Abstract
PIWIL2, a member of PIWI/AGO family, is expressed in germline stem cells and precancerous stem cells, but not in adult somatic cells. PIWIL2 plays an important role in tumor development. It is considered as a cancer–testis antigen (CT80). It has been reported that the spliced fragment of PIWIL2, PL2L60, was widely expressed in cancer cell lines. In this study, HLA-A2-restricted epitopes from PL2L60 were predicted by online tools. To improve the activity of the native epitope, a candidate peptide P281 with potent binding affinity was chosen to investigate the modification strategy. A series of aromatic amino acids were introduced to substitute the first residue of P281. Then, we tested the binding affinity and stability of the peptide analogs and their ability to elicit specific immune responses both in vitro and in vivo. Our results indicated that the cytotoxic T lymphocytes (CTLs) induced by [4-Cl-Phe1]P281 could elicit more potent activities than that of P281 and other analogs. The CTLs induced by this analog could lyze target cells in HLA-A2-restricted and antigen-specific manners. [4-Cl-Phe1]P281 also showed the best resistance against degradation in human serum. In conclusion, the introduction of the unnatural amino acid, 4-Cl-Phe, into the first position could enhance the activity of the native epitope to induce cytotoxic T lymphocytes. It might be a good strategy to modify other promising native epitopes. The novel epitopes identified in this study could be used as novel candidates to the immunotherapy of HLA-A2 positive patients with tumors expressing PL2L60.
Electronic supplementary material
The online version of this article (doi:10.1007/s00262-013-1478-7) contains supplementary material, which is available to authorized users.
Keywords: PIWIL2, Cytotoxic T lymphocyte, Epitope, Cancer, Unnatural amino acid
Introduction
Immunotherapy is considered as an ideal choice to the treatment of cancer patients. Among all the immunotherapy methods, the activation of cytotoxic T lymphocyte (CTL) by epitopes from tumor-specific antigens could kill the cancer cells in an antigen-specific manner [1]. Epitopes recognized by CD8+ T cells were generally thought to be derived from full-length proteins by degradation in the cytosol and presented on MHC class I molecules [2, 3]. In the past two decades, great advances were made in tumor immunology and immunotherapy, and a lot of tumor antigens and epitopes were identified [4, 5]. The identification of CTL epitopes from different kinds of tumor antigens has contributed significantly to the development of therapeutic vaccines for the treatment of cancer [6]. The functional tumor antigens with tissue-restricted expression profiles have been considered as potential candidates because of the less opportunity to cause side effects and to be down-regulated by cancer cells. Precancerous stem cells (pCSCs) constitutively express PIWIL2 (Piwi-like 2, CT80) transcripts, which belong to the cancer–testis antigen family. PIWIL2 was reported to be associated with the proliferation, damage repair, and differentiation of cancer cells. Among the PIWIL2 family members, the 60 kD spliced fragment PL2L60 rather than other fragments or full length of PIWIL2 is widely expressed in various kinds of human and mouse cancer cells [7]. This is the first report to identify cytotoxic T lymphocyte epitopes from such an ideal tumor antigen, PL2L60.
The immunogenicity of the CTL epitopes derived from tumor antigens could be improved by proper modification strategies. The modification of these epitopes could enhance the binding affinity and stability, and overcome the immune tolerance existed in the patients [8, 9]. To activate the immune system during the disease state, peptides with lower binding affinity are often selected to be modified at the anchor residues, which are correlated with both the affinity of the peptides toward MHC and the stability of the peptide/MHC complex [10, 11]. This approach has been successfully used to modify epitopes derived from several tumor antigens including MART-1 and gp100 [12, 13]. Ruppert et al. [14] reported that HLA-A2.1 binding motif could be defined as a leucine (L) or methione (M) at position 2 and a leucine (L), valine (V), or isoleucine (I) at position 9. Tourdot et al. [15] reported that substitution of position 1 by a tyrosine could be a general strategy to enhance the immunogenicity of HLA-A2-restricted epitopes. But one of the disadvantages of modification of CTL epitopes is that it might bring unexpected effects such as totally changing the antigen specificity and T cell receptor (TCR) reactivity, which might bring the side effects. Therefore, we tried to find a strategy to slightly modify the native peptides but get significant improvement of the immune activity.
It was reported that incorporation of unnatural amino acids, retro-inversion, and cyclization could improve bioavailability [8]. A heteroclitic epitope describes an altered peptide that is a better agonist for inducing T cell responses than the native peptide. This terminology has been applied to other altered peptides with a higher immunological potency than their unaltered counterparts. Heteroclitic peptides have increased potency either due to increased binding to MHC molecules or to increased agonist properties to stimulate TCR [16]. Unnatural amino acids are resistant to proteolytic degradation and can provide enhanced stability compared to natural amino acids [17]. In the present study, we combined the strategies of introduction of aromatic and unnatural amino acids together. After the selection of the candidate native peptide, a series of unnatural aromatic amino acids were introduced to substitute the position 1. The in vitro and in vivo activities of the native peptide and its analogs were investigated.
Materials and methods
Peptides
The HLA-A2-restricted native epitopes in PL2L60 were predicted by online tools. Native peptides and all substituted peptides were synthesized by a standard solid phase Fmoc strategy and were purified to more than 95 % purity by reverse-phase high-performance liquid chromatography (RP-HPLC), and their molecular weights were confirmed by electrospray ionization–mass spectrometry (ESI–MS). COX-2_P321 (ILIGETIKI) was used as a positive control in T2 binding assay [18].
Animals, cell lines, and blood samples
HLA-A2.1/Kb transgenic mice were kindly supplied by Professor Xue-tao Cao (Second Military Medical University, China). Mice were bred and maintained in specific pathogen-free facilities. For experimental purposes, mice were used at 6–8 weeks of age.
The transporter associated with antigen processing (TAP)-deficient T2 cell line (transfected with HLA-A*0201 molecule) was kindly supplied by Professor Yu-Zhang Wu (Third Military Medical University, China). T2 cells cannot present the endogenous peptides onto its surface. The breast cancer cell line MCF-7 (HLA-A2+, PL2L60+), colorectal cancer cell line HT-29 (HLA-A2−, PL2L60+), and esophageal cancer cell line EC-9706 (HLA-A2+, PL2L60−) were maintained in our laboratory. The T2 and cancer cells were cultured in RPMI 1640 medium (Invitrogen, US) supplemented with 10 % fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin in an incubator with a humidified atmosphere containing 5 % CO2.
Peripheral blood samples were obtained from HLA-A2+ healthy donors. The sample collection was approved by the Ethics Committee of Zhengzhou University.
T2 binding assay
The addition of exogenous binding peptide could stabilize the expression of HLA-A*0201 molecules on the T2 cell surface. To determine whether the synthetic peptides could bind to HLA-A*0201 molecules, peptide-induced HLA-A*0201 up-regulation on T2 cells was examined. Briefly, T2 cells were incubated with 50 μM candidate peptides and 3 μg/ml human β2-microglobulin (β2-M, Merck, Germany) in serum-free RPMI 1640 medium for 18 h at 37 °C in 5 % CO2 atmosphere. The expression of HLA-A*0201 on T2 cells was determined by being stained with FITC-conjugated anti-HLA-A2 mAb BB7.2 (Santa Cruz, US) [19]. The data were analyzed by using a FACSCalibur flow cytometer (Becton–Dickinson, US) with a CellQuest software (Becton–Dickinson, US). The fluorescence index (FI) was calculated as follows: FI = (mean FITC fluorescence with the given peptide—mean FITC fluorescence without peptide)/(mean FITC fluorescence without peptide). Peptides with FI more than 1.5 were regarded as high-affinity candidates.
T2 stabilization assay
Cell surface stabilization of HLA-A*0201 molecules was also examined by using the T2 cell line. T2 cells (1 × 106 cells/ml) were incubated with peptide (100 μM) and β2-M (100 ng/ml) for 18 h at 37 °C in serum-free RPMI 1640 medium. Thereafter, cells were washed twice to remove free peptides, then incubated with brefeldin A (to block transport of newly synthesized empty class I MHC molecules to the cell surface) (10 μg/ml, Sigma, US) for 1 h, washed, and incubated at 37 °C for 0, 2, 4, and 6 h. Cells were then washed twice, stained, and analyzed by a flow cytometry [20]. The dissociation constant 50 (DC50) was defined as the time required for 50 % dissociation of the HLA-A*0201 ⁄ peptide complex stabilized at t = 0 h.
RT-PCR analysis
Total RNA was extracted from cancer cell lines. The cDNA was generated by reverse-transcribed with Avian myoblastosis virus (AMV) reverse transcriptase and oligo dT (Clontech, US). Quality of the cDNA was confirmed by polymerase chain reaction (PCR) of GAPDH. Gene-specific PCR primers used to amplify PL2L60 and GAPDH were designed (PL2L60 sense: 5′-CCC AGG TTG TCA ATG TTC G-3′, antisense: 5′-CAG GCT GTC CAC AAT CTC C-3′, size: 278 bp; GAPDH sense: 5′-GAA GGT GAA GGT CGG AGT C-3′, antisense: 5′-GAA GAT GGT GAT GGG ATT TC-3′, size: 226 bp). Briefly, an aliquot of 0.5 μl cDNA was used in each 20 μl PCR reaction, using PCR Master Mix (Promega, US). The following conditions were used: an initial denaturation at 95 °C for 5 min followed by denaturation at 94 °C for 30 s, annealing at 65 °C for 1 min, touchdown 1 °C per cycle, and extension at 72 °C for 1 min for a total of 10 cycles. Then, the condition was fixed for 25 cycles of denaturation at 94 °C for 30 s, annealing at 50 °C for 1 min, and extension at 72 °C for 1 min with a final extension at 72 °C for 10 min. PCR products were analyzed by 1.5 % agarose gel containing 0.01 mg/ml ethidium bromide [7].
Western blot analysis
For Western blot analysis, cell pellets were lyzed in electrophoresis sample buffer. Cell lysates were separated on a 10 % SDS-PAGE gel and transferred to a nitrocellulose membrane. Membranes were blocked in TBS-T buffer (50 mM Tris–HCl, 0.1 % Tween-20, pH 7.4) supplemented with 5 % nonfat dry milk and probed with the specific antibodies. The membranes were incubated overnight at 4 °C with the indicated primary antibody: rabbit polyclonal anti-PL2L60 (Bioss, China), or rabbit polyclonal anti-β-actin antibody (Bioss, China), followed by 5-min washing in TBS-T for three times and incubation with appropriate secondary antibody, and the bands were visualized with the ECL plus system [21].
Induction of CTLs from human PBMCs
Cytotoxic T lymphocytes (CTLs) induction in vitro was performed in accordance with the procedure described previously [22]. Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood of HLA-A2+ healthy volunteer donors by a Ficoll–Paque density gradient centrifugation and then cultured in RPMI 1640 supplemented with 10 % FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. Then, these cells were stimulated once a week with the synthetic peptides and the β2-M at the final concentration of 10 μg/ml. Human recombinant IL-2 was added to the culture medium at a concentration of 50 U/ml on day 3 and the day after each stimulation. The enzyme-linked immunospot (ELISPOT) assay and cytotoxic assay were performed on day 21.
Generation of CTLs from HLA-A2.1/Kb transgenic mice
HLA-A2.1/Kb transgenic mice were immunized with 100 μg of various peptides and 100 μg of the IAb-restricted HBVcore antigen-derived T helper epitope (sequence 128–140: TPPAYRPPNAPIL) prepared in incomplete Freund’s adjuvant (IFA) on days 0, 5, and 10. Group of mice received IFA containing PBS or T helper peptide was used as negative control. After 11 days, spleen lymphocytes (5 × 107 cells in 10 ml) were stimulated in vitro with peptide (10 μg). At day 6 of the stimulation, the specific cytotoxicity and enzyme-linked immunospot (ELISPOT) assays were performed. Cytotoxic activity was tested based on the measurement of LDH release using the nonradioactive cytotoxicity assay kit (Promega, US) at E:T ratios, 20:1, 40:1, and 80:1.
Mouse serum IFN-γ ELISA assay
Serum was obtained from immunized mice after the immunization. Mouse blood samples were allowed to clot at 37 °C for 2 h, and then, the serum was collected. The assays were performed according to the manufacturer’s guidelines of the mouse IFN-γ ELISA kits (R&D, US). The absorbance was read at 450 nm in an ELISA microplate reader (Bio-Rad, US) [23].
IFN-γ ELISPOT assay
Enzyme-linked immunospot assays were performed using a commercially available kit (Dakewe, China). T2 cells, pulsed with the indicated concentration of synthetic peptides, were used as stimulator cells. Effector cells (1 × 105) and peptide-pulsed T2 cells (1 × 105) were seeded into 96-well microplates coated with IFN-γ antibody. After incubation at 37 °C for 18 h, cells were removed and plates were processed. The number of spots was determined automatically by using a computer-assisted spot analyzer (Dakewe, China).
Cytotoxicity assay
Cytotoxic activity was tested based on the measurement of lactate dehydrogenase (LDH) release using the nonradioactive cytotoxicity assay kit (Promega, US) at various E:T ratios (12.5:1, 25:1, and 50:1, CTLs from the PBMCs of healthy donors; 20:1, 40:1, and 80:1, CTLs from the spleen lymphocytes of transgenic mice) [24]. For CTLs-generated form PBMCs of healthy donors, target cells were T2 cells loaded with or without peptides, cancer cells MCF-7, HT-29, and EC-9706. In order to determine the HLA restriction, monoclonal antibody of HLA-A2 (BB7.2) was added 30 min before the addition of effector cells [25]. For CTLs generated from HLA-A2.1/Kb transgenic mice, MCF-7 cells and T2 cells loaded with peptides were served as target cells. Target cells (5 × 103/well) were co-cultured with various number of effector cells at 37 °C for 5 h. The percentage of specific lysis of the target cells was determined as follows: percentage of specific lysis = (experimental release − effector spontaneous release − target spontaneous release)/(target maximum release − target spontaneous release) ×100.
Digestion of peptides in human serum
Human serum was obtained from the whole blood of healthy donors by a Ficoll–Paque density gradient centrifugation [26]. Each peptide was dissolved in PBS (pH 7.2) to a concentration of 1 mg/ml. Same peptide solution without serum was used as control. At the starting time point, 10 μl of peptide solution was mixed with 190 μl of 10 % (v/v) human serum. The mixture was incubated at 37 °C, and samples were taken out at 0, 30, 60, 120, 240, and 480 min. The reaction was stopped by the addition of ice-cold acetonitrile and acetic acid. Samples were centrifuged at 13,000×g for 5 min at −4 °C, and the concentration of the intact peptide in the supernatant was determined by RP-HPLC [27]. The experiment was repeated three times.
Statistical analysis
All data were expressed as mean ± SD. Significances were analyzed by one-way ANOVA. P < 0.05, P < 0.01, and P < 0.001 were considered as significant differences.
Results
Selection, modification, and binding capacity of candidate epitopes derived from PL2L60
Based on the four prediction programs, SYFPEITHI, BIMAS, IEDB, and NetCTL [28–31], nine native peptides with higher prediction scores were selected (Table S1). These peptides were synthesized with a purity of over 95 %. The molecular weights of all the peptides were confirmed by ESI–MS (Table 1 and S2).
Table 1.
The analysis data, HLA-A*0201 binding affinity, and stability of the candidate peptides
| Peptides | Sequence | ESI–MS [M + H]+ | Puritya (%) | FIb | DCc50 | |
|---|---|---|---|---|---|---|
| Calculated | Observed | |||||
| P281 | KLGGELWGV | 958.1 | 958.3 | 98 | 2.42 | >4 h |
| P281_1Y | YLGGELWGV | 993.1 | 993.1 | 98 | 3.38 | <2 h |
| P281_1y | [D-Tyr1]LGGELWGV | 993.2 | 993.2 | 95 | 1.77 | <2 h |
| P281_1Nal | [Nal1]LGGELWGV | 1026.2 | 1027.6 | 98 | 2.94 | >6 h |
| P281_1NF | [3-NO2-Tyr1]LGGELWGV | 1038.0 | 1038.6 | 96 | 3.72 | <2 h |
| P281_1MY | [4-OMe-Phe1]LGGELWGV | 1007.2 | 1007.0 | 95 | 3.91 | <2 h |
| P281_1CF | [4-Cl-Phe1]LGGELWGV | 1011.5 | 1011.0 | 98 | 4.73 | >4 h |
| P281_1F | FLGGELWGV | 977.1 | 977.6 | 95 | 3.01 | >6 h |
| P281_1Dmt | [Dmt1]LGGELWGV | 1021.1 | 1021.6 | 98 | 4.22 | >4 h |
| HBcAg18-27 | FLPSDFFPSV | 1155.3 | 1155.6 | 96 | 1.20 | >6 h |
aEstimated by the peak area analyzed by RP-HPLC
b FI = [mean fluorescence intensity (MFI) of the peptide—MFI background]/[MFI background]
c DC50 was defined as the time (h) required for 50 % dissociation of the HLA-A*0201/peptide complex stabilized at t = 0 h
To evaluate the binding affinity of these peptides to HLA-A*0201 molecule in vitro, a binding assay in T2 cells was used [32]. As shown in Table S2, among the nine candidate peptides, the native peptides P281 showed the most potent binding affinity to HLA-A*0201. Because of the poor immunogenicity of P281 in the preliminary experiment to induce cytotoxic T lymphocytes in human PBMCs, it was selected as the candidate native peptide to carry out the modification strategy. Eight analogs with aromatic amino acids substitution at position 1 of P281 were also synthesized (Fig. 1 and Table 1). The peptides with potent binding affinity to HLA-A*0201 were tested for the peptide/HLA-A*0201 complex stability, and the binding stability of these peptides was shown as DC50 [33]. As shown in Table 1, all the analogs of P281 showed potent binding affinity with FI > 1.5. Four analogs of P281 showed potent binding stability with the DC50 > 4 h.
Fig. 1.
The chemical structure of the aromatic amino acids
Expression of PL2L60 in cancer cell lines
The expression profiles of PL2L60 in cancer cell lines were determined by RT-PCR and Western blot. As shown in Fig. 2, PL2L60 mRNA expression was observed in MCF-7 and HT-29 cells, but not in EC-9706 cells. The expression of PL2L60 protein was examined by Western blot, and the results were consistent with the RT-PCR.
Fig. 2.
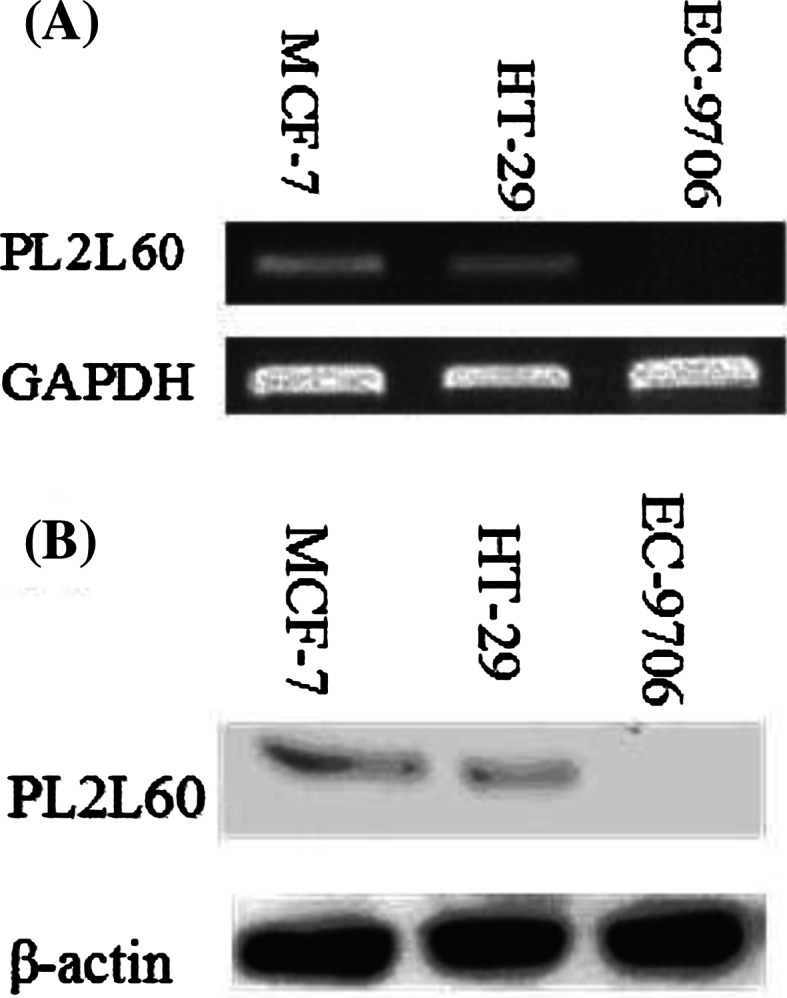
Expression of PL2L60 in cancer cell lines. a mRNA detected by RT-PCR. PCR products were tested by electrophoresis on 1.5 % agarose gel with ethidium bromide staining. GAPDH was used as positive control. b protein was analyzed by Western blot. Each lane contains 30 μg protein extract from the cancer cells. β-actin was used as positive control
IFN-γ ELISPOT assay
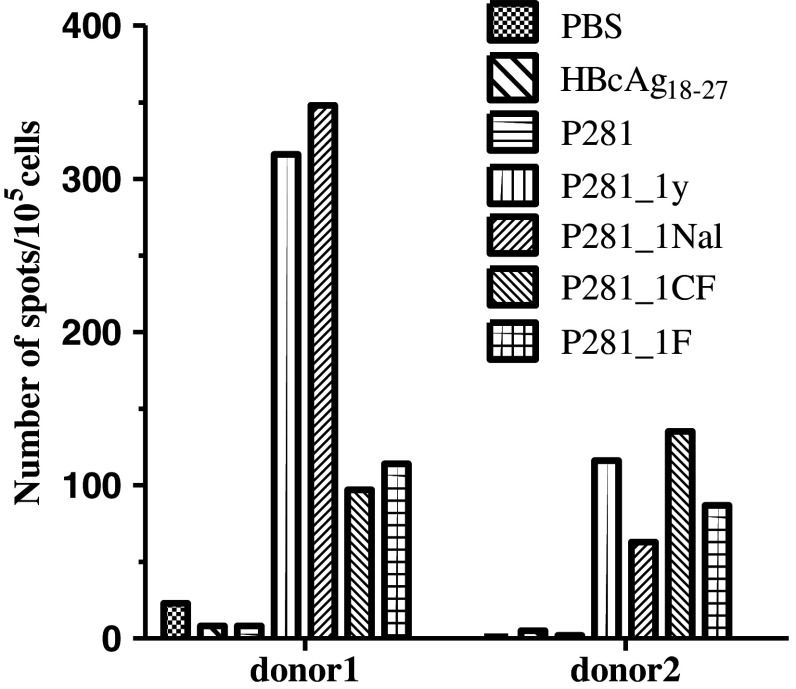
Based on the results of the T2 binding assay and peptide/HLA-A*0201 stability assay, P281 and the eight analogs were selected to investigate their ability to induce T cell response by using IFN-γ release ELISPOT assay. The CTLs were induced from the PBMCs of the HLA-A2+ healthy donors. Our results showed that the CTLs induced by the analogs, P281_1y, P281_1Nal, P281_1CF, and P281_1F, showed more potent activity which could induce more amounts of IFN-γ in two donors (Fig. 3).
Fig. 3.
ELISPOT assay to measure IFN-γ release by CTLs induced from PBMCs of healthy donors. PBMCs from healthy donors were separated and stimulated with synthetic peptides and IL-2 in RPMI 1640 supplemented with 10 % FCS for 21 days. Then, these PBMCs were collected, and ELISPOT assay was performed to determine the IFN-γ production by these cells. CTLs induced by PBS and irrelevant peptide HBcAg18–27 were taken as negative controls. To each donor, the experiment was repeated twice
Cytolytic activity of epitope-specific CTLs
To investigate whether the CTLs induced from the PBMCs of healthy donors could lyze target cells, an LDH cytotoxicity assay was performed. MCF-7 cells (HLA-A2+, PL2L60+) were used as positive targets, while HT-29 (HLA-A2−, PL2L60+) and EC-9706 (HLA-A2+, PL2L60−) cells as negative targets. In the preliminary experiment, two analogs, P281_1CF and P281_1F, but not others could widely induce potent CTL responses in HLA-A2+ healthy donors, so they were chosen for further investigation. As shown in Fig. 4, the CTLs induced by these two analogs, especially P281_1CF, could lyze MCF-7 cells, but not HT-29 and EC-9706 cells. The anti-HLA-A2 mAb BB7.2 could totally block the cytotoxic activity of the CTLs (induced by P281_1CF and P281_1F) on MCF-7 cells. The results indicated that the two modified analogs, P281_1CF and P281_1F, could induce potent CTL responses in HLA-A2-restricted and antigen-specific manners. The CTLs induced by them in PBMCs could recognize the endogenously processed and presented epitope to the surface of cancer cells. Similar results were observed in the additional CFSE assay to confirm the specific lysis of the CTLs induced by these two analogs (Fig. S1). The analog P281_1CF showed more potent activity.
Fig. 4.
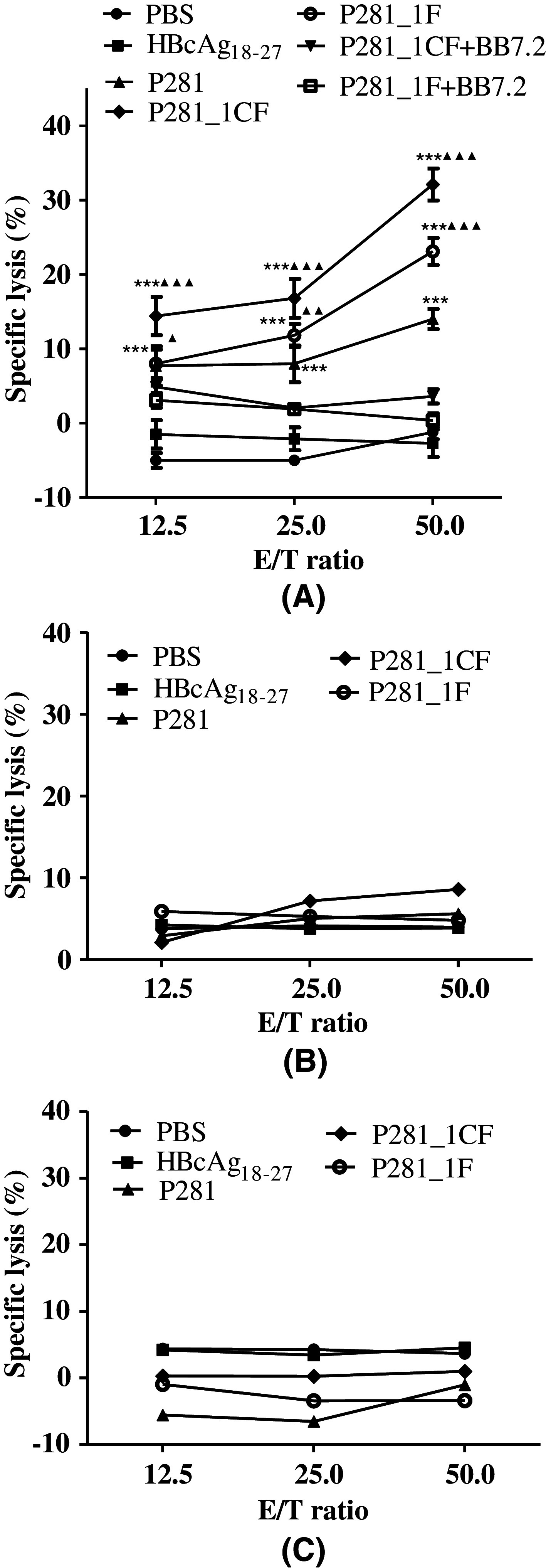
The cytotoxic activity of the CTLs induced by P281 and its analogs P281_1CF and P281_1F from human PBMCs. PBMCs from healthy HLA-A2+ donors were stimulated with peptides (10 μg/ml) for three times. After 5 days of the final stimulation, the stimulated PBMCs were used as effector cells to detect their cytotoxic activity against tumor cells in a LDH release assay. a MCF-7 (HLA-A2+, PL2L60+) as target cells. Anti-HLA-A2 mAb BB7.2 was used to block the HLA-A2 molecules on MCF-7 cells. b HT-29 (HLA-A2−, PL2L60+) as target cells. c EC-9706 (HLA-A2+, PL2L60−) as target cells. CTLs induced by PBS and irrelevant peptide HBcAg18–27 were taken as negative controls. ***P < 0.001 represented the significances versus HBc18–27 group. ▲ P < 0.05, ▲▲ P < 0.01, and ▲▲▲ P < 0.001 represented the significances versus P281 group, respectively. Data were represented as mean ± SD (n = 3)
Cross-recognition of P281 and its analogs by specific CTLs
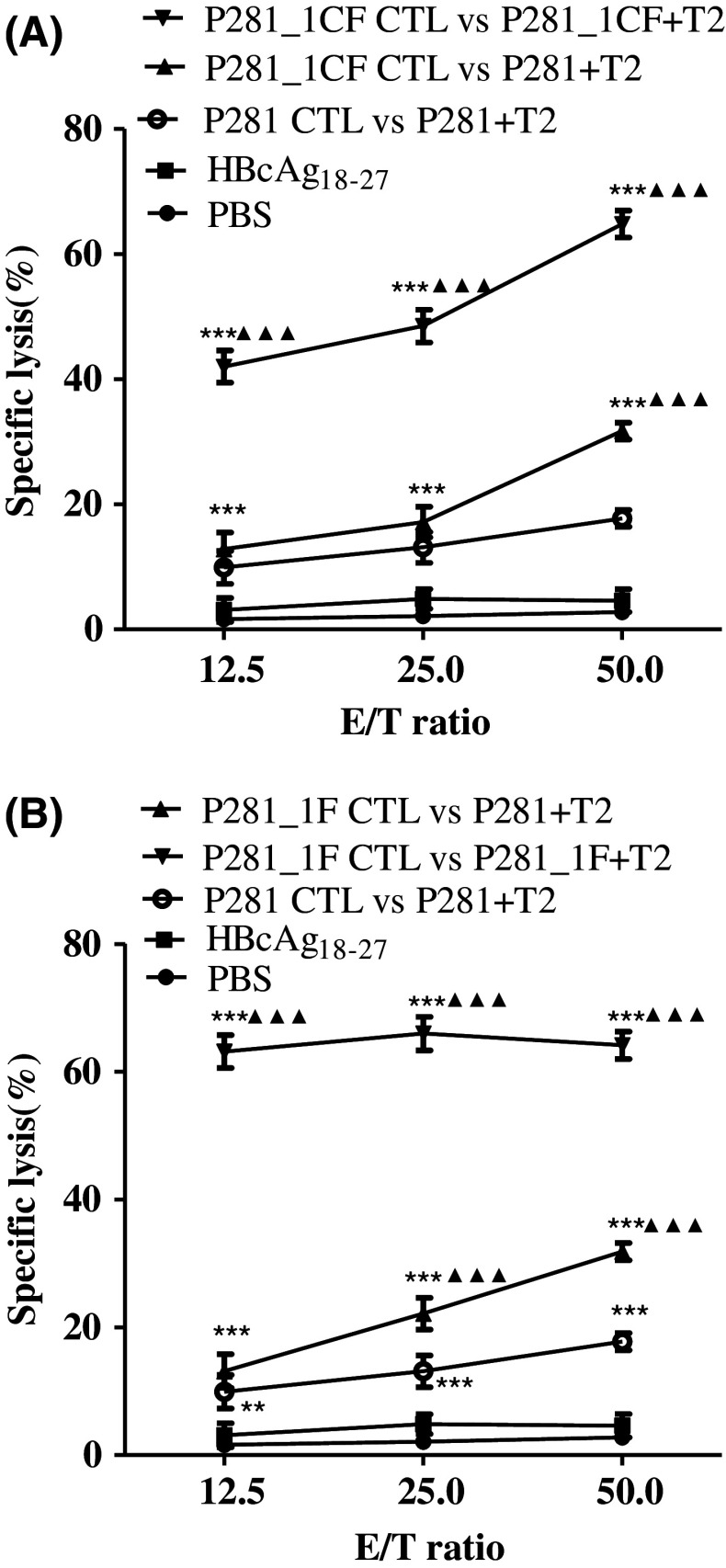
To further verify whether the specific CTLs could recognize the native peptide P281 and its analogs, we examined the cytolytic activity of CTLs induced by P281_1CF or P281_1F. As shown in Fig. 5, the CTLs induced by P281_1CF could lyze T2 cells loaded with both native peptide P281 and analog P281_1CF. Similar results were observed to the CTLs induced by P281_1F. The results indicated that the modification of the native peptide might not alter the antigenic specificity and confirmed that the CTLs induced by the analogs P281_1CF and P281_1F could recognize and kill target cells that presenting the native peptide.
Fig. 5.
Recognition of native peptide P281 by CTLs induced by two analogs. CTLs induced by P281_1CF (a) or P281_1F (b) from human PBMCs were tested for their capacity to lyze T2 cells loaded with or without each peptide. **P < 0.01, and ***P < 0.001 represented the significances versus HBc18–27 group, respectively. ▲▲▲ P < 0.001 represented the significances versus P281 group. Data were represented as mean ± SD (n = 3)
Cytotoxic and IFN-γ release activities of peptide-specific CTLs induced from HLA-A2.1/Kb transgenic mice
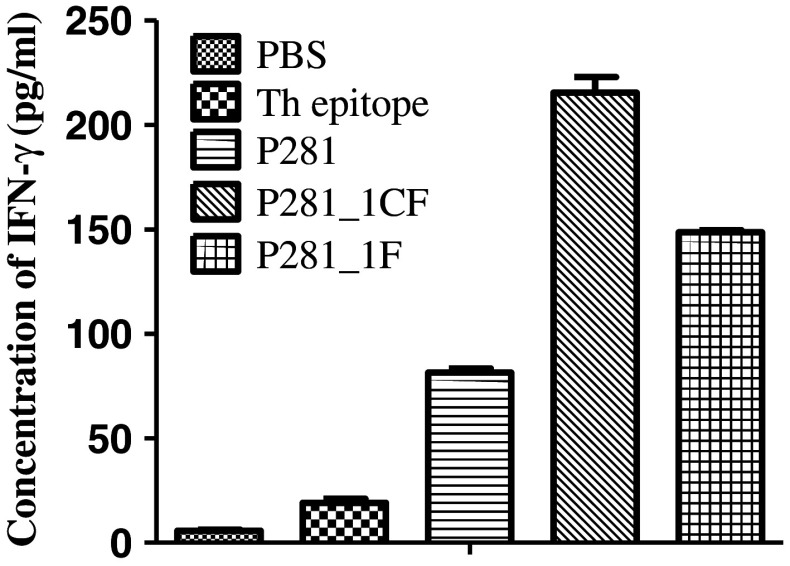
To investigate whether the peptides could induce specific CTLs in vivo, HLA-A2.1/Kb transgenic mice were immunized with P281_1CF and P281_1F emulsified in IFA in the presence of HBV core 128 T helper epitope. After immunization three times, spleen lymphocytes were pooled and re-stimulated in vitro with the related peptides, respectively. Then, IFN-γ ELISA and LDH release assays were carried out to test the cytotoxic activity of the CTLs induced by P281, P281_1CF, and P281_1F, respectively. P281-loaded T2 cells, MCF-7 cells, or MCF-7 cells blocked by anti-HLA-A2 mAb were used as target cells, and the effector/target ratios were 20:1, 40:1, and 80:1. The results showed that when the targets were MCF-7 cells and P281-loaded T2 cells, the peptide-specific CTLs induced by P281_1CF and P281_1F showed more potent cytotoxic activity than that of P281 (Fig. 6). In addition, the results from the ELISA assay also showed that the two analogs, especially P281_1CF, could produce more IFN-γ than that of P281 (Fig. 7).
Fig. 6.
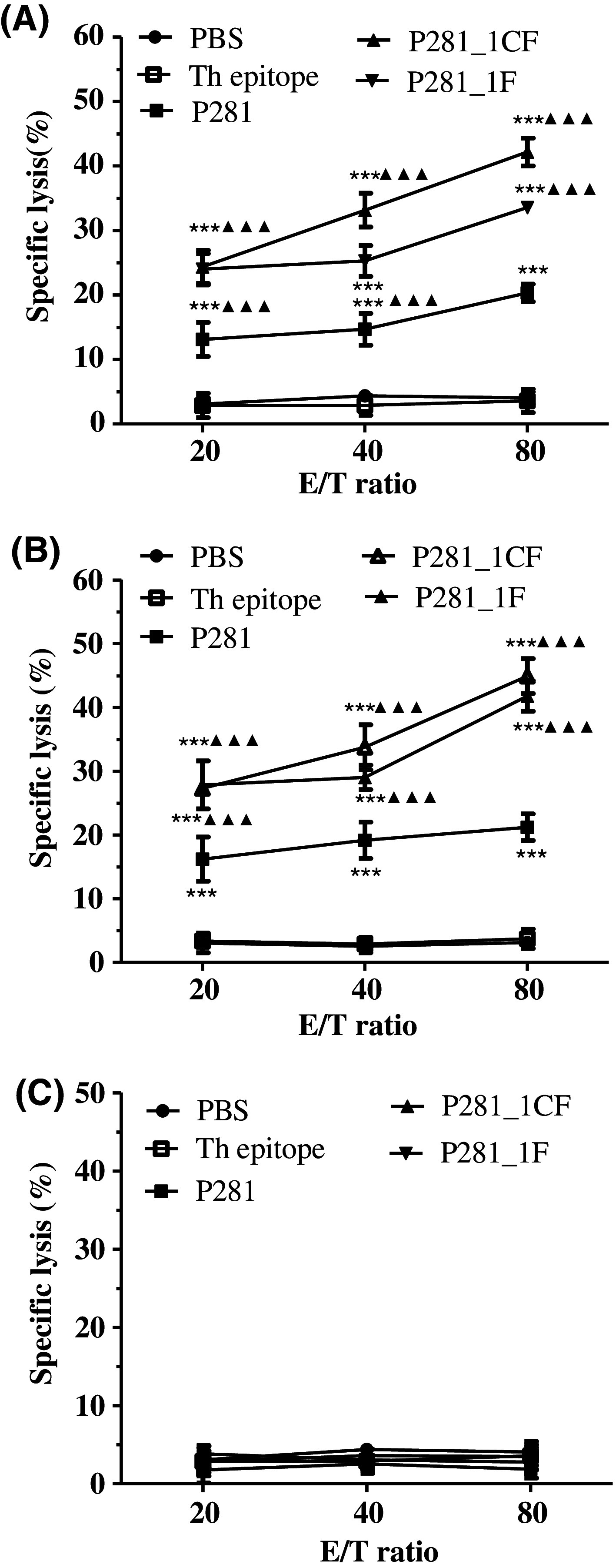
Specific lysis of cancer cell lines by the CTLs generated from immunized HLA-A2.1⁄ Kb transgenic mice. The HLA-A2.1⁄Kb transgenic mice were immunized with peptide P281, P281_1CF, or P281_1F emulsified in IFA in the presence of 100 μg of the HBVcore128 T helper epitope on days 0, 5, and 10. Group of mice received IFA containing PBS or T helper peptide was used as negative control. About 11 days after the first immunization, the animals were killed, and the splenocytes were re-stimulated in vitro for an additional 6 days with 10 μg/ml peptide and 10 units/ml rmIL-2. LDH cytotoxicity assay was used to evaluate the lysis of a MCF-7 cells (HLA-A2+, PL2L60+), b T2 cells pulsed with P281, and c MCF-7 cells on which surface the HLA-A2 molecules were blocked with anti-HLA-A2 mAb. ***P < 0.001 represented the significances versus Th group. ▲▲▲ P < 0.001 represented the significances versus P281 group. Data were represented as mean ± SD (n = 5)
Fig. 7.
IFN-γ release by the CTLs generated from immunized HLA-A2.1⁄Kb transgenic mice. Serum was obtained from immunized mice after the immunization, and the concentration of IFN-γ was evaluated by ELISA. Data were represented as mean ± SD (n = 3)
Combined with the results both in vitro and in vivo, substitution of position 1 with aromatic amino acids could enhance the binding ability and immunogenicity of the native peptide P281. The analog with the unnatural amino acid 4-Cl-Phe showed the most potent activity.
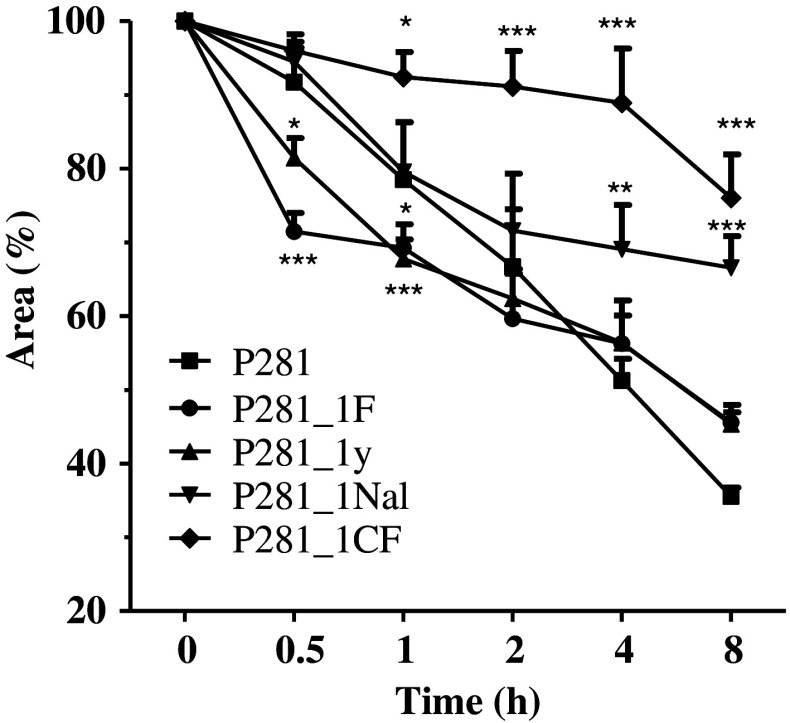
Stability in human serum
To investigate if the introduction of unnatural amino acids enhanced the stability of the analogs, their stability in human serum was tested. As shown in Fig. 8, each peptide with unnatural amino acid was more stable than the native one. P281_1CF showed the most potent ability against the degradation of the human serum.
Fig. 8.
Digest of the synthetic peptides in 10 % human serum. Data were presented as percentages of peptide remained which was calculated from the area of the corresponding peak obtained by RP-HPLC. *P < 0.05, **P < 0.01, and ***P < 0.001 represented the significances versus P281 group. Data were represented as mean ± SD (n = 5)
Application of this strategy to other epitopes
To investigate whether this modification strategy can be performed to other epitopes, additional experiment was done. Two putative native epitopes, MAGE-3_P271 (FLWGPRALV) and gp100_P154 (KTWGQYWQV), were chosen as candidates. LDH release assay was carried out to test the cytotoxic activity of the CTLs induced by the related peptides. As shown in Fig. S2, the introduction of 4-Cl-Phe could enhance the immune response of the two native peptides. The results indicated that introduction of the unnatural aromatic amino acid 4-Cl-Phe to substitute the position 1 of the CTL epitope could be a general strategy to enhance the immune response of the native peptide.
Discussion
T cell epitopes play an important role in the cellular immunity against cancer. Immunizing cancer patients with synthetic epitopes and vaccines has been elaborated since the identification of a defined tumor-specific CTL epitope [34]. Numerous clinical trials have been conducted with promising results. The effectiveness of T cell-mediated cancer immunotherapy depends on both an optimal immunostimulatory strategy and the selection of the T cell epitopes in promising tumor antigens [1]. The identification of T cell epitopes is important not only to the development of new vaccine, but also to help us understanding the antigen processing [35, 36].
However, several problems to the native epitope are still unsolved, such as low immunogenicity and rapid proteolytic degradation in vivo. Design of peptidomimetics through chemical alteration of epitope structure could be helpful to improve immunogenicity and stability [37]. During the modification of native epitope, it was essential to maintain the basic hydrogen bonding properties of peptide/MHC and peptide/TCR. This would help the CTLs induced by the modified epitope to maintain the antigen specificity to recognize the native peptide presented by the cancer cells. Therefore, it is important for us to find a strategy to modify native peptide slightly and efficiently.
Ruppert et al. [14] determined the HLA-A2.1 binding motif with a leucine (L) or methione (M) at position 2 and a leucine (L), valine (V), or isoleucine (I) at position 9. To a 9-mer epitope, the positions 2 and 9 are considered as the anchor residues, which contribute most to the binding to the MHC molecule. Tourdot et al. [15] reported that the substitution of position 1 by a tyrosine was a general strategy to enhance immunogenicity of HLA-A2-restricted epitopes. Considering that most of the peptides composed of natural amino acids could be easily degraded from the terminal residues, we tried to introduce a series of unnatural aromatic amino acids to modify the native epitope. Then, we tested if these modifications could improve the binding activity and maintain the immune property of the native epitope.
Our results showed that P281 and their analogs are the high-affinity peptide, with potent binding affinity and stability to HLA-A*0201 molecule. Subsequent IFN-γ release and LDH release assays by using PBMCs from HLA-A2+ healthy donors and spleen lymphocytes from immunized mice showed that the analog P281_1CF could induce the most potent T cell response. The CTLs induced by P281_1CF could specifically lyze the cancer cells in antigen-specific and HLA-A2-restricted manners. The substitution of the native peptide P281 with a 4-Cl-Phe in position 1 could prolong its half-life in human serum, which might be one of the reasons why this analog showed the more potent activity than that of the tyrosine-substituted analog. In addition, we discovered that the strategy we presented here could also be performed to the modification of native CTL epitopes from other tumor antigens.
In conclusion, we firstly identified a potential CTL epitope from the promising tumor antigen PL2L60 and proposed a novel modification strategy by substituting the position 1 of the native peptide by unnatural aromatic amino acids. Our results indicated that the substitution of the residue in position 1 by 4-Cl-Phe could maintain the antigen specificity and enhance the binding affinity, stability, and immunogenicity of the native CTL epitope. This strategy could be used as a general method to improve the activity of the native epitope. The novel epitopes reported here could serve as good candidates to develop peptide vaccines against PL2L60-positive cancers.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (Nos. 81373228, 81172893) and the National Science and Technology Major Projects of New Drugs (2012ZX09103301-023).
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Kessler JH, Melief CJ. Identification of T-cell epitopes for cancer immunotherapy. Leukemia. 2007;21:1859–1874. doi: 10.1038/sj.leu.2404787. [DOI] [PubMed] [Google Scholar]
- 2.Uenaka A, Hirano Y, Hata H, et al. Cryptic CTL epitope on a murine sarcoma Meth A generated by exon extension as a novel mechanism. J Immunol. 2003;170:4862–4868. doi: 10.4049/jimmunol.170.9.4862. [DOI] [PubMed] [Google Scholar]
- 3.Shen H, Shao HW, Chen XH, et al. Identification of a novel HLA-A2-restricted mutated Survivin epitope and induction of specific anti-HCC CTLs that could effectively cross-recognize wild-type Survivin antigen. Cancer Immunol Immunother. 2012 doi: 10.1007/s00262-012-1323-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenberg SA, Restifo NP, Yang JC, Morgan RA, Dudley ME. Adoptive cell transfer: a clinical path to effective cancer immunotherapy. Nat Rev Cancer. 2008;8:299–308. doi: 10.1038/nrc2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blattman JN, Greenberg PD. Cancer immunotherapy: a treatment for the masses. Science. 2004;305:200–205. doi: 10.1126/science.1100369. [DOI] [PubMed] [Google Scholar]
- 6.Bredenbeck A, Losch FO, Sharav T, Eichler-Mertens M, Filter M, Givehchi A, Sterry W, Wrede P, Walden P. Identification of noncanonical melanoma-associated T cell epitopes for cancer immunotherapy. J Immunol. 2005;174:6716–6724. doi: 10.4049/jimmunol.174.11.6716. [DOI] [PubMed] [Google Scholar]
- 7.Ye Y, Yin DT, Chen L, et al. Identification of Piwil2-like (PL2L) proteins that promote tumorigenesis. PLoS One. 2010;5:e13406. doi: 10.1371/journal.pone.0013406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lazoura E, Apostolopoulos V. Rational Peptide-based vaccine design for cancer immunotherapeutic applications. Curr Med Chem. 2005;12:629–639. doi: 10.2174/0929867053202188. [DOI] [PubMed] [Google Scholar]
- 9.Cole DK, Edwards ES, Wynn KK, et al. Modification of MHC anchor residues generates heteroclitic peptides that alter TCR binding and T cell recognition. J Immunol. 2010;185:2600–2610. doi: 10.4049/jimmunol.1000629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tirosh B, el-Shami K, Vaisman N, Carmon L, Bar-Haim E, Vadai E, Feldman M, Fridkin M, Eisenbach L. Immunogenicity of H-2 Kb-low affinity, high affinity, and covalently-bound peptides in anti-tumor vaccination. Immunol Lett. 1999;70:21–28. doi: 10.1016/S0165-2478(99)00115-7. [DOI] [PubMed] [Google Scholar]
- 11.Tu SH, Huang HI, Lin SI, et al. A novel HLA-A2-restricted CTL epitope of tumor-associated antigen L6 can inhibit tumor growth in vivo. J Immunother. 2012;35:235–244. doi: 10.1097/CJI.0b013e318248f2ae. [DOI] [PubMed] [Google Scholar]
- 12.Valmori D, Fonteneau JF, Lizana CM, et al. Enhanced generation of specific tumor-reactive CTL in vitro by selected Melan-A/MART-1 immunodominant peptide analogues. J Immunol. 1998;160:1750–1758. [PubMed] [Google Scholar]
- 13.van Stipdonk MJ, Badia-Martinez D, Sluijter M, Offringa R, van Hall T, Achour A. Design of agonistic altered peptides for the robust induction of CTL directed towards H-2Db in complex with the melanoma-associated epitope gp100. Cancer Res. 2009;69:7784–7792. doi: 10.1158/0008-5472.CAN-09-1724. [DOI] [PubMed] [Google Scholar]
- 14.Ruppert J, Sidney J, Celis E, Kubo RT, Grey HM, Sette A. Prominent role of secondary anchor residues in peptide binding to HLA-A2.1 molecules. Cell. 1993;74:929–937. doi: 10.1016/0092-8674(93)90472-3. [DOI] [PubMed] [Google Scholar]
- 15.Tourdot S, Scardino A, Saloustrou E, Gross DA, Pascolo S, Cordopatis P, Lemonnier FA, Kosmatopoulos K. A general strategy to enhance immunogenicity of low-affinity HLA-A2. 1-associated peptides: implication in the identification of cryptic tumor epitopes. Eur J Immunol. 2000;30:3411–3421. doi: 10.1002/1521-4141(2000012)30:12<3411::AID-IMMU3411>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 16.Gold JS, Ferrone CR, Guevara-Patino JA, Hawkins WG, Dyall R, Engelhorn ME, Wolchok JD, Lewis JJ, Houghton AN. A single heteroclitic epitope determines cancer immunity after xenogeneic DNA immunization against a tumor differentiation antigen. J Immunol. 2003;170:5188–5194. doi: 10.4049/jimmunol.170.10.5188. [DOI] [PubMed] [Google Scholar]
- 17.Fischer PM. The design, synthesis and application of stereochemical and directional peptide isomers: a critical review. Curr Protein Pept Sci. 2003;4:339–356. doi: 10.2174/1389203033487054. [DOI] [PubMed] [Google Scholar]
- 18.Gao YF, Sun ZQ, Qi F, Qi YM, Zhai MX, Lou HP, Chen LX, Li YX, Wang XY. Identification of a new broad-spectrum CD8 + T cell epitope from over-expressed antigen COX-2 in esophageal carcinoma. Cancer Lett. 2009;284:55–61. doi: 10.1016/j.canlet.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 19.Gritzapis AD, Voutsas IF, Lekka E, Tsavaris N, Missitzis I, Sotiropoulou P, Perez S, Papamichail M, Baxevanis CN. Identification of a novel immunogenic HLA-A*0201-binding epitope of HER-2/neu with potent antitumor properties. J Immunol. 2008;181:146–154. doi: 10.4049/jimmunol.181.1.146. [DOI] [PubMed] [Google Scholar]
- 20.Chen F, Zhai MX, Zhu YH, Qi YM, Zhai WJ, Gao YF. In vitro and in vivo identification of a novel cytotoxic T lymphocyte epitope from Rv3425 of Mycobacterium tuberculosis. Microbiol Immunol. 2012;56:548–553. doi: 10.1111/j.1348-0421.2012.00470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hui X, Chen H, Zhang S, Ma X, Wang X, Huang B. Antitumor activities of recombinant human interferon (IFN)-lambda1 in vitro and in xenograft models in vivo for colon cancer. Cancer Lett. 2011;311:141–151. doi: 10.1016/j.canlet.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Zhu B, Chen Z, Cheng X, et al. Identification of HLA-A*0201-restricted cytotoxic T lymphocyte epitope from TRAG-3 antigen. Clin Cancer Res. 2003;9:1850–1857. [PubMed] [Google Scholar]
- 23.Wang RN, Wang YB, Geng JW, Guo DH, Liu F, Chen HY, Zhang HY, Cui BA, Wei ZY. Enhancing immune responses to inactivated porcine parvovirus oil emulsion vaccine by co-inoculating porcine transfer factor in mice. Vaccine. 2012;30:5246–5252. doi: 10.1016/j.vaccine.2012.05.077. [DOI] [PubMed] [Google Scholar]
- 24.Ding FX, Wang F, Lu YM, Li K, Wang KH, He XW, Sun SH. Multiepitope peptide-loaded virus-like particles as a vaccine against hepatitis B virus-related hepatocellular carcinoma. Hepatology. 2009;49:1492–1502. doi: 10.1002/hep.22816. [DOI] [PubMed] [Google Scholar]
- 25.Liu W, Zhai M, Wu Z, Qi Y, Wu Y, Dai C, Sun M, Li L, Gao Y. Identification of a novel HLA-A2-restricted cytotoxic T lymphocyte epitope from cancer-testis antigen PLAC1 in breast cancer. Amino Acids. 2011 doi: 10.1007/s00726-011-0966-3. [DOI] [PubMed] [Google Scholar]
- 26.Zhu YH, Gao YF, Chen F, Liu W, Zhai MX, Zhai WJ, Qi YM, Ye Y. Identification of novel T cell epitopes from efflux pumps of Mycobacterium tuberculosis. Immunol Lett. 2011;140:68–73. doi: 10.1016/j.imlet.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 27.Tugyi R, Uray K, Ivan D, Fellinger E, Perkins A, Hudecz F. Partial D-amino acid substitution: improved enzymatic stability and preserved Ab recognition of a MUC2 epitope peptide. Proc Natl Acad Sci USA. 2005;102:413–418. doi: 10.1073/pnas.0407677102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 29.Parker KC, Bednarek MA, Coligan JE. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J Immunol. 1994;152:163–175. [PubMed] [Google Scholar]
- 30.Nielsen M, Lundegaard C, Worning P, Lauemøller SL, Lamberth K, Buus S, Brunak S, Lund O. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12:1007–1017. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larsen MV, Lundegaard C, Lamberth K, Buus S, Lund O, Nielsen M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinformatics. 2007;8:424. doi: 10.1186/1471-2105-8-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schalich J, Vytvytska O, Zauner W, Fischer MB, Buschle M, Aichinger G, Klade CS. Analysis of the human cytomegalovirus pp65-directed T-cell response in healthy HLA-A2-positive individuals. Biol Chem. 2008;389:551–559. doi: 10.1515/BC.2008.065. [DOI] [PubMed] [Google Scholar]
- 33.Wu ZY, Gao YF, Wu YH, Liu W, Sun M, Zhai MX, Qi YM, Ye Y. Identification of a novel CD8 + T cell epitope derived from cancer-testis antigen MAGE-4 in oesophageal carcinoma. Scand J Immunol. 2011;74:561–567. doi: 10.1111/j.1365-3083.2011.02606.x. [DOI] [PubMed] [Google Scholar]
- 34.Traversari C, van der Bruggen P, Luescher IF, Lurquin C, Chomez P, Van Pel A, De Plaen E, Amar-Costesec A, Boon T. A nonapeptide encoded by human gene MAGE-1 is recognized on HLA-A1 by cytolytic T lymphocytes directed against tumor antigen MZ2-E. J Exp Med. 1992;176:1453–1457. doi: 10.1084/jem.176.5.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goonetilleke N, Moore S, Dally L, et al. Induction of multifunctional human immunodeficiency virus type 1 (HIV-1)-specific T cells capable of proliferation in healthy subjects by using a prime-boost regimen of DNA- and modified vaccinia virus Ankara-vectored vaccines expressing HIV-1 Gag coupled to CD8 + T-cell epitopes. J Virol. 2006;80:4717–4728. doi: 10.1128/JVI.80.10.4717-4728.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salmon-Ceron D, Durier C, Desaint C, et al. Immunogenicity and safety of an HIV-1 lipopeptide vaccine in healthy adults: a phase 2 placebo-controlled ANRS trial. AIDS. 2010;24:2211–2223. doi: 10.1097/QAD.0b013e32833ce566. [DOI] [PubMed] [Google Scholar]
- 37.Croft NP, Purcell AW. Peptidomimetics: modifying peptides in the pursuit of better vaccines. Expert Rev Vaccines. 2011;10:211–226. doi: 10.1586/erv.10.161. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.