Abstract
Rapidly detectable and easily accessible markers of tumor cell death are needed for evaluating early therapeutic efficacy for immunotherapy and chemotherapy so that patients and their physicians can decide whether to remain with a given therapeutic strategy. Currently, image-based tests such as computed tomography scans and magnetic resonance imaging are used to visualize the response of a patient’s tumor, but often these evaluations are not conducted for weeks to months after treatment begins. While serum levels of secreted proteins such as carcinoembryonic antigen and prostate specific antigen are commonly monitored to gauge tumor status during therapy and between image evaluations, the levels of these proteins do not always correlate well with the actual tumor response. In laboratory studies, it has been shown that tumor cells undergoing apoptosis can release cellular components into cell culture media such as cytochrome c, nucleosomes, cleaved cytokeratin-18 and E-cadherin. Studies of patient sera have found that these and other macromolecules can be found in circulation during cancer therapy, providing a potential source of material for monitoring treatment efficacy. In the future, analysis of biofluids from severe combined immunodeficiency mice bearing patient tumor specimens treated with a targeted therapy such as Apo2L/tumor necrosis factor-related apoptosis-inducing ligand will be useful in the preclinical identification of therapy response markers. In this review, the current status of the identification of serum markers of tumor cell apoptosis is provided, as well as a discussion of critical research questions that must be addressed and the considerations necessary when identifying a marker that reflects true clinical outcome.
Keywords: Apoptosis, Treatment response markers, Treatment efficacy markers, Monitoring cancer therapy
Introduction
Apoptosis in normal and malignant cells
A novel use of the word, “apoptosis,” was proposed in 1972 when Kerr et al. [43] used it to describe a controlled mechanism of cell death, suggesting that it was a process that complemented mitosis. Now it is accepted that apoptosis is a type of programmed cell death that is highly regulated; a process during which chromatin condenses, caspases are activated, and DNA is systematically fragmented. Furthermore, unlike necrosis, an immune response is typically not thought to be generated during apoptosis, however, some studies have shown that apoptotic T cells can induce proinflammatory cytokine secretion from phagocytes [95] and that treatment of resident macrophages in vivo with FasL leads to the production of proinflammatory cytokines [35].
Apoptosis is an important part of normal cellular homeostasis and regulation in humans. Specifically, apoptosis plays a significant role in lymphocyte homeostasis including the eliminating B cells that express auto-antibodies against self antigens as well as controlling the duration of T cell activation through the FasL-Fas mediated apoptotic pathway [77]. The ability to induce apoptosis in virally infected or malignant cells is an essential feature of immune cells like cytotoxic T lymphocytes and natural killer cells that are crucial to the healthy survival of an organism [70, 80]. When the normal balance of populations of apoptotic and proliferating cells is disturbed, this can lead to the development of diseases like cancer and autoimmunity.
In tumors compared to normal tissue, spontaneous apoptosis is increased and often associated with tumor cell turnover [92]. Higher baseline apoptotic indices in (untreated) tumors are associated with undifferentiated malignancies and lower survival rates [59]. Upon treatment of a tumor with a therapy that induces killing, tumor size regresses and this can be attributed to apoptosis or senescence and seems to be tumor type specific [82]. Restoration of p53 leads to tumor regression primarily by apoptosis in lymphomas, and senescence, although delayed, in sarcomas [98]. In vivo imaging of annexin-V+ breast tumor cells correlates with a reduction in tumor size following treatment with Herceptin® and paclitaxel while therapy resistant cells do not exhibit tumor regression or increased annexin-V staining [27]. Patient colon tumors grown as xenografts in severe combined immunodeficiency (SCID) mice and treated with Apo2L/tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) and CPT-11 experienced tumor regression that correlated to increased numbers of tumor cell apoptosis as quantified with the terminal deoxynucleotidyl transferase biotin-dUTP nick end ligand (TUNEL) assay [64]. Although apoptosis is not the only mechanism by which tumor cells die in response to therapy, there is evidence that apoptosis significantly contributes to the death of the tumor and because of this, the quantification of this process may have potential as a surrogate marker of response.
In this review, we discuss the difficulties with the current methods available for monitoring patients’ responses to cancer therapy as well as the need for optimally measuring treatment-induced responses in real time using easily obtainable bodily fluids. Preclinical and clinical studies have already begun to elucidate which molecules are indicative of tumor cell apoptosis and these candidate molecules, summarized below, will need further validation before they are utilized by clinicians. These recent discoveries expose several challenges to identifying markers of therapeutic efficacy, including: the component of blood most likely to contain the information of interest, the preclinical model system best suited for study and the functional relevance of a tumor cell to release molecular ‘signals’ during apoptosis, to name a few.
Obstacles to monitoring tumor response to therapy
Major efforts by large numbers of researchers have led to the identification of novel therapies for cancer and this success has raised hopes for increased survival and improved patient selection. The identification of targeted therapies in addition to improvements in the use of more standard therapies has provided a substantial increase in the number of Phase I and II trials that are available for advanced cancer patients. However, the availability of new cancer drugs serves to highlight the existence of a major concern in the treatment of cancer: how to accurately and continuously measure a patient’s tumor response to therapy so that patients and their clinicians know as early as possible whether a particular therapeutic choice is actually working, and if it is not, when to choose an alternative approach as early as possible, before a tumor develops additional protective escape mechanisms. Many times, the tumor response is not evaluated until after an entire course of therapy, often months after the initial treatment, and even at that time, it is difficult to measure with current methods. In the clinic, X-rays, ultrasound, and computed tomography (CT) scans are commonly used to assess treatment efficacy by imaging the tumor [105]. These tests are not only costly, but provide the clinician with little information about the molecular response of the tumor to the treatment. There is a need in the clinic to be able to measure a tumor’s response to cancer therapy in real time by sampling the tumor throughout the course of treatment, beginning hours or days after the first therapeutic administration. For example, CPT-11 (camptothecin-11, camptosar®, irinotecan), a potent drug that many patients with metastatic carcinoma of the colon and rectum receive, inhibits topoisomerase I, causing cells to die by G2/M cell cycle arrest and apoptosis. But even for this commonly used treatment (one whose mechanism of action is understood at the molecular level), it is difficult to know whether it is actually resulting in apoptosis of tumor cells. If tumors could be ‘sampled’ or biopsied repeatedly from patients during drug treatments, evidence of this specific DNA damage and/or dead cells could be used to evaluate drug efficacy. However, in patients, repeated invasive procedures to obtain biopsies are not feasible for several important reasons, including the fact that in advanced cancer, tumors may be disseminated and located in organs such as the lung, liver or bone marrow, making them difficult to biopsy. Instead, patients are examined by CT scans at the end of a given course of therapy, and unfortunately, during this time interval many patients’ tumors may develop drug resistance and no longer respond favorably. In this case, continued inappropriate use of an ineffective therapeutic on tumor cells that are not induced to undergo apoptosis and have infinite potential to proliferate [31] may actually lengthen the period of time for tumor progression to occur. When an effective drug is selected, the disease may be more advanced, making treatment more difficult.
In current practice, surgical resection is the initial treatment for early stage colon cancer; adjuvant chemotherapy is used for stages III and IV [11]. Patient follow-up for colon cancer usually involves monitoring progress with a colonoscopy one year post-treatment [1] while further evaluation may involve a chest X-ray or CT scan to check the patient for possible metastasis. These methods are relatively insensitive, costly methods used to evaluate the patient months after treatment for changes in morphology that can be recognized by eye. However, by the time the imaging tests are carried out, the tumor may be quite large, making therapy all the more difficult. Currently, response evaluation criteria in solid tumors (RECIST) and the World Health Organization (WHO) provide guidelines for measuring (unidimensional and bidimensional, respectively) and quantifying patient tumor response to therapy [40]. After tumor dimensions are obtained with electronic calipers from CT or magnetic resonance imaging (MRI) images, patient responses are categorized into one of four groups: progressive disease, stable disease, partial response or complete response. While measuring a tumor that has smooth, defined boundaries is possible, accurately quantifying a tumor that has an irregular structure or has indistinguishable margins that merge with nearby organs can be technically challenging [40] and tumors that have spread to bone are “nonmeasureable” according to RECIST criteria (as stated in [50]). Clinically accessible molecular markers of therapeutic response that could be used in combination with these methods would provide an additional way to validate a decision to continue with the current therapy or change it in order to improve efficacy. Markers of on-going response could be invaluable especially in the case of “stable” disease when tumor cells could be proliferating as quickly as they are being killed by chemotherapy, although such markers may not be relevant in the case of treatment with tyrosine kinase inhibitors, such as Tarceva®, whose main mechanism of action is to slow tumor cell proliferation. If the number of dying cells were equal to the number of proliferating cells, this would be an important fact to know, a quantification which is currently not feasible in the clinic.
Although there are some blood-sampling methods to assess efficacy and the possible recurrence of colon cancer, they cannot be relied on solely. For example, carcinoembryonic antigen (CEA), the preferred tumor antigen used to monitor metastatic colorectal cancer following chemotherapy, often does not provide useful information about the response of the tumor to therapy. A decrease in serum CEA levels is expected with a response to chemotherapy, however, it has been reported that CEA is shed from 5-fluorouracil-treated colon tumor cells in vitro [4] and when treating patients with oxaliplatin, a significant increase can occur within 4 weeks from the start of treatment that does not correlate with disease progression [88, 89]. An earlier study reported that 16% of patients that did not have a colon cancer recurrence had a false-positive CEA test [62]. Further, tests for markers like CEA are usually performed every 1–3 months following therapy [57, National Comprehensive Cancer Network (NCCN) Practice Guidelines] when a test that could evaluate therapeutic efficacy within hours or days of the start of treatment would be much more useful. Other potential biomarkers that could be used to monitor therapeutic efficacy, such as CA 19-9, just do not have enough supportive clinical data to justify their use in these situations [57]. Even the use of common markers of other types of cancer for treating monitoring, like prostate specific antigen (PSA) that has been widely used in the diagnosis of prostate cancer, remains controversial [26, 100].
One exception to the types of biomarkers mentioned above is one that has been very effective in detecting cancer and monitoring therapy is in pediatric oncology. Alpha-fetoprotein (AFP) has shown to be very sensitive in the detection of hepatoblastoma and upon treatment with chemotherapy, sera levels decrease in patients [79]. The weekly monitoring of AFP serum levels has also shown promise as a tool that can guide therapy protocols in order to diminish toxic side effects by selecting an alternative therapy at the appropriate time.
Currently used biomarkers
The importance of detecting cancer as early as possible is evident when the survival data for cancers such as breast, colorectal, and prostate are examined by the stage of diagnosis. There is an increase in 5 or 10-year survival for these diseases when cancer is detected at an earlier stage surveillance epidemiology and end results (SEER) data (as discussed in [22]) mostly because of the lack of curative treatments available for late-stage cancer patients, although survival rates are increasing in some late stage cancers, mainly due to new, effective targeted therapies [69]. Because detecting and diagnosing cancer early usually correlates with a positive survival outcome, a large body of work has focused on the discovery and validation of biomarkers for detection of cancer, including biomarkers for the detection of prostate, colon and ovarian cancers (as reviewed in [9, 11, 24]). Besides detection or disease biomarkers, there are several other categories of molecules used as predictive tests in medicine- from surrogate endpoints of heart disease such as cholesterol levels to toxicity markers such as the induction of cytochrome P-450 which can be monitored for indication of adverse effects of a therapy [6] as well as markers of drug sensitivity. Different types of biomarkers seem to have their own niche for aiding in the process of diagnosis, drug sensitivity prediction, as well as in monitoring treatment. Not only is it beneficial to detect a patient’s disease in the earliest stage and to choose a therapy based on genomic or proteomic data that would imply sensitivity, but these approaches should also be useful to confirm that the tumor is responding as predicted and as a direct effect of the initial treatment. Espina et al. [21] as part of suggestion for a new paradigm for the detection and treatment of cancer, emphasizes the need for monitoring a patient’s tumor response to therapy in real-time. This group also pointed out that a panel of biomarkers, not just a single one, may be necessary, especially when delivering combination therapy to patients.
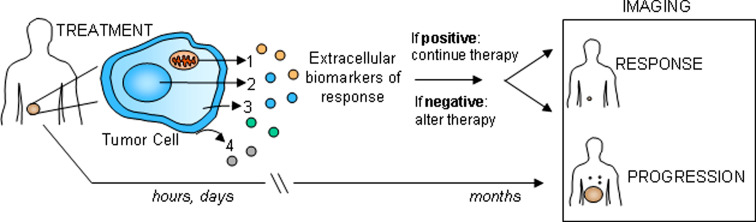
Thus, there is increasing interest in identifying markers of therapeutic efficacy in cancer, also referred to as apoptotic or outcome markers [6, 37]. A blood test is a simple, non-invasive method to collect information about a patient’s tumor, especially for a malignancy that cannot be easily sampled by biopsy because of its location. The proposed use of these markers is outlined in Fig. 1. Ideally, a tumor could be monitored during the first hours and days following therapy to determine whether or not treatment is effective. If markers of treatment efficacy are present, the patient would remain on the initial therapy, however, if the markers are absent, therapy would be altered accordingly in order to optimize the disease outcome in the most time efficient way. Alternatively, patients monitored solely with imaging methods weeks to months after therapy are potentially at risk for advanced disease progression, and by that time, there may be few treatment options available for improving outcome.
Fig. 1.
Strategy for monitoring tumor response in the clinic. After detection, a first-line cancer therapy is chosen and traditionally, follow-up is conducted months later with imaging techniques such as X-ray, computed tomography (CT) scan or magnetic resonance imaging (MRI). This time delay could result in the progression if the tumor is not sensitive to the initial therapy. With therapeutic efficacy or response markers, blood could be sampled from patients within hours to days following the administration of treatment. These biomarkers may have several origins: 1 mitochondria, 2 nucleus, 3 cytosol or 4 membrane (see the text and table for details). At this time the absence or presence of biomarkers could guide the course of therapy by providing more information about the immediate response of the tumor. When response markers are not detected, the therapy would be adjusted to another available, alternative therapeutic agent. Once again, blood would be sampled and the presence of biomarkers assessed
Inducing and monitoring tumor cell apoptosis
The major mechanisms of cell kill of many chemotherapeutic agents are to induce apoptosis in rapidly proliferating cells [42]. Doxorubicin, an intercalating agent that inhibits topoisomerase II, cisplatin, an interstrand DNA cross-linking agent, and 5-fluorouracil, anti-metabolite, are all commonly used chemotherapeutics that have been shown to cause apoptosis in cells cultured in vitro [85]. After the initial chemotherapeutic insult to the DNA, cellular damage is sensed by p53 and then a decision is made to repair the damage or undergo apoptosis through the mitochondrial pathway [33]. It has recently been shown that DNA damaging agents can induce PUMA in a p53-dependent manner to initiate apoptosis in tumor cells [109]. Although therapy-induced apoptosis is the focus of this review, we acknowledge that anticancer treatments can induce other forms of cellular death including autophagy, extrinsic senescence, and necrosis (as reviewed in [29, 44]) and that these processes may occur simultaneously with apoptosis and could significantly contribute to tumor responses.
Other targets of cancer therapy are directed towards activating the extrinsic pathway, mainly the members of the tumor necrosis factor (TNF) superfamily of receptors, including: DR4 (TRAIL-R1), DR5 (TRAIL-R2), Fas (CD95), and TNF-R1. While Apo2L/TRAIL has shown to induce apoptosis in malignant cells but not normal cells [5] [102] through the activation of DR4 and DR5, TNF alpha has demonstrated severe toxic side effects in the clinic including manifestation of pro-inflammatory responses and organ failure [34]. Additionally, antibodies to Fas have led to lethality in mice due to the destruction of the liver [67]. During activation of the extrinsic pathway, FasL or Apo2L/TRAIL binds to a trimerized death receptor and recruits Fas-associated death domain (FADD) to the cytoplasmic region of the receptor. Caspase-8 or -10, initiator caspases, are then recruited to FADD as part of the death-inducing signaling complex (DISC) where they undergo autoproteolysis to take an activated form [13, 45]. Activated initiator caspases are then able to prompt cleavage of effector caspases-3, 6, and 7, inducing apoptosis [3]. Crosstalk between the extrinsic and intrinsic pathways occurs when activated caspase-8 cleaves Bid, activating the mitochondrial pathway [54, 58]. Through the activation of pro-apoptotic molecules Bax and Bak in the mitochondria, the transmembrane mitochondrial potential is disrupted, causing the release of cytochrome c into the cytosol. Cytochrome c then interacts with Apaf-1, assembling the apoptosome where caspase-9 is activated [30]. Subsequent activation of the effector caspases brings the cell closer to completion of the apoptotic signaling cascade and its demise.
Non-invasive imaging technologies to detect apoptosis in vivo
The process of apoptosis is easily detected in vitro, relying on the measurement of intracellular or surface molecules; however, the measurement of apoptosis in vivo can be much more challenging. In vitro, apoptosis is readily observed when phosphatidylserine is translocated to the outer leaflet of the plasma membrane [99]. Other biochemical features characteristic of apoptosis are: activated caspases, chromatin condensation, cytoplasmic shrinkage, membrane blebbing, formation of apoptotic bodies, DNA fragmentation and laddering [81].
Other techniques are in development for the detection of apoptosis in vivo when patient tumor specimens are not accessible. Besides using CT scans or X-rays to monitor a patient’s response to treatment by measuring tumor size, newer imaging techniques are available to provide more sensitive monitoring in vivo by monitoring cellular processes. In fact, radiopharmaceuticals like 18F-deoxyglucose, a glucose analogue, is can be utilized to monitor glucose uptake or tumor cell proliferation during a course of treatment like radiotherapy (as reviewed in [97]). Although 18F-deoxyglucose (FDG)–positron emission tomography (PET) is a sensitive method to measure individual tumor cell response, the resolution is only 3–7 mm (as stated in [65]) and it does not identify the dying cells, only those that continue to undergo active metabolism during treatment. In addition, it is also possible that tumor-infiltrating immune cells, also requiring energy to function, could take up FDG and misconstrue the signal perceived to come from the tumor. While this is a method to measure tumor response, it would be useful to quantify cell death due to cancer therapy.
New technology uses 99mTc-radiolabeled annexin-V to locate apoptotic cells in vivo and visualize the location with single photon emission computerized tomography (SPECT) imaging (as reviewed in [97]). In a study performed by Belhocine et al. [10], fifteen patients diagnosed with lung cancer, lymphoma or breast cancer were monitored for 99mTc-radiolabeled annexin-V uptake before and in the hours following the first course of chemotherapy. Seven patients were observed to have an increased uptake of 99mTc-radiolabeled annexin-V at the site of the tumor as compared with pre-treatment analyses. Upon follow-up all seven patients had either a complete or partial response to chemotherapy. These exciting results are promising for the development of a new non-invasive measurement of apoptosis; however, the study also identified patients that although showed no significant tracer uptake following chemotherapy, went on to have complete or partial responses to treatment. Other methods of assaying apoptosis in vivo are in development. A nanoprobe that utilizes the C2A domain of synaptotagmin I, a protein that also binds to phosphatidylserine, carries a biotin tag that allows for its binding to Gd3+-labeled avidin for image contrast in tumors during detection with MRI [66].
The FDG-PET technique has shown promise in preclinical models as a measure of cell viability based on glucose metabolism. In a treatment response study, human non-small cell lung cancer cell lines that were sensitive or resistant to gefitinib, an epithelial growth factor receptor (EGFR) kinase inhibitor, were studied [93]. Cellular uptake of FDG measurements were established in vitro after treatment with gefitinib then the responses were confirmed in a SCID mouse xenograft model where FDG uptake was calculated with microPET. Within 2 days of the start of treatment, treated mice bearing the EGFR kinase inhibitor-sensitive cell lines demonstrated about a 50% decrease in FDG uptake as compared with mice bearing the resistant cell lines where no change was observed [93]. The authors of this study cautioned that even though dramatic results were found with the SCID mouse model, it may be more difficult to measure responses in humans, especially since there is much more heterogeneity in a patient tumor as compared with that of a cell line. In fact, Gillham et al. [28] reported that pathological response of patients with esophageal cancer who underwent FDG-PET imaging seven days after the start of chemotherapy and radiation could not be correlated with the metabolic response of the tumor. Results of FDG-PET imaging were compared with histological data for classifying responses. This study also cited that an inflammatory response, resulting from radiation treatment, could have confounded the ability to measure the metabolic response of the tumor cells alone to treatment.
Because of the possibility that FDG-PET measures tumor cell proliferation as well as any cells involved in an inflammatory response, the use of 18F-fluorothymidine–PET has been explored. Once in the cytosol, [18F]-18F-fluorothymidine (FLT) is phosphorylated by the cell cycle regulated enzyme, thymidine kinase 1 [84]. Used as a measure of thymidine kinase activity and to identify cells in S-phase, [18 F]-FLT has been shown to correlate better with tumor cell proliferation than FDG-PET when RIF-1 tumors were treated with cisplatin in vivo. Assessments were made by comparing the imaging methods to immunohistochemical studies of the tumors for localization of the cell proliferation marker, PCNA [53].
Other types of imaging utilize the fluorescent properties of molecules to localize tumor cells in mice and, in other studies, to monitor targeted therapies. Transfection of tumor cells with red fluorescent protein for visualization of the cytoplasm and with green fluorescent protein linked to histone H2B allowed for differential visualization of the cytoplasm and nucleus when the tumor cells were injected intravenously in mice and tracked with a whole-mouse imaging system [108]. It is clear that using fluorescent or bioluminescent imaging methods for tumor cell detection could be used to monitor a tumor’s response to therapy in mice, measuring tumor burden in real-time [20], certainly more sensitive than characterizing a therapeutic response with a series of tumor growth measurements, as well as providing information about tumor cell extravasation and metastasis. These methods are very useful in preclinical evaluations of drug efficacy on tumor metastasis, but are not directly translational for monitoring therapy responses in patients. However, with diffusion MRI, a more clinically relevant approach can be used to monitor tumors. Once tumor cell viability is compromised, movement of water occurs and this change can be quantified with MRI and functional diffusion mapping [50].
Using a novel construct, a study has based therapeutic response detection on the cleavage of caspase 3 during photodynamic therapy (PDT)- induced apoptosis. A photosensitizer for PDT was engineered for targeting tumor cells that overexpress the folate receptor [90]. In addition to the folate molecule for targeting to the receptor, the construct includes the following elements: a photosensitizer, a fluorescent quencher and a caspase-3 cleavage sequence. This agent can be added to cell cultures or delivered by intravenous injection to mice bearing tumors. Once inside the tumor cell, when light is applied in the presence of oxygen, the photosensitizer is activated causing the targeted cell to undergo apoptosis. During this process, caspase-3 is activated and it can then cleave the target sequence on the construct, thereby releasing the quencher and producing a fluorescent signal [90]. Although this type of therapeutic coupled with an apoptotic detection agent is still under development and has some limitations (e.g. peptide degradation when delivered in vivo and photobleaching) [90], the design of targeted therapies that possess multifaceted domains could provide some of the most promising methods for detecting tumor cell apoptosis.
While the above mentioned features are characteristic to or can be correlated with apoptosis, they lack the high throughput capabilities that are conducive to classifying patient response quickly in the clinic. The discovery of reliable markers of tumor response that are shed or released from apoptotic tumor cells, released into circulation and then quantified are needed for closely monitoring and guiding cancer therapy. Several ongoing investigations have identified potential extracellular markers of treatment response.
Tumor-derived markers of apoptosis found extracellularly or in circulation
As demonstrated by the number of studies conducted to identify a variety of components that are released from tumor cells during treatment-induced apoptosis, this is an area that has potentially important implications for the noninvasive, real-time measurement of therapeutic response. Unfortunately, it is not yet clear how cellular constituents make their way outside of the cell, especially in the case for components that are not normally found on or near the cell membrane. Of functional interest is the biological role that proteins, DNA, lipids and others may have as they reach the extracellular space. The section below provides a summary of the current literature that identifies potential candidate markers of a response to cancer therapy. A summary of molecules and other cellular components discussed in this section that are released extracellularly can be found in Table 1.
Table 1.
Cellular components or molecules whose levels are altered in the extracellular environment during treatment-induced apoptosis
| Origin | Marker | Released into | Inducer | Tumor | Timecourse | Reference |
|---|---|---|---|---|---|---|
| Mitochondria | Cytochrome c | Culture media | Anti-CD95, staurosporine, eptoposide, doxorubicin | Jurkat | Hours | [8, 78] |
| Serum | Multidrug chemotherapy | Hematological cancer | Days | [8, 78] | ||
| Nucleus | Nucleosomes | Cell surface | Camptothecin | Jurkat | Hours | [75] |
| Culture media | Cisplatin | HeLa | Hours | [94] | ||
| Serum | Cisplatin | HeLa in BALB/c nu/nu mice | 1 day | [94] | ||
| Serum | Gemcitabine oxaliplatin | Cervical carcinoma | Hours–days | [94] | ||
| Serum | Chemotherapy | NSCLC | Days | [38] | ||
| Serum | Radiochemotherapy | Pancreatic cancer | Hours–days | [49] | ||
| Serum | Radiochemotherapy | Colorectal cancer | Hours–days | [48] | ||
| Serum | 5- Fluorouracil + folinic acid ± irinotecan | Colorectal cancer | Days | [36] | ||
| DNA | Serum | Multidrug chemotherapy | Acute myeloid leukemia | Days | [63] | |
| Culture media | Staurosporine ± oligomycin | Jurkat | Hours | [41] | ||
| Plasma | Anti-CD95 or acetaminophen | Normal liver in BALB/c mice | Hours | [41] | ||
| Serum | Tamoxifen | Breast cancer | 1 year | [25] | ||
| Cytosol | Cleaved CK18 | Culture media | Mitomycin C | MCF-7 | Hours | [83] |
| Serum | Docetaxel or cyclophosphamide + epirubicine + 5-fluorouacil | Recurrent breast cancer | Days | [12] | ||
| Culture media | Cisplatin | MDA-MB-231 | Days | [46] | ||
| Serum | Estramustine phosphate + docetaxel or vinorelbine | Hormone refractory prostate carcinoma | Days | [47] | ||
| Membrane | MMP-2 | Serum | Chemotherapy | Metastatic colorectal cancer | Weeks | [32] |
| sFas | Serum | Chemotherapy | Various | Hours–days | [73] | |
| Lipids | Culture media | Vincristine, several others | ALL-697 | Hours | [110] | |
| E-cadherin | Culture media | Staurosporine | H184A1 | Hours | [91] | |
| Culture media | Apo2L/TRAIL | Colo 205 | Hours | see Fig. 2 |
Cytochrome c
Cytochrome c, a 14.5 kDa protein, is located in the inner mitochondria membrane in healthy cells and is a key component of the electron transport chain, functioning as an electron shuttle. During apoptosis, cytochrome c is released from the mitochondria into the cytosol where it can bind to Apaf-1 [111]. It has been shown that soluble cytochrome c is rapidly released into the cell culture media after the triggering of apoptosis in tumor cells with anti-CD95, staurosporine, eptoposide and doxorubicin in vitro [78]. Upon inducing necrosis, cytochrome c remained cellular and was not found in the media, while during apoptosis, cytochrome c was found as early as 1 h in the media following treatment [78]. Lactate dehydrogenase (LDH), already known to be secreted during cell death, was released much later after an apoptotic stimulus was given than cytochrome c, suggesting that the two proteins exit the cell via a different mechanism [78].
These in vitro findings prompted investigation of cytochrome c in patients who had received chemotherapy. Sera from patients with hematological malignancies were analyzed for cytochrome c levels during the first eight days of therapy. Although the response was variable, most patients experienced an increase in cytochrome c concentration and then a decrease during the later stages of the course of chemotherapy [78]. The patient sera studied described here included only 17 patient samples and other than measuring sera LDH levels, there was no correlation to the cytochrome c levels and patient response.
In 2005, Barczyk et al. [8] observed that patients undergoing chemotherapy for their disease generally had an increase in levels of cytochrome c during the first couple of days following the start of therapy [8]. However, those patients whose tumors responded well to the therapy and were alive three years later, had much lower sera levels of cytochrome c (<25 ng/ml) overall. Sudden spikes and variations in levels of cytochrome c could be attributed to chemotherapy-related liver toxicity or an inflammatory response in 2 of the patients and perhaps variable kidney clearance rates of the protein among patients. Because of the small patient sample sizes in these studies and without image or histological analyzes performed at the same time points as the levels of cytochrome c were measured, it is difficult to correlate an early patient response with cytochrome c levels thus far.
Although the biological significance of the extracellular release of cytochrome c is not fully understood, there has been speculation that its presence in the extracellular space could have anti-inflammatory effects, or perhaps guide phagocytes to the site of injury for corpse clearance [78]. Others have suggested that this protein could contribute to the bystander effect since exogenous cytochrome c added to the cell culture media of neurons increased their apoptosis as compared with controls [2]. There is also evidence that cytosolic cytochrome c may contribute to the cell-shrinkage associated with apoptosis by interacting with K+ channels in the cell membrane to induce K+ efflux from the cell [74].
Nucleosomes and DNA
Nucleosomes provide a mechanism for chromatin compaction and consist of four pairs of histones, encircled by DNA. During apoptosis, DNA is systematically cleaved between the histones/nucleosomes by endonucleases [107]. As evidence that nucleosomes can be exposed to the extracellular environment, autoimmune responses can be generated against nuclear antigens; autoantibodies to nucleosomes are found in systemic lupus erythematosus patients [16]. Normally positioned in the nucleus, nucleosomes were exposed on the surface of apoptotic Jurkat T cells suggesting that there is a mechanism by which they are directed out of the nucleus and to the cell membrane during cell death [75]. As pointed out by this group, it is possible that the display of nucleosomes on the cell surface may provide a signal to phagocytes for clearance of these cells. However, the function of nucleosomes that are shed extracellularly is less clear. In the studies examined in this review, while lower baseline and post treatment nucleosome sera levels in vivo tended to correlate with better patient response in general, the kinetics of the nucleosome levels varied based on the treatment and tumor type and therefore, the utility of nucleosomes as a biomarker of therapeutic response remains questionable.
Trejo-Bercerril et al. [94] begin to elucidate the release of nucleosomes from tumor cells in vitro and then in vivo in rodent models and in patients. When HeLa cells were treated with cisplatin in vitro, this group reported that nucleosome levels increased in the cell culture supernatant through 24 h. In BALB/C nu/nu mice bearing HeLa tumors, a decrease in nucleosome levels were found 24 h after treatment with cisplatin when compared to untreated mice with a tumor. This group also addressed the issue of the extent to which nucleosome measurements were due to the secretion of these molecules from dying, non-malignant cells during chemotherapy. They found that at therapeutic doses in untreated non-tumor-bearing mice, there was no difference in nucleosome levels as compared with non-tumor-bearing mice at 24 h post-treatment with cisplatin. However, when rats without tumors were administered chemotherapy at toxic doses above normal therapeutic levels, nucleosome levels increased [94]. Five of six cervical carcinoma patients that experienced a decrease in nucleosome levels 24 h following chemotherapy were responders as calculated by tumor size using the two longest perpendicular diameters from CT images [94]. Among the five patients that had an increase in nucleosome sera levels, two patients responded and these patients had only slight increased in nucleosomes compared with the rest of the group. However, when blood was drawn from responders just a few hours after the initiation of treatment, there was an increase in these levels before it eventually tapered off at 24 h, suggesting that the nucleosomes are rapidly cleared from circulation [94].
In a study of 212 non-small cell lung cancer patients undergoing chemotherapy, a prompt increase in serum nucleosomes followed by a decrease in these levels during periods without treatment was experienced and the degree of the response differed between responders and non-responders [38]. Responders or patients with remission based on CT image measurement classified by the WHO guidelines experienced less of an initial increase in nucleosomes followed by a significant decrease in levels until day eight [38]. In comparison, non-responders (patients classified as having progression or no change) experienced a dramatically greater initial increase, meaning that these patients experienced an increase in nucleosome levels and these levels did not drop as quickly as responders [38]. Some of the reasons suggested by the authors for the difference between response groups are the variable rates of cell death and proliferation in the aggressive tumors in the non-responder group as well as increased access to blood vasculature in patients with metastasis.
Kremer et al. [49] found that nucleosome levels increased within hours of radio- and chemotherapy treatment in pancreatic cancer patients but that these levels dropped by 6 h and often rose again before falling for a second time. As stated, the delay in increased levels after 6 h could be attributed to damaged tumor cells attempting to repair themselves unsuccessfully and then finally surrendering to apoptosis. After analyzing the area under the curve for nucleosome concentration at several time points, the observation was made that in general, patients with no progression had lower overall concentrations of nucleosomes during days 1–3, while patients with progression had higher levels in their sera during this time [49]. Patient responses in this study were categorized based on sera levels of CA 19-9. Similar results were obtained in a study of colorectal cancer patients receiving radiochemotherapy [48].
In a study of colorectal cancer patients undergoing a variety of chemotherapy regimens, a combined group of adjuvant and palliative patients experienced increases in nucleosomes in sera during the therapy, followed by decreases in periods of no treatment [36]. Patients were assessed for their treatment response based on CT scans and sonography using the WHO method. Baseline nucleosome values measured before each treatment cycle declined in the adjuvant group while the palliative group did not demonstrate this trend [36]. Palliative therapy patients with disease progression had increasing baseline values and high increases in nucleosomes during the treatment cycles while patients without progression demonstrated mostly decreasing nucleosome levels [36].
While many studies examined only the nucleosome levels in the sera, Mueller et al. [63] studied circulating nucleosomal DNA in acute myeloid leukemia patients during their first week of chemotherapy. Response was assessed in these patients by quantifying the number of blasts in the bone marrow and neutrophils and platelets in the blood [63]. In general, the 18 patients with complete remission experienced a slight increase in circulating nucleosomal DNA by day 2 of treatment, followed by a drop in levels over the next few days. Overall, patients without complete remission (n = 7) had lower levels of nucleosomes (as measured by the area under the curve for days 2–4 of treatment) than did those who were classified as complete remission [63]. These results are in contrast to the results of work previously mentioned when patients with solid tumors and lower nucleosome levels experienced better outcomes [36, 49]. Explanations are offered that non-responsive patients with solid tumors may have a large tumor load with rapidly proliferating cells during therapy and perhaps these patients have defective clearance mechanisms as a result of a deficient immune system [63].
These nucleosome studies emphasize the importance of studying the kinetics of a therapeutic response when monitoring molecules that are released from apoptotic cells. Future kinetic studies with a larger number of patient samples from different histological types of tumors and a variety of treatment modalities are needed to elucidate the significance of nucleosome levels for monitoring therapy in the clinic.
There have also been reports of DNA fragments found in the serum of cancer patients [52] presumably because apoptosis and necrosis are occurring in and around the tumor and while macrophages can normally clear this debris, circulatory access to this area might be limited [41]. Increases in soluble DNA fragments were identified in media supernatants when Jurkat T cells were induced to undergo apoptosis or necrosis in vitro [41]. Mice were injected with acetaminophen or anti-CD95 to induce necrosis or apoptosis of liver tissue, respectively [41]. In both in vitro and in vivo models, DNA was released as cells died by either necrosis or apoptosis, however, the most DNA was released from cells undergoing necrosis. Necrosis results in the release of large pieces of DNA but apoptosis, as a regulated process, was characterized by cleaved DNA into multiples of 180 bp [41].
Tumor-specific cell-free DNA has been studied as a surrogate marker of the presence of tumor cells. An epigenetic alteration of RASSF1A is common in cancers, especially in breast cancer [14]. In a study of 148 patients with breast cancer who received tamoxifen as adjuvant therapy, patients who were relapse-free or had the best overall survival manifested a disappearance of DNA methylation of gene RASSF1A in sera [25]. DNA from sera obtained one year after of treatment was analyzed, however, it would be useful to examine the kinetics of the response to identify the earliest possible time that prediction of survival could be determined.
Caspase-cleaved cytokeratin 18
Recently, other groups have turned their focus towards the examination of cleaved cytokeratin that may be released outside of the cell during the process of apoptosis. Intermediate filament proteins such as cytokeratins, specific to epithelial cells and found in most carcinomas, provide a supportive structure for the cell by linking the surface of the nucleus to the cell membrane. Cytokeratin-18 is located in the cytosol, however, its presence has been reported on the cell surface of heptoma cells [104]. Cytokeratin-18 is cleaved at Asp396 by caspases during cell death [51], exposing a neo-epitope that can be detected with the M30 antibody [12, 51]. This is an event that occurs during the early stages of apoptosis since the neo-epitope can be found before cells will react with annexin V [51]. Intracellular cytokeratin-18 is cleaved within 24 h of mitomycin C treatment of MCF-7 cells and this correlates with the release of the Cytokeratin-18 (CK18) fragments into the cell culture media while caspase inhibitors abrogate this effect [83]. Ueno et al. [96] were the first to show the presence of an apoptosis-specific marker in the sera of cancer patients using the M30 antibody, although sera obtained at several time points during therapy was not included in this study.
The presence of CK18-Asp396 is easily detectable in patient sera. As enumerated by Biven et al. [12], inducing apoptosis in only 10% of cells in a tumor with 109 cells should yield 103 U of the CK18 fragment, or a concentration of 330 U/l in plasma when healthy controls contain an estimated 150 U/l. This group also found that in a cohort of patients receiving chemotherapy for recurrent breast cancer with a baseline value of cleaved CK18 lower than 200 U/l, responders had larger increases over those of non-responders. How response was quantified was not outlined in this study.
Kramer et al. [46] reported that caspase-cleaved CK18 comprises only 1% of cytokeratin 18 detected in the media from necrotic MDA-MB-231 in vitro. When MDA-MB-231 was treated with cisplatin to initiate apoptosis, CK18 fragments cleaved at Asp396 (CK18-Asp396) were found in the media at 24 and 48 h. Addition of a caspase inhibitor to these cultures impeded release of CK18-Asp396 but not sCK18 into the media. In patients with endometrial cancer, both sCK18 and CK18-Asp396 were found in increased levels in the sera collected from local pelvic blood during surgery as compared with patients who had benign conditions, but not in increased levels in sera from peripheral blood in the same groups of patients [46]. As the authors pointed out, this observation suggests that the cytokeratin originates from the tumor. In the same study, prostate cancer patients were monitored for soluble CK18 and CK18-Asp396 during second and third line chemotherapy. Variable results were found among patients and response (as measured by change in PSA levels) did not correlate well with CK18-Asp396 [46]. The authors suggest that because of circumstances where PSA levels fall but CK18-Asp396 remains unchanged, it is possible that apoptosis is not the main mechanism of cell death and that perhaps, a measurement for total cell death would be more useful than just measuring apoptosis.
More recently, it has been noted that measuring CK18-Asp396 in patient serum during therapy may be beneficial because it originates from epithelial cells while nucleosomes and cytochrome c, however, can be released from a variety of cell types, including chemotherapy and radiation-sensitive bone marrow cells [12, 47]. In a study of patients with hormone refractory prostate carcinoma, a significant increase in CK18-Asp396 was found in the serum of patients between days 5 and 7 who received estramustine phosphate on days 1–3 and then docetaxel on days 3–5 of the treatment cycle [47]. However, statistically significant increases in CK18-Asp396 were not found after patients received estramustine phosphate or estramustine phosphate and then vinorelbine. The study also reported that baseline levels of PSA and total soluble CK18 correlated with increases in CK18-Asp396 in patients who received docetaxel but a similar association did not exist for those who were treated with vinorelbine, perhaps suggesting that vinorelbine was toxic to non-malignant cells [47]. One patient demonstrated an impressive reduction of liver lesion diameter on a CT scan with a decrease in PSA levels that correlated with spiking CK18-Asp396 levels during third-line therapy with estramustine phosphate/docetaxel. However, the patients in this study had advanced prostate cancer, many of them with metastases and correlations were only made to PSA levels and not to responses as measured by CT images. In a study of breast cancer patients who received neoadjuvant chemotherapy, total CK18 sera levels correlated positively with treatment response and patient survival [68]. Importantly, CK18 from frozen sera samples demonstrated stability even after a series of freeze-thaw cycles [68].
Not only in cancer, but in other diseases, such as hepatitis C virus infection where the cell death of hepatocytes correlates with the severity of the disease, it would be beneficial to be able to monitor the amount of apoptosis with a noninvasive test [23]. Bantel et al. [7] reported an increase in the presence of cleaved CK18 in the sera from HCV-infected patients over healthy controls. This may be a much needed measure of the early stages of apoptosis in this disease, providing another way to detect disease other than liver biopsy or serum alanine aminotransferase levels which are not always elevated in HCV patients (as discussed in [7]).
While apoptotic tumor cell release of CK18-Asp396 in vitro corresponds with therapy-induced death, the conclusions are not as clear in vivo. It appears that there may be a correlation between response to therapy and the level of CK18 fragment, however, more studies are needed that correlate data from a large number of patients to a currently accepted measure of tumor response.
MMPs/TIMPs
Matrix metalloproteinases (MMPs) are known for their enzymatic activities of digesting the basement membranes in tissues. In a study of patients diagnosed with metastatic colorectal cancer that spread to the liver, MMP-2 and tissue inhibitor of MMPs (TIMP)-1 were assayed for in sera collected during various treatments with chemotherapy [32]. Patients were classified based on spiral CT results and according to the WHO standards. The ratio of MMP-2/TIMP-1 increased in patients who experienced tumor control as compared to prechemotherapy levels and a cohort that succumbed to tumor progression [32]. This result seems counterintuitive because increased MMP activity is often correlated with growth of the tumor and invasion, while decreased TIMP would imply a lack of control over metastasis potential. The authors offer the explanation that cell death that occurs during chemotherapy may free MMPs that are located on the cell surface thereby binding TIMP [32].
sFas
A recent preliminary study of cancer patients of various tumor types investigated the plasma concentrations of chemotherapy, and sera concentrations of sFas and the CK18-Asp 396 neo-epitope during the first 48 h of therapy [73]. The results indicated that there was a correlation between the concentrations of sFas and 5-fluorouracil or cisplatin but not with anthracyclines or ifosfamide. The time it took to reach a maximum increase in sFas seemed to be therapy-dependent [73]. Patients with baseline sFas concentrations below the median value had an increased overall survival. However, levels of sFas during chemotherapy provided little information about survival data. It is significant that in these patients, an increase in CK18 neo-epitope concentrations above the median correlated with better survival than those patients with lower values during treatment. Because of the small number of patients studied and the variety of tumor types included, it is difficult to come to conclusive results about the data.
Lipids
A study of cellular membranes found that lipids are released during the process of apoptosis. Zhang et al. [110], labeled cell membrane lipids with 3H and measured their release into the cell culture media from tumor cells in vitro during apoptosis. The released radioactive lipids were mostly neutral lipids, such as triacylglycerol, and phospholipids, like phosphatidylcholine. Electron microscopy revealed the release of fragments from cell membranes, mitochondria and organelles just 6 h after vincristine treatment. Overexpression of Bcl-2, and inhibitor of apoptosis, protein kinase C activation and inhibition of caspases all resulted in prevention release of lipids from the cells. The release of lipids was a general occurrence; it did not depend on the cell line or the inducer of apoptosis.
E-cadherin
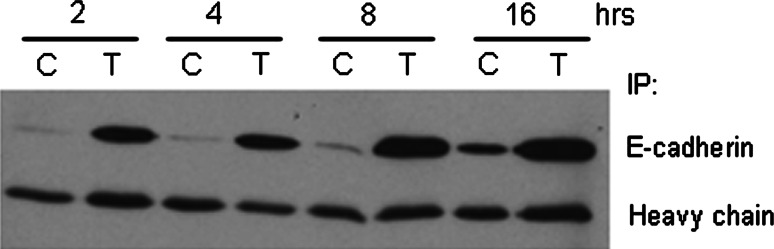
During apoptosis, transmembrane E-cadherin is cleaved in the intracellular domain by activated caspase-3 and in the extracellular domain by metalloproteinases [91]. As a result of the extracellular cleavage, an 84-kDa product of E-cadherin is shed into the surrounding environment within 6 h of staurosporine treatment and this shedding is reduced when cells are treated with an MMP inhibitor [91]. We have also detected soluble E-cadherin in the cell culture media of human colon adenocarcinoma cells within hours after treatment with recombinant human Apo2L/TRAIL (Fig. 2). The extracellular cleavage of E-cadherin releases the cell from contacts with neighboring cells, perhaps a mechanism by which an apoptotic cell makes its exodus from the tissue.
Fig. 2.
E-cadherin is released from Colo 205 cells as they undergo Apo2L/tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis. Cells were treated with 250 ng/ml Apo2L/TRAIL for 2, 4, 8, and 16 h. Cell culture media was collected and E-cadherin was immunoprecipitated from the samples and then detected on a western blot with the HECD-1 clone. C control, T Apo2L/TRAIL-treated
IL-6
Proteomic strategies have also been employed to identify markers of early clinical response in patients undergoing radiation therapy for cancer. Tandem mass spectrometry was used to identify several low abundant serum proteins that appeared in the serum following radiotherapy, perhaps coming from necrotic or apoptotic cells [61]. One of the proteins identified in this study was a fragment of IL-6, a proinflammatory cytokine that has also been found in increased levels in patient sera after radiation treatment of liver cancer [106]. However, it is unclear whether the source of the secreted IL-6 is from dying normal or tumor cells in the liver.
Discussion
Many challenges arise when investigators consider the use of real-time markers of therapeutic efficacy during cancer therapy just as there is for the discovery and validation of biomarkers used to initially detect cancer. False negative and false positive results are of concern, especially when an apoptotic marker could be used as a tool to direct or modulate the type of therapy a patient receives—a patient’s therapy could only be changed once the initial therapy was proven ineffective. In fact, relying on the discovery or use of a single marker for this purpose may not be practical, especially during the years of early discovery and validation, when the use of markers of therapeutic efficacy in the clinic will only be complementary to conventional X-ray, CT, MRI or histological evaluations. It is quite possible that the use of apoptotic markers will arise as a test that surveys several proteins or other molecules, and that perhaps a combination of some of the circulating markers in serum mentioned in this review and others not yet identified will prove to be better predictors of therapeutic outcome than any single marker alone. Often attributed to the heterogeneity of tumors, the inability to identify a marker with high specificity and sensitivity is a common problem in tumor-specific biomarker research. It is thought that by including several markers as a screening test, it is possible to encompass more of the patient population, thus increasing the probability of an efficacious assay for the clinic (as reviewed in [72]).
In some metabonomic studies, instead of identifying one or two biomarkers, complex samples are examined by interpreting patterns of proteins or lower molecular weight molecules [76] for the detection of cancer. Although criticized for identifying patterns of unknown molecules that may be irrelevant to a biological response or disease state [18, 87], studies like these [71] have been an important step in trying to understand the enormously complex response of the body to a diseased state or in the context of this review, a response to cancer therapy. The challenge then becomes comprehending the number of molecules involved in the perturbation of a system and why they are released from tumor cells induced to undergo apoptosis. The function of the proteins and other cellular components that are released extracellularly remains to be fully understood. Do these molecules interact with other neighboring cell types to trigger differential responses? Are they involved in the activation and recruitment of immune cells to mount an anti-tumor response or, conversely, is their function to initiate immune evasion by signaling nearby tumor cells to proliferate? Perhaps it is neither of these hypotheses, and that the release of cellular molecules into the surrounding environment is simply an effect of secondary necrosis that occurs when the phagocytes cannot sustain the necessary clearance rate in conditions of massive apoptosis. In any case, observations of extracellular release during apoptosis suggest that there may be other molecules- proteins, lipids, metabolites-shed into the external environment during this process. Perhaps a more comprehensive description of apoptosis aims to include the many events that are known to occur within a cell as well as the possible interactions or signals that can occur outside the cell during this same process.
Which subsets of extracellular components are most important for monitoring treatment is not yet known. In fact, proteomic studies of complex samples like serum have created a forum for discussion of the contribution of peptides, rather than proteins, as relevant molecules in other biomarker studies [19, 72, 101]. Additionally, albumin has been reconsidered—its high abundance once thought to distract from the proteins of interest—now possibly a very valuable carrier of biomarkers throughout the circulation, protecting its cargo from clearance through the kidney [56, 60]. Because biofluids can contain numerous molecules, making analysis difficult, the discovery of apoptotic markers in vivo may be best assessed by initially simplifying the dataset and examining a fraction of the sample-like a specific molecular weight range or only those proteins bound to albumin.
Another relevant question to be addressed in future apoptosis studies is whether or not the molecules that are released are dependent upon the type of therapy used, especially since each drug has a different target and mechanism for inducing cell death as mentioned earlier. Is it possible that for each therapy, a different set of response markers is needed or is there a general signature that all apoptotic tumor cells release irrespective of the inducer?
Tumor heterogeneity is a concern when attempting to understand the kinetics of how a therapy affects the malignant tissue. Time course studies are needed for each therapy to decipher how quickly a drug can reach the tumor, often dependent upon vascular access of the tumor, to penetrate it so that a significant number of cells die. Similarly, because apoptotic cells are cleared by macrophages and other cells surrounding the site of insult, samples would need to be collected in vivo during this process to determine when the capacity of phagocytic cells is overwhelmed, providing an opportunity for secreted molecules to find their way into circulation [55] and suggesting an optimal time frame for biofluid collection. For small proteins and metabolites that are released, it is likely that with their circulation time in the blood being limited by the filtration of the kidneys, studies of urine could be complementary to those of other biofluid components. Another issue that arises is whether the decrease of a biomarker is just as significant as observing an increase, as suggested by preliminary studies in the literature. Decreases in serum nucleosomes were found in mice with tumors 1 day after cisplatin treatment [94] and Apo2L/TRAIL induced a significant reduction of vascular endothelial growth factor secreted by several tumor cell lines in vitro [15]. Once an optimal window of time for sample collection is determined, methods for detecting biomarkers and the sensitivity of these assays need to be optimized for clinical use.
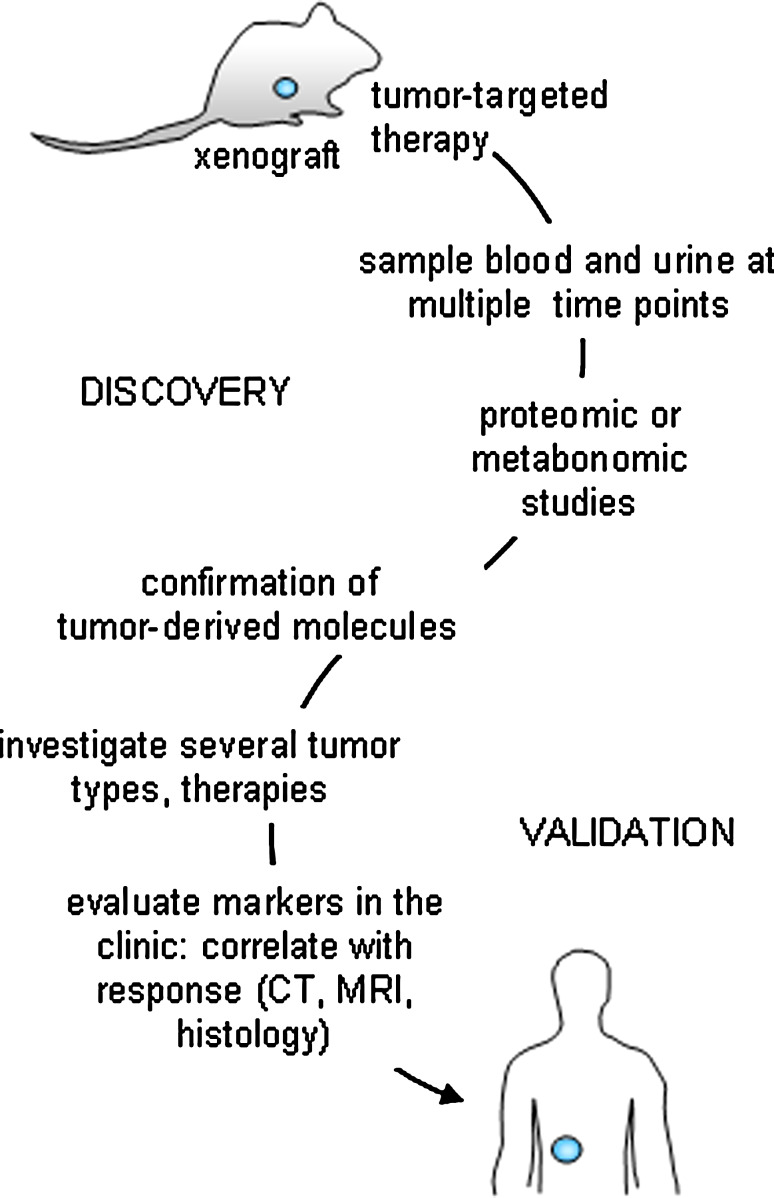
Another hurdle to overcome in the field of response markers is to ascertain that the molecular markers of interest originate from the tumor itself and are not off target effects of susceptible, non-tumor tissue (as so many chemotherapy regimens do affect). In preclinical studies, an appropriate model system is of utmost importance when deciphering malignant signals from benign ones. The SCID mouse-human tumor xenograft model is a system where a human tumor can be targeted with a tumor specific therapy, such as Apo2L/TRAIL [64]. Apo2L/TRAIL kills malignant cells and spares normal cells [5, 102], providing a therapy ideal for studying molecules that are released specifically from the tumor as it undergoes apoptosis. Once proteins or other molecules are identified as released from the tumor in a targeted therapy model, studies can be conducted in a more complex system where chemotherapies attack rapidly proliferating cells. In this model, it is also important to note that the human material is a freshly obtained patient specimen, passaged in SCID mice, and does not originate from an injection of a tumor cell line. The patient specimen xenograft maintains similar architecture and heterogeneity to that of the original patient specimen, a characteristic that is not consistent with observations of a tumor cell line [103], an important property that would most closely mimic drug access to a tumor in the clinic. In fact, models such as these provide a critical starting point where potential candidate molecules can be identified (Fig. 3). Once biomarkers from targeted therapy-induced apoptotic tumor cells are recognized, studies can be expanded to include other cytotoxic therapies (e.g. chemotherapy) as well as a variety of different tumor types, stages and grades.
Fig. 3.
Scheme for validating response markers as measures of true clinical outcome. Initially, the study of human tumor specimens engrafted into immunocompromised mice will be most useful for the identification of candidate markers of tumor response when treated with a targeted therapy (e.g. Apo2L/TRAIL, Herceptin®). Once identified and confirmed with proteomic and/or metabonomic studies, other more non-specific therapies can be investigated (e.g. chemotherapy, radiation). Inquiries into whether the biomarkers are tumor or treatment specific will be elucidated. Once established, clinical studies will identified whether the same molecules are detectable in patients during therapy. Importantly, cohorts of patients will need to be followed for assessment of tumor response with traditional techniques such as tumor imaging and histology and these evaluations will need to be correlated with the newly identified therapeutic efficacy markers to assess their measurement of true clinical response
Adding to the complexity of the situation, the question may be asked whether or not it is sufficient to detect extracellular apoptotic signals from tumor cells in general, or is it more meaningful—or even possible—to measure and identify those molecules that originate from the more elusive and allegedly drug resistant cancer stem cells? Recent research has suggested that these tumor cell subpopulations, however few in number, are what we should be most concerned with in terms of progression free or disease-free survival [39, 86]. It is also very likely that biomarkers of treatment response will be released from tumor cells undergoing other types of cell death such as autophagy, extrinsic senescence, and necrosis, however, these scenarios have not yet been studied. Besides the tumor cells themselves, perhaps other cellular components like tumor endothelial cells may provide distinct extracellular signatures when treated with antiangiogenic agents. Recently, the number of circulating tumor cells in the blood prior to and during treatment was useful in predicting survival rates for patients with metastatic breast cancer [17].
It is also important that the appropriate controls are used in studies that aim to identify biomarkers. Petricoin et al. [72] emphasize that in the early phases of biomarker discovery, it is necessary to include groups that not only represent healthy patients and those with cancer, but that also other conditions, such as benign and inflammatory diseases are also studied because of their influence on a change of pathology that could be common to the more malignant phenotype. Especially in preclinical models, it would be more convincing to validate a marker of therapeutic response by demonstrating that a sensitive tumor releases a protein into circulation while a tumor that has resistance to the same therapy does not manifest the marker similarly.
Summary
The discovery of biomarkers of a therapeutic response may enlighten our understanding of fundamental aspects of apoptosis, in particular, extracellular events associated with this process. Whether or not extracellular markers of apoptosis are dependent upon, and specific to, the inducer of cell death and tumor type is still not clear. Additionally, the timing of when a blood sample is obtained for analysis will be essential in order to collect meaningful data before potential biomarkers are cleared from circulation. It is likely that circulating molecular markers of apoptosis will be used in conjunction with accepted methods of monitoring tumor response, such as imaging and histology, with the goal of improving patient outcomes and survival while minimizing time spent on ineffective cancer treatments. In fact, an ideal treatment regimen would include pre-selection of the initial therapy based upon known tumor markers (e.g. HER2+ breast cancer patients would receive Herceptin®) followed by monitoring of response with the consideration of an adjustment to an alternate therapy based on biomarkers of therapeutic efficacy and the other conventional methods mentioned above. Most importantly, the validation of these markers is critical when using them as surrogate endpoints that measure the true clinical outcome. The future holds considerable promise for utilization of novel markers of therapeutic efficacy in combination with other steps toward pre-selection of patients to receive the best therapy for their cancer.
Abbreviations
- CEA
Carcinoembryonic antigen
- CK18
Cytokeratin-18
- CT
Computed tomography
- DISC
Death-inducing signaling complex
- EGFR
Epithelial growth factor receptor
- FADD
Fas-associated death domain
- FDG
18F-deoxyglucose
- FLT
18F-fluorothymidine
- LDH
Lactate dehydrogenase
- MMP
Matrix metalloproteinase
- MRI
Magnetic resonance imaging
- NCCN
National comprehensive cancer network
- PDT
Photodynamic therapy
- PET
Positron emission tomography
- PSA
Prostate specific antigen
- RECIST
Response evaluation criteria in solid tumors
- SEER
Surveillance epidemiology and end results
- SCID
Severe combined immunodeficiency
- SPECT
Single photon emission computerized tomography
- TIMP
Tissue inhibitor of MMPs
- TNF
Tumor necrosis factor
- TRAIL
Tumor necrosis factor-related apoptosis-inducing ligand
- TUNEL
Terminal deoxynucleotidyl transferase biotin-dUTP nick end labeling
- WHO
World Health Organization
References
- 1.Abraham J, Allegra CJ (2001) (eds) Bethesda Handbook of Clinical Oncology. Lippincott Williams and Wilkins, Philadelphia
- 2.Ahlemeyer B, Klumpp S, Krieglstein J. Release of cytochrome c into the extracellular space contributes to neuronal apoptosis induced by staurosporine. Brain Res. 2002;934:107–116. doi: 10.1016/s0006-8993(02)02365-x. [DOI] [PubMed] [Google Scholar]
- 3.Almasan A, Ashkenazi A. Apo2L/TRAIL: apoptosis signaling, biology, and potential for cancer therapy. Cytokine Growth Factor Rev. 2003;14:337–348. doi: 10.1016/s1359-6101(03)00029-7. [DOI] [PubMed] [Google Scholar]
- 4.Aquino A, Prete SP, Guadagni F, Greiner JW, Giuliani A, Orlando L, Masci G, De Santis S, Bonmassar E, Graziani G. Effect of 5-fluorouracil on carcinoembryonic antigen expression and shedding at clonal level in colon cancer cells. Anticancer Res. 2000;20:3475–3484. [PubMed] [Google Scholar]
- 5.Ashkenazi A, Pai RC, Fong S, Leung S, Lawrence DA, Marsters SA, Blackie C, Chang L, McMurtrey AE, Hebert A, DeForge L, Koumenis IL, Lewis D, Harris L, Bussiere J, Koeppen H, Shahrokh Z, Schwall RH. Safety and antitumor activity of recombinant soluble Apo2 ligand. J Clin Invest. 1999;104:155–162. doi: 10.1172/JCI6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker M. In biomarkers we trust? Nat Biotechnol. 2005;23:297–304. doi: 10.1038/nbt0305-297. [DOI] [PubMed] [Google Scholar]
- 7.Bantel H, Lugering A, Heidemann J, Volkmann X, Poremba C, Strassburg CP, Manns MP, Schulze-Osthoff K. Detection of apoptotic caspase activation in sera from patients with chronic HCV infection is associated with fibrotic liver injury. Hepatology. 2004;40:1078–1087. doi: 10.1002/hep.20411. [DOI] [PubMed] [Google Scholar]
- 8.Barczyk K, Kreuter M, Pryjma J, Booy EP, Maddika S, Ghavami S, Berdel WE, Roth J, Los M. Serum cytochrome c indicates in vivo apoptosis and can serve as a prognostic marker during cancer therapy. Int J Cancer. 2005;116:167–173. doi: 10.1002/ijc.21037. [DOI] [PubMed] [Google Scholar]
- 9.Bast RC, Jr, Badgwell D, Lu Z, Marquez R, Rosen D, Liu J, Baggerly KA, Atkinson EN, Skates S, Zhang Z, Lokshin A, Menon U, Jacobs I, Lu K. New tumor markers: CA125 and beyond. Int J Gynecol Cancer. 2005;15(Suppl 3):274–281. doi: 10.1111/j.1525-1438.2005.00441.x. [DOI] [PubMed] [Google Scholar]
- 10.Belhocine T, Steinmetz N, Hustinx R, Bartsch P, Jerusalem G, Seidel L, Rigo P, Green A. Increased uptake of the apoptosis-imaging agent (99m)Tc recombinant human Annexin V in human tumors after one course of chemotherapy as a predictor of tumor response and patient prognosis. Clin Cancer Res. 2002;8:2766–2774. [PubMed] [Google Scholar]
- 11.Benson AB, III, Choti MA, Cohen AM, Doroshow JH, Fuchs C, Kiel K, Martin EW, Jr, McGinn C, Petrelli NJ, Posey JA, Skibber JM, Venook A, Yeatman TJ. NCCN practice guidelines for colorectal cancer. Oncol (Williston Park) 2000;14:203–212. [PubMed] [Google Scholar]
- 12.Biven K, Erdal H, Hagg M, Ueno T, Zhou R, Lynch M, Rowley B, Wood J, Zhang C, Toi M, Shoshan MC, Linder S. A novel assay for discovery and characterization of pro-apoptotic drugs and for monitoring apoptosis in patient sera. Apoptosis. 2003;8:263–268. doi: 10.1023/a:1023672805949. [DOI] [PubMed] [Google Scholar]
- 13.Bodmer JL, Holler N, Reynard S, Vinciguerra P, Schneider P, Juo P, Blenis J, Tschopp J. TRAIL receptor-2 signals apoptosis through FADD and caspase-8. Nat Cell Biol. 2000;2:241–243. doi: 10.1038/35008667. [DOI] [PubMed] [Google Scholar]
- 14.Burbee DG, Forgacs E, Zochbauer-Muller S, Shivakumar L, Fong K, Gao B, Randle D, Kondo M, Virmani A, Bader S, Sekido Y, Latif F, Milchgrub S, Toyooka S, Gazdar AF, Lerman MI, Zabarovsky E, White M, Minna JD. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst. 2001;93:691–699. doi: 10.1093/jnci/93.9.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cantarella G, Risuglia N, Dell’eva R, Lempereur L, Albini A, Pennisi G, Scoto GM, Noonan DN, Bernardini R. TRAIL inhibits angiogenesis stimulated by VEGF expression in human glioblastoma cells. Br J Cancer. 2006;94:1428–1435. doi: 10.1038/sj.bjc.6603092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chabre H, Amoura Z, Piette JC, Godeau P, Bach JF, Koutouzov S. Presence of nucleosome-restricted antibodies in patients with systemic lupus erythematosus. Arthritis Rheum. 1995;38:1485–1491. doi: 10.1002/art.1780381015. [DOI] [PubMed] [Google Scholar]
- 17.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, Hayes DF. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 18.Diamandis EP. Analysis of serum proteomic patterns for early cancer diagnosis: drawing attention to potential problems. J Natl Cancer Inst. 2004;96:353–356. doi: 10.1093/jnci/djh056. [DOI] [PubMed] [Google Scholar]
- 19.Diamandis EP. Peptidomics for cancer diagnosis: present and future. J Proteome Res. 2006;5:2079–2082. doi: 10.1021/pr060225u. [DOI] [PubMed] [Google Scholar]
- 20.Dickson PV, Hamner JB, Cauthen LA, Ng CY, McCarville MB, Davidoff AM. Efficacy of zoledronate against neuroblastoma. Surgery. 2006;140:227–235. doi: 10.1016/j.surg.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 21.Espina V, Dettloff KA, Cowherd S, Petricoin EF, III, Liotta LA. Use of proteomic analysis to monitor responses to biological therapies. Expert Opin Biol Ther. 2004;4:83–93. doi: 10.1517/14712598.4.1.83. [DOI] [PubMed] [Google Scholar]
- 22.Etzioni R, Urban N, Ramsey S, McIntosh M, Schwartz S, Reid B, Radich J, Anderson G, Hartwell L. The case for early detection. Nat Rev Cancer. 2003;3:243–252. doi: 10.1038/nrc1041. [DOI] [PubMed] [Google Scholar]
- 23.Feldstein AE, Gores GJ. An apoptosis biomarker goes to the HCV clinic. Hepatology. 2004;40:1044–1046. doi: 10.1002/hep.20479. [DOI] [PubMed] [Google Scholar]
- 24.Feneley MR, Partin AW. Diagnosis of localized prostate cancer: 10 years of progress. Curr Opin Urol. 2000;10:319–327. doi: 10.1097/00042307-200007000-00006. [DOI] [PubMed] [Google Scholar]
- 25.Fiegl H, Millinger S, Mueller-Holzner E, Marth C, Ensinger C, Berger A, Klocker H, Goebel G, Widschwendter M. Circulating tumor-specific DNA: a marker for monitoring efficacy of adjuvant therapy in cancer patients. Cancer Res. 2005;65:1141–1145. doi: 10.1158/0008-5472.CAN-04-2438. [DOI] [PubMed] [Google Scholar]
- 26.Fleming TR, DeMets DL. Surrogate end points in clinical trials: are we being misled? Ann Intern Med. 1996;125:605–613. doi: 10.7326/0003-4819-125-7-199610010-00011. [DOI] [PubMed] [Google Scholar]
- 27.Gee MS, Upadhyay R, Bergquist H, Weissleder R, Josephson L, Mahmood U. Multiparameter noninvasive assessment of treatment susceptibility, drug target inhibition and tumor response guides cancer treatment. Int J Cancer. 2007;121:2492–2500. doi: 10.1002/ijc.22995. [DOI] [PubMed] [Google Scholar]
- 28.Gillham CM, Lucey JA, Keogan M, Duffy GJ, Malik V, Raouf AA, O’Byrne K, Hollywood D, Muldoon C, Reynolds JV. (18)FDG uptake during induction chemoradiation for oesophageal cancer fails to predict histomorphological tumour response. Br J Cancer. 2006;95:1174–1179. doi: 10.1038/sj.bjc.6603412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gonzalez VM, Fuertes MA, Alonso C, Perez JM. Is cisplatin-induced cell death always produced by apoptosis? Mol Pharmacol. 2001;59:657–663. doi: 10.1124/mol.59.4.657. [DOI] [PubMed] [Google Scholar]
- 30.Green DR. Apoptotic pathways: paper wraps stone blunts scissors. Cell. 2000;102:1–4. doi: 10.1016/s0092-8674(00)00003-9. [DOI] [PubMed] [Google Scholar]
- 31.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 32.Hanke B, Wein A, Martus P, Riedel C, Voelker M, Hahn EG, Schuppan D. Serum markers of matrix turnover as predictors for the evolution of colorectal cancer metastasis under chemotherapy. Br J Cancer. 2003;88:1248–1250. doi: 10.1038/sj.bjc.6600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hannun YA. Apoptosis and the dilemma of cancer chemotherapy. Blood. 1997;89:1845–1853. [PubMed] [Google Scholar]
- 34.Hersh EM, Metch BS, Muggia FM, Brown TD, Whitehead RP, Budd GT, Rinehart JJ, Crawford ED, Bonnet JD, Behrens BC. Phase II studies of recombinant human tumor necrosis factor alpha in patients with malignant disease: a summary of the Southwest Oncology Group experience. J Immunother. 1991;10:426–431. doi: 10.1097/00002371-199112000-00006. [DOI] [PubMed] [Google Scholar]
- 35.Hohlbaum AM, Gregory MS, Ju ST, Marshak-Rothstein A. Fas ligand engagement of resident peritoneal macrophages in vivo induces apoptosis and the production of neutrophil chemotactic factors. J Immunol. 2001;167:6217–6224. doi: 10.4049/jimmunol.167.11.6217. [DOI] [PubMed] [Google Scholar]
- 36.Holdenrieder S, Holubec L, Jr, Topolcan O, Finek J, Stieber P. Circulating nucleosomes and cytokeratin 19-fragments in patients with colorectal cancer during chemotherapy. Anticancer Res. 2005;25:1795–1801. [PubMed] [Google Scholar]
- 37.Holdenrieder S, Stieber P. Apoptotic markers in cancer. Clin Biochem. 2004;37:605–617. doi: 10.1016/j.clinbiochem.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 38.Holdenrieder S, Stieber P, von Pawel J, Raith H, Nagel D, Feldmann K, Seidel D. Circulating nucleosomes predict the response to chemotherapy in patients with advanced non-small cell lung cancer. Clin Cancer Res. 2004;10:5981–5987. doi: 10.1158/1078-0432.CCR-04-0625. [DOI] [PubMed] [Google Scholar]
- 39.Huntly BJ, Gilliland DG. Leukaemia stem cells and the evolution of cancer-stem-cell research. Nat Rev Cancer. 2005;5:311–321. doi: 10.1038/nrc1592. [DOI] [PubMed] [Google Scholar]
- 40.Jaffe CC. Measures of response: RECIST, WHO, and new alternatives. J Clin Oncol. 2006;24:3245–3251. doi: 10.1200/JCO.2006.06.5599. [DOI] [PubMed] [Google Scholar]
- 41.Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, Knippers R. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659–1665. [PubMed] [Google Scholar]
- 42.Kaufmann SH, Earnshaw WC. Induction of apoptosis by cancer chemotherapy. Exp Cell Res. 2000;256:42–49. doi: 10.1006/excr.2000.4838. [DOI] [PubMed] [Google Scholar]
- 43.Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim R, Emi M, Tanabe K. The role of apoptosis in cancer cell survival and therapeutic outcome. Cancer Biol Ther. 2006;5:1429–1442. doi: 10.4161/cbt.5.11.3456. [DOI] [PubMed] [Google Scholar]
- 45.Kischkel FC, Lawrence DA, Chuntharapai A, Schow P, Kim KJ, Ashkenazi A. Apo2L/TRAIL-dependent recruitment of endogenous FADD and caspase-8 to death receptors 4 and 5. Immunity. 2000;12:611–620. doi: 10.1016/s1074-7613(00)80212-5. [DOI] [PubMed] [Google Scholar]
- 46.Kramer G, Erdal H, Mertens HJ, Nap M, Mauermann J, Steiner G, Marberger M, Biven K, Shoshan MC, Linder S. Differentiation between cell death modes using measurements of different soluble forms of extracellular cytokeratin 18. Cancer Res. 2004;64:1751–1756. doi: 10.1158/0008-5472.can-03-2455. [DOI] [PubMed] [Google Scholar]
- 47.Kramer G, Schwarz S, Hagg M, Havelka AM, Linder S. Docetaxel induces apoptosis in hormone refractory prostate carcinomas during multiple treatment cycles. Br J Cancer. 2006;94:1592–1598. doi: 10.1038/sj.bjc.6603129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kremer A, Holdenrieder S, Stieber P, Wilkowski R, Nagel D, Seidel D. Nucleosomes in colorectal cancer patients during radiochemotherapy. Tumour Biol. 2006;27:235–242. doi: 10.1159/000094694. [DOI] [PubMed] [Google Scholar]
- 49.Kremer A, Wilkowski R, Holdenrieder S, Nagel D, Stieber P, Seidel D. Nucleosomes in pancreatic cancer patients during radiochemotherapy. Tumour Biol. 2005;26:44–49. doi: 10.1159/000084339. [DOI] [PubMed] [Google Scholar]
- 50.Lee KC, Sud S, Meyer CR, Moffat BA, Chenevert TL, Rehemtulla A, Pienta KJ, Ross BD. An imaging biomarker of early treatment response in prostate cancer that has metastasized to the bone. Cancer Res. 2007;67:3524–3529. doi: 10.1158/0008-5472.CAN-06-4236. [DOI] [PubMed] [Google Scholar]
- 51.Leers MP, Kolgen W, Bjorklund V, Bergman T, Tribbick G, Persson B, Bjorklund P, Ramaekers FC, Bjorklund B, Nap M, Jornvall H, Schutte B. Immunocytochemical detection and mapping of a cytokeratin 18 neo-epitope exposed during early apoptosis. J Pathol. 1999;187:567–572. doi: 10.1002/(SICI)1096-9896(199904)187:5<567::AID-PATH288>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 52.Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646–650. [PubMed] [Google Scholar]
- 53.Leyton J, Latigo JR, Perumal M, Dhaliwal H, He Q, Aboagye EO. Early detection of tumor response to chemotherapy by 3′-deoxy-3′-[18F]fluorothymidine positron emission tomography: the effect of cisplatin on a fibrosarcoma tumor model in vivo. Cancer Res. 2005;65:4202–4210. doi: 10.1158/0008-5472.CAN-04-4008. [DOI] [PubMed] [Google Scholar]
- 54.Li H, Zhu H, Xu CJ, Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- 55.Linder S, Havelka AM, Ueno T, Shoshan MC. Determining tumor apoptosis and necrosis in patient serum using cytokeratin 18 as a biomarker. Cancer Lett. 2004;214:1–9. doi: 10.1016/j.canlet.2004.06.032. [DOI] [PubMed] [Google Scholar]
- 56.Liotta LA, Petricoin EF. Serum peptidome for cancer detection: spinning biologic trash into diagnostic gold. J Clin Invest. 2006;116:26–30. doi: 10.1172/JCI27467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Locker GY, Hamilton S, Harris J, Jessup JM, Kemeny N, Macdonald JS, Somerfield MR, Hayes DF, Bast RC., Jr ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol. 2006;24:5313–5327. doi: 10.1200/JCO.2006.08.2644. [DOI] [PubMed] [Google Scholar]
- 58.Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 1998;94:481–490. doi: 10.1016/s0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- 59.Meggiato T, Calabrese F, Valente M, Favaretto E, Baliello E, Del Favero G. Spontaneous apoptosis and proliferation in human pancreatic cancer. Pancreas. 2000;20:117–122. doi: 10.1097/00006676-200003000-00002. [DOI] [PubMed] [Google Scholar]
- 60.Mehta AI, Ross S, Lowenthal MS, Fusaro V, Fishman DA, Petricoin EF, III, Liotta LA. Biomarker amplification by serum carrier protein binding. Dis Markers. 2003;19:1–10. doi: 10.1155/2003/104879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Menard C, Johann D, Lowenthal M, Muanza T, Sproull M, Ross S, Gulley J, Petricoin E, Coleman CN, Whiteley G, Liotta L, Camphausen K. Discovering clinical biomarkers of ionizing radiation exposure with serum proteomic analysis. Cancer Res. 2006;66:1844–1850. doi: 10.1158/0008-5472.CAN-05-3466. [DOI] [PubMed] [Google Scholar]
- 62.Moertel CG, Fleming TR, Macdonald JS, Haller DG, Laurie JA, Tangen C. An evaluation of the carcinoembryonic antigen (CEA) test for monitoring patients with resected colon cancer. JAMA. 1993;270:943–947. [PubMed] [Google Scholar]
- 63.Mueller S, Holdenrieder S, Stieber P, Haferlach T, Schalhorn A, Braess J, Nagel D, Seidel D. Early prediction of therapy response in patients with acute myeloid leukaemia by nucleosomal DNA fragments. BMC Cancer. 2006;6:143. doi: 10.1186/1471-2407-6-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Naka T, Sugamura K, Hylander BL, Widmer MB, Rustum YM, Repasky EA. Effects of tumor necrosis factor-related apoptosis-inducing ligand alone and in combination with chemotherapeutic agents on patients’ colon tumors grown in SCID mice. Cancer Res. 2002;62:5800–5806. [PubMed] [Google Scholar]
- 65.Neves AA, Brindle KM. Assessing responses to cancer therapy using molecular imaging. Biochim Biophys Acta. 2006;1766:242–261. doi: 10.1016/j.bbcan.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 66.Neves AA, Krishnan AS, Kettunen MI, Hu DE, Backer MM, Davletov B, Brindle KM. A paramagnetic nanoprobe to detect tumor cell death using magnetic resonance imaging. Nano Lett. 2007;7:1419–1423. doi: 10.1021/nl070126v. [DOI] [PubMed] [Google Scholar]
- 67.Ogasawara J, Watanabe-Fukunaga R, Adachi M, Matsuzawa A, Kasugai T, Kitamura Y, Itoh N, Suda T, Nagata S. Lethal effect of the anti-Fas antibody in mice. Nature. 1993;364:806–809. doi: 10.1038/364806a0. [DOI] [PubMed] [Google Scholar]
- 68.Olofsson MH, Ueno T, Pan Y, Xu R, Cai F, van der Kuip H, Muerdter TE, Sonnenberg M, Aulitzky WE, Schwarz S, Andersson E, Shoshan MC, Havelka AM, Toi M, Linder S. Cytokeratin-18 is a useful serum biomarker for early determination of response of breast carcinomas to chemotherapy. Clin Cancer Res. 2007;13:3198–3206. doi: 10.1158/1078-0432.CCR-07-0009. [DOI] [PubMed] [Google Scholar]
- 69.Ozols RF, Herbst RS, Colson YL, Gralow J, Bonner J, Curran WJ, Jr, Eisenberg BL, Ganz PA, Kramer BS, Kris MG, Markman M, Mayer RJ, Raghavan D, Reaman GH, Sawaya R, Schilsky RL, Schuchter LM, Sweetenham JW, Vahdat LT, Winn RJ. Clinical cancer advances 2006: major research advances in cancer treatment, prevention, and screening—a report from the American Society of Clinical Oncology. J Clin Oncol. 2007;25:146–162. doi: 10.1200/JCO.2006.09.7030. [DOI] [PubMed] [Google Scholar]
- 70.Pardo J, Bosque A, Brehm R, Wallich R, Naval J, Mullbacher A, Anel A, Simon MM. Apoptotic pathways are selectively activated by granzyme A and/or granzyme B in CTL-mediated target cell lysis. J Cell Biol. 2004;167:457–468. doi: 10.1083/jcb.200406115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Petricoin EF, Ardekani AM, Hitt BA, Levine PJ, Fusaro VA, Steinberg SM, Mills GB, Simone C, Fishman DA, Kohn EC, Liotta LA. Use of proteomic patterns in serum to identify ovarian cancer. Lancet. 2002;359:572–577. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- 72.Petricoin EF, Belluco C, Araujo RP, Liotta LA. The blood peptidome: a higher dimension of information content for cancer biomarker discovery. Nat Rev Cancer. 2006;6:961–967. doi: 10.1038/nrc2011. [DOI] [PubMed] [Google Scholar]
- 73.Pichon MF, Labroquere M, Rezai K, Lokiec F. Variations of soluble fas and cytokeratin 18-Asp 396 neo-epitope in different cancers during chemotherapy. Anticancer Res. 2006;26:2387–2392. [PubMed] [Google Scholar]
- 74.Platoshyn O, Zhang S, McDaniel SS, Yuan JX. Cytochrome c activates K+ channels before inducing apoptosis. Am J Physiol Cell Physiol. 2002;283:C1298–C1305. doi: 10.1152/ajpcell.00592.2001. [DOI] [PubMed] [Google Scholar]
- 75.Radic M, Marion T, Monestier M. Nucleosomes are exposed at the cell surface in apoptosis. J Immunol. 2004;172:6692–6700. doi: 10.4049/jimmunol.172.11.6692. [DOI] [PubMed] [Google Scholar]
- 76.Rantalainen M, Cloarec O, Beckonert O, Wilson ID, Jackson D, Tonge R, Rowlinson R, Rayner S, Nickson J, Wilkinson RW, Mills JD, Trygg J, Nicholson JK, Holmes E. Statistically integrated metabonomic-proteomic studies on a human prostate cancer xenograft model in mice. J Proteome Res. 2006;5:2642–2655. doi: 10.1021/pr060124w. [DOI] [PubMed] [Google Scholar]
- 77.Rathmell JC, Thompson CB. Pathways of apoptosis in lymphocyte development, homeostasis, and disease. Cell. 2002;109(Suppl):S97–S107. doi: 10.1016/s0092-8674(02)00704-3. [DOI] [PubMed] [Google Scholar]
- 78.Renz A, Berdel WE, Kreuter M, Belka C, Schulze-Osthoff K, Los M. Rapid extracellular release of cytochrome c is specific for apoptosis and marks cell death in vivo. Blood. 2001;98:1542–1548. doi: 10.1182/blood.v98.5.1542. [DOI] [PubMed] [Google Scholar]
- 79.Sayar D, Yaniv I, Goshen Y, Cohen IJ. Treatment of alpha-fetoprotein secreting hepatoblastoma by response of serum alpha-fetoprotein levels: a new concept. Pediatr Hematol Oncol. 2001;18:509–518. doi: 10.1080/088800101753328475. [DOI] [PubMed] [Google Scholar]
- 80.Screpanti V, Wallin RP, Grandien A, Ljunggren HG. Impact of FASL-induced apoptosis in the elimination of tumor cells by NK cells. Mol Immunol. 2005;42:495–499. doi: 10.1016/j.molimm.2004.07.033. [DOI] [PubMed] [Google Scholar]
- 81.Sellins KS, Cohen JJ. Gene induction by gamma-irradiation leads to DNA fragmentation in lymphocytes. J Immunol. 1987;139:3199–3206. [PubMed] [Google Scholar]
- 82.Serrano M. Cancer regression by senescence. N Engl J Med. 2007;356:1996–1997. doi: 10.1056/NEJMcibr071461. [DOI] [PubMed] [Google Scholar]
- 83.Sheard MA, Vojtesek B, Simickova M, Valik D. Release of cytokeratin-18 and -19 fragments (TPS and CYFRA 21-1) into the extracellular space during apoptosis. J Cell Biochem. 2002;85:670–677. doi: 10.1002/jcb.10173. [DOI] [PubMed] [Google Scholar]
- 84.Sherley JL, Kelly TJ. Regulation of human thymidine kinase during the cell cycle. J Biol Chem. 1988;263:8350–8358. [PubMed] [Google Scholar]
- 85.Skladanowski A, Konopa J. Adriamycin and daunomycin induce programmed cell death (apoptosis) in tumour cells. Biochem Pharmacol. 1993;46:375–382. doi: 10.1016/0006-2952(93)90512-u. [DOI] [PubMed] [Google Scholar]
- 86.Smalley M, Ashworth A. Stem cells and breast cancer: a field in transit. Nat Rev Cancer. 2003;3:832–844. doi: 10.1038/nrc1212. [DOI] [PubMed] [Google Scholar]
- 87.Sorace JM, Zhan M. A data review and re-assessment of ovarian cancer serum proteomic profiling. BMC Bioinf. 2003;4:24. doi: 10.1186/1471-2105-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sorbye H, Dahl O. Carcinoembryonic antigen surge in metastatic colorectal cancer patients responding to oxaliplatin combination chemotherapy: implications for tumor marker monitoring and guidelines. J Clin Oncol. 2003;21:4466–4467. doi: 10.1200/JCO.2003.99.200. [DOI] [PubMed] [Google Scholar]
- 89.Sorbye H, Dahl O. Transient CEA increase at start of oxaliplatin combination therapy for metastatic colorectal cancer. Acta Oncol. 2004;43:495–498. doi: 10.1080/02841860410032380. [DOI] [PubMed] [Google Scholar]
- 90.Stefflova K, Chen J, Li H, Zheng G. Targeted photodynamic therapy agent with a built-in apoptosis sensor for in vivo near-infrared imaging of tumor apoptosis triggered by its photosensitization in situ. Mol Imaging. 2006;5:520–532. [PubMed] [Google Scholar]
- 91.Steinhusen U, Weiske J, Badock V, Tauber R, Bommert K, Huber O. Cleavage and shedding of E-cadherin after induction of apoptosis. J Biol Chem. 2001;276:4972–4980. doi: 10.1074/jbc.M006102200. [DOI] [PubMed] [Google Scholar]
- 92.Strater J, Koretz K, Gunthert AR, Moller P. In situ detection of enterocytic apoptosis in normal colonic mucosa and in familial adenomatous polyposis. Gut. 1995;37:819–825. doi: 10.1136/gut.37.6.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Su H, Bodenstein C, Dumont RA, Seimbille Y, Dubinett S, Phelps ME, Herschman H, Czernin J, Weber W. Monitoring tumor glucose utilization by positron emission tomography for the prediction of treatment response to epidermal growth factor receptor kinase inhibitors. Clin Cancer Res. 2006;12:5659–5667. doi: 10.1158/1078-0432.CCR-06-0368. [DOI] [PubMed] [Google Scholar]
- 94.Trejo-Becerril C, Perez-Cardenas E, Trevino-Cuevas H, Taja-Chayeb L, Garcia-Lopez P, Segura-Pacheco B, Chavez-Blanco A, Lizano-Soberon M, Gonzalez-Fierro A, Mariscal I, Wegman-Ostrosky T, Duenas-Gonzalez A. Circulating nucleosomes and response to chemotherapy: an in vitro, in vivo and clinical study on cervical cancer patients. Int J Cancer. 2003;104:663–668. doi: 10.1002/ijc.11003. [DOI] [PubMed] [Google Scholar]
- 95.Uchimura E, Kodaira T, Kurosaka K, Yang D, Watanabe N, Kobayashi Y. Interaction of phagocytes with apoptotic cells leads to production of pro-inflammatory cytokines. Biochem Biophys Res Commun. 1997;239:799–803. doi: 10.1006/bbrc.1997.7556. [DOI] [PubMed] [Google Scholar]
- 96.Ueno T, Toi M, Biven K, Bando H, Ogawa T, Linder S. Measurement of an apoptotic product in the sera of breast cancer patients. Eur J Cancer. 2003;39:769–774. doi: 10.1016/s0959-8049(02)00865-1. [DOI] [PubMed] [Google Scholar]
- 97.Van de Wiele C, Lahorte C, Oyen W, Boerman O, Goethals I, Slegers G, Dierckx RA. Nuclear medicine imaging to predict response to radiotherapy: a review. Int J Radiat Oncol Biol Phys. 2003;55:5–15. doi: 10.1016/s0360-3016(02)04122-6. [DOI] [PubMed] [Google Scholar]
- 98.Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 99.Vermes I, Haanen C, Steffens-Nakken H, Reutelingsperger C. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J Immunol Methods. 1995;184:39–51. doi: 10.1016/0022-1759(95)00072-i. [DOI] [PubMed] [Google Scholar]
- 100.Vicini FA, Vargas C, Abner A, Kestin L, Horwitz E, Martinez A. Limitations in the use of serum prostate specific antigen levels to monitor patients after treatment for prostate cancer. J Urol. 2005;173:1456–1462. doi: 10.1097/01.ju.0000157323.55611.23. [DOI] [PubMed] [Google Scholar]
- 101.Villanueva J, Shaffer DR, Philip J, Chaparro CA, Erdjument-Bromage H, Olshen AB, Fleisher M, Lilja H, Brogi E, Boyd J, Sanchez-Carbayo M, Holland EC, Cordon-Cardo C, Scher HI, Tempst P. Differential exoprotease activities confer tumor-specific serum peptidome patterns. J Clin Invest. 2006;116:271–284. doi: 10.1172/JCI26022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Walczak H, Miller RE, Ariail K, Gliniak B, Griffith TS, Kubin M, Chin W, Jones J, Woodward A, Le T, Smith C, Smolak P, Goodwin RG, Rauch CT, Schuh JC, Lynch DH. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat Med. 1999;5:157–163. doi: 10.1038/5517. [DOI] [PubMed] [Google Scholar]
- 103.Weinberg RA (2007) (eds) The biology of cancer. Garland Science, Taylor and Francis Group, LLC, New York
- 104.Wells MJ, Hatton MW, Hewlett B, Podor TJ, Sheffield WP, Blajchman MA. Cytokeratin 18 is expressed on the hepatocyte plasma membrane surface and interacts with thrombin–antithrombin complexes. J Biol Chem. 1997;272:28574–28581. doi: 10.1074/jbc.272.45.28574. [DOI] [PubMed] [Google Scholar]
- 105.Werner-Wasik M, Xiao Y, Pequignot E, Curran WJ, Hauck W. Assessment of lung cancer response after nonoperative therapy: tumor diameter, bidimensional product, and volume. A serial CT scan-based study. Int J Radiat Oncol Biol Phys. 2001;51:56–61. doi: 10.1016/s0360-3016(01)01615-7. [DOI] [PubMed] [Google Scholar]
- 106.Wickremesekera JK, Chen W, Cannan RJ, Stubbs RS. Serum proinflammatory cytokine response in patients with advanced liver tumors following selective internal radiation therapy (SIRT) with (90)Yttrium microspheres. Int J Radiat Oncol Biol Phys. 2001;49:1015–1021. doi: 10.1016/s0360-3016(00)01420-6. [DOI] [PubMed] [Google Scholar]
- 107.Wyllie AH. Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature. 1980;284:555–556. doi: 10.1038/284555a0. [DOI] [PubMed] [Google Scholar]
- 108.Yamauchi K, Yang M, Jiang P, Xu M, Yamamoto N, Tsuchiya H, Tomita K, Moossa AR, Bouvet M, Hoffman RM. Development of real-time subcellular dynamic multicolor imaging of cancer-cell trafficking in live mice with a variable-magnification whole-mouse imaging system. Cancer Res. 2006;66:4208–4214. doi: 10.1158/0008-5472.CAN-05-3927. [DOI] [PubMed] [Google Scholar]
- 109.Yu J, Yue W, Wu B, Zhang L. PUMA sensitizes lung cancer cells to chemotherapeutic agents and irradiation. Clin Cancer Res. 2006;12:2928–2936. doi: 10.1158/1078-0432.CCR-05-2429. [DOI] [PubMed] [Google Scholar]
- 110.Zhang J, Driscoll TA, Hannun YA, Obeid LM. Regulation of membrane release in apoptosis. Biochem J. 1998;334(Pt 2):479–485. doi: 10.1042/bj3340479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zou H, Henzel WJ, Liu X, Lutschg A, Wang X. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]