased, resulting in increased NK and CD8+ T cell infiltration and the establishment of an immunostimulatory microenvironment [36, 37].
Tumor cell differentiation has been associated with the activation of cellular senescence [38]. Treatments that induce senescence and tumor cell differentiation may need to be accompanied by chemotherapeutic drugs or radiation. Treatment-induced senescence was shown to be one of the key determinants of tumor response to therapy both in vitro and in vivo. Clinical and preclinical studies have shown that the expression of different biological classes of senescence-associated genes in tumor cells has significant prognostic implications [39–41]. Although senescent cells do not proliferate, they remain metabolically active and secrete proteins with either tumor-suppressing or tumor-promoting activities [42, 43]. A role has been reported for the innate immune system in eliminating senescent cells from tumors upon p53 reexpression, and CD4+ T cells have been shown to mediate anti-tumor effects through Myc-induced cellular senescence [44, 45]. We have shown that mouse shMaml1-B16 cells and human shMaml1-M537 cells undergoing senescence expressed multiple senescence-associated immune cytokines and chemokines, both in culture and in tumor microenvironments. Consequently, more NK and CD8+ T cell infiltrations occurred, demonstrating that the disruption of Maml1- and Notch-induced transcription repression may be a possible strategy for cancer treatment. Maml1 may participate in the Notch and Wnt signaling pathways during normal tissue development and cell differentiation; the knockdown of Maml1 blocked the transcriptional repression of Hes1 and affected the activation of STAT1, STAT3, and NF-κB. This specific knockdown of Maml1 in tumor cells has tremendous therapeutic potential; therefore, further studies of the involvement of Maml1 in the Notch and Wnt pathways are necessary.
Acknowledgments
This work was supported by the Province of Guangdong Science Grant (10251051501000008).
Conflict of interest
The authors declare that they have no conflict of interest.
Abbreviations
- Maml1
Mastermind-like protein-1
- siMaml1
Small interfering Maml1 RNA
- Hes1
Hairy/enhancer-of-split 1
- Hey1
Hairy/enhancer-of-split related with YRPW motif 1
- CCL2
MCP-1, monocyte chemoattractant-1
- CCL3
Macrophage inflammatory protein-1α (MIP-1α)
- CCL5
Regulated upon activation normal T cell expressed and secreted (RANTES)
- CCL18
Macrophage inflammatory protein-4 (MIP-4, PARC)
- CXCL9
Monokine induced by gamma interferon (Mig)
- CXCL10
Interferon gamma-induced protein-10 (IP-10)
- CXCL11
Interferon-inducible T cell alpha chemoattractant (I-TAC, IP-9)
References
- 1.Takebe N, Harris PJ, Warren RQ, Ivy SP. Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nat Rev Clin Oncol. 2011;8:97–106. doi: 10.1038/nrclinonc.2010.196. [DOI] [PubMed] [Google Scholar]
- 2.Zhou BB, Zhang H, Damelin M, Geles KG, Grindley JC, Dirks PB. Tumour-initiating cells: challenges and opportunities for anticancer drug discovery. Nat Rev Drug Discov. 2009;8:806–823. doi: 10.1038/nrd2137. [DOI] [PubMed] [Google Scholar]
- 3.Crea F, Duhagon MA, Farrar WL, Danesi R. Pharmacogenomics and cancer stem cells: a changing landscape? Trends Pharmacol Sci. 2011;32:487–494. doi: 10.1016/j.tips.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Radtke F, Raj K. The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat Rev Cancer. 2003;3:756–767. doi: 10.1038/nrc1186. [DOI] [PubMed] [Google Scholar]
- 5.Hansson EM, Lendahl U, Chapman G. Notch signaling in development and disease. Semin Cancer Biol. 2004;14:320–328. doi: 10.1016/j.semcancer.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 6.Nickoloff BJ, Osborne BA, Miele L. Notch signaling as a therapeutic target in cancer: a new approach to the development of cell fate modifying agents. Oncogene. 2003;22:6598–6608. doi: 10.1038/sj.onc.1206758. [DOI] [PubMed] [Google Scholar]
- 7.Kang S, Yang C, Luo R. Induction of CCL2 by siMAML1 through upregulation of TweakR in melanoma cells. Biochem Biophys Res Commun. 2008;372:629–633. doi: 10.1016/j.bbrc.2008.05.079. [DOI] [PubMed] [Google Scholar]
- 8.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alves-Guerra MC, Ronchini C, Capobianco AJ. Mastermind-like 1 Is a specific coactivator of beta-catenin transcription activation and is essential for colon carcinoma cell survival. Cancer Res. 2007;67:8690–8698. doi: 10.1158/0008-5472.CAN-07-1720. [DOI] [PubMed] [Google Scholar]
- 10.Shimizu T, Kagawa T, Inoue T, Nonaka A, Takada S, Aburatani H, Taga T. Stabilized beta-catenin functions through TCF/LEF proteins and the Notch/RBP-Jkappa complex to promote proliferation and suppress differentiation of neural precursor cells. Mol Cell Biol. 2008;28:7427–7441. doi: 10.1128/MCB.01962-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ingram WJ, McCue KI, Tran TH, Hallahan AR, Wainwright BJ. Sonic Hedgehog regulates Hes1 through a novel mechanism that is independent of canonical Notch pathway signalling. Oncogene. 2008;27:1489–1500. doi: 10.1038/sj.onc.1210767. [DOI] [PubMed] [Google Scholar]
- 12.Wall DS, Mears AJ, McNeill B, Mazerolle C, Thurig S, Wang Y, Kageyama R, Wallace VA. Progenitor cell proliferation in the retina is dependent on Notch-independent Sonic hedgehog/Hes1 activity. J Cell Biol. 2009;184:101–112. doi: 10.1083/jcb.200805155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatton BA, Villavicencio EH, Pritchard J, LeBlanc M, Hansen S, Ulrich M, Ditzler S, Pullar B, Stroud MR, Olson JM. Notch signaling is not essential in sonic hedgehog-activated medulloblastoma. Oncogene. 2010;29:3865–3872. doi: 10.1038/onc.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sang L, Roberts JM, Coller HA. Hijacking HES1: how tumors co-opt the anti-differentiation strategies of quiescent cells. Trends Mol Med. 2010;16:17–26. doi: 10.1016/j.molmed.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schreck KC, Taylor P, Marchionni L, Gopalakrishnan V, Bar EE, Gaiano N, Eberhart CG. The Notch target Hes1 directly modulates Gli1 expression and Hedgehog signaling: a potential mechanism of therapeutic resistance. Clin Cancer Res. 2010;16:6060–6070. doi: 10.1158/1078-0432.CCR-10-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sang L, Coller HA. Fear of commitment: Hes1 protects quiescent fibroblasts from irreversible cellular fates. Cell Cycle. 2009;8:2161–2167. doi: 10.4161/cc.8.14.9104. [DOI] [PubMed] [Google Scholar]
- 17.Sang L, Coller HA, Roberts JM. Control of the reversibility of cellular quiescence by the transcriptional repressor HES1. Science. 2008;321:1095–1100. doi: 10.1126/science.1155998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu L, Kobayashi K, Sun T, Gao P, Liu J, Nakamura M, Weisberg E, Mukhopadhyay NK, Griffin JD. Cloning and functional characterization of the murine mastermind-like 1 (Maml1) gene. Gene. 2004;328:153–165. doi: 10.1016/j.gene.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 19.Jin B, Shen H, Lin S, Li JL, Chen Z, Griffin JD, Wu L. The mastermind-like 1 (MAML1) co-activator regulates constitutive NF-kappaB signaling and cell survival. J Biol Chem. 2010;285:14356–14365. doi: 10.1074/jbc.M109.078865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 21.Moellering RE, Cornejo M, Davis TN, Del Bianco C, Aster JC, Blacklow SC, Kung AL, Gilliland DG, Verdine GL, Bradner JE. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462:182–188. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartlett DW, Davis ME. Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live-animal bioluminescent imaging. Nucleic Acids Res. 2006;34:322–333. doi: 10.1093/nar/gkj439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang S, Xie J, Ma S, Liao W, Zhang J, Luo R. Targeted knock down of CCL22 and CCL17 by siRNA during DC differentiation and maturation affects the recruitment of T subsets. Immunobiology. 2010;215:153–162. doi: 10.1016/j.imbio.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 24.Hiroi M, Ohmori Y. Constitutive nuclear factor kappaB activity is required to elicit interferon-gamma-induced expression of chemokine CXC ligand 9 (CXCL9) and CXCL10 in human tumour cell lines. Biochem J. 2003;376:393–402. doi: 10.1042/BJ20030842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biswas SK, Gangi L, Paul S, Schioppa T, Saccani A, Sironi M, Bottazzi B, Doni A, Vincenzo B, Pasqualini F, Vago L, Nebuloni M, Mantovani A, Sica A. A distinct and unique transcriptional program expressed by tumor-associated macrophages (defective NF-kappaB and enhanced IRF-3/STAT1 activation) Blood. 2006;107:2112–2122. doi: 10.1182/blood-2005-01-0428. [DOI] [PubMed] [Google Scholar]
- 26.Riccio O, van Gijn ME, Bezdek AC, Pellegrinet L, van Es JH, Zimber-Strobl U, Strobl LJ, Honjo T, Clevers H, Radtke F. Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO Rep. 2008;9:377–383. doi: 10.1038/embor.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kavanagh E, Joseph B. The hallmarks of CDKN1C (p57, KIP2) in cancer. Biochim Biophys Acta. 2011;1816:50–56. doi: 10.1016/j.bbcan.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 28.Jin B, Shen H, Lin S, Li JL, Chen Z, Griffin JD, Wu L. The mastermind-like 1 (MAML1) co-activator regulates constitutive NF-kappaB signaling and cell survival. J Biol Chem. 2010;285:14356–14365. doi: 10.1074/jbc.M109.078865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao Q, Kaur C, Wu CY, Lu J, Ling EA. Nuclear factor-kappa beta regulates Notch signaling in production of proinflammatory cytokines and nitric oxide in murine BV-2 microglial cells. Neuroscience. 2011;192:140–154. doi: 10.1016/j.neuroscience.2011.06.060. [DOI] [PubMed] [Google Scholar]
- 30.Bhoopathi P, Chetty C, Dontula R, Gujrati M, Dinh DH, Rao JS, Lakka SS. SPARC stimulates neuronal differentiation of medulloblastoma cells via the Notch1/STAT3 pathway. Cancer Res. 2011;71:4908–4919. doi: 10.1158/0008-5472.CAN-10-3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee JH, Suk J, Park J, Kim SB, Kwak SS, Kim JW, Lee CH, Byun B, Ahn JK, Joe CO. Notch signal activates hypoxia pathway through HES1-dependent SRC/signal transducers and activators of transcription 3 pathway. Mol Cancer Res. 2009;7:1663–1671. doi: 10.1158/1541-7786.MCR-09-0191. [DOI] [PubMed] [Google Scholar]
- 32.Ma J, Meng Y, Kwiatkowski DJ, Chen X, Peng H, Sun Q, Zha X, Wang F, Wang Y, Jing Y, Zhang S, Chen R, Wang L, Wu E, Cai G, Malinowska-Kolodziej I, Liao Q, Liu Y, Zhao Y, Xu K, Dai J, Han J, Wu L, Zhao RC, Shen H, Zhang H. Mammalian target of rapamycin regulates murine and human cell differentiation through STAT3/p63/Jagged/Notch cascade. J Clin Invest. 2010;120:103–114. doi: 10.1172/JCI37964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee H, Deng J, Xin H, Liu Y, Pardoll D, Yu H. A requirement of STAT3 DNA binding precludes Th-1 immunostimulatory gene expression by NF-kappaB in tumors. Cancer Res. 2011;71:3772–3780. doi: 10.1158/0008-5472.CAN-10-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee H, Herrmann A, Deng JH, Kujawski M, Niu G, Li Z, Forman S, Jove R, Pardoll DM, Yu H. Persistently activated Stat3 maintains constitutive NF-kappaB activity in tumors. Cancer Cell. 2009;15:283–293. doi: 10.1016/j.ccr.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee H, Pal SK, Reckamp K, Figlin RA, Yu H. STAT3: a target to enhance antitumor immune response. Curr Top Microbiol Immunol. 2011;344:41–59. doi: 10.1007/82_2010_51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Regis G, Pensa S, Boselli D, Novelli F, Poli V. Ups and downs: the STAT1:STAT3 seesaw of Interferon and gp130 receptor signalling. Semin Cell Dev Biol. 2008;19:351–359. doi: 10.1016/j.semcdb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 38.Pinho AV, Rooman I, Reichert M, De Medts N, Bouwens L, Rustgi AK, Real FX. Adult pancreatic acinar cells dedifferentiate to an embryonic progenitor phenotype with concomitant activation of a senescence programme that is present in chronic pancreatitis. Gut. 2011;60:958–966. doi: 10.1136/gut.2010.225920. [DOI] [PubMed] [Google Scholar]
- 39.Lleonart ME, Artero-Castro A, Kondoh H. Senescence induction; a possible cancer therapy. Mol Cancer. 2009;8:3. doi: 10.1186/1476-4598-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roninson IB. Tumor cell senescence in cancer treatment. Cancer Res. 2003;63:2705–2715. [PubMed] [Google Scholar]
- 41.Schmitt CA. Cellular senescence and cancer treatment. Biochim Biophys Acta. 2007;1775:5–20. doi: 10.1016/j.bbcan.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 42.Kahlem P, Dorken B, Schmitt CA. Cellular senescence in cancer treatment: friend or foe? J Clin Invest. 2004;113:169–174. doi: 10.1172/JCI20784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Freund A, Orjalo AV, Desprez PY, Campisi J. Inflammatory networks during cellular senescence: causes and consequences. Trends Mol Med. 2010;16:238–246. doi: 10.1016/j.molmed.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rakhra K, Bachireddy P, Zabuawala T, Zeiser R, Xu L, Kopelman A, Fan AC, Yang Q, Braunstein L, Crosby E, Ryeom S, Felsher DW. CD4(+) T cells contribute to the remodeling of the microenvironment required for sustained tumor regression upon oncogene inactivation. Cancer Cell. 2010;18:485–498. doi: 10.1016/j.ccr.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]