Abstract
The European Searchable Tumour line Database (ESTDAB) (http://www.ebi.ac.uk/ipd/estdab) is a freely available and fully searchable database of melanoma-derived cell lines, which have been characterised for over 250 immunologically relevant markers by a consortium of European scientists. The database is linked to a cell bank, which can provide melanoma cell lines to non-profit investigators for a nominal handling charge. All cells are fully HLA typed at the genomic and surface expression levels. The expression of a number of surface antigens, apoptotic markers, tumour-associated antigens and extracellular matrix proteins has also been determined. Cytokine secretion has been tested and polymorphisms in cytokine genes have been identified. Glycans at the cell surface were identified and glycosyltransferase activity quantified. Cell lines with a particular constellation of these parameters can be sought online via the ESTDAB interface, which is included as part of the Immuno-Polymorphism Database (IPD) section of the European Bioinformatics Institute’s (EBI) website.
Keywords: Cell bank, Melanoma, Searchable database, Infrastructure resource, Immunological characterisation
Introduction
While many cell banks like ATCC (http://www.lgcpromochem-atcc.com/) or ECACC (http://www.ecacc.org.uk/) make immortal cell line materials available to the scientific community, these services have seldom, until this point, made available data on the nature and characteristics of the cells contained in the cell bank. The European Searchable Tumour line Database (ESTDAB) project [1], part of the European Commission’s fifth framework infrastructures (FP5) program (contract no. QLRI-CT-2001-01325) is unique in that it links a cell bank of more than 170 melanoma-derived cell lines to a database of information about the available cells. Melanoma cell lines are used extensively in immunology and immunotherapy studies. They have a number of uses including monitoring the presence of tumour-reactive antibodies and cells, and they could also be used as a source of tumour-associated antigens in vaccination and as well as screening agents for anti-cancer activity.
The ESTDAB consortium [1] was made up of seven laboratories from countries around the EU, with each laboratory responsible for the production of data on a prearranged set of immunological markers that reflected that laboratory’s interests and technical specialities. The central facility and physical location of the cell bank was established at the Centre for Medical Research (ZMF) of the University of Tübingen in Germany, where cells lines were gathered from a variety of sources around Europe, Australia and the United States of America. Since the project began, cells have been acquired as they became available, this has meant in particular that a number of cell lines have been sourced from melanoma samples collected from patients entered into clinical immunotherapy trials associated with ESTDAB’s sister project, OISTER (Outcome and Impact of Specific Treatment in European Research on Melanoma, http://www.dkfz.de/oister/). As a consequence of the central facility’s efforts to gather cell lines continuously, we now believe that ESTDAB provides access to several times more cell lines of melanoma origin than any other cell bank currently available.
Data collection
Each melanoma cell line held in the ESTDAB database and cell bank has been characterised for a large number of markers, with many of the analyses detailing the expression levels of various markers both under resting conditions and following stimulation with Interferon Gamma (IFN)-γ for 48 h.
High-resolution HLA typing of the six major HLA genes was performed by Reference Strand mediated Conformational Analysis (RSCA) [2, 3] or by sequencing-based typing (SBT) [4, 5]. Surface expression of 11 HLA and 20+ other surface antigens was analysed using the FACsort flow cytometer (Becton Dickinson, Oxford, UK) and a panel of primary monoclonal antibodies specific for the antigen being tested and FITC-conjugated goat/rabbit–anti-mouse Ig F(ab)2 secondary antibodies. Levels of secretion of the cytokines IL-6, -8 and -10, TGF-β, TNF-α and IFN-γ were measured in whole cell medium and in BD Matrigel™ Basement Membrane Matrix (Becton Dickinson, Oxford, UK.) In the same set of cytokines, genetic polymorphisms were identified at 29 polymorphic positions by polymerase chain reaction (PCR) using sequence-specific primers (PCR-SSP) and One Lambda’s cytokine genotyping tray (One Lambda, Canoga Park, CA). Expression of 19 tumour-associated antigens was assessed by reverse transcriptase PCR (RT-PCR) as previously described [6]. Glycan chain analysis was performed using a modified version of the Glycan Differentiation Kit (Roche Diagnostics, Nutley, NJ) protocol and a panel of five plant lectins (DSA, GNA, MAA, PHA-L and SNA). Five cadherin family proteins, gp100 and Survivin were detected by immunohistochemistry methods. RNase protection assay or Western Blot were used to quantify the expression of 28 apoptosis-related genes, including several constituents of the Caspase-8 pathway and members of the Bcl family. The activity of five glycosyltransferase enzymes at the cell surface was measured by RT-PCR and direct binding assays were performed for Collagen IV, Fibronectin and Laminin. Chromium release assays were used to characterise binding of HLA by peptides of MART-1 and gp100.
Data production was quality-controlled by a system of hidden duplication. Further crucial quality control was that all lines were DNA fingerprinted prior to entering the cell bank and periodically retested to assure fidelity in the system. This DNA fingerprint is derived from nine short tandem repeat (STR) loci and the amelogenin gene by PCR (AmpFlSTR Profiler PCR Amplification Kit, Applied Biosystems, Foster City, CA, USA). Mycoplasma testing was also carried out on a routine basis. Patient anonymity was maintained throughout the project; this is an additional reason why neither clinical data nor autologous normal tissue is available for any of the ESTDAB cell lines. ESTDAB is updated periodically and the first release contained more than 170 cell lines, each of which has been characterised for up to 250 separate markers.
Organisation and content of the online ESTDAB database
The ESTDAB database is based on programming and tools used in the design of the IMGT/HLA Database (http://www.ebi.ac.uk/imgt/hla/) [7, 8] which maintains a searchable database of the source material used in defining all new and confirmatory HLA allele sequences. While not as complex as the ESTDAB data, this model could be expanded to provide a suitable platform around which to base the ESTDAB tools. The model also incorporates well-used web-based search facilities that are suitable for such a dataset.
The current data are stored in an Oracle® 9i relational database (Oracle Corporation, Redwood Shores, CA, USA), in collaboration with the European Bioinformatics Institute (EBI). A simplified schematic of the database is shown in Fig. 1. The data are then made available to the scientific community through the ESTDAB pages on the EBI’s web server. The ESTDAB pages are contained within the Immuno-Polymorphism Database (IPD) project alongside other databases of immunological interest. The IPD was developed in 2003 to provide a centralised system for the study of polymorphisms in genes of the immune system. IPD currently consists of four databases: IPD-KIR contains the allelic sequences of killer-cell immunoglobulin-like receptors; IPD-MHC is a database of sequences of the major histocompatibility complex of different species; IPD-HPA lists alloantigens expressed only on platelets; and IPD-ESTAB. The project works with specialist groups or nomenclature committees who provide and curate individual sections before they are submitted to IPD for online publication. The IPD project was established by the HLA Informatics Group of the Anthony Nolan Research Institute in close collaboration with the EBI [9].
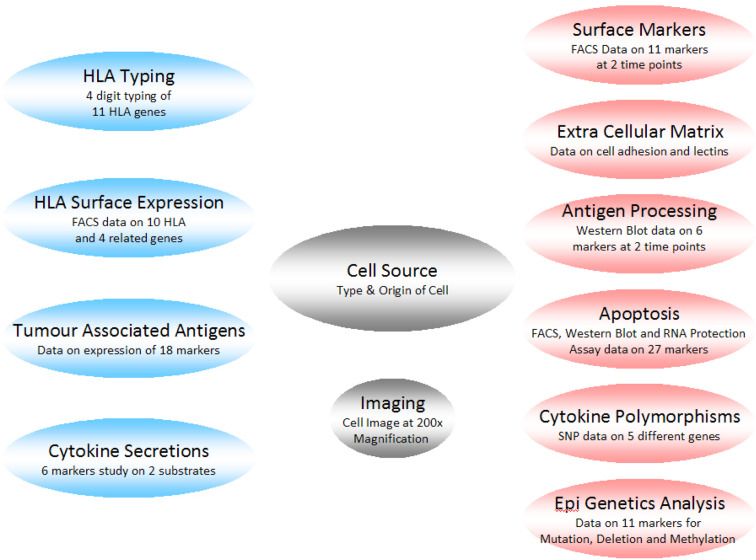
Fig. 1.
Simplified schematic diagram of the ESTDAB data. The grey boxes represent the mandatory data, the blue boxes the primary search data and the red boxes the secondary search data
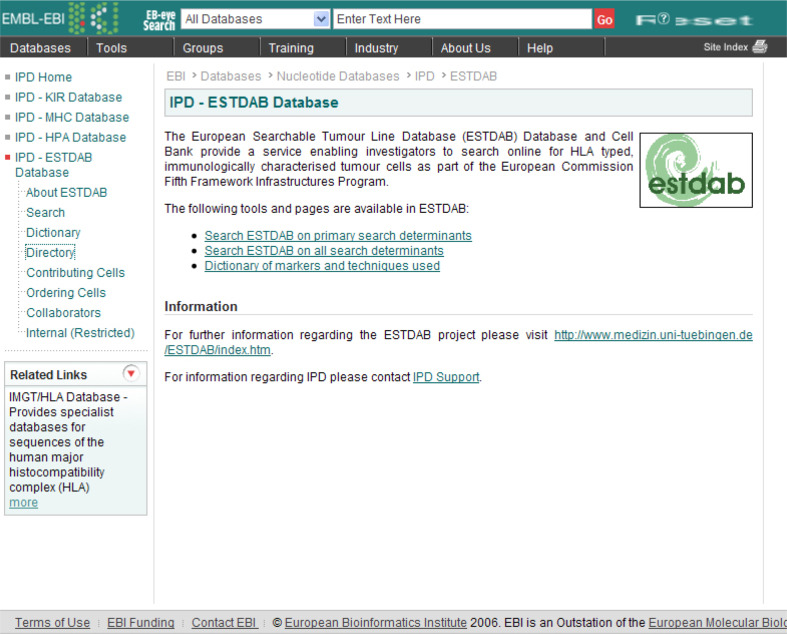
Access to all tools is made available through the ESTDAB website (see Fig. 2); there are several ways to search the ESTDAB database for cells of interest. The different searches are available from the ESTDAB homepage, which also provides links to supporting documentation including protocols and/or information sufficient to reproduce the experimental conditions applied in the characterisation of the ESTDAB cells.
Fig. 2.
The ESTDAB homepage, which has links to the different search tools and support documentation
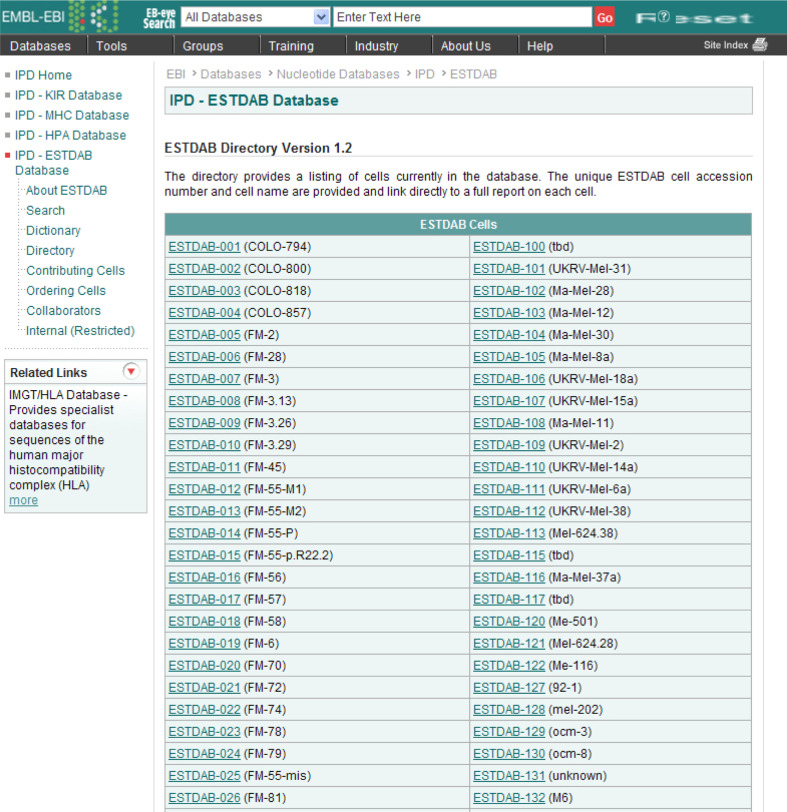
The simplest method of obtaining a cell′s profile is to go the ESTDAB directory (http://www.ebi.ac.uk/ipd/estdab/directory.html), which provides a link to each cell in the directory based on its unique ESTDAB identification code and name. This simple and easy-to-use page (see Fig. 2) only requires the user to know the name of the melanoma cell line of interest in order to access its full profile.
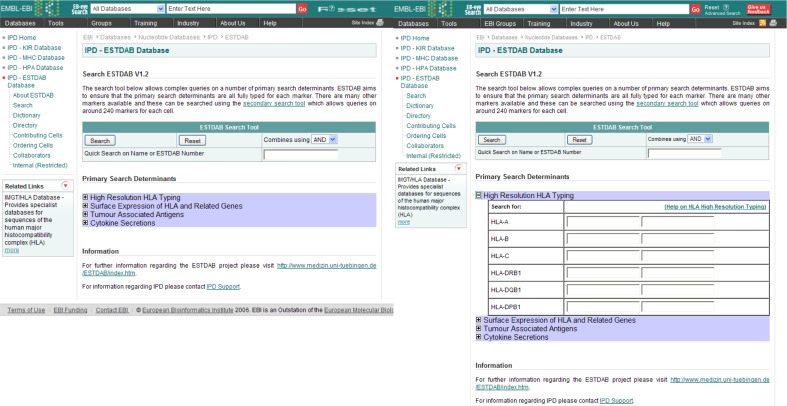
The advanced search tool is split into two versions because of the number of markers studied by the ESTDAB consortium. The primary search tool (http://www.ebi.ac.uk/ipd/estdab/primary_search.html) allows users to find cells with a particular HLA genotype or surface expression pattern, or otherwise according to the cells’ expression of tumour associated antigens or secretion of cytokines. These are the most well-defined markers in the dataset and as such are more likely to return positive hits than those factors included in the secondary search tool (see Fig. 3). If the user desires to search for a more specific factor, the ESTDAB database can be searched according to any marker by using the secondary search tool (http://www.ebi.ac.uk/ipd/estdab/secondary_search.html).
Fig. 3.
The ESTDAB directory, which provides a list of all cells in the database, and direct links to their profiles
Whichever search tool is chosen the same methods apply when searching for a cell. Through a series of expanding menus, the user can define the marker required and select or enter the type of interest. The search tool will then display the results of these queries as a list of links to the cells that match. The tool can be used to define simple queries based on a single marker or more complex queries using Boolean algebra (AND, OR) to search for more complex patterns found in several markers. There is also the option to simply search on the cell name or unique ESTDAB identification code.
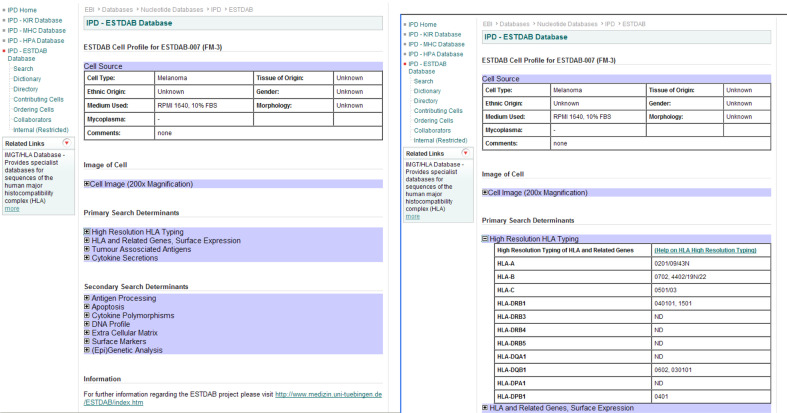
From the results the user can easily follow a link to the full profile of any cell. The main page displays the basic data for each cell. The same series of expanding menus is used to allow the large dataset to be easily displayed on a single page (see Fig. 4). Simply expanding the relevant section will provide the full information on the particular set of markers. A number of the lines also have images of the cells available to view. The output (see Fig. 5) can therefore be tailored to the user and the data from as many markers as necessary can be displayed or printed by the user.
Fig. 4.
The ESTDAB primary search tool, allows the user to perform complex queries on the most well-defined markers. The information on individual markers can be viewed by expanding each section using the small “plus” button. The figure illustrates the expanded high-resolution HLA typing section
Fig. 5.
An ESTDAB cell profile. The cell profile contains a large number of markers, which can be viewed by expanding the entry for each section. The right hand image shows the detailed information available for high-resolution HLA typing
Once cells of interest have been identified using the online search tools, samples of those cells can be obtained from the ESTDAB cell bank for a nominal charge to cover handling and shipping on completion of a standard Material Transfer Agreement. Since the release of ESTDAB there have been a number of publications that have utilised ESTDAB cell lines, for example in the study of glycosylation characterisation [10], the direct mechanisms of loss of IFN-gamma mediated HLA class I inducibility [11] and the characterisation of HLA class I altered phenotypes [12].
The ESTDAB database and cell bank provide a freely available and fully searchable database of highly characterised melanoma-derived cell lines. Users can easily search for cells suitable for use in their research, which can then be ordered from the cell bank. This service should be of use primarily to those interested in melanoma biology, but will also be of interest to anyone involved in cancer biology.
Acknowledgments
This project was supported by the European Commission within the Fifth Framework Infrastructures Program (contract no. QLRI-CT-2001-01325). ESTDAB was a continuation of EUCAPS (European Concerted Action on Peptide Sensitization. http://www.medizin.uni-tuebingen.de/eucaps/home/). The characterisation of the lines used in ESTDAB was provided by Raja Choudhury, Federico Garrido, Rolf Kiessling, Piotr Laidler, Elissaveta Naumova, Per thor Straten and Wolfgang Wagner.
Footnotes
This paper is a focussed research review from the meeting which took place on the 28th–29th May 2008 in Nottingham, UK, celebrating the contribution of Prof. I.A. “Tony” Dodi (29.1.2008) to the EU project “Network for the identification and validation of antigens and biomarkers in cancer and their application in clinical tumour immunology (ENACT).”
References
- 1.Pawelec G, Marsh SGE. ESTDAB: a collection of immunologically characterised melanoma cell lines and searchable databank. Cancer Immunol Immunother. 2006;55:623–627. doi: 10.1007/s00262-005-0117-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arguello JR, Little AM, Bohan E, Goldman JM, Marsh SGE, Madrigal JA. High resolution HLA class I typing by reference strand mediated conformation analysis (RSCA) Tissue Antigens. 1998;52:57–66. doi: 10.1111/j.1399-0039.1998.tb03024.x. [DOI] [PubMed] [Google Scholar]
- 3.Arguello JR, Little AM, Pay AL, Gallardo D, Rojas I, Marsh SGE, Goldman JM, Madrigal JA. Mutation detection and typing of polymorphic loci through double-strand conformation analysis. Nat Genet. 1998;18:192–194. doi: 10.1038/ng0298-192. [DOI] [PubMed] [Google Scholar]
- 4.Rozemuller EH, Bouwens AG, van Oort E, Versluis LF, Marsh SGE, Bodmer JG, Tilanus MG. Sequencing-based typing reveals new insight in HLA-DPA1 polymorphism. Tissue Antigens. 1995;45:57–62. doi: 10.1111/j.1399-0039.1995.tb02415.x. [DOI] [PubMed] [Google Scholar]
- 5.Santamaria P, Lindstrom AL, Boyce-Jacino MT, Myster SH, Barbosa JJ, Faras AJ, Rich SS. HLA class I sequence-based typing. Hum Immunol. 1993;37:39–50. doi: 10.1016/0198-8859(93)90141-M. [DOI] [PubMed] [Google Scholar]
- 6.thor Straten P, Guldberg P, Gronbaek K, Hansen MR, Kirkin AF, Seremet T, Zeuthen J, Becker JC. In situ T cell responses against melanoma comprise high numbers of locally expanded T cell clonotypes. J Immunol. 1999;163:443–447. [PubMed] [Google Scholar]
- 7.Robinson J, Malik A, Parham P, Bodmer JG, Marsh SGE. IMGT/HLA database—a sequence database for the human major histocompatibility complex. Tissue Antigens. 2000;55:280–287. doi: 10.1034/j.1399-0039.2000.550314.x. [DOI] [PubMed] [Google Scholar]
- 8.Robinson J, Waller MJ, Parham P, Bodmer JG, Marsh SGE. IMGT/HLA database—a sequence database for the human major histocompatibility complex. Nucleic Acids Res. 2001;29:210–213. doi: 10.1093/nar/29.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robinson J, Waller MJ, Stoehr P, Marsh SGE. IPD—the immuno polymorphism database. Nucleic Acids Res. 2005;33:D523–D526. doi: 10.1093/nar/gki032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laidler P, Litynska A, Hoja-Lukowicz D, Labedz M, Przybylo M, Ciolczyk-Wierzbicka D, Pochec E, Trebacz E, Kremser E. Characterization of glycosylation and adherent properties of melanoma cell lines. Cancer Immunol Immunother. 2006;55:112–118. doi: 10.1007/s00262-005-0019-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez T, Mendez R, Del Campo A, Jimenez P, Aptsiauri N, Garrido F, Ruiz-Cabello F. Distinct mechanisms of loss of IFN-gamma mediated HLA class I inducibility in two melanoma cell lines. BMC Cancer. 2007;7:34. doi: 10.1186/1471-2407-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendez R, Rodriguez T, Del Campo A, Monge E, Maleno I, Aptsiauri N, Jimenez P, Pedrinaci S, Pawelec G, Ruiz-Cabello F, Garrido F. Characterization of HLA class I altered phenotypes in a panel of human melanoma cell lines. Cancer Immunol Immunother. 2008;57:719–729. doi: 10.1007/s00262-007-0411-3. [DOI] [PMC free article] [PubMed] [Google Scholar]