Abstract
Our genome consists to about 8% of human endogenous retroviral (HERV) sequences. These HERVs have been discussed to be linked to human diseases for decades. Recently, a detailed analysis of a HERV-H sequence located on chromosome Xp22.3 revealed a strong expression in a subset of gastrointestinal cancers whereas expression in normal tissues and in other cancer entities was low. In the present study, we used the reverse immunology approach to test the immunological potential of this HERV-H ORF on Xp22.3. A total of ten peptides displaying HLA-A2.1-binding motifs were selected from the predicted env protein sequence. Stimulation of peripheral T cells with retroviral peptides (RVPs) presented by autologous antigen-presenting cells clearly resulted in sustained proliferation of predominantly CD8+ T cells. High numbers of IFN-γ-secreting T cells were detectable after several weekly stimulations with RVP mixes. Reactivity observed in RVP-Mix–stimulated cultures was attributable to RVP03, RVP09 and to a lower extend to RVP08, suggesting those to be highly immunogenic epitopes. Besides killing of RVP-loaded target cells, up to 40% specific lysis of colorectal carcinoma cell lines endogenously expressing this HERV-H Xp22.3 ORF was achieved. These data demonstrate that human T cells can be sensitized toward HERV peptides and moreover posses a high lytic potential toward HERV-H expressing CRC cells. Additionally, these data hint toward endogenous ENV protein expression followed by proteasomal degradation and presentation in the context of HLA molecules. Finally, our data strengthen the view that HERV-encoded sequences should be considered as a new class of tumor-specific antigens.
Electronic supplementary material
The online version of this article (doi:10.1007/s00262-011-1183-3) contains supplementary material, which is available to authorized users.
Keywords: HERV-H, Endogenous retrovirus, Tumor-specific antigen, T cell epitope, Reverse immunology
Introduction
A substantial part of the human genome is derived from transposable elements—remnants of ancient retroviral infections. Conservative estimates set the percentage of human endogenous retroviruses (HERVs) in the genome at 8% [1]. The process of endogenization is dependant on the viruses’ ability to withstand restrictions common to all cells, their potential to infect reproductive tissues, that is, cells of the germ line and finally to proceed to fixation in the human genome. All these processes must happen without loss of fitness of the host, thus excluding highly pathogenic ERV [2]. Generally, HERVs are classified into three groups: class I (gamma (like) retroviruses), class II (beta (like) retroviruses) and class III (spuma (like) retroviruses) [2, 3]. The most common nomenclature utilizes the single-letter amino acid code corresponding to the tRNA primer that is used for reverse transcription of the HERV genome [4]. HERVs posses a single open reading frame containing the genes gag, pol and env, which are flanked by LTRs at the 5′ and 3′ ends [5, 6]. For the most part, the interplay between mutations [3], epigenetic mechanisms and posttranscriptional regulations silence HERV expression [2, 7]. Yet, there are some examples of HERVs executing preserved functions as do the proviruses of their exogenous ancestors, making them something between parasites and symbionts. Among the cellular functions influenced by HERVs are enhancement and promotion of gene expression, splicing and polyadenylation, protein degradation, RNA synthesis and processing as well as membrane fusion [reviewed in 2].
Not only because of their above-mentioned roles in human cells, but also for their transforming ability, HERVs have been suggested to be linked to human diseases for a long time. They seem to play a role in neurological diseases such as multiple sclerosis [8] or schizophrenia [9] and also have repeatedly been found to be associated with a variety of cancer entities [7, 10, 11].
In a recent study, a detailed analysis of a HERV-H sequence located on chromosome Xp22.3 has been performed [12]. This sequence was found to be strongly expressed in a subset of gastrointestinal cancers whereas expression in normal tissues (including matched epithelium) and in other cancer entities like lung and cervical carcinoma was low. Besides, expression was correlated with demethylation of 5′LTR CpG-rich regions. These data demonstrate tumor-restricted activation of HERV-H Xp22.3 ORF expression.
This led us to hypothesize that HERV-H Xp22.3 ORF is an interesting candidate tumor-specific antigen similar to HERV-K env, which has been described as trigger of specific immune responses in breast cancer patients [13]. In the present study, we used the reverse immunology approach to test the immunological potential of HERV-H Xp22.3 ORF.
Materials and methods
Cell culture
Human colorectal carcinoma (CRC) cell lines (Colo60H, Colo94H, CaCo2 and LS174T) and T2 cells (174xCEM.T2 hybridoma, expressing empty HLA-A2.1 molecules on their surface because of TAP1 and TAP2 deficiency) were cultured in DMEM/Ham’s F12 (1:1) supplemented with 10% fetal calf serum, 2 mmol/l l-glutamine and 1% penicillin–streptomycin and incubated at 37°C and 5% CO2. All cell lines were obtained from the DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen) or the tumor bank of the German cancer research center; media and supplements were purchased from PAA (Cölbe, Germany).
CD40 ligand system for the culture of normal peripheral blood B cells
CD40 ligand-activated B cells (CD40 B cells) were obtained and cultured as described [14]. In brief, B cells from peripheral blood mononuclear cells (PBMC) were stimulated via NIH/3T3 cells stably expressing human CD154. These cells were irradiated lethally (60 Gy), plated on 6-well plates (0.4 × 105 cells/well) and cultured overnight. After rinsing with PBS, PBMC (2 × 106 cells/ml) in Iscove’s MEM supplemented with 10% human AB serum, 5 μg/ml insulin, 50 μg/ml transferrin and 15 μg/ml gentamicin were added and cultured in the presence of IL-4 (2 ng/ml; Cellgenix, Freiburg, Germany) and cyclosporin A (5.5 × 10−7 M). At intervals of 3–5 days, cells were transferred to new plates containing fresh irradiated stimulator cells.
Peptides and HLA-A2.1-binding assay
With the specific computer prediction program SYFPEITHI [15] (access via: http://www.syfpeithi.de), peptides displaying HLA-A2.1-binding motifs from the HERV-H Xp22.3 ORF sequence (Accession number: CAE84557) were selected. Peptides (retroviral peptides (RVPs); see Table 1 for details) were purchased from the peptide synthesis unit of the German cancer research center. Stock solutions (5 mg/ml in DMSO) were prepared and stored at −70°C; a dilution of 500 μg/ml in PBS was prepared before use. T2 cells were pulsed with 10 μg/ml peptide and 5 μg/ml β2-microglobulin (Sigma, Deisenhofen, Germany) overnight at 37°C. HLA-A2.1 expression was analyzed by flow cytometry using MAb BB7.2 (specific for pan HLA-A2) followed by incubation with a FITC-conjugated goat Ab binding anti-mouse Ig (Dako, Hamburg, Germany).
Table 1.
Retroviral and control peptides used in the present study
| Protein | Accession numbera | Name | Peptideb | SYFPEITHI scorec | Fluorescence indexd |
|---|---|---|---|---|---|
| Influenza matrix protein | AAA43682 | MP | 57-GILGFVFTL | 30 | 0.80 |
| Growth-regulated protein P68 | 226021 | P68 | 128-YLLPAIVHI | 30 | 0.88 |
| HERV-H Xp22.3 | CAE84557 | RVP01 | 171-LLSVSLPLL | 27 | 0.13 |
| RVP02 | 192-SLRVSTPSL | 27 | 0.11 | ||
| RVP03 | 215-CLYPFSAFL | 26 | 1.70 | ||
| RVP04 | 59-FLLSSPTSL | 25 | 0.28 | ||
| RVP05 | 53-NLVSSLFLL | 24 | 0.09 | ||
| RVP06 | 21-ILRPPALCSL | 30 | 0.26 | ||
| RVP07 | 112-ALVTDWEGSL | 24 | 0.00 | ||
| RVP08 | 170-PLLSVSLPLL | 24 | 0.05 | ||
| RVP09 | 87-SLNFNSFHFL | 23 | 0.49 | ||
| RVP10 | 59-FLLSSPTSLT | 17 | 0.00 |
aProtein or nucleotide accession numbers are indicated
bPosition of the start amino acid in the protein is indicated
cPredicted binding scores to HLA-A2.1 using computer assisted analysis (http://www.syfpeithi.de)
d(Mean fluorescence with peptide − mean fluorescence without peptide)/(mean fluorescence without peptide). Results are representative of two experiments
T cell purification and induction of peptide-specific cytotoxic T lymphocytes (CTL)
PBMC were depleted of non-T cells with the MACS Pan T Cell Isolation Kit II (Miltenyi-Biotec, Bergisch Gladbach, Germany) according to the manufacturer’s instructions. Preparations containing at least 97% of CD3+ cells were used in subsequent analyses. CD40 B cells of a healthy HLA-A2.1+ donor were incubated with different RVP mixes (10 μg/ml; Table 1) in serum-free Iscove’s MEM for 1 h at room temperature, followed by two wash steps to remove excess peptide, irradiated (30 Gy) and added to purified CD3+ autologous T cells at a ratio of 1:4 (T cells: CD40 B cells) in Iscove’s MEM containing 10% human AB serum, supplements (1:100) and hIL-7 (10 ng/ml, Cellgenix). T cells were distributed in 24-well plates at a density of 2 × 106 T cells/well in 1 ml medium. After a 3-day culture period 1 ml complete medium was added. T cells were harvested once a week and stained with trypan blue to distinguish between live and dead cells. Vital (unstained) T cells were then counted by a person blinded to the stimulation conditions using a Neubauer chamber. The growth of the T cell cultures was calculated for the whole stimulation period assuming that all T cells would always have been restimulated. The procedure was repeated on a weekly basis for T cell restimulation. IL-2 was added at days 21 (10 IU/ml, Proleukine®) and 24, and from day 28 on, only hIL-2 was used.
Enzyme-linked immunospot (ELISpot) assay
ELISpot assays were performed as described [16]. In short, nitrocellulose 96-well plates (Multiscreen; Millipore, Bedford, MA) were covered with mouse anti-human IFN-γ MAb (Mabtech, Nacka Strand, Sweden) and blocked with medium containing serum. Varying numbers of effector cells were plated in triplicates, and 3.5 × 104 peptide-loaded T2 cells per well served as targets. The incubation period was 16 h; thereafter, plates were washed, incubated with biotinylated rabbit anti-human INF-γ secondary antibody, washed a second time, incubated with streptavidin-coupled alkaline phosphatase and washed again. INF-γ-secreting cells were visualized by incubation with NBT/BCIP (Sigma) for 45 min to 1 h; the reaction was stopped with tap water. Spots were counted after drying of the plates.
Cytotoxicity assay
Standard chromium-release cytotoxicity assays were carried out as described before [14]. Roughly, effector T cells were incubated in triplicate in 96-well plates with 51Cr-labeled target cells (100 μCi) at a ratio of 2–100:1 (E:T). The cells were incubated for 4 h (T2 cell targets) or 8 h (CRC cell targets) at 37°C and 5% CO2. Plates were then centrifuged, aliquots of the supernatants (100 μl) were harvested and analyzed in a γ-counter. Percent cytotoxicity was calculated as follows: 100 × (cpm spontaneous release)/(cpm total release − cpm spontaneous release).
Results
Retroviral peptides (RVPs)
A total of ten peptides displaying HLA-A2.1-binding motifs (RVPs; see Table 1 for details) were selected from the predicted protein sequence of the open reading frame of a newly identified HERV-H copy (HERV-H Xp22.3 ORF; accession number: CAE84557). In a first step, binding capacity to HLA-A2.1 was assessed in a functional binding assay using T2 cells. These cells posses a defect in the protein processing machinery that prevents endogenous peptide presentation in their class I MHC molecules and additionally, they express HLA-A2.1 as the sole MHC-molecule on their surface. In summary, T2 cells express empty HLA-A2.1 molecules, which exhibit a rapid turnover, finally resulting in low levels of HLA-A2.1 on T2 cells’ surface. Exogenously added peptides can stabilize the HLA-A2.1 molecules and thus augment the HLA-A2.1 expression levels. Subsequent flow cytometric measurement of HLA-A2.1 levels is a reliable functional way of measuring the HLA-A2.1 binding capacity of candidate peptides [17]. As expected, the ten RVPs displayed varying HLA-A2.1-binding capacity with RVP03 and RVP09 showing the highest levels. Details can be depicted from Table 1.
Stimulation with RVPs leads to T cell proliferation
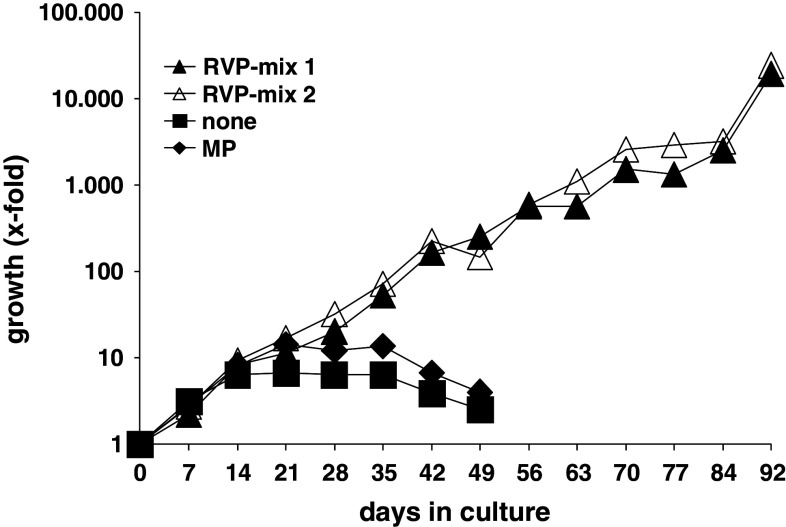
The feasibility of induction and antigen recognition of RVP-specific CTL was tested next. Therefore, T cells were isolated and CD40 B cells were generated from PBMC of healthy donors. The latter were loaded with RVPs in two different mixes of five RVPs each and used for stimulation of autologous T cells. We prefer CD40 B cells as antigen-presenting cells for in vitro T cell stimulation instead of the more classical dendritic cells (DCs). The percentage of DCs in PBMCs is only 0.1–0.5%; furthermore, the expansion of those DCs bears difficulties and is costly; the same is true for the differentiation of monocytes to DCs. Lastly, after 2–3 weeks DCs cease to proliferate and lose their high T cells stimulating potential [18, 19]. T cell proliferation rates were determined weekly by trypan blue cell counting, and the total number of T cells was calculated for the whole stimulation period. Stimulation with RVPs clearly resulted in sustained T cell proliferation (Fig. 1), whereas stimulation with the control peptide MP or without any peptide did not result in sustained T cell proliferation (Fig. 1). Phenotypic analysis of outgrowing T cell cultures revealed the predominance of CD8+ T cells (Supplementary material, available online).
Fig. 1.
Growth of RVP-Mix–stimulated T cells. T cells from a healthy HLA-A2+ donor were stimulated with the nonamer RVP-mix 1 (dark triangles), the decamer RVP-mix 2 (light triangles), the irrelevant peptide MP (diamonds) or no peptide at all (squares). The outgrowth of RVP-stimulated T cells was assessed by counting the number of viable T cells weekly and calculating an accumulated growth factor
INFγ producing CTL are induced by RVP stimulation
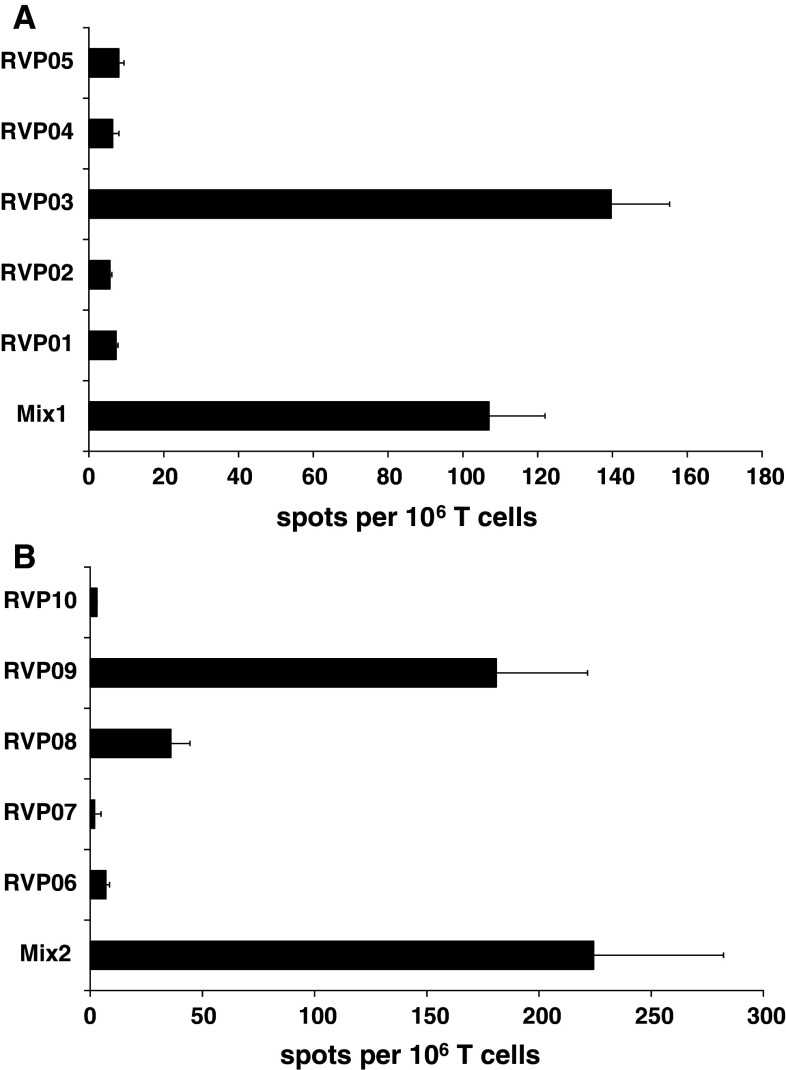
The frequency of peptide-specific stimulation was analyzed by IFN-γ-ELISpot analyses. Here, T2 cells were loaded with the respective peptides used for T cell stimulation (either single peptides or as mixes) and served as targets in IFN-γ-ELISpot tests. Significant numbers of IFN-γ-secreting T cells were detectable and therefore hint toward a strong reactivity against several RVPs (Fig. 2a, b). Of note, after several additional stimulation rounds, RVP03 was the only remaining reactivity observed in the RVP-mix 1 and RVP09 in the RVP-mix 2. Minor reactivity was still observed against RVP08 (data not shown).
Fig. 2.
ELISpot analysis of RVP-specific IFN-γ release. The number of specific spots derived from T cells, of a healthy HLA-A2+ donor, secreting IFN-γ in response to RVP-loaded target cells was determined in a series of ELISpot experiments. Reactivity of RVP-stimulated T cells on day 77 is given for the nonamer RVP-mix 1 (a) and the decamer RVP-mix 2 (b). Analysis was performed in triplicates with 1,000 effector and 10,000 target cells per well. The ELISpot results are representative for three individual analyses
RVP-specific T cells have strong cytolytic potential
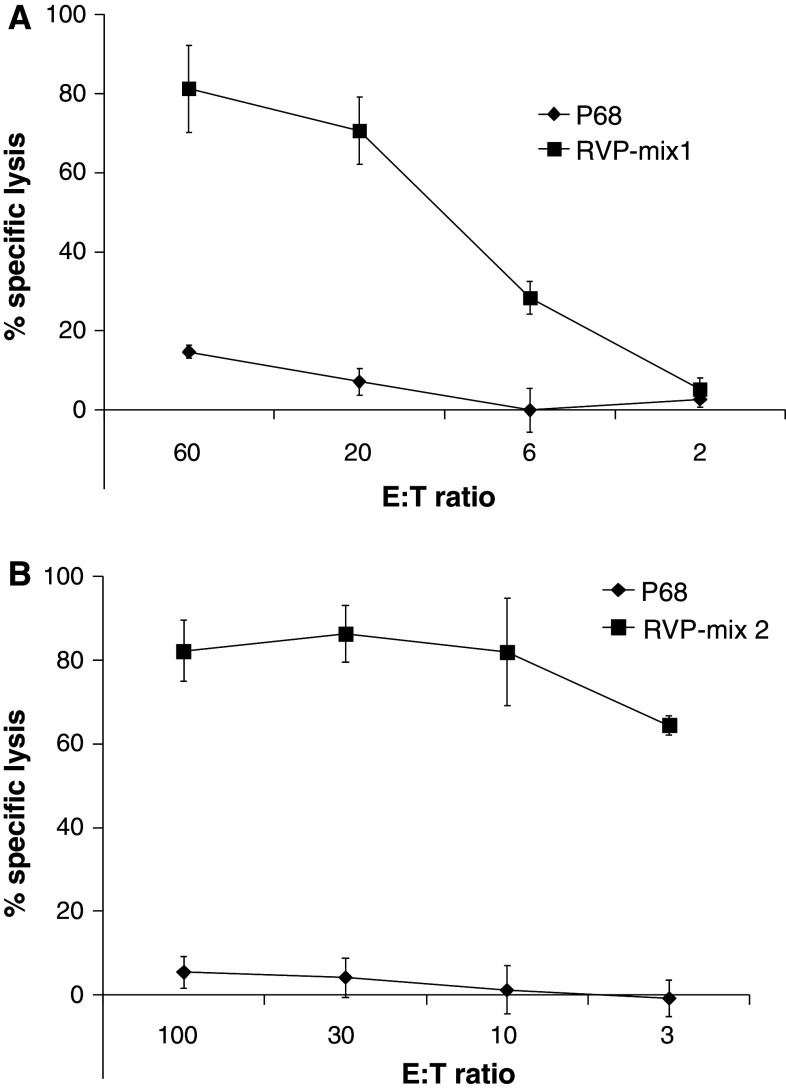
IFN-γ production is a clear sign of specific activation. It does, however, not prove cytotoxic ability. Therefore, we next tested the potential of the peptide-specific CTL to kill peptide-loaded T2 target cells. Here, we observed high efficiency in target cell killing when peptides used for T cell stimulation and target cell loading matched (Fig. 3). Both CTL lines’ reactivity toward RVP-loaded T2 targets was titrated but the mix-1 stimulated T cells showed higher background recognition of T2 loaded with irrelevant P68 compared to the mix-2 stimulated ones. In line with the results of the ELISpot-analysis, reactivity in the mix-2 stimulated CTL line was mostly due to CTL reactive against RVP09 (Fig. 3b). Accordingly, reactivity of the mix-1-stimulated CTL could be attributed to recognition of RVP03 (Fig. 3a). Since there is virtually only one peptide recognized in both T cell cultures at a later time point of culture, these cultures can then be considered as specific for RVP03 (mix 1) and RVP09 (mix 2). Thus, reactivity most likely can be attributed to these two distinct epitopes, which is why we continued using mix-stimulated CTLs for the assays.
Fig. 3.
Analysis of RVP-specific CTL activity against peptide-sensitized T2 target cells. Cytotoxic activity of RVP-specific CTL after at least three rounds of restimulation with the indicated RVP mixes was analyzed in standard 51Cr-release assays. RVP-specific reactivity of T cell bulk cultures was tested against T2 target cells loaded with the indicated RVP mixes. Effector T cells were added at different effector-to-target cell (E:T) ratios. A representative experiment with CTL stimulated with either the nonamer RVP-mix 1 (a) or decamer RVP-mix 2 (b) is given. The analysis was performed in triplicates with 1,000 effector cells per well and after a 4-h incubation period. The results of the cytotoxicity assays are representative for three individual analyses
RVP-specific CTL lyse tumor cells
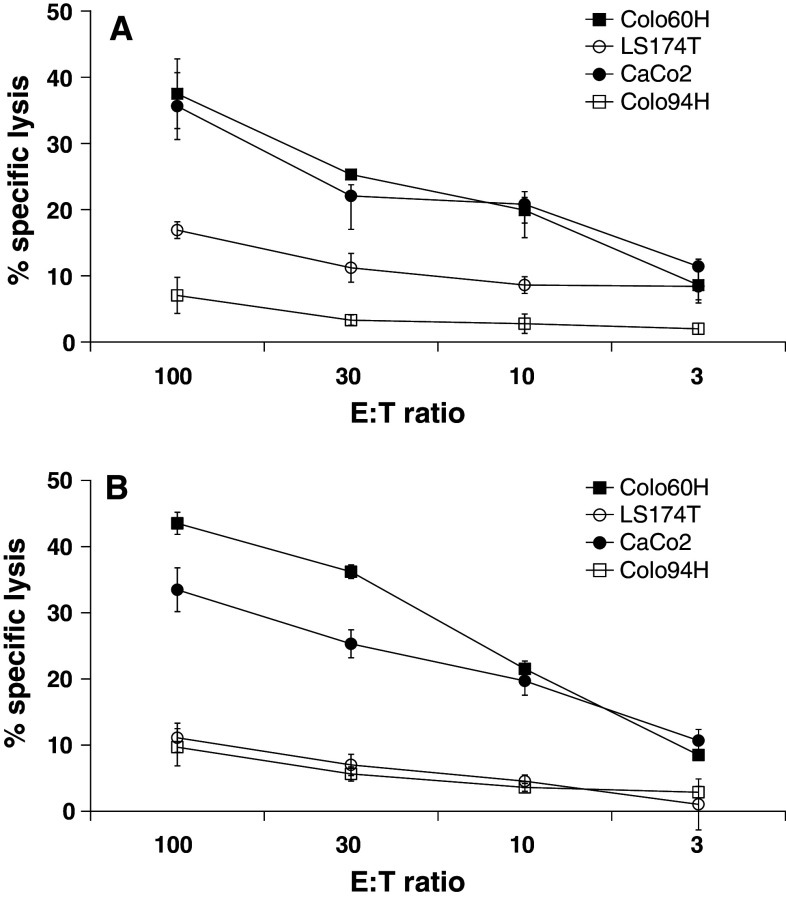
The ability to kill tumor cells endogenously expressing the target peptides used to induce the CTLs is the ultimate goal of each reverse immunology approach. We thus analyzed, in further cytotoxicity tests, whether RVP-specific CTL have the potential to attack CRC cells. As targets, we used Colo60H and CaCo2 (both HLA-A2.1+, HERV-H Xp22.3 ORF+), Colo94H (HLA-A2.1+, HERV-H Xp22.3 ORFneg) and LS174T (HLA-A2.1neg, HERV-H Xp22.3 ORF+). Approximately 40% specific lysis of Colo60H and CaCo2 cells was observed with the highest effector-to-target cell ratio used (100:1; Fig. 4a, b). The most effective CTL were the mix-2 stimulated T cells followed by mix-1 stimulated CTL. The tumor cell lysis was titrated, and the control cells Colo94H expressing HLA-A2.1 but no HERV-H Xp22.3 ORF and LS174T expressing HERV-H Xp22.3 ORF but no HLA-A2.1 were only marginally affected by both CTL cultures tested. In summary, these data hint toward a specific recognition of endogenously processed HERV-H Xp22.3 ORF-derived peptides presented in HLA molecules on surface of CRC cells.
Fig. 4.
Analysis of RVP-specific CTL activity against CRC cell lines. Standard 51Cr-release cytotoxicity assays were performed with RVP-specific CTL using CRC cell lines as target cells: Colo60H and CaCo2 (both HLA-A2.1+, HERV-H Xp22.3 ORF+), Colo94H (HLA-A2.1+, HERV-H Xp22.3 ORFneg) and LS174T (HLA-A2.1neg, HERV-H Xp22.3 ORF+). Effector T cells were added at different effector-to-target cell (E:T) ratios. A representative experiment with T cells stimulated with the nonamer RVP-mix 1 (a) and decamer RVP-mix 2 (b) is shown. Analysis was performed in triplicates with 1,000 effector cells per well and after an 8-h incubation period. The cytotoxicity results are representative for three individual analyses on different days
Discussion
Activation of members of several HERV families has been associated with tumor development. For instance, HERV-K expression is detectable in melanoma [11], breast cancer [10], testicular cancer [20] as well as ovarian cancer [21]. HERV-H seems to be associated with colorectal carcinoma [5, 12], HERV-E with prostate, kidney, ovarian and uterus cancer [7, 10] and HERV-W with testicular cancer [7]. Finally, HERV-R is associated with tumors of the liver and lung [20]. Expression of HERVs has been recurrently observed during cellular transformation processes in both cell culture and in vivo [7].
Increase in HERVs transcription in cancer cells has been linked to the liberation of HERV LTRs from epigenetic constraints via demethylation [3]. This is also in agreement with recent data showing that 5–8% of repetitive elements demonstrate cancer-related DNA methylation patterns [22]. Over-representation of the U5 regions of differentially expressed LTR loci suggests that activation of HERVs, predominantly found in tumors, is essentially autonomous [7]. Wentzensen and colleagues analyzed the methylation status of the HERV-H Xp22.3 LTR at length. Bisulfite sequencing of the respective LTR revealed CpG methylation patterns with 11–23 methylated CpGs for CRC cell lines lacking expression of the HERV-H Xp22.3 sequence, whereas there was no methylation detectable in the HERV-H expressing CRC cell lines [12]. This strongly supports the hypotheses that changes in the methylation status are tumor specific.
Repetitive elements might impose detrimental effects on the host genome by mediating ectopic recombination. HERVs may offer a better opportunity to mediate illegitimate crossover than non-repetitive fractions of the genome [3]. Consequently, their activation may directly contribute to chromosomal instability observed in most human tumors [23].
If this finding is true, one may hypothesize that chromosomally stable tumors must exhibit lower levels of HERVs expression than the majority of tumors, which show a high-level chromosomal instability. This would thus be especially interesting for microsatellite-instable and CpG-island methylation-high CRCs, both with typically low levels of chromosomal instability [24]. Respective analyses are currently being performed in our laboratory using a panel of primary CRC cell lines of the different molecular classes.
Transcripts from X-chromosome-linked members of the HERV-H family are frequently (and very selectively) expressed in CRC but not in normal tissue. Recently, a 93AA-long gag ORF of a HERV-H copy on Xp22 has been suggested as specific target for immunological interventions against CRC [5]. Wentzensen and coworkers identified an HERV-H env ORF on Xp22 with specific expression in CRC encoding for a putative protein of 273AA in length [12]. We decided to concentrate on the latter HERV-H Xp22.3 ORF for this pilot immunological analysis simply because of the length of the encoded putative ENV protein and for the relatively high number of theoretically strong HLA-A2.1-binding peptides.
We provide clear evidence for (1) easy induction of RVP-specific CTLs out of healthy donors’ peripheral blood, (2) strong recognition of RVP-loaded target cells in both ELISpot and cytotoxicity assays, (3) functional recognition of CRC tumor cells in cytotoxicity assays when expressing HERV-H Xp22.3 ORF and HLA-A2.1, thus demonstrating that (4) endogenous HERV-H Xp22.3 ENV protein must be synthesized, processed by proteasomal pathways and presented in the context of HLA-A2.1 on the cell surface.
Since HERV-H env genes exhibit abundant expression in liver, lung and testicular tumor tissues [20] our data on their immunogenic potential implies that they may be suited not only as CRC-specific but more universal tumor-associated antigens. In another immunological analysis, Takahashi and coworkers traced the regression of a human kidney cancer after allogeneic stem cell transplantation back to a specific recognition of a HERV-E-encoded and HLA-A11-restricted 10mer peptide [25]. It was the first report on the potential clinical relevance and usefulness of HERV-encoded proteins. In a very recent study, Wang-Johanning and coworkers provided convincing evidence of naturally occurring cellular as well as serological immune reactions against a HERV-K env protein in a high percentage of breast carcinoma patients [13].
Supposing that the latter observation would be of more general character, HERV ORFs indeed represent another very interesting source of both shared and individual tumor-specific antigens with even the potential to surpass entity boundaries. The development of tumor immunotherapies is a very active field, and true tumor-specific antigens are of outstanding interest since they minimize the risk of autoimmunity. The data presented in this manuscript provide the rationale for inclusion of HERV-H Xp22.3 into clinical vaccination protocols targeting colorectal tumors expressing the underlying ORF. Finally, when considering the recent groundbreaking work of SenGupta and coworkers, which suggested spontaneous anti-HERV T cell responses for control of chronic HIV-1 infections [26], one may even hypothesize that HERV-specific vaccination strategies have the potential to generally improve the human health situation and thus should be investigated with strong emphasis.
Another potentially relevant link to the immune system is highlighted by the fact that Parseval and coworkers showed the murine retrovirus Mo-MLV to encode an immunosuppressive protein, which allows tumor cells to escape immune rejection in immunocompetent mice [27]. HERV-H env contains a sequence very closely related to the immunosuppressive domains of this Mo-MLV protein. It will therefore be very interesting to analyze HERV ORF-specific T cell responses in patients in detail since it may shed light into the immunological balance between specific tumor antigen recognition and immune escape mechanisms potentially originating from the very same protein.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
Thanks to Michael Mullins for his editing advice as a native speaker and proofreading of the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
The authors declare that all experiments on human subjects were conducted in accordance with the Declaration of Helsinki and that all procedures were carried out with the adequate understanding and written consent of the subjects.
References
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Doylen M, FitzHugh W, Funke R, Gage D, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Blikstad V, Benachenhou F, Sperber GO, Blomberg J. Evolution of human endogenous retroviral sequences: a conceptual account. Cell Mol Life Sci. 2008;65(21):3348–3365. doi: 10.1007/s00018-008-8495-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Romanish MT, Cohen CJ, Mager DL. Potential mechanisms of endogenous retroviral-mediated genomic instability in human cancer. Semin Cancer Biol. 2010;20(4):246–253. doi: 10.1016/j.semcancer.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Cohen M, Larsson E. Human endogenous retroviruses. Bioessays. 1988;9(6):191–196. doi: 10.1002/bies.950090603. [DOI] [PubMed] [Google Scholar]
- 5.Alves PM, Lévy N, Stevenson BJ, Bouzourene H, Theiler G, Bricard G, Viatte S, Ayyoub M, Vuilleumier H, Givel JC, Rimoldi D, Speiser DE, Jongeneel CV, Romero PJ, Lévy F. Identification of tumor-associated antigens by large-scale analysis of genes expressed in human colorectal cancer. Cancer Immun. 2008;8:11. [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen T. HERVs in neuropathogenesis. J Neuroimmune Pharmacol. 2010;5(3):326–335. doi: 10.1007/s11481-010-9214-y. [DOI] [PubMed] [Google Scholar]
- 7.Gimenez J, Montgiraud C, Pichon JP, Bonnaud B, Arsac M, Ruel K, Bouton O, Mallet F. Custom human endogenous retroviruses dedicated microarray identifies self-induced HERV-W family elements reactivated in testicular cancer upon methylation control. Nucleic Acids Res. 2010;38(7):2229–2246. doi: 10.1093/nar/gkp1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen T. Association of human endogenous retroviruses with multiple sclerosis and possible interactions with herpes viruses. Rev Med Virol. 2005;15(3):179–211. doi: 10.1002/rmv.465. [DOI] [PubMed] [Google Scholar]
- 9.Karlsson H, Bachmann S, Schröder J, McArthur J, Torrey EF, Yolken RH. Retroviral RNA identified in the cerebrospinal fluids and brains of individuals with schizophrenia. Proc Natl Acad Sci USA. 2001;98(8):4634–4639. doi: 10.1073/pnas.061021998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang-Johanning F, Frost AR, Jian B, Epp L, Lu DW, Johanning GL. Quantitation of HERV-K env gene expression and splicing in human breast cancer. Oncogene. 2003;22(10):1528–1535. doi: 10.1038/sj.onc.1206241. [DOI] [PubMed] [Google Scholar]
- 11.Büscher K, Hahn S, Hofmann M, Trefzer U, Ozel M, Sterry W, Löwer J, Löwer R, Kurth R, Denner J. Expression of the human endogenous retrovirus-K transmembrane envelope, Rec and Np9 proteins in melanomas and melanoma cell lines. Melanoma Res. 2006;16(3):223–234. doi: 10.1097/01.cmr.0000215031.07941.ca. [DOI] [PubMed] [Google Scholar]
- 12.Wentzensen N, Coy JF, Knaebel HP, Linnebacher M, Wilz B, Gebert J, von Knebel Doeberitz M. Expression of an endogenous retroviral sequence from the HERV-H group in gastrointestinal cancers. Int J Cancer. 2007;121(7):1417–1423. doi: 10.1002/ijc.22826. [DOI] [PubMed] [Google Scholar]
- 13.Wang-Johanning F, Radvanyi L, Rycaj K, Plummer JB, Yan P, Sastry KJ, Piyathilake CJ, Hunt KK, Johanning GL. Human endogenous retrovirus K triggers an antigen-specific immune response in breast cancer patients. Cancer Res. 2008;68(14):5869–5877. doi: 10.1158/0008-5472.CAN-07-6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linnebacher M, Gebert J, Rudy W, Woerner S, Yuan YP, Bork P, von Knebel Doeberitz M. Frameshift peptide-derived T-cell epitopes: a source of novel tumor-specific antigens. Int J Cancer. 2001;93(1):6–11. doi: 10.1002/ijc.1298. [DOI] [PubMed] [Google Scholar]
- 15.Rammensee H-G, Bachmann J, Emmerich NN, Bachor OA, Stevanovic Stefan. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 16.Linnebacher M, Wienck A, Boeck I, Klar E. Identification of an MSI-H tumor-specific cytotoxic T cell epitope generated by the (−1) frame of U79260(FTO) J Biomed Biotechnol. 2010;2010:841451. doi: 10.1155/2010/841451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vonderheide RH, Hahn WC, Schultze JL, Nadler LM. The telomerase catalytic subunit is a widely expressed tumor-associated antigen recognized by cytotoxic T lymphocytes. Immunity. 1999;10:673–679. doi: 10.1016/S1074-7613(00)80066-7. [DOI] [PubMed] [Google Scholar]
- 18.Schultze JL, Michalak S, Seamon MJ, Dranoff G, Jung K, Daley J, Delgado JC, Gribben JG, Nadler LM. CD40-activated human B cells: an alternative source of highly efficient antigen presenting cells to generate autologous antigen-specific T cells for adoptive immunotherapy. J Clin Invest. 1997;100(11):2757–2765. doi: 10.1172/JCI119822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Bergwelt-Baildon MS, Vonderheide RH, Maecker B, Hirano N, Anderson KS, Butler MO, Xia Z, Zeng WY, Wucherpfennig KW, Nadler LM, Schultze JL. Human primary and memory cytotoxic T lymphocyte responses are efficiently induced by means of CD40-activated B cells as antigen-presenting cells: potential for clinical application. Blood. 2002;99(9):3319–3325. doi: 10.1182/blood.V99.9.3319. [DOI] [PubMed] [Google Scholar]
- 20.Ahn K, Kim HS. Structural and quantitative expression analyses of HERV gene family in human tissues. Mol Cells. 2009;28(2):99–103. doi: 10.1007/s10059-009-0107-y. [DOI] [PubMed] [Google Scholar]
- 21.Wang-Johanning F, Liu J, Rycaj K, Huang M, Tsai K, Rosen DG, Chen DT, Lu DW, Barnhart KF, Johanning GL. Expression of multiple human endogenous retrovirus surface envelope proteins in ovarian cancer. Int J Cancer. 2007;120(1):81–90. doi: 10.1002/ijc.22256. [DOI] [PubMed] [Google Scholar]
- 22.Szpakowski S, Sun X, Lage JM, Dyer A, Rubinstein J, Kowalski D, Sasaki C, Costa J, Lizardi PM. Loss of epigenetic silencing in tumors preferentially affects primate-specific retroelements. Gene. 2009;448(2):151–167. doi: 10.1016/j.gene.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schulz WA, Steinhoff C, Florl AR. Methylation of endogenous human retroelements in health and disease. Curr Top Microbiol Immunol. 2006;310:211–250. doi: 10.1007/3-540-31181-5_11. [DOI] [PubMed] [Google Scholar]
- 24.Ostwald C, Linnebacher M, Weirich V, Prall F. Chromosomally and microsatellite stable colorectal carcinomas without the CpG island methylator phenotype in a molecular classification. Int J Oncol. 2009;35(2):321–327. [PubMed] [Google Scholar]
- 25.Takahashi Y, Harashima N, Kajigaya S, Yokoyama H, Cherkasova E, McCoy JP, Hanada K, Mena O, Kurlander R, Tawab A, Srinivasan R, Lundqvist A, Malinzak E, Geller N, Lerman MI, Childs RW. Regression of human kidney cancer following allogeneic stem cell transplantation is associated with recognition of an HERV-E antigen by T cells. J Clin Invest. 2008;118(3):1099–1109. doi: 10.1172/JCI34409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.SenGupta D, Tandon R, Vieira RG, Ndhlovu LC, Lown-Hecht R, Ormsby CE, Loh L, Jones RB, Garrison KE, Martin JN, York VA, Spotts G, Reyes-Terán G, Ostrowski MA, Hecht FM, Deeks SG, Nixon DF. Strong human endogenous retrovirus-specific T cell responses are associated with control of HIV-1 in chronic infection. J Virol. 2011;85(14):6977–6985. doi: 10.1128/JVI.00179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Parseval N, Casella J, Gressin L, Heidmann T. Characterization of the three HERV-H proviruses with an open envelope reading frame encompassing the immunosuppressive domain and evolutionary history in primates. Virology. 2001;279(2):558–569. doi: 10.1006/viro.2000.0737. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.