Abstract
The role of the cap-binding complex, eIF4F, in the translation of vaccinia virus mRNAs has been analyzed within infected cells. Plasmid DNAs, which express dicistronic mRNAs containing a picornavirus internal ribosome entry site, produced within vaccinia virus-infected cells both β-glucuronidase and a cell surface-targeted single-chain antibody (sFv). Cells expressing sFv were selected from nonexpressing cells, enabling analysis of protein synthesis specifically within the transfected cells. Coexpression of poliovirus 2A or foot-and-mouth disease virus Lb proteases, which cleaved translation initiation factor eIF4G, greatly inhibited cap-dependent protein (β-glucuronidase) synthesis. Under these conditions, internal ribosome entry site-directed expression of sFv continued and cell selection was maintained. Furthermore, vaccinia virus protein synthesis persisted in the selected cells containing cleaved eIF4G. Thus, late vaccinia virus protein synthesis has a low requirement for the intact cap-binding complex eIF4F. This may be attributed to the short unstructured 5′ noncoding regions of the vaccinia virus mRNAs, possibly aided by the presence of poly(A) at both 5′ and 3′ termini.
Vaccinia virus infection of cells results in the inhibition of host cell protein synthesis and a switch to the production of virus-encoded polypeptides (reviewed in reference 23). The mechanism underlying this effect is not established. Vaccinia virus replicates in the cytoplasm of the cell and encodes its own enzymes for DNA replication and RNA production. The viral mRNAs are capped (at their 5′ terminus) by the virus-encoded capping enzyme and polyadenylated (at the 3′ terminus) and hence have a structure similar to the host cell cytoplasmic mRNAs. The initiation of protein synthesis is generally considered to be the key regulatory stage of polypeptide formation (reviewed in reference 22). This step involves the recognition of the 5′-terminal cap structure by the translation initiation complex eIF4F. This factor is a heterotrimer consisting of eIF4E (which recognizes the cap structure, m7GpppN…), eIF4A (an RNA helicase), and eIF4G (believed to act as a scaffold for the other proteins). eIF4F, probably in association with the 40S ribosomal subunit, is believed to migrate along the mRNA, unwinding the secondary structure, until an AUG codon in a suitable context is encountered (18). At this point the 60S ribosomal subunit joins and polypeptide formation can commence.
In contrast to cellular mRNAs, the translation of vaccinia virus mRNAs has been shown to be relatively resistant to inhibition by the cap analogue m7GTP in vitro, suggesting that the initiation of protein synthesis on the viral mRNAs is relatively cap independent (2). An alternative strategy for analyzing the mechanism of initiation of protein synthesis in vaccinia virus-infected cells has also been described (11). These authors coexpressed, in a transient assay, the poliovirus (PV) 2A protease within vaccinia virus-infected cells and reported a major reduction in the level of viral protein synthesis. The PV 2A protease induces cleavage of the eIF4G component of the cap-binding complex eIF4F. This cleavage results in the inhibition of cap-dependent protein synthesis without affecting cap-independent translation directed by the picornavirus internal ribosome entry site (IRES) elements (reviewed in reference 5). These data are also consistent with the observation that it has been impossible to introduce the PV 2A protease coding region into the genome of vaccinia virus (16, 33). Furthermore, a similar incompatibility was observed between vaccinia virus and the foot-and-mouth disease virus (FMDV) L coding sequence, which also specifies a protease which cleaves eIF4G (4). These results appear to suggest that the inhibition of cap-dependent protein synthesis induced by cleavage of eIF4F is deleterious to vaccinia virus.
Recently, the isolation of temperature-sensitive (ts) mutants in the vaccinia virus capping enzyme has been reported (15). The mutant protein is defective at the nonpermissive temperature in at least two activities of this enzyme, namely, the guanyltransferase and methyltransferase activities. The mutant virus is also markedly ts, as production of virus was reduced several hundredfold at the nonpermissive temperature. However, it was observed that little difference in the production of viral proteins could be detected between the permissive and nonpermissive temperatures. Thus, it appeared that the viral protein synthesis was relatively independent of the capping of the viral transcripts.
Although there have been a number of studies in which the vaccinia virus-T7 RNA polymerase transient-expression system has been used to express, from plasmids, the PV 2A protease or the FMDV Lb protease, these studies (e.g., references 3, 21, and 25) have not attempted to analyze the effect of these proteases on the vaccinia virus itself. The studies do show, however, that these proteases very efficiently inhibit cap-dependent translation of mRNA transcripts generated from cotransfected plasmids within vaccinia virus-infected cells. The study of the effects of these proteases on the vaccinia virus mRNA translation itself is complicated by the fact that while all the cells can be readily infected with vaccinia virus, only a proportion of the cells are transfected by plasmids. Hence, the analysis is hampered by the background of cells that are not expressing the picornavirus proteases. This background may mask other effects that these proteases may be causing.
In order to assess specifically the influence of PV 2A and FMDV Lb on vaccinia virus protein synthesis compared to the cap-dependent translation of a reporter gene construct, we have utilized a recently developed system which selects only those cells which have been transfected. In these selected vaccinia virus-infected cells we have analyzed, in parallel, the effects of these proteases on reporter gene expression and also on the vaccinia virus protein synthesis.
MATERIALS AND METHODS
Plasmids.
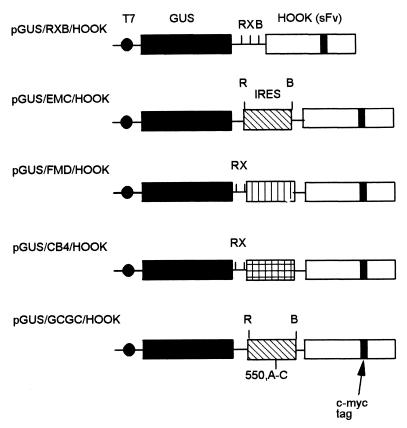
The plasmids in this study were constructed by standard methods as described previously (28) and by the manufacturers. Plasmid DNA was amplified in Escherichia coli DH5α and purified by using a Bio 101 Maxi Prep kit (Anachem). pHOOK-1 was obtained from Invitrogen. The construction of the dicistronic vector pGUS/RXB/HOOK (Fig. 1) will be described elsewhere (26). Derivatives of this construct that contain picornavirus IRES elements from FMDV, encephalomyocarditis virus (EMCV), coxsackie B4 virus (CB4) were produced and are also illustrated in Fig. 1. An inactive mutant form of the EMCV IRES (termed GCGC [see reference 24]) containing a single A-C change at nucleotide 550 within a conserved GNRA motif was also used. The EMCV plasmids contain EcoRI-BamHI fragments of EMCV cDNA derived from pSKEMCRB and its mutant derivative (24) between the unique EcoRI and BamHI sites of pGUS/RXB/HOOK. The FMDV IRES cDNA (as an EcoRI-ClaI fragment from pKSRCla [9]) and the CB4 IRES cDNA (as a HindIII-SstI fragment from pSKCB4 [31]) were blunt ended and inserted upstream of the HOOK sequence into the blunt-ended BamHI site (see Fig. 1). Plasmids of the correct structure were identified from miniprep DNA and amplified.
FIG. 1.
Structure of reporter plasmids used in this study. IRES elements from FMDV, CB4, and both wild-type and mutant forms of EMCV were positioned between the two cistrons of the parental GUS/RXB/HOOK. The T7 promoter (filled circle) and unique restriction sites in the parental vector for EcoRI (R), XbaI (X), and BamHI (B) are indicated.
Transient-expression assay and cell selection.
COS-7 cells (35-mm dishes) were infected with the recombinant vaccinia virus vTF7-3 (12), which expresses the T7 RNA polymerase. The infected cells were transfected with the pGUS/IRES/HOOK plasmid DNA (2.5 μg) alone or also with pAΔ802 (0.5 μg) (which expresses the PV 2A protease [17]) or pLb (0.5 μg) (which expresses the FMDV Lb protease [21]), or other plasmids as indicated in the Figure legends by using Lipofectin (8 μg; Life Technologies) in Optimem as described previously (see reference 6). After about 20 h, cells were harvested for analysis of gene expression directly by using buffer C (20 mM Tris-HCl, pH 8.0; 0.135 M NaCl; Nonidet P-40, 0.5%), or else cells expressing the sFv encoded by the HOOK coding sequence were selected. The cell selection method will be described in detail elsewhere (26). Briefly, the cells were removed from the dish by washing in Ca-Mg-free phosphate-buffered saline (PBS), with an aliquot retained for analysis of the total cell extracts, and then incubated with the mouse monoclonal antibody (MAb) 9E10 (specific for the c-myc tag [10]) for 1 h on ice. After being washed, the cells were incubated with sheep anti-mouse immunoglobulin G (IgG)-coated magnetic beads (Dynabeads M-450; Dynal) for 45 min on a rotating wheel at 4°C. Beads were captured on a magnetic stand (Dynal) and washed, and the selected cells were extracted in buffer C.
Cell extracts were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (6 or 10%) (20) and, where appropriate, by immunoblotting with rabbit anti-β-glucuronidase (GUS) (5prime-3prime, Inc.), rabbit anti-actin (Sigma), rabbit anti-eIF4G (a gift from N. Sonenberg, McGill University, Montreal, Quebec, Canada), mouse anti-myc tag (9E10 [10]), or rat monoclonal antibody 15B6 anti-VVp37 (29), followed by peroxidase-linked anti-rabbit, anti-mouse IgG antibodies (Amersham) or anti-rat IgG (Dako), as appropriate, with detection by using chemiluminescent reagents (Pierce).
Protein synthesis was monitored by metabolic labeling with [35S]EXPRESS (NEN) (50 μCi/dish) in methionine- and cysteine-free medium for 1 h prior to cell selection.
RESULTS
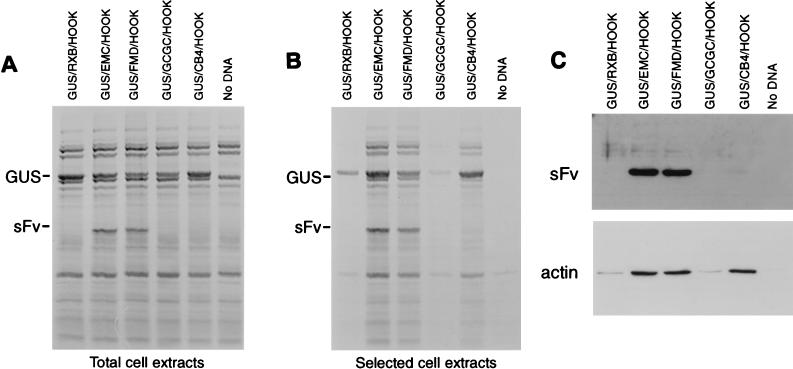
A system has been developed which permits the isolation of transfected cells away from untransfected cells dependent on the expression of a cell surface targeted single-chain antibody (sFv) encoded by the plasmid pHOOK-1 (Invitrogen). In order for this system to function when cap-dependent protein synthesis is blocked, the coding sequence for the myc-tagged sFv has been placed downstream of picornavirus IRES elements (Fig. 1). The picornavirus IRES elements direct cap-independent internal initiation of protein synthesis, which continues in the presence of cleaved eIF4G. Upstream of the IRES sequence there is a reporter gene sequence encoding GUS. Thus, plasmids expressing from the T7 promoter mRNAs of the form GUS/IRES/HOOK have been constructed. A number of versions have been made (see Fig. 1) with either no IRES element (GUS/RXB/HOOK), a variety of different picornavirus IRES elements (from EMCV, FMDV, and CB4), or a severely defective mutant EMCV IRES called GCGC (see reference 24). Transfection of these plasmids into cells infected with the recombinant vaccinia virus vTF7-3 (12) results in the transcription of these plasmids. In each case the expression of GUS was readily observed by 20 h after infection by direct analysis of [35S]methionine- and [35S]cysteine-labeled total cell extracts (Fig. 2A) and was then confirmed by immunoblotting with anti-GUS antibodies (data not shown). The polypeptide synthesis observed in these cell extracts is a combination of that directed by vaccinia virus plus that produced from the T7 transcripts derived from the plasmids transfected into the cells (host cell protein synthesis is inhibited by vaccinia virus). Expression of the sFv was dependent on the IRES element. In the absence of the IRES or with a mutant form of the EMCV IRES (GCGC), no expression of the sFv was detectable in these labeled cell extracts (Fig. 2A). However, strong expression of the sFv from the plasmids expressing dicistronic mRNAs containing the EMCV or FMDV IRES was observed. With the plasmid expressing the CB4 IRES element, a much-lower expression of the sFv was obtained (Fig. 2A; see also Fig. 2C).
FIG. 2.
IRES-dependent cell selection. The indicated plasmids were transfected into vTF7-3-infected COS-7 cells as described in Materials and Methods. Cells were harvested (after metabolic labeling with [35S]EXPRESS) in Ca-Mg-free PBS, and an aliquot was retained and analyzed as the total extract (A). Of the remaining cells those expressing the sFv were selected with MAb 9E10 and anti-mouse IgG-coated magnetic beads, and the selected cell extracts (panel B) were analyzed by SDS-PAGE (10%) and autoradiography. The positions of the GUS and sFv proteins are indicated. Alternatively, selected cell extracts were analyzed by immunoblotting (panel C) for the presence of cellular actin (as a measure of cell recovery) and for myc tagged sFv expression (as a measure of IRES activity).
Expression of the sFv permitted efficient selection of the transfected cells. Intact cells were harvested in the absence of trypsin or detergent by using Ca-Mg-free PBS and incubated with the MAb 9E10 which recognizes the myc tag on the sFv displayed on the cell surface. After being washed, the cells which bound the antibody were isolated by using magnetic beads coated with anti-mouse IgG (as described in Materials and Methods), and cell extracts were analyzed by SDS-PAGE (Fig. 2B). In the absence of a functional IRES (GUS/RXB/HOOK and GUS/GCGC/HOOK) within the construct or when untransfected cells were used, only a low level of cellular protein was isolated on the magnetic beads (actin was monitored by Western blot analysis [Fig. 2C]; the actin represents stable accumulated cellular protein, since all cellular protein synthesis is inhibited by the vaccinia virus infection). However, all three constructs encoding a functional IRES element expressed the sFv, and efficient selection of cells was observed (Fig. 2B and C). Thus, the lower level of sFv achieved by the CB4 IRES was still sufficient to achieve efficient selection. Within the selected cell extracts (Fig. 2B), the expression of the upstream cistron, GUS, was even more evident than in the total cell extracts, while the relatively low level expression of the sFv directed by the CB4 IRES was demonstrated by Western blot analysis, using the anti-myc-tag MAb 9E10 (Fig. 2C). Thus, in these assays, the vaccinia virus-directed protein synthesis, as well as the cap-dependent and the IRES-directed (cap-independent) translation of the dicistronic mRNA expressed from the plasmids, could each be separately assayed.
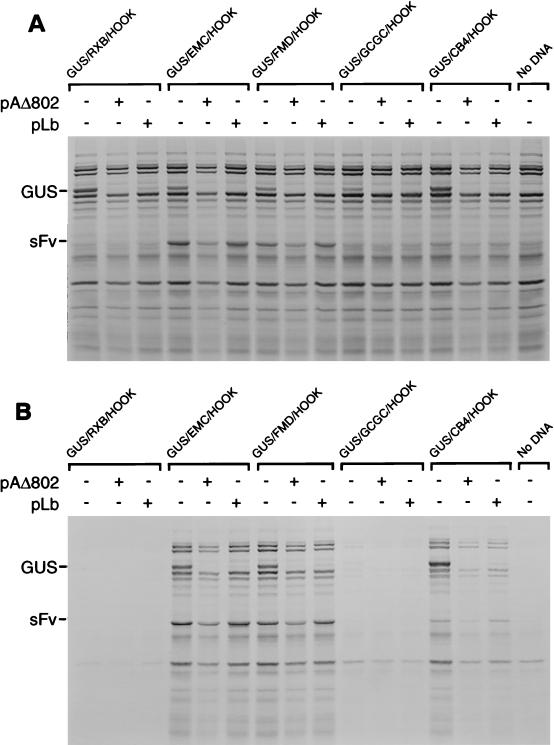
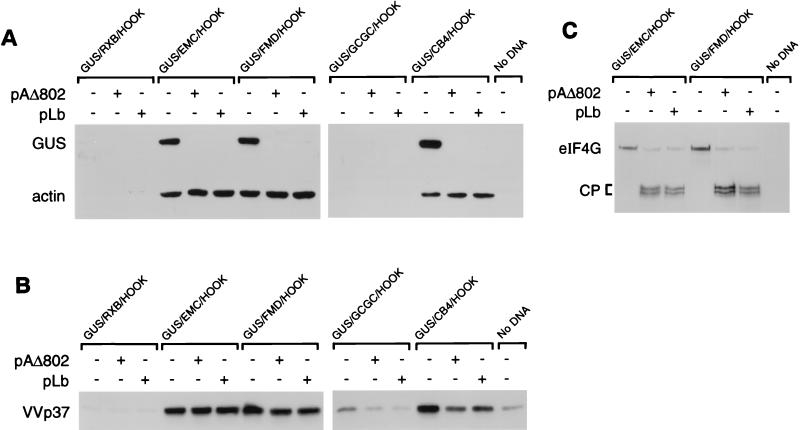
To assess the effect of the PV 2A protein or the FMDV Lb protein on the expression of the reporter gene products and the vaccinia virus proteins, the GUS/HOOK plasmids were cotransfected with plasmid pAΔ802 (17), which expresses PV 2A, or with pLb (21), which expresses FMDV Lb. Cell extracts were analyzed either as total cell extracts or after cell selection with MAb 9E10 and the anti-mouse IgG-coated magnetic beads as described above. In the total cell extracts, the expression of the GUS open reading frame was readily detected in all of the cells transfected with the GUS/HOOK constructs alone (Fig. 3A). However, when cotransfected with plasmids expressing PV 2A (from pAΔ802) or FMDV Lb (from pLb), the expression of GUS was greatly inhibited. As expected, no significant change in the IRES-directed expression of the sFv was observed (Fig. 3A), since the IRES-directed translation occurs when eIF4G is cleaved. Furthermore, no difference in the pattern of vaccinia polypeptide synthesis was seen between cell extracts from dishes transfected with the reporter constructs alone or in combination with the viral proteases. In these total cell extracts this may be attributed to the fact that many of the cells will not express the plasmid-encoded proteases since they have not taken up the DNA. To overcome this background effect, the infected and transfected cells were selected and cell extracts were analyzed (Fig. 3B). The IRES-dependent isolation of transfected cells was still achieved in the presence of either PV 2A or FMDV Lb as anticipated. In the selected cell extracts, obtained from cells expressing the GUS/FMD/HOOK, GUS/EMC/HOOK, and GUS/CB4/HOOK plasmids, the inhibition of GUS expression in the presence of PV 2A or FMDV Lb was readily apparent (Fig. 3B). This effect on GUS expression was confirmed by immunoblotting analysis (Fig. 4A), although no change in the level of actin isolated was apparent (reflecting the efficiency of selection and not the pattern of actin synthesis, which is inhibited in each case by the vaccinia virus infection). It was also clear that, in contrast to the dramatic drop in GUS expression, little effect on the pattern of expression of the sFv or of the vaccinia virus proteins occurred in these extracts (Fig. 3B). To demonstrate that no change in the accumulation of a vaccinia virus protein (p37) occurred in the presence of PV 2A or FMDV Lb, immunoblotting analysis was performed with an anti-VVp37 MAb (MAb 15B6 [29]), revealing very similar levels of this protein in cell extracts obtained from cells with or without the viral proteases (Fig. 4B). Thus, in these selected cell extracts the vast majority of the cells must have contained the GUS/IRES/HOOK transcript (or else they would not be selected) and, where appropriate, the PV 2A or FMDV Lb (to cause the inhibition of GUS expression). Analysis of the eIF4G within these extracts demonstrated that, in the presence of the PV 2A or the FMDV Lb proteases, the eIF4G was present predominantly as the cleaved form (Fig. 4C). The low level of intact eIF4G may be derived from a small number of untransfected cells which may be aggregated with the selected cells. Clearly, there is little background of cells selected in the absence of the selection plasmid (Fig. 4C).
FIG. 3.
Differential regulation of cap-dependent, IRES-dependent, and vaccinia virus protein synthesis. COS-7 cells were infected with vTF7-3 and transfected with the indicated GUS/HOOK plasmids alone or with pAΔ802 (encodes PV 2A) or pLb (encodes FMDV Lb). After metabolic labeling with [35S]EXPRESS, cells were harvested and both total cell extracts (A) and selected cell extracts (B) were analyzed by SDS-PAGE (10%) and autoradiography as described in Fig. 2. The positions of the GUS and sFv proteins are indicated.
FIG. 4.
Vaccinia virus protein synthesis is maintained following cleavage of eIF4G. COS-7 cells were infected with vTF7-3 and transfected with the indicated GUS/HOOK plasmids alone or with pAΔ802 (encodes PV 2A) or pLb (encodes FMDV Lb). Selected cell extracts were prepared and analyzed by immunoblotting for GUS and actin (A), VVp37 (B), and eIF4G (C). The cleavage products (CP) of eIF4G are indicated.
Our conclusion from these data is that, in general, the translation of the vaccinia virus mRNAs is rather insensitive to the loss of intact eIF4G and hence has only a limited requirement for the cap-binding complex eIF4F. Since this conclusion is at variance with the results obtained by Feduchi et al. (11), we sought to investigate the basis for this disparity. A significant difference between their studies and those reported here is in the plasmids used to induce eIF4G cleavage. Feduchi et al. (11) used a derivative of pTM1 which includes an IRES element from EMCV to express the PV 2A, and this may be expected to yield large amounts of the PV 2A protein. In contrast, we have used plasmids that lack an IRES element to express PV 2A and FMDV Lb and, furthermore, in the case of the PV 2A plasmid the initiation codon is the second AUG codon (see reference 17); hence, the expression level obtained is likely to be much lower than that achieved by Feduchi et al. (11). The yield of PV 2A and FMDV Lb in our studies is clearly sufficient to block GUS expression (and to cleave eIF4G), but it may be insufficient to cause other undefined effects that high-level expression of these proteases may be able to achieve within cells.
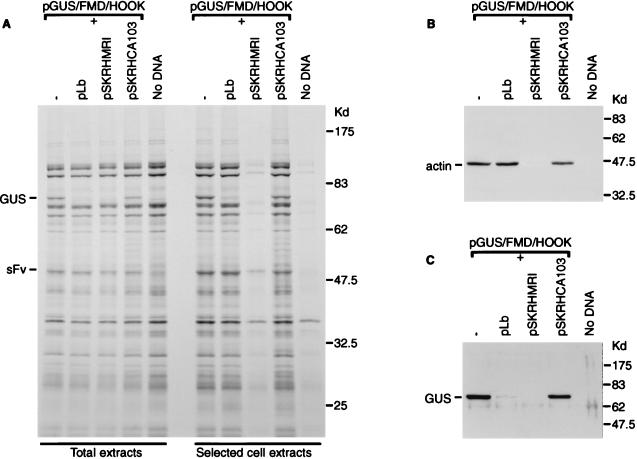
By cotransfecting the GUS/FMD/HOOK selection vector with plasmids expressing the FMDV L protease with or without an IRES element, we have sought to test whether this hypothesis may be correct. In Fig. 5, it is shown that significantly fewer cells (as monitored by the level of virus protein synthesis (Fig. 5A) and the yield of cellular actin (Fig. 5B) are recovered when the selection vector is cotransfected with a plasmid expressing FMDV L (from pSKRHMR1 [3]) protease under the control of an IRES. In contrast, no significant effect on cell selection or GUS expression (Fig. 5C) was observed in the presence of a very similar plasmid (pSKRHCA103 [3]) which has the L gene specifically inactivated. As seen previously (Fig. 4B), the coexpression of pLb (which lacks an IRES) had no inhibitory effect on the cell selection, but it did inhibit GUS expression. Similar results were also obtained in a parallel experiment with the pGUS/EMC/HOOK vector (data not shown). These results are consistent with the view that high-level expression of the viral proteases has a detrimental effect on protein expression within the vaccinia virus-infected cells that is unrelated to the cleavage of eIF4G.
FIG. 5.
IRES-directed expression of FMDV L inhibits protein expression in vaccinia virus-infected cells. COS-7 cells were infected with vTF7-3 alone (no DNA) or also transfected with pGUS/FMD/HOOK alone or with one of the following plasmids as indicated: pLb (FMDV Lb), pSKRHMR1 (FMDV IRES-L-P1-2A+3C) (3), or pSKRHCA103 (FMDV IRES-ΔL-P1-2A+3C) (3). Cells were labeled with [35S]EXPRESS and selected as described in Materials and Methods. Samples of the total cell extracts and extracts of the selected cells were analyzed by SDS-PAGE and autoradiography (A). The selected cell extracts were also analyzed by immunoblotting with antibodies specific for actin (B) or GUS (C) and detected by chemiluminescence.
DISCUSSION
In these studies we have assayed at a gross level the effect of PV 2A and FMDV Lb, which induce cleavage of the eIF4G component of the cap-binding complex eIF4F, on the translation of reporter gene mRNAs and vaccinia virus mRNAs in vaccinia virus-infected cells. It is clear that the cap-dependent expression of GUS from the plasmid construct was greatly inhibited by the coexpression of the viral proteases, whereas IRES-directed translation of the sFv and the vaccinia virus mRNA translation continued efficiently. We have not examined all of the individual mRNAs produced by vaccinia virus, and it is certainly possible that particular mRNAs may be more sensitive to the loss of eIF4G than was indicated by this general analysis. However, since the pattern of viral protein synthesis observed was identical in each case, it would appear that the picornavirus proteases had no adverse effect on any essential early virus function. Our results are consistent with the relative insensitivity of vaccinia virus mRNA translation in the rabbit reticulocyte lysate to inhibition by cap analogue (2). Furthermore, a recent report (15) showed that a ts lesion in the vaccinia capping enzyme had no significant effect on the translation of vaccinia virus mRNAs within infected cells, even at the nonpermissive temperature. These results suggest that translation of vaccinia virus mRNAs has a low requirement for the cap-binding complex. These findings are analogous to observations on the translation of late adenovirus mRNAs containing the tripartite leader, which also have a low requirement for eIF4F (8, 30). The adenovirus late mRNAs are relatively unstructured in the 5′-proximal portion of their 5′-untranslated region (UTR), and this may facilitate ribosome attachment in the absence of efficient recognition of the mRNA by eIF4F. Vaccinia virus mRNAs also have relatively short and unstructured 5′ UTRs; indeed, late mRNAs have a capped 5′ terminus followed by a heterogeneous length of poly(A) generated by the viral RNA polymerase upstream of the initiation codon (23). This contrasts with the extensive secondary structure and large size (ca. 450 bases) of the picornavirus IRES elements, which also function without the requirement for intact eIF4F (5). Thus, two distinct mechanisms of translation initiation with little or no dependence on the intact eIF4F complex are operating in these assays. The function of the cap structure on vaccinia virus mRNAs may be to help stabilize the mRNAs, as decapping of mRNAs leads to rapid degradation (7).
Our results and conclusions differ from those reported previously (11), where it was shown that expression of the PV 2A protein in vaccinia virus-infected cells produced a major inhibition of vaccinia virus protein synthesis. We have demonstrated that the high-level expression of FMDV L, achieved by the use of an IRES-containing vector to express this protease, is capable of achieving effects on the vaccinia virus that exceed those resulting from the loss of eIF4G and cap-dependent protein synthesis (see Fig. 5). Thus, we believe that the loss of vaccinia virus translation observed earlier (11) was an artifact of that experimental system.
Very recently, a functional homologue of eIF4G, termed eIF4GII, has been reported (14). Since the cap-dependent expression of GUS is clearly inhibited when either FMDV L or PV 2A proteases are expressed, it is apparent that no functional homologue of eIF4G can persist under these conditions. Furthermore, we have now demonstrated that eIF4GII is also cleaved in the vaccinia-T7 RNA polymerase transient-expression system by using the plasmids that express FMDV Lb and PV 2A (6a). Thus, the vaccinia virus mRNAs are translated when both forms of eIF4G are cleaved.
Earlier reports had indicated that constitutive expression of PV 2A or FMDV Lb within recombinant vaccinia virus was not tolerated (4, 16, 33), since such recombinant viruses could not be isolated. Interruption or elimination of the FMDV L coding sequence within FMDV cassettes greatly facilitated the production of recombinant FMDV-vaccinia viruses (1, 4). It should be noted, however, that even when only low-level expression of the PV 2A or FMDV L proteases could be expected (i.e., when the coding sequence was not specifically positioned under the control of a vaccinia virus promoter [4, 33]), it was still not possible to isolate recombinant vaccinia viruses, suggesting that the toxic effects of these proteases is apparent even at low levels. The incompatibility of vaccinia virus with PV 2A or FMDV Lb may represent effects on a minor population of mRNAs or, alternatively, may reflect only a small diminution in replication efficiency that is amplified by the multiple cycles of virus growth required to generate plaques.
In conclusion, the results presented here clearly demonstrate that vaccinia virus protein synthesis proceeds efficiently when the cap-binding complex (eIF4F) is inactivated through the cleavage of the eIF4G component. Thus, initiation of protein synthesis on these viral mRNAs appears to be accomplished with a low requirement for this translation initiation complex; this is probably as a result of the short, unstructured nature of the 5′ UTRs of the vaccinia virus mRNAs. It is interesting to note that, since late vaccinia virus mRNAs have a stretch of poly(A) at both their 5′ and 3′ termini, interaction with a protein which recognizes poly(A) could bring the two termini into close proximity. This may facilitate the recycling of ribosomes from the 3′ terminus to the 5′ end with little requirement for eIF4F. This is similar to the model for poly(A)-stimulated translation initiation within yeast cells, as described previously (27), suggesting circularization of mRNA resulting from the interaction between the poly(A) binding protein (PABP) and eIF4G (32). Within mammalian cells no evidence for a direct interaction between these two proteins has been presented. However, a PABP-interacting protein that interacts with both PABP and eIF4A has very recently been characterized (6b) that can also bridge the two mRNA termini. In vaccinia virus-infected cells, circularization could also be achieved by the vaccinia virus poly(A) polymerase, a dimer of VP55 and VP39 (23) that has affinity for both poly(A) and the 5′ cap structure. Alternatively, perhaps the abundant cellular PABP could achieve this function alone on vaccinia virus mRNAs, since it has four RNA recognition motifs and has the ability to multimerize (13, 19).
ACKNOWLEDGMENTS
We thank Tom Wileman (Pirbright) for the anti-c-myc and anti-VVp37 antibodies, Nahum Sonenberg (Montreal) for the anti-eIF4G antibodies, and Ann Kaminski (Cambridge) for the plasmids pAΔ802.
M.E.M.R. was supported by a BBSRC studentship.
REFERENCES
- 1.Abrams C C, King A M Q, Belsham G J. Assembly of foot-and-mouth disease virus empty capsids synthesized by a vaccinia virus expression system. J Gen Virol. 1995;76:3089–3098. doi: 10.1099/0022-1317-76-12-3089. [DOI] [PubMed] [Google Scholar]
- 2.Bablanian R, Goswami S K, Esteban M, Banerjee A K, Merrick W C. Mechanism of selective translation of vaccinia virus mRNAs: differential role of poly(A) and initiation factors in the translation of viral and cellular mRNAs. J Virol. 1991;65:4449–4460. doi: 10.1128/jvi.65.8.4449-4460.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belsham G J, Brangwyn J K. A region of the 5′ non-coding region of foot-and-mouth disease virus RNA directs efficient internal initiation of protein synthesis within cells: involvement with the role of L protease in translational control. J Virol. 1990;64:5389–5395. doi: 10.1128/jvi.64.11.5389-5395.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belsham G J, Brangwyn J K, Ryan M D, Abrams C C, King A M Q. Intracellular expression and processing of foot-and-mouth disease virus capsid precursors using vaccinia virus vectors: influence of the L protease. Virology. 1990;176:524–530. doi: 10.1016/0042-6822(90)90022-j. [DOI] [PubMed] [Google Scholar]
- 5.Belsham G J, Sonenberg N. RNA protein interactions in regulation of picornavirus RNA translation. Microbiol Rev. 1996;60:499–511. doi: 10.1128/mr.60.3.499-511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belsham G J. Analysis of picornavirus internal ribosome entry site function in vivo. In: Richter J, editor. mRNA formation and function. New York, N.Y: Academic Press, Inc.; 1997. pp. 323–340. [Google Scholar]
- 6a.Belsham, G. J., and N. Sonenberg. Unpublished results.
- 6b.Craig A, Haghighat A, Yu A T K, Sonenberg N. Interaction of polyadenylate-binding protein with eIF4G homologue PAIP enhances translation. Nature. 1998;392:520–523. doi: 10.1038/33198. [DOI] [PubMed] [Google Scholar]
- 7.Decker C J, Parker R. Mechanism of mRNA degradation in eukaryotes. Trends Biochem Sci. 1994;19:336–340. doi: 10.1016/0968-0004(94)90073-6. [DOI] [PubMed] [Google Scholar]
- 8.Dolph P J, Huang J, Schneider R J. Translation by the adenovirus tripartite leader: elements which determine independence from cap-binding protein complex. J Virol. 1990;64:2669–2677. doi: 10.1128/jvi.64.6.2669-2677.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drew J, Belsham G J. trans complementation by RNA of defective foot-and-mouth disease virus internal ribosome entry site elements. J Virol. 1994;68:697–703. doi: 10.1128/jvi.68.2.697-703.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evan G I, Lewis G K, Ramsay G, Bishop K M. Isolation of monoclonal antibodies specific for c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feduchi E, Aldabe R, Novoa I, Carrasco L. Effect of poliovirus 2Apro on vaccinia virus gene expression. Eur J Biochem. 1995;234:849–854. doi: 10.1111/j.1432-1033.1995.849_a.x. [DOI] [PubMed] [Google Scholar]
- 12.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorlach M, Burd C G, Dreyfuss G. The mRNA poly(A)-binding protein: localization, abundance, and RNA-binding specificity. Exp Cell Res. 1994;211:400–407. doi: 10.1006/excr.1994.1104. [DOI] [PubMed] [Google Scholar]
- 14.Gradi A, Imataka H, Svitkin Y V, Rom E, Raught B, Sonenberg N. A novel functional human eukaryotic translation initiation factor 4G. Mol Cell Biol. 1998;18:334–342. doi: 10.1128/mcb.18.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hassett D E, Lewis J I, Xing X, DeLange L, Condit R C. Analysis of a temperature-sensitive vaccinia virus mutant in the viral capping enzyme isolated by clustered charge-to-alanine mutagenesis and transient dominant selection. Virology. 1997;238:391–409. doi: 10.1006/viro.1997.8820. [DOI] [PubMed] [Google Scholar]
- 16.Jewell J E, Ball L A, Rueckert R R. Limited expression of poliovirus by vaccinia virus recombinants due to inhibition of the vector by proteinase 2A. J Virol. 1990;64:1388–1393. doi: 10.1128/jvi.64.3.1388-1393.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaminski A, Howell M T, Jackson R J. Initiation of encephalomyocarditis virus RNA translation: the authentic initiation site is not selected by a scanning mechanism. EMBO J. 1990;9:3753–3759. doi: 10.1002/j.1460-2075.1990.tb07588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuhn U, Pieler T. Xenopus poly(A) binding protein: functional domains in RNA binding and protein-protein interaction. J Mol Biol. 1996;256:20–30. doi: 10.1006/jmbi.1996.0065. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Medina M, Domingo E, Brangwyn J K, Belsham G J. The two species of the foot-and-mouth disease virus leader protein, expressed individually, exhibit the same activities. Virology. 1993;194:355–359. doi: 10.1006/viro.1993.1267. [DOI] [PubMed] [Google Scholar]
- 22.Merrick W C, Hershey J W B. The pathway and mechanism of eukaryotic protein synthesis. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 31–69. [Google Scholar]
- 23.Moss B. Poxviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2637–2671. [Google Scholar]
- 24.Roberts L O, Belsham G J. Complementation of defective picornavirus internal ribosome entry site (IRES) elements by the coexpression of fragments of the IRES. Virology. 1997;227:53–62. doi: 10.1006/viro.1996.8312. [DOI] [PubMed] [Google Scholar]
- 25.Roberts L O, Seamons R A, Belsham G J. Recognition of picornavirus internal ribosome entry sites within cells: influence of cellular and viral proteins. RNA. 1998;4:520–529. doi: 10.1017/s1355838298971989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robertson, M. E. M., R. A. Seamons, and G. J. Belsham. Submitted for publication.
- 27.Sachs A B, Sarnow P, Hentze M W. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Schmelz M, Sodeik B, Ericsson M, Wolffe E J, Shida H, Hiller G, Griffiths G. Assembly of vaccinia virus—the second wrapping cisterna is derived from the trans-golgi network. J Virol. 1994;68:130–147. doi: 10.1128/jvi.68.1.130-147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneider R J. Cap-independent translation in adenovirus-infected cells. Curr Top Microbiol Immunol. 1995;203:117–129. doi: 10.1007/978-3-642-79663-0_6. [DOI] [PubMed] [Google Scholar]
- 31.Stone D M, Almond J W, Brangwyn J K, Belsham G J. trans complementation of cap-independent translation directed by poliovirus 5′ noncoding region deletion mutants: evidence for RNA-RNA interactions. J Virol. 1993;67:6215–6223. doi: 10.1128/jvi.67.10.6215-6223.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tarun S Z, Sachs A B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 33.Turner P C, Young D C, Flanegan J B, Moyer R W. Interference with vaccinia virus growth caused by insertion of the coding sequence for poliovirus protease 2A. Virology. 1989;173:509–521. doi: 10.1016/0042-6822(89)90563-1. [DOI] [PubMed] [Google Scholar]