Abstract
Multiple determinant factors are involved in the occurrence and progression of esophageal squamous cell carcinoma (ESCC). Human papillomavirus (HPV) and human leukocyte antigen (HLA) polymorphism were identified as important factors. This study examined the associations between the development of Kazakh ESCC and the determinant factors including HLA-DRB1*0901, 1501; DQB1*0301, 0602; high-risk HPV infection in the area of Xinjiang, China. 200 Kazakh patients with ESCC and 150 controls were recruited, and polymerase chain reaction (PCR) was performed to detect HLA-DRB1*0901, 1501 and DQB1*0301,0602 using sequence-specific primers (SSPs). HPV16 was detected in esophageal specimens using PCR. HPV16 infection rate in Kazakh ESCC case group was 41 %, significantly higher than that of control group 14 % (OR = 3.62; 95 % CI, 2.15–6.09; P < 0.001). A positive association between ESCC and HLA-DRB1*1501 (OR = 2.46, P < 0.0125) or HLA-DQB1*0301 (OR = 3.34, P < 0.0125) alleles was observed. Similar tendencies were observed for HLA-DRB1*1501 (OR = 3.095, P < 0.0125) and HLA-DQB1*0301 (OR = 2.410, P < 0.0125) alleles with HPV16-positive ESCC. HLA-DRB1*1501, HLA-DQB1*0301 and DQB1*0602 were significantly associated with ESCC when the age was ≥55 years (P < 0.0125 for all), whereas only HLA-DQB1*0301 was significantly associated with ESCC when the age was <55 years (P < 0.0125). HLA-DRB1*1501 and HLA-DQB1*0301 were significantly associated with an increase in ESCC occurrence in females (P < 0.0125), whereas only HLA-DQB1*0301 was significantly associated with ESCC in males. Moreover, the occurrence of HLA-DQB1*0602 gene in poorly differentiated ESCC group (68.8 %) was slightly higher than that of well-differentiated squamous cell carcinoma group (31.2 %). The difference was not statistically significant (P > 0.0125). The study suggests that HLA-DRB1*1501 and HLA-DQB1*0301 may influence the immune response to specific tumor and HPV-encoded epitopes and affect the risk of Kazakh ESCC in XinJiang, China.
Keywords: HLA, Esophageal squamous cell carcinoma, Human papillomavirus, PCR-SSP, Kazakh
Introduction
Esophageal carcinoma is one of the top ten commonest malignant tumors in the world. The incidence rate varies in different physiographic regions, nations and races [1]. China has high incidence and mortality rate of esophageal carcinoma [2]. Kazakh national minority (ethnic) living in Xinjiang (northwest of China) has been reported to be one of the ethnics with the highest incidence of esophageal Carcinoma [3]. The esophageal carcinoma mortality rate within Kazakh reaches 155.9/100,000 which is higher than the average chinese rate of 15.2/100,000 [4]. The occurrence of Esophageal Carcinoma is a complex process involving multifactors, multistages and multiway interactions [5]. It has been documented that the HPV infection could be the one important attribute of esophagus carcinogenesis [6, 7]. However, previous studies are not definitive due to the use of insensitive laboratory approaches, different HPV detection techniques and different study populations [8–12]. In our previous study, we have observed a significantly high prevalence of HPV infection [8]. Even though the HPV infections are common, only a small fraction progresses into persistent infection and cancer, suggesting that other factors are also involved in the pathogenesis [13]. One potential cofactor may be the host cellular immune response to HPV infection and cancer cells mediated by the HLA-restricted T lymphocytes [14].
HLA-II alleles, which are involved in presenting foreign antigens to the immune system cells, are important in host immune responses to viruses and other pathogens. Among the most polymorphic human genes, HLA-II polymorphism results in variations of the peptide-binding cleft, thus influencing the bound and presentation of antigens to T cells [15]. Therefore, given that host immune response to HPV is thought to be an important determinant of HPV persistence and progression to high-grade lesions and cancer, [16, 17] it is possible that HLA-II variations, which are also related to ethnicity, may affect esophageal carcinoma pathogenesis through immunologic control of HPV.
HLA encodes molecules essential for immune clearance of virus-infected cells. HLA class II molecules encoded by DR, DQ and DP genes are expressed in immune cells and are responsible for presenting antigenic peptides to CD4+ T cells. A number of studies have examined the association between HLA class II allele polymorphism, HPV infection and the development of tumors, especially for those with a high-degree polymorphism of the DRB1 and DQB1 [18–20]. However, few studies have examined HPV-related esophageal carcinoma and HLA class II allele polymorphism to date. We, therefore, examined whether the four HLA alleles (HLA-DRB1*0901, 1501 and DQB1*0301, 0602) are associated with HPV infection, and the occurrence and progression of ESCC were analyzed in 200 patient and 150 control groups.
Materials and methods
Study population
Two hundred Kazakans specimens were collected between 2000 and 2007. The patients were aged 34–79 years (121 men and 79 women) who were diagnosed with ESCC and did not received radiotherapy or chemotherapy before surgery. At the same time, 150 specimens were collected from normal Kazakans esophageal biopsy tissues as controls. The normal patients were aged 30 to 73 years (82 men and 68 women). All specimens were recruited from the Department of Yili friendship hospital, XinJiang, China, where the biopsy paraffin-embedded tissues were prepared. Each participant provided written informed consent, and the study was approved by the participating hospital.
All ESCC specimens obtained in the surgery were embedded in paraffin, subsequently sectioned into 5-μm-thick slices, and subjected to conventional H&E staining. The diagnosis of ESCCs was confirmed by two pathologists according to WHO histological tumor classification criteria [21]: 76 cases of well-differentiated ESCC and 124 cases of poorly differentiated ESCC. Control group specimens were gastroscope biopsy tissues, preserved in paraffin and confirmed as normal esophageal mucosa.
DNA preparation
We used some methods to minimize the possibility of tissues contamination for patient and control groups. Briefly, each formalin-fixed and paraffin-embedded sample was cut into 7-μm-thick sections; 10–15 slides were placed into a new high-pressure EP tube, disinfected blade with 75 % medicine alcohol before the sample cut. And for every 5 samples, we used no tissues paraffin-embedded sample to cut for no contamination control. Then, genomic DNA was isolated from paraffin-embedded tissues by phenol–chloroform method [22] and dissolved in sterile double-distilled water at −80 °C in the refrigerator for 12–24 h to preserve it.
HPV16 typing
Before HPV16 detection, we used β-globin signals to confirm sample DNA quality and chose strong β-globin samples to genotype. HPV DNA was detected using a nested primer PCR. Each reaction includes a positive and negative control. The positive control was CaSki cell DNA (generously provided by Professor Yang Ke, Beijing Institute for Cancer Prevention Laboratory). Primers for HPV16 E6 were the following: FOR 5′- GACCCAGAAAGTTACCACAG -3′, RER 5′—CACAACGGTTTGTT GTATTG -3′ (primer synthesis from Shanghai Health Industrial Co., Ltd). PCRs were carried out under the following conditions: predegeneration, 94 °C 3 min; degeneration, 94 °C 45 s; annealing, 55 °C 45 s; an extension of 72 °C 45 s, a total of 35 cycles.
HLA alleles polymorphism identification
Previous studies have found that HLA-II alleles are closely related to carcinoma risks, [23, 24], but the studies on the relationship of ESCC with HLA-II are very limited. Lin [25] genotyped 23 HLA-II alleles in ESCC among 42 Han ethnic adults living in Hubei, China, and found that only HLA-DRB1*0901 was significantly associated with the risk of ESCC. Further, throughout the paper review, we found that the three HLA-II alleles (HLA DRB1*1501, DQB1*0301, *0602) are closely related to HPV16 infection and other tumors [24, 26]. However, whether these four HLA-II alleles are related to HPV infection and esophageal carcinoma has not been reported to date. Thus, we selected these four HLA-II alleles to examine whether the four alleles were associated with the occurrence and progression of ESCC.
We used SSP based on polymerase chain reaction (PCR-SSP) method to genotype the HLA polymorphisms. PCRs are set each positive control and negative control. We deleted the false-positive and false-negative results. The sequence-specific primers refer to O.Olerup’s article [27, 28]. PCR reactions were carried out as described above. Electrophoresis was performed on a 2 % agarose gel, which was then imaged under ultraviolet light and photographed(Fig. 1). PCR products were purified and sequenced at the Shanghai Health Industrial Co., Ltd, and the results subjected to homology analysis using BLAST.
Fig. 1.

a HLA-DRB1*0901PCR gel electrophoresis. 2: HLA-DRB1*0901 positive control; 1,4,5,6,8,9,10,11,12: HLA-DRB1*0901 negative samples; 3, 7: HLA-DRB1*0901-positive samples; 13: negative control; 14: blank control; b HLA-DQB1*0301PCR gel electrophoresis. 14: HLA-DQB1*0301 positive control; 3,4,5,7, 8,9: HLA-DRB1*0301 negative samples; 2,6,11,12,13: HLA-DRB1*0301 positive samples; 1: negative control; 15: blank control; c HLA-DRB1*1501 PCR electrophoresis Fig. 15: HLA-DRB1*1501 positive control; 1-10: HLA-DRB1*1501 negative samples; 11,12,13: HLA-DRB1*1501 positive samples; 14: negative control; 16: blank control; d HLA-DQB1*0602PCR gel electrophoresis. 14: HLA-DQB1*0602 positive control; 2,4,5,6,7,9,11: HLA-DRB1*0602 negative samples; 3,8,10,12: HLA-DRB1*0602 positive samples; 1: negative control; 15: blank control; M: DNA Marker; 501-bp pUC19 DNA/MspI
Quality control
DNA preparation, PCR and PCR product detection were carried out in separated spaces. Using ultraviolet light to monitor environmental contamination before experiments, new sterile gloves and supplies were used to avoid contamination. Using β-globin signals to confirm sample DNA quality, DNA of sufficient quality was chosen for PCR. At last, 30 % samples were randomly chosen to repeat the test, and same results were obtained.
Statistical analysis
The software SPSS version 13.0 was employed for all statistical analyses. The t-test was used for continuous variables, and Mantel–Haenszel χ2 test was used for categorical variables. Odds ratios (OR) and 95 % CI were calculated by logistic regression to estimate the relative likelihood between four HLA-II polymorphisms and the risk of ESCC across genotypes. OR, CI, and P values were calculated by the Epi-Info program. P values less than 0.0125 (0.05/4) were considered significant.
Results
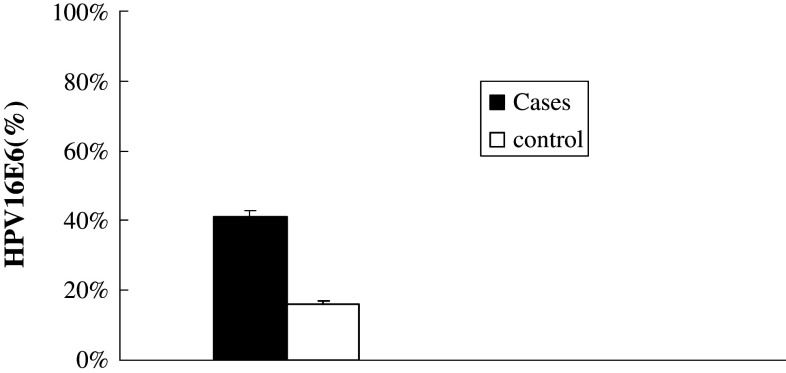
Case and control groups had similar age range and distribution of sex (P > 0.05 for both) (Table 1). The β-globin PCR results show that all subjects had specimens with sufficient quality for HLA and HPV analyses. Among cases, 41 % were HPV16 positive, significantly higher than 16.1 % of control group (OR = 3.62; 95 % CI, 2.15–6.09; P < 0.001) (Fig. 2).
Table 1.
Sex distribution of case and control groups
| Group | Cases (n) | Sex n (%) | χ 2 | P | |
|---|---|---|---|---|---|
| Male | Female | ||||
| Case | 200 | 121 (60.5) | 79 (39.5) | 2.0 | 0.157 |
| Control | 150 | 82 (54.7) | 68 (45.3) | ||
Fig. 2.
The distribution of HPV16E6 in case and control groups
We detect the distribution of HLA class II alleles (HLA-DRB1* 0901, 1501 and DQB1*0301,0602) among HPV16-positive and HPV16-negative healthy group(no cancer group), which was showed in Table 2. We found the frequencies distribution of HLA-DRB1*0901 and HLA-DRB1*1501 allele in HPV16-positive group are higher than that of the HPV16-negative ones (0.125 vs. 0.087 and 0.292 vs. 0.246), and the frequency distribution of HLA-DQB1*0301 and HLA-DQB1*0602 allele in HPV16-positive group is lower than that of the HPV16-negative ones (0.458 vs. 0.492 and 0.548 vs. 0.625). However, these differences are not statistically significant (P > 0.0125 for all).
Table 2.
The distribution of HLA-DRB1*0901, 1501 and DQB1*0301, 0602 in HPV16 positive and HPV16 negative in control group (no cancer patients)
| HLA | HPV16 positive | HPV16 negative | |||||
|---|---|---|---|---|---|---|---|
| (n = 24) | AF | (n = 126) | AF | X2 | P | OR (95 % CI) | |
| DRB1*0901 | 3 | 0.125 | 11 | 0.087 | 0.339 | 0.561 | 1.494 (0.384–5.811) |
| DRB1*1501 | 7 | 0.292 | 31 | 0.246 | 0.222 | 0.638 | 1.262 (0.479–3.326) |
| DQB1*0301 | 11 | 0.458 | 62 | 0.492 | 0.092 | 0.762 | 0.873 (0.364–2.097) |
| DQB1*0602 | 15 | 0.548 | 69 | 0.625 | 0.490 | 0.484 | 1.377 (0.561–3.379) |
The distribution of HLA-DRB1*0901, 1501 and DQB1*0301, 0602 alleles among case and control groups was showed in Table 2. We found that HLA-DQB1*0301 and HLA-DRB1*1501 were significantly associated with an increase in the risk of ESCC (ORs ranged 2.5–3.3; P < 0.0125 for all). The comparison results of four HLA-II alleles in HPV16-infected ESCC with healthy controls were also showed Table 3. HLA-DRB1*1501 has most significant association with the risk of HPV16 positive Kazakh ESCC, which was also closely associated with HLA-DQB1*0301 (ORs ranged 2.4–3.1, P < 0.0125 for all).
Table 3.
The distribution of HLA-DRB1*0901, 1501 and DQB1*0301, 0602 in patients and control group
| HLA | ECC | HPV16ECC | Control | ECC | HPV16ECC | ||||
|---|---|---|---|---|---|---|---|---|---|
| (n = 200) | (n = 30) | (n = 150) | P | OR | 95 % CI | P | OR | 95 % CI | |
| DRB1*0901 | 21 | 11 | 14 | 0.857 | 1.14 | 0.559–2.323 | 0.461 | 1.505 | 0.650–3.487 |
| DRB1*1501 | 91 | 42 | 38 | 0.000 | 2.46 | 1.551–3.903 | 0.000 | 3.095 | 1.753–5.463 |
| DQB1*0301 | 152 | 57 | 73 | 0.000 | 3.34 | 2.117–5.269 | 0.004 | 2.410 | 1.362–4.248 |
| DQB1*0602 | 138 | 58 | 84 | 0.017 | 1.75 | 1.126–2.716 | 0.039 | 1.899 | 1.069–3.373 |
With age control, we found when the age was ≥55 years, HLA-DRB1*1501, HLA-DQB1*0301 and DQB1*0602 were significantly associated with an increase in ESCC (P < 0.0125 for all). When the age was <55, HLA-DQB1*0301 was significantly associated with an increase in ESCC (P < 0.0125) (Table 4).
Table 4.
The distribution of HLA-DRB1*0901, 1501 and DQB1*0301, 0602 in patients and control group (age stratification)
| Characteristic | Age <55 | P | Age ≥55 | P | ||
|---|---|---|---|---|---|---|
| ESCC | Control | ESCC | Control | |||
| HLA-DRB1*0901 | 11 (10.9 %) | 10 (10.3 %) | 1.000 | 10 (10.1 %) | 4 (7.5 %) | 0.604 |
| HLA-DRB1*1501 | 44 (43.6 %) | 25 (25.8 %) | 0.013 | 47 (47.5 %) | 13 (24.5 %) | 0.010 |
| HLA-DQB1*0301 | 74 (73.3 %) | 40 (51.3 %) | 0.002 | 78 (78.8 %) | 24 (45.3 %) | 0.000 |
| HLA-DQB1*0602 | 62 (61.4 %) | 49 (50.5 %) | 1.000 | 76 (76.8 %) | 24 (45.3 %) | 0.000 |
With sex control, we found HLA-DQB1*0301 was significantly associated with an increase in male ESCC (P < 0.0125).HLA-DRB1*1501 and HLA-DQB1*0301 was significantly associated with an increase in female esophageal squamous cell carcinoma (P < 0.0125 for all) (Table 5).
Table 5.
The distribution of HLA-DRB1*0901, 1501 and DQB1*0301,0602 in patients and control group (male and female)
| Characteristics | Male | P | Female | P | ||
|---|---|---|---|---|---|---|
| ESCC | Control | ESCC | Control | |||
| HLA-DRB1*0901 | 13 (10.7 %) | 5 (6.4 %) | 0.431 | 8 (10.1 %) | 9 (12.5 %) | 0.839 |
| HLA-DRB1*1501 | 59 (48.8 %) | 24 (30.8 %) | 0.018 | 32 (40.5 %) | 14 (19.4 %) | 0.008 |
| HLA-DQB1*0301 | 91 (75.2 %) | 40 (51.3 %) | 0.001 | 61 (77.2 %) | 33 (45.8 %) | 0.000 |
| HLA-DQB1*0602 | 83 (68.6 %) | 43 (55.1 %) | 0.076 | 55 (69.6 %) | 41 (56.9 %) | 0.148 |
To determine the difference between well-differentiated squamous cell carcinoma and poorly differentiated squamous cell carcinoma in the distribution of HLA-DRB1*0901, 1501and DQB1*0301, 0602 alleles, statistical analysis was performed. We found that the frequency distribution of HLA-DQB1*0602 allele in poorly differentiated squamous cell carcinoma group was higher than that of the well-differentiated squamous cell carcinoma group (0.688 vs. 0.312), but the difference was not statistically significant (P > 0.0125) Table 6.
Table 6.
The distribution of HLA allele in different histopathological grade esophageal squamous cell carcinoma
| HLA alleles | W-ECC | P-ECC | |||
|---|---|---|---|---|---|
| N1 | AF | N2 | AF | P | |
| DRB1*0901 | 4 | 0.56 | 17 | 0.133 | 0.087 |
| DRB1*1501 | 38 | 0.528 | 53 | 0.414 | 0.121 |
| DQB1*0301 | 53 | 0.736 | 99 | 0.773 | 0.553 |
| DQB1*0602 | 43 | 0.597 | 95 | 0.742 | 0.033 |
Discussion
Many reports regarding high-risk HPV infection in esophageal carcinoma were inconsistence [6–8, 10–12]. To explain these marked differences in the reports, sampling methods, demographic ethnic factors, disease status, and sensitivity of detection methods have been cited as potential causes of inconsistency. In our study, HPV16 virus infection rate in the patients of ethnic Kazakh ESCC was significantly higher than that of the normal ethnic Kazakh population. Therefore, it has been proposed that HPV16 infection may play a role in esophageal carcinogenesis. A similar observation was also revealed in previous studies [9]. However, some studies suggest that HPV infection was not related to esophageal carcinoma, which may reflect discrepancy of different race, different ethnic and different region. In our study, we have performed strict quality controls on the object and method of the study. Therefore, we can confirm that HPV infection is one of the important factors in high incidence of Xinjiang ethnic Kazakh esophageal carcinoma. It was considered by some investigators that high polymorphism of HLA-II type alleles was closely related to HPV infection in early studies [17, 19] HLA-II genes are the main functions of identifying and presenting exogenous antigen, such as the HPV. There are different HLA-II alleles with different capacity of identifying as well as combining HPV and presenting antigenic peptides to CD4+ T cells. HLA-II alleles that are hard to be identified and cleared by HPV virus are called HPV virus susceptibility alleles, whereas those are easy to be identified and cleared by HPV virus can prevent the infection of HPV alleles. Those HPV susceptibility alleles cause HPV persistent infection and maintained in the body, subsequently causing tumor.
At present, published data on the distribution of HLA-II alleles among ESCC are very limited, and no data are available for HPV-related ESCC. However, many studies on the distribution of HLA-II alleles refer to some tumors, especially cervical cancer. Because ESCC has many similar properties with cervical squamous cell carcinoma, so the studies on cervical carcinoma are worthy using for references. Allele frequencies were calculated for cases and controls of HLA-DRB1*0901, 1501, DQB1*0301, 0602 alleles whose association with increased or decreased risk of cervical disease was previously reported in different populations by other investigators: HLA-DRB1*0901, [29, 30] HLA-DQB1*0301, [26, 31–33] HLA-DRB1*1501, [24, 34–36] and HLA-DQB1*0602 [36, 37].
In this study, HLA-DQB1*0301 is associated not only with increasing risk of ESCC but also with the increasing risk of the HPV16-positive ESCC. The same phenomenon was observed in other tumors, especially in cervical cancer. Madeleine MM study of American women squamous cell cervical cancer (SCC) demonstrated that HLA-DQB1*0301 increased the risk of SCC, which is associated with HPV16-positive SCC. This association was also observed in other area women, including Norway, [38] Brazil [39] and British women [26]. When do age and sex stratification, we found that HLA-DQB1*0301 had similar result that increased the risk of ESCC in different groups (age less than or greater than 55 in male and female).
The statistical analysis of HLA-DRB1*1501 distribution among cases and controls showed that the allele was more frequent in ESCC than in control population. Similar tendencies were observed for HLA-DRB1*1501 with HPV16-positive ESCC. It is consistent with cervical cancer reports of others. Beskow AH [35] examined the distribution of HLA-DRB1 and DQB1 allele frequencies in HPV infection in cervical cancer and found that HLA-DRB1*1501 was related to long-term HPV infection, especially closely related to HPV16-related cervical cancer. Cuzick [26] have detected cervical squamous cell carcinoma in the women of the United Kingdom and found that frequency distribution of HLA DRB1*1501 and HLA-DQB1*0602 in all patients with cervical squamous cell carcinoma did not increase, but is associated with the HPV16-positive cervical squamous cell carcinoma. Similar reports included the studies on Sweden women [35] and Brazilian women [36]. In our study, although HLA-DQB1*0602 allele frequency in patients with ESCC and HPV16-positive ESCC was higher than that of control group, it was not statistical significance. On the other hand, it is also considered by some investigators that HLA DRB1*1501 and HLA-DQB1*0602 were associated with a decreased risk of cervical cancer [29, 30]. To explain these differences in the reports, sampling methods, geographic area and ethnic factors, disease status, and sensitivity of detection methods have been cited as potential causes of inconsistency. By age control, we found that HLA-DRB1*1501 increased the risk of ESCC in the patient of >55 years, but not in the patient of less than 55 years. And when we study >55-year-old ESCC patients, we found HLA-DQB1*0602 increased the risk of ESCC. The result suggests that HLA-DQB1*0602 was susceptible alleles at >55-year-old patients with esophageal carcinoma. By sex control, we found HLA-DRB1*1501 increased the risk of female ESCC, indicating that HLA-DRB1*1501 was close related to female ESCC.
We found that HLA-DRB1*1501 and HLA-DQB1*0301 were closely related to HPV16-positive ESCC, but not to HPV16-positive healthy group (no cancer group). It may be prompt that HLA-DRB1*1501 and HLA-DQB1*0301 has to do with lack of defense against HPV16 infection, but for ESCC, especially HPV16 persistent infection ESCC, these two alleles have important roles.
We found the distribution of HLA-DRB1*0901 in ESCC was higher than that of control group. However, the difference was not statistically significant. This result was different with the study by LinJun on ESCC of Han ethnic in China [25]. It may be that the samples were from different ethnic regions.
In this study, phenol–chloroform extraction method was used to extract DNA from paraffin-embedded tissues of ESCC and esophageal mucosa, and the frequency distribution of HLA alleles with the DNA was then detected. This method is slightly different with the traditional methods that detect distribution of HLA allele frequencies. The traditional way for the detection usually uses peripheral blood DNA, whereas our study used organization of DNA. In cancer research, it is important to select same part of tissue to prepare DNA for case and control groups because it is more comparable and will more real show the distribution of allele frequency. At the same time, organization of DNA can be used for detecting HPV, which can a comprehensive study of genetic and environmental factors for esophageal carcinoma.
A limitation in our works is the relatively small number of samples collected. A larger cohort of samples might substantiate the data. Another limitation is the loss of other epidemiology information, such as alcohol use, smoking status and eating habits, which may help us to assess the interaction between high-risk HPV infection and other environment factors in esophageal carcinoma.
Acknowledgments
The authors are grateful to Dr. Xiang Gao in Department of Nutrition, Harvard School of Public Health, Boston, Massachusetts, and Dr. Mei Wan in Department of Orthopaedic Surgery, Johns Hopkins University School of Medicine, Baltimore, USA, for carefully revising the manuscript. This work was supported by National Key Technology R&D Program of China [2009BAI82B03], International S&T Cooperation Program of China [2010DFB34100], and The National Natural Science Foundation, China [81160301].
Conflict of interest
The authors disclose no conflict of interest.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Bray F, Pisani P, Parkin DM. GLOBOCAN (2000) cancer incidence, mortality and prevalence worldwide. Lyon: IARC Press; 2001. [Google Scholar]
- 3.Zhang Y. The distribution of esophageal cancer in Xinjiang. J Xinjiang Med Univ (China) 1988;11:139–144. [Google Scholar]
- 4.Hou C, Chen ZF, He YT. Esophageal cancer epidemiology. Hebei Work J Med. 2000;17:27–29. [Google Scholar]
- 5.Cheung WY, Liu G. Genetic variations in esophageal cancer risk and prognosis. Gastroenterol Clin North Am. 2009;38(1):75–91. doi: 10.1016/j.gtc.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Syrjanen KJ. HPV infections and oesophageal cancer. J Clin Pathol. 2002;55:721–728. doi: 10.1136/jcp.55.10.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Tian X, Liu F, Zhao Y, Sun M, Chen D, Lu C, Wang Z, Shi X, Zhang Q, Zhang D, Shen Z, Li F, Harris CC, Cai H, Ke Y. Detection of HPV DNA in esophageal cancerspecimens from different regions and ethnic groups: a descriptive study. BMC Cancer. 2010;10:19. doi: 10.1186/1471-2407-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang L, Chen L, Sun ZZ, Zhang HY, Tian XY, Qi Y, Zhu J, Qin JM, Li F. Analysis of human papillomavirus DNA infection in Kazakh esophageal carcinomas in Xinjiang province. World Chin J Digestol. 2007;15(19):2114–2119. [Google Scholar]
- 9.Gu LY, Sai LM, Hou XL, Chen G, Li F, Liu CX, Chen YZ, Chang B, Yang L, Liang WH, Hu JM. Correlation between HPV16 infection and esophagus carcinoma of Kazakh people in Xinjiang. China Oncol. 2010;20(9):668–672. [Google Scholar]
- 10.Antonsson A, Nancarrow DJ, Brown IS, Green AC, Drew PA, Watson DI, Hayward NK, Whiteman DC. High-risk human papillomavirus in esophageal squamous cell carcinoma. Cancer Epidem Biomark Prev. 2010;19(8):2080–2087. doi: 10.1158/1055-9965.EPI-10-0033. [DOI] [PubMed] [Google Scholar]
- 11.Koshiol J, Wei WQ, Kreimer AR, Chen W, Gravitt P, Ren JS, Abnet CC, Wang JB, Kamangar F, Lin DM, von Knebel-Doeberitz M, Zhang Y, Viscidi R, Wang GQ, Gillison ML, Roth MJ, Dong ZW, Kim E, Taylor PR, Qiao YL, Dawsey SM. No role for human papillomavirus in esophageal squamous cell carcinoma in China. Int J Cancer. 2010;127(1):93–100. doi: 10.1002/ijc.25023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herrera-Goepfert R, Lizano M, Akiba S, Carrillo-García A, Becker-D’Acosta M. Human papilloma virus and esophageal carcinoma in a Latin-American region [J] World J Gastroenterol. 2009;7(25):3142–3147. doi: 10.3748/wjg.15.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arias-Pulido H, Joste N, Wheeler CM. Loss of heterozygosity on chromosome 6 in HPV-16 positive cervical carcinomas carrying the DRB1*1501–DQB1*0602 haplotype. Genes Chromosom Cancer. 2004;40(4):277–284. doi: 10.1002/gcc.20048. [DOI] [PubMed] [Google Scholar]
- 14.Mota F, Rayment N, Chong S, Singer A, Chain B. Theantigen-presenting environment in normal and humanpapillomavirus (HPV)-related premalignant cervical epithelium. Clin Exp Immunol. 1999;116(1):33–40. doi: 10.1046/j.1365-2249.1999.00826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breitburd F, Ramoz N, Salmon J, Orth G. HLA control in the progression of human papillomavirus infections. Semin Cancer Biol. 1996;7(6):359–371. doi: 10.1006/scbi.1996.0045. [DOI] [PubMed] [Google Scholar]
- 16.Clerici M, Merola M, Ferrario E, Trabattoni D, Villa ML, Stefanon B, Venzon DJ, Shearer GM, De Palo G, Clerici E. Cytokine production patterns in cervical intraepithelial neoplasia: association with human papillomavirus infection. J Natl Cancer Inst. 1997;89(3):245–250. doi: 10.1093/jnci/89.3.245. [DOI] [PubMed] [Google Scholar]
- 17.Hildesheim A, Schiffman MH, Tsukui T, Swanson CA, Lucci J, Scott DR, Glass AG, Rush BB, Lorincz AT, Corrigan A, Burk RD, Helgesen K, Houghten RA, Sherman ME, Kurman RJ, Berzofsky JA, Kramer TR. Immune activation in cervical neoplasia: crosssectional association between plasma soluble interleukin 2 receptor levels and disease. Cancer Epidemiol Biomark Prev. 1997;6(10):807–813. [PubMed] [Google Scholar]
- 18.De Araujo Souza PS, Maciag PC, Ribeiro KB, Petzl-Erler ML, Franco EL, Villa LL (2008) Interaction between polymorphisms of the human leukocyte antigen and HPV-16 variants on the risk of invasive cervical cancer. BMC Cancer 22;8:246. [PMID:18721466] [DOI] [PMC free article] [PubMed]
- 19.Chan PK, Cheung JL, Cheung TH, Lin CK, Siu SS, Yu MM, Tang JW, Lo KW, Yim SF, Wong YF, To KF, Ng HK. Chung TK.HLA-DQB1 polymorphisms and risk for cervical cancer: A case-control study in a southern Chinese population. Gynecol Oncol. 2007;105(3):736–741. doi: 10.1016/j.ygyno.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 20.El-Chennawi FA, Auf FA, Metwally SS, Mosaad YM, El-Wahab MA. Tawhid ZE.HLA-class II alleles in Egyptian patients with hepatocellular carcinoma. Immunol Invest. 2008;37(7):661–674. doi: 10.1080/08820130802111605. [DOI] [PubMed] [Google Scholar]
- 21.Hamilton SR, Aaltonen LA. WHO classification tumours of the digestive system. Lyon: IARC Press; 2000. pp. 9–11. [Google Scholar]
- 22.Shi SR, Cote RJ, Wu L, Liu C, Datar R, Shi Y, Liu D, Lim H. Taylor CR.DNA extraction from archival formalin-fixed, paraffin-embedded tissue sections based on the antigen retrieval principle: heating under the influence of pH. J Histochem Cytochem. 2002;50(8):1005–1011. doi: 10.1177/002215540205000802. [DOI] [PubMed] [Google Scholar]
- 23.Yang L, Wang LJ, Shi GL, Ni L, Song CX, Zhang ZX, Xu SF. Analysis of HLA-A, HLA-B and HLA-DRB1 alleles in Chinese patients with lung cancer. Genet Mol Res. 2010;9(2):750–755. doi: 10.4238/vol9-2gmr735. [DOI] [PubMed] [Google Scholar]
- 24.Wu Y, Liu B, Lin W, Xu Y, Li L, Zhang Y, Chen S, Lin Z, Xu A. Human leukocyte antigen class II alleles and risk of cervical cancer in China. Hum Immunol. 2007;68(3):192–200. doi: 10.1016/j.humimm.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 25.Lin J, Deng CS, Sun J, Zheng XG, Huang X, Zhou Y, Xiong P, Wang YP. HLA-DRB1 allele polymorphism in genetic susceptibility to esophages carcinoma[J] World J Gasunenterol. 2003;9(3):412–416. doi: 10.3748/wjg.v9.i3.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cuzick J, Terry G, Ho L, Monaghan J, Lopes A, Clarkson P, Duncan I. Association between high-risk HPV types, HLA DRB1* and DQB1*alleles and cervical cancer in British women. Br J Cancer. 2000;82(7):1348–1352. doi: 10.1054/bjoc.1999.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olerup O, Aldener A, Fogdell A. HLA-DQB1 and DQA1 typing by PCR amplification with sequence-specific primers(PCR-SSP) in 2 hours. Tissue Antigens. 1993;41(3):119–134. doi: 10.1111/j.1399-0039.1993.tb01991.x. [DOI] [PubMed] [Google Scholar]
- 28.Olerup O, Zetterquist H. HLA-DR typing by PCR amplification with sequence-specific primers(PCR-SSP) in 2 h: An alternative to serological DR typing in clinical practice including donor-recipient matching in cadaveric transplantations. Tissue Antigens. 1992;39(5):225–235. doi: 10.1111/j.1399-0039.1992.tb01940.x. [DOI] [PubMed] [Google Scholar]
- 29.Yang YC, Chang TY, Lee YJ, Su TH, Dang CW, Wu CC, Liu HF, Chu CC, Lin M. HLA-DRB1 alleles and cervical squamous cell carcinoma: experimental study and meta-analysis. Hum Immunol. 2006;67(4–5):331–340. doi: 10.1016/j.humimm.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 30.Saito M, Okubo M, Hirata R, Takeda S, Maeda H. Association of human leukocyte antigen and T cell message with human papillomavirus 16-positive cervical neoplasia in Japanese women. Int J Gynecol Cancer. 2007;17(6):1314–1321. doi: 10.1111/j.1525-1438.2007.00938.x. [DOI] [PubMed] [Google Scholar]
- 31.Ferrera A, Olivo A, Alaez C, Melchers WJ, Gorodezky C. HLA DQA1 and DQB1 loci in Honduran women with cervical dysplasia and invasive cervical carcinoma and their relationship to human papillomavirus infection. Hum Biol. 1999;71(3):367–379. [PubMed] [Google Scholar]
- 32.Madeleine MM, Johnson LG, Smith AG, Hansen JA, Nisperos BB, Li S, Zhao LP, Daling JR, Schwartz SM, Galloway DA. Comprehensive analysis of HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 loci and squamous cell cervical cancer risk. Cancer Res. 2008;68(9):3532–3539. doi: 10.1158/0008-5472.CAN-07-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madeleine MM, Brumback B, Cushing-Haugen KL, Schwartz SM, Daling JR, Smith AG, Nelson JL, Porter P, Shera KA, McDougall JK, Galloway DA. Human leukocyte antigen class II and cervical cancer risk: a population-based study. J Infect Dis. 2002;186(11):1565–1574. doi: 10.1086/345285. [DOI] [PubMed] [Google Scholar]
- 34.Hildesheim A, Schiffman M, Scott DR, Marti D, Kissner T, Sherman ME, Glass AG, Manos MM, Lorincz AT, Kurman RJ, Buckland J, Rush BB, Carrington M. Human leukocyte antigen class I/II alleles and development of human papillomavirus-related cervical neoplasia: results from a case-control study conducted in the United States. Cancer Epidemiol Biomark Prev. 1998;7(11):1035–1041. [PubMed] [Google Scholar]
- 35.Beskow AH, Josefsson AM, Gyllensten UB. HLA class II alleles associated with infection by HPV16 in cervical cancer in situ. Int J Cancer. 2001;93(6):817–822. doi: 10.1002/ijc.1412. [DOI] [PubMed] [Google Scholar]
- 36.Maciag PC, Schlecht NF, Souza PS, Franco EL, Villa LL, Petzl-Erler ML. Major histocompatibility complex class II polymorphisms and risk of cervical cancer and human papillomavirus infection in Brazilian women. Cancer Epidemiol Biomark Prev. 2000;9(11):1183–1191. [PubMed] [Google Scholar]
- 37.Beskow AH, Gyllensten UB. Host genetic control of HPV 16 titer in carcinoma in situ of the cervix uteri. Int J Cancer. 2002;101(6):526–531. doi: 10.1002/ijc.90010. [DOI] [PubMed] [Google Scholar]
- 38.Lie AK, Skarsvåg S, Haugen OA, Skjeldestad FE, Olsen AO, Skovlund E, Rønningen KS. Association between the HLA DQB1*0301 gene and human papillomavirus infection in high-grade cervical intraepithelial neoplasia. Int J Gynecol Pathol. 1999;18(3):206–210. doi: 10.1097/00004347-199907000-00003. [DOI] [PubMed] [Google Scholar]
- 39.Maciag PC, Schlecht NF, Souza PS, Rohan TE, Franco EL, Villa LL. Polymorphisms of the human leukocyte antigen DRB1 and DQB1 genes and the natural history of human papillomavirus infection. J Infect Dis. 2002;15(2):164–172. doi: 10.1086/341080. [DOI] [PubMed] [Google Scholar]