Abstract
The cytotoxic activity of T cells selects the outgrowth of tumor cells that escape from immune surveillance by different strategies. The different mechanisms that interfere with immune recognition and limit vaccination efficiency are still poorly understood. We analysed six cell lines established from different metastases of melanoma patient UKRV-Mel-20 for specific characteristics known to have an impact on the tumor-T cell interaction: (1) alterations in the HLA class I phenotype, (2) expression of Fas/CD95, and (3) expression of specific cytokines and chemokines. One of the cell lines, UKRV-Mel-20f, exhibited an HLA class I haplotype loss and just this cell line was also characterised by the expression of Fas/CD95 and of relatively high levels of proinflammatory chemokines suggesting that the cytotoxic activity of tumor-infiltrating T cells might have selected the outgrowth of this tumor cell variant. All other cell lines analysed showed no alterations in HLA class I expression, but, in contrast to UKRV-Mel-20f, expressed much lower levels of Fas/CD95 and of proinflammatory chemokines and some of them produced high levels of immunosuppressive TGF-β1. These results suggest that in patient UKRV-Mel-20, tumor cells interfere with T cell recognition by different strategies which might partially explain why this patient did not have a clinical response to an autologous tumor cell vaccine.
Keywords: Immune escape, HLA loss, LOH
Introduction
Evidence that melanoma-associated antigens can be specifically recognised by the immune system led to the development and clinical application of several tumor-specific vaccination strategies. But so far clinical responses have only been observed infrequently. Successful immunotherapy of cancer will ultimately require an understanding of the natural relationship between the immune system and tumors as they transform, invade and metastasize. Recent evidence suggests that tumors defeat specific immunity through a variety of mechanisms that account for the infrequent clinical immune responses [8, 18]. Thus, tumors acquire resistance to CTLs by loss of tumor antigen expression and due to the lack of components of the antigen processing and presentation machinery [4], e.g. tumor cells can partially or totally lose the expression of major histocompatibility complex (MHC) class I antigens during progression [17]. Beside defects in antigen presentation other immune suppressive mechanisms have been characterised: tumor cells have been demonstrated to loose expression of specific molecules, e.g. Fas/CD95 protecting them from T cell mediated apoptosis [6, 11].
Furthermore, they can secrete immunosuppressive factors like TGF-β-1, IL-10, and other cytokines, that interfere not only with T cell activity but also with activation and differentiation of dendritic cells infiltrating the tumor [33]. On the other hand trafficking of lymphocytes to the tumor is prevented by the inhibition of proinflammatory cytokine and chemokine production due to a constitutive activation of the STAT-3 protein within the tumor cells [30]. But recent research has also emphasised the importance of active suppressor mechanisms mediated by the immune system itself: CD4+CD25+T lymphocytes have been demonstrated to suppress both the proliferation and effector functions of other immune cells [10, 22] and these cells can be detected with higher frequency in the peripheral blood and tumor tissue of tumor patients [31].
In this study we analysed the pattern of MHC class I, CD95/Fas, chemokine, and cytokine expression in melanoma cell lines derived from six metastasis obtained at different time points from late stage disease patient UKRV-Mel-20. Interestingly, in one metastatic cell line that produced the highest levels of pro-inflammatory cytokines an haplotype HLA loss was observed suggesting that the microenvironment of this tumor favoured the activity of MHC-class I restricted T cells which then selected the outgrowth of this loss variant.
Material and methods
Cell lines
Melanoma cell lines established from different metastasis of patient UKRV-Mel-20 designated as UKRV-Mel-20b, UKRV-Mel-20c, UKRV-Mel-20d, UKRV-Mel-20e, UKRV-Mel-20f, and UKRV-Mel-20g (Fig. 1) were maintained in RPMI 1640/HEPES/2 mM glutamine (PAA Laboratories, Cölbe, Germany) supplemented with 10% FCS (PAA Laboratories), 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C and 5% CO2.
Fig. 1.
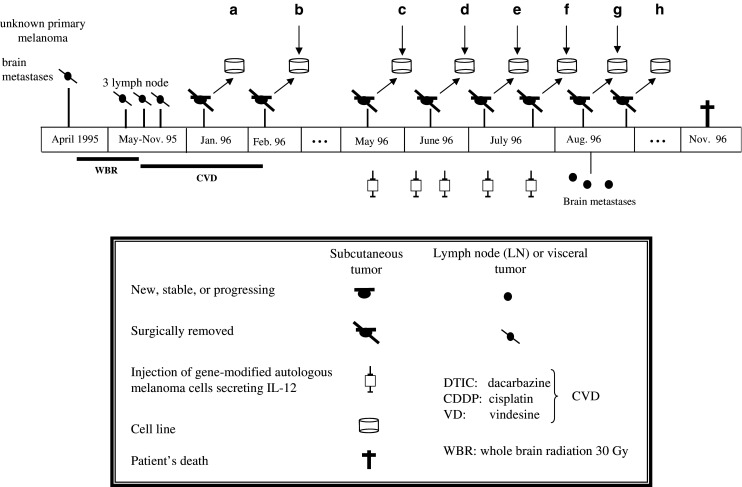
Scheme of the clinical evolution of melanoma patient UKRV-Mel-20
Flow cytometry analysis
Surface HLA class I expression on cultured melanoma cells was determined by indirect immunofluorescence using an appropriate anti-HLA-class-I monoclonal antibody (mAb) and FITC-labelled rabbit anti-mouse Ig (Fab2) fragments (Sigma Chemical CO, St. Louis, USA). The following mAbs were used: W6/32 against HLA class I heavy chain/β2m complex [1]; GRH1 against free β2m; A131 defining an HLA-A locus-specific determinant [25]; and YTH-76 defining an HLA-B locus-specific determinant [3]. To increase MHC antigen expression, cell lines were treated with recombinant interferon-γ (800 U/ml) (Amershan, Aylesbury, UK) for 48 h. Surface expression of CD95/Fas was determined by an anti-CD95 PE-conjugated monoclonal antibody (BD Biosciences, San Jose, CA, USA). A total of 104 cells were analysed for each immunofluorescence profile. As negative controls, cells were incubated with irrelevant mAb. Fluorescence was analysed with a FACSort flow cytometer (Becton Dickinson, Mountain View, CA, USA) and the CellQUEST software (BD Biosciences) was used for data acquisition and analysis.
Microsatellite analysis
Loss of heterozygosity (LOH) analysis was performed by PCR amplification of five highly polymorphic microsatellite sequences. The markers were chosen on the basis of their heterozygosity (PIC value greater than 0.7) and their location in the HLA region of chromosome 6 [20]. For fluorescent microsatellite assay PCR reactions were performed in a total volume of 15 μl containing 60 ng of each DNA sample, 1×PCR buffer, 5 μM each of unlabelled primer and 5′ end primers labelled with fluorescent dyes, 0.5 units Taq DNA polymerase, and 250 μM of each dideoxynucleotide. Specific Genescan and Genotyper® software was used to determine the size and quantity of the PCR products and to compare normal and tumor amplicon patterns for each marker.
Real-time PCR
Melanoma cells were analysed for expression of several target genes (IP-10, ITAC-1, RANTES, MCP-1, MIP1α, SDF-1, IFN-γ, TGF-β-1, VEGF-c, and IL-10) by quantitative real-time PCR. As a control, expression of the G6PDH housekeeping gene was tested. All PCR reactions were performed in a Light Cycler instrument using the LC-FastStart DNA Master SYBR Green I Kit (Roche Diagnostics, Manheim, Germany), with the exception of G6PDH for which the Housekeeping Gene Set Kit was used (Roche Diagnostics). The primers for the cytokine and chemokine amplification reactions were used from LightCycler-Primer Set (Search LC GmbH, Heidelberg, Germany).
Thermocycling for each reaction was performed in a final volume of 20 μl containing 2 μl cDNA sample, 4 mmol/l MgCl2, 0.5 μmol/l of each primer, and 2 μl LC-FastStart DNA Master SYBR Green. After 10 min of initial denaturation at 95°C, the cycling conditions consisted of denaturation at 95°C for 6 s, annealing at 68°C for 10 s, and elongation at 72°C. Elongation periods varied depending on the length of the product (1 s/25 bp). After amplification the temperature was slowly raised above the melting point of the PCR product to measure the fluorescence for the melting curve, which allows the identification of specific transcripts. Expression levels of target genes were given relative to the expression levels of G6PDH. All PCR products were checked by melting point analysis and by gel electrophoresis to verify that the products were of the correct size.
Results
Analysis of the HLA class I phenotype of melanoma cell lines from patient UKRV-Mel-20
In April 1995 patient UKRV-Mel-20 was presented with melanoma and brain metastasis. After radiation and subsequent polychemotherapy the patient was included in an immunotherapy trial, during which he received irradiated autologous gene-modified tumor cells secreting IL-12 [26]. Vaccination was started in May 1996. The vaccine was prepared from two cell lines, UKRV-Mel-20a, and -20b established from cutaneous metastatic lesions excised in January and February 1996, respectively. From May to July the patient received five autologous vaccines. Vaccination was abrogated in August when new brain metastases were diagnosed and no objective response of any lesion was detected. Under vaccination different cutaneous tumors were successively excised, leading to the establishment of the cell lines UKRV-Mel-20c, -Mel-20d, -Mel-20e, -Mel-20f, -Mel-20g, and -Mel-20h.
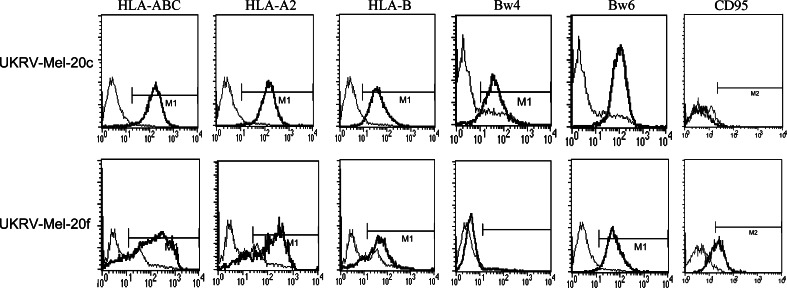
Due to the non responsiveness during vaccination we analysed the different tumor cell lines for specific characteristics that might have interfered with tumor cell recognition by cytotoxic T cells, such as the alteration of the HLA phenotype, the loss of CD95/Fas expression or the secretion of immunosuppressive cytokines. To answer this we first determined the HLA class I phenotype of UKRV-Mel-20b, -Mel-20c, -Mel-20d, -Mel-20e, -Mel-20f, and -Mel-20g cells by indirect immunofluorescence. Representative results of the cytofluorometric analysis of UKRV-Mel-20c cells are presented in Fig. 2. These melanoma cells were positively stained by anti-HLA-ABC, anti-β2m and by locus-specific antibodies for HLA-A and HLA-B antigens, suggesting that no alteration in HLA class I expression occurred. Comparable results were also obtained for four additional cell lines: UKRV-Mel-20d, -Mel-20e, -Mel-20g, and -Mel-20h (data not shown). In contrast, UKRV-Mel-20f cells, which exhibited apparently normal HLA-ABC expression were clearly HLA-Bw4-negative, corresponding to HLA-B*3801 of the patients genotype (HLA-A*02011, -A*0204, -B*40011, -B*3801, HLA-Cw*03) (Fig. 2). This specificity also remained undetectable after treatment of melanoma cells with IFN-γ.
Fig. 2.
Flow cytometric analysis of HLA class I and CD95 surface expression on melanoma cells from patient UKRV-Mel-20. Cells were labelled with monomorphic, locus-specific and allele-specific anti-HLA class I mAbs. Additionally staining with an anti-CD95 antibody was performed. Thin line represents profiles of the isotypic Ig used as a negative control. Representative results for two (UKRV-Mel-20c, UKRV-Mel-20f) of six cell lines are given. Expression of HLA-Bw4 could be detected on UKRV-Mel-20c and for UKRV-Mel-20b, UKRV-Mel-20d, UKRV-Mel-20e, and UKRV-Mel-20g cells (data not shown) but not for UKRV-Mel-20f cells. In contrast, a strong expression of CD95/Fas could only be detected on UKRV-Mel-20f cells
UKRV-Mel-20f exhibit loss of heterozygosity in the HLA-region
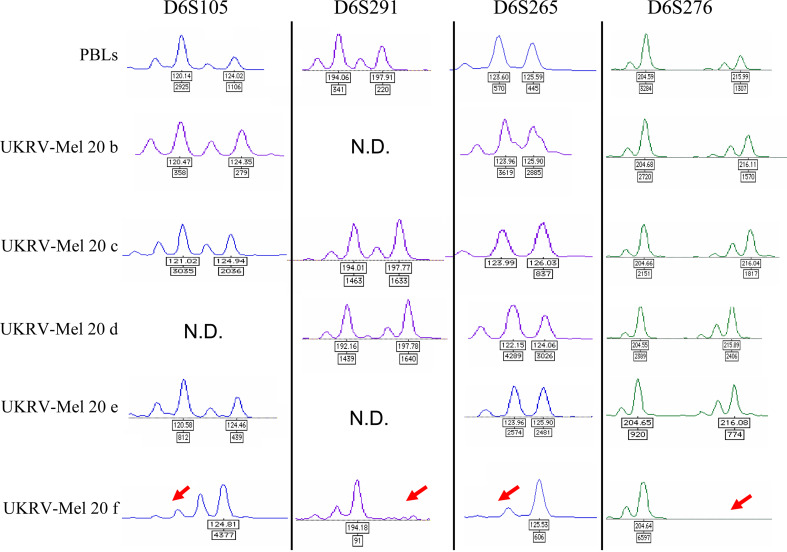
The studies performed at the protein level demonstrated that UKRV-Mel-20f cells lost HLA-B*3801 expression. However, this analysis did not allow the detection of alterations at the HLA-A level since both alleles of patient UKRV-Mel-20 (HLA-A*0201, A*0204) were stained by the allele-specific HLA-A2 antibody. To determine if alterations in UKRV-Mel-20f cells affected additional genes of the HLA-region we analysed the status of heterozygosity of five microsatellite markers spanning chromosome 6p. PCR analysis on the genomic DNA from autologous peripheral blood lymphocytes of patient UKRV-Mel-20 revealed heterozygosity for four of the five markers, with the exception of marker D6S273. Comparison of normal and autologous tumor DNA demonstrated that the cell lines UKRV-Mel-20b, -20c, -20d, and -20e exhibited retention of heterozygosity for all informative markers employed, while UKRV-Mel-20f cells presented loss heterozygosity for all these markers (Fig. 3). These data demonstrated that an extended loss of chromosome 6p material occurred within these cells most probably associated with a complete loss of an HLA haplotype.
Fig. 3.
Pattern of polymorphic markers on chromosome 6. These examples illustrate the status of heterozygosity of several microsatellites markers on chromosome 6p in melanoma cell lines and PBLs of patient UKRV-Mel-20. Microsatellite analysis was performed on DNA from UKRV-Mel-20 melanoma cells compared to DNA obtained form autologous PBLs. The scan showed a pattern of allelic loss (LOH) for all markers employed for the UKRV-Mel-20f melanoma cells
Expression of Fas on the UKRV-Mel 20 cell lines
Beside presentation of HLA molecules we analysed the cell lines for the presence of the CD95/Fas receptor at their cell surface. Cytofluorometric analysis revealed a significant variation in CD95/Fas surface expression. Whereas UKRV-Mel-20f cells showed a strong CD95/Fas expression, all of the other metastatic cell lines, exhibited low or undetectable expression comparable to UKRV-Mel-20c (Fig. 2 and data not shown). These results suggested that except for UKRV-Mel-20f cells all other cell lines might be resistant to Fas ligand mediated killing by cytotoxic CD8+T cells.
Chemokine and cytokine gene expression in UKRV-Mel-20 cell lines
In addition to alterations in the HLA class I phenotype, immune-modulating factors secreted by tumor cells, such as cytokines and chemokines, influence the activity of T cells and professional antigen presenting cells infiltrating the tumor. By Real-time RT-PCR we compared the expression pattern of different cytokines (IL-10, IFN-γ, TGF-β1), chemokines (MCP-1, MIP-1α, IP-10, ITAC-1, RANTES, SDF-1) and of the vascular endothelial growth factor (VEFG-c), by the different melanoma cell lines. Expression analysis revealed strong variations between the different UKRV-Mel-20 cell lines (Fig. 4).
Fig. 4.
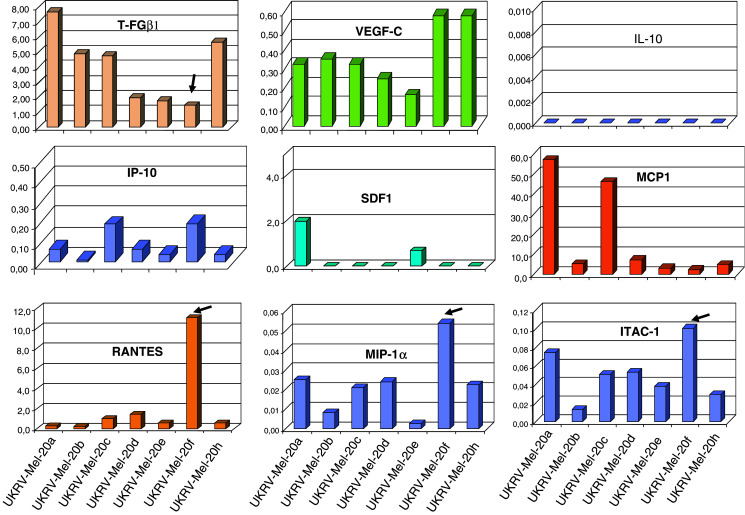
Comparison of the expression pattern of specific cytokines and chemokines by real-time RT-PCR. The proinflammatory chemokines ITAC-1, IP10, MIP-1α, RANTES, were significantly increased in UKRV-Mel-20f cells compared to the other melanoma cell lines from this patient. TGF-β-1 was detected clearly and with a tendency towards decreased levels in all cell lines through cell line UKRV-Mel-20f and VEGF-c was expressed in all cell lines at a comparable level. MCP-1 and SDF-1 were expressed variably between the cell lines
VEGF-c was expressed in all cell lines at a comparable level. The immunosuppressive cytokine TGF-β-1 was detected clearly with a tendency towards a decrease in the RNA level from cell line UKRV-Mel-20a through to cell line -Mel-20f. In contrast, expression of IL-10 and IFN-γ was undetectable. Interestingly, UKRV-Mel-20f cells were characterised by an elevated expression level of certain chemokines e.g. ITAC-1, IP-10, MIP-1α, RANTES, most of them were less abundantly expressed in all other cell lines.
Discussion
Recent studies have presented evidence that the immune system influences the development of some spontaneous malignancies, shaping the resulting repertoire of tumor-cell phenotypes [24]. Thus, cancer progression and metastases are the result of a balance between tumor immunosurveillance and tumor escape. It is assumed that the microenvironment exerts an evolutionary pressure that promotes the selection of tumor cell clones [23]. Based on the divergent evolution of tumor cells, this most probably leads to the outgrowth of subclones characterised by a heterogeneous pattern of immune escape mechanisms. To test this proposition, patterns of HLA class I, CD95/Fas, cytokine, chemokine, and growth factor expression were determined in several melanoma cell lines from various metastases obtained at different time points from a melanoma patient (UKRV-Mel-20) (Fig. 1). One cell line, UKRV-Mel-20f was characterised by an HLA haplotype loss, and a distinct pattern of CD95/Fas, cytokine and chemokine expression (Figs. 2, 4) as compared to other cell lines. Proliferation of the tumor cell clones with haplotype loss (e.g. UKRV-Mel-20f cells) might be due to selective immunoreactivity of tumor-infiltrating antigen-specific T cells that favour the outgrowth of malignant cells that are less immunogenic [7, 8, 13, 15]. The infiltration of T lymphocytes into tumor UKRV-Mel-20f might have been supported by the expression of specific chemokines, since we detected high levels of RANTES, MIP-1α and CXCR3 ligands ITAC-1 and IP-10. Chemokine receptors CXCR3 and CCR5 are preferentially expressed on cytotoxic T cells and helper T cells of the Th1 phenotype, which is abundant in inflammatory diseases [16, 19]. Accordingly, this pattern of chemokines and the relatively low expression of the immunosuppressive cytokine TGF-β-1 might have enforced T cell activity, leading to the outgrowth of a tumor cell escape variant characterised by an HLA haplotype loss. However, tumor cells that lack expression of certain MHC class I alleles become more susceptible to lysis by NK cells [12]. NK cells are potently attracted by the chemokine SDF-1 [9]. Interestingly, we detected no SDF-1 expression by UKRV-Mel 20f cells. Cytokines and growth factors such as IL-10 and VEGF-c, which suppress or attenuate an antitumor immune response, interfering with DC activation and differentiation, were also found in melanoma cell lines established from patient UKRV-mel-20 [32]. IL-10 expression was not detected in these melanoma cell lines, while VEGF-c expression varied among these cell lines.
Based on these results, it could be speculated that the production of specific chemokines involved in tumor development and spread might contribute indirectly to the immune escape of malignant cells. Interestingly, unlike any of the other cell lines analysed, UKRV-Mel-20f cells exhibited a high surface expression of CD95/Fas, indicating their sensitivity against the apoptosis inducing activity of Fas ligand secreted by cytotoxic tumor-infiltrating T cells.
In comparison with UKRV-Mel-20f cells, the other cell lines all expressed higher levels of the immunosuppressive cytokine TGF-β-1 which is known to impair CTL and NK cell activity [5, 14]. Elevated concentration of TGF-β-1 has been correlated with a CD3 zeta loss in tumor infiltrating lymphocytes (TILs) [29], a mechanism that prevents direct TIL/tumor cell contact and inhibits TILs activation [33]. TGF-β-1 also acts on cytotoxic T cells lymphocytes to specifically inhibit the expression of genes encoding perforin, granzyme A, granzyme B, IFN-γ and CD95L, which are effectors of CTL-mediated cytotoxicity [28]. Furthermore, these melanoma cells (except for UKRV-Mel-20f) were characterised by a reduction or loss of Fas/CD95 expression on the cell surface, as reported on other melanoma cells [2] which protects them from Fas ligand-induced cell death exerted by cytotoxic T cells.
We postulate, based on our data, that each tumor in a given patient is characterised by a specific microenvironment that promotes a status of immune tolerance, and that the mechanisms underlying this status are heterogeneous. This agrees with our previous report of the coexistence of multiple mechanisms of tumor immune evasion in a single patient [21]. It is believed that different cellular variants with increased metastatic ability are the result of genomic instability. The coexistence of different phenotypes in one patient supports a selection model, in which cells that possesses advantageous characteristics are selected [27].
Acknowledgements
We would like to thank Carmen Amezcua, M. Jose Rivas and Antje Sucker for expert technical assistance. This work was partially supported by the Fondo de Investigaciones Sanitarias, the plan Andaluz de Investigacion, Instituto de Salud Carlos III-Red de Centros de Cancer-RTICCC-contract CO3/10 and the Plan Nacional, Spain as well as the EU 6th framework project ENACT (contract No. 553306).
Footnotes
This article is a symposium paper from the conference “Progress in Vaccination against Cancer 2005 (PIVAC 5),” held in Athens, Greece, on 20–21 September 2005.
References
- 1.Barnstable CJ, Bodmer WF, Brown G, Galfre G, Milstein C, Williams AF, Ziegler A. Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens-new tools for genetic analysis. Cell. 1978;14:9. doi: 10.1016/0092-8674(78)90296-9. [DOI] [PubMed] [Google Scholar]
- 2.Bullani RR, Wehrli P, Viard_Leveugle I, Rimoldi D, Cerottini JC, Saurat JH, Tschopp J, French LE. Frequent downregulation of Fas (CD95) expression and function in melanoma. Melanoma Res. 2002;12:263. doi: 10.1097/00008390-200206000-00010. [DOI] [PubMed] [Google Scholar]
- 3.Burrone OR, Kefford RF, Gilmore D, Milstein C. Stimulation of HLA-A,B,C by IFN-alpha. The derivation of Molt 4 variants and the differential expression of HLA-A,B,C subsets. EMBO J. 1985;4:2855. doi: 10.1002/j.1460-2075.1985.tb04014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cabrera CM, Jimenez P, Cabrera T, Esparza C, Ruiz-Cabello F, Garrido F. Total loss of MHC class I in colorectal tumors can be explained by two molecular pathways: beta2-microglobulin inactivation in MSI-positive tumors and LMP7/TAP2 downregulation in MSI-negative tumors. Tissue Antigens. 2003;61:211. doi: 10.1034/j.1399-0039.2003.00020.x. [DOI] [PubMed] [Google Scholar]
- 5.Castriconi R, Cantoni C, Della Chiesa M, Vitale M, Marcenaro E, Conte R, Biassoni R, Bottino C, Moretta L, Moretta A. Transforming growth factor beta 1 inhibits expression of NKp30 and NKG2D receptors: consequences for the NK-mediated killing of dendritic cells. Proc Natl Acad Sci USA. 2003;100:4120. doi: 10.1073/pnas.0730640100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong H, Strome SE, Salomao DR, Tamura H, Hirano F, Flies DB, Roche PC, Lu J, Zhu G, Tamada K, Lennon VA, Celis E, Chen L. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med. 2002;8:793. doi: 10.1038/nm730. [DOI] [PubMed] [Google Scholar]
- 7.Garrido F, Algarra I. MHC antigens and tumor escape from immune surveillance. Adv Cancer Res. 2001;83:117. doi: 10.1016/S0065-230X(01)83005-0. [DOI] [PubMed] [Google Scholar]
- 8.Garrido F, Ruiz-Cabello F, Cabrera T, Perez-Villar JJ, Lopez-Botet M, Duggan-Keen M, Stern PL. Implications for immunosurveillance of altered HLA class I phenotypes in human tumours. Immunol Today. 1997;18:89. doi: 10.1016/S0167-5699(96)10075-X. [DOI] [PubMed] [Google Scholar]
- 9.Hanna J, Wald O, Goldman-Wohl D, Prus D, Markel G, Gazit R, Katz G, Haimov-Kochman R, Fujii N, Yagel S, Peled A, Mandelboim O, Hanna J, Wald O, Goldman-Wohl D, Prus D, Markel G, Gazit R, Katz G, Haimov-Kochman R, Fujii N, Yagel S, Peled A, Mandelboim O. CXCL12 expression by invasive trophoblasts induces the specific migration of CD16-human natural killer cells. Blood. 2003;102:1569. doi: 10.1182/blood-2003-02-0517. [DOI] [PubMed] [Google Scholar]
- 10.Hori S, Takahashi T, Sakaguchi S. Control of autoimmunity by naturally arising regulatory CD4+T cells. Adv Immunol. 2003;81:331. doi: 10.1016/s0065-2776(03)81008-8. [DOI] [PubMed] [Google Scholar]
- 11.Igney FH, Krammer PH. Death and anti-death: tumour resistance to apoptosis. Nat Rev Cancer. 2002;2:277. doi: 10.1038/nrc776. [DOI] [PubMed] [Google Scholar]
- 12.Karre K. NK cells, MHC class I molecules and the missing self. Scand J Immunol. 2002;55:221. doi: 10.1046/j.1365-3083.2002.01053.x. [DOI] [PubMed] [Google Scholar]
- 13.Khong HT, Restifo NP. Natural selection of tumor variants in the generation of “tumor escape” phenotypes. Nat Immunol. 2002;3:999. doi: 10.1038/ni1102-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JC, Lee KM, Kim DW, Heo DS. Elevated TGF-beta1 secretion and down-modulation of NKG2D underlies impaired NK cytotoxicity in cancer patients. J Immunol. 2004;172:7335. doi: 10.4049/jimmunol.172.12.7335. [DOI] [PubMed] [Google Scholar]
- 15.Lehmann F, Marchand M, Hainaut P, Pouillart P, Sastre X, Ikeda H, Boon T, Coulie PG. Differences in the antigens recognized by cytolytic T cells on two successive metastases of a melanoma patient are consistent with immune selection. Eur J Immunol. 1995;25:340. doi: 10.1002/eji.1830250206. [DOI] [PubMed] [Google Scholar]
- 16.Loetscher P, Uguccioni M, Bordoli L, Baggiolini M, Moser B, Chizzolini C, Dayer JM. CCR5 is characteristic of Th1 lymphocytes. Nature. 1998;391:344. doi: 10.1038/34814. [DOI] [PubMed] [Google Scholar]
- 17.Paschen A, Mendez RM, Jimenez P, Sucker A, Ruiz-Cabello F, Song M, Garrido F, Schadendorf D. Complete loss of HLA class I antigen expression on melanoma cells: a result of successive mutational events. Int J Cancer. 2003;103:759. doi: 10.1002/ijc.10906. [DOI] [PubMed] [Google Scholar]
- 18.Pawelec G, Zeuthen J, Kiessling R. Escape from host-antitumor immunity. Crit Rev Oncog. 1997;8:111. doi: 10.1615/critrevoncog.v8.i2-3.10. [DOI] [PubMed] [Google Scholar]
- 19.Qin S, Rottman JB, Myers P, Kassam N, Weinblatt M, Loetscher M, Koch AE, Moser B, Mackay CR. The chemokine receptors CXCR3 and CCR5 mark subsets of T cells associated with certain inflammatory reactions. J Clin Invest. 1998;101:746. doi: 10.1172/JCI1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramal LM, Feenstra M, van der Zwan AW, Collado A, Lopez-Nevot MA, Tilanus M, Garrido F. Criteria to define HLA haplotype loss in human solid tumors. Tissue Antigens. 2000;55:443. doi: 10.1034/j.1399-0039.2000.550507.x. [DOI] [PubMed] [Google Scholar]
- 21.Real LM, Jimenez P, Kirkin A, Serrano A, Garcia A, Canton J, Zeuthen J, Garrido F, Ruiz-Cabello F. Multiple mechanisms of immune evasion can coexist in melanoma tumor cell lines derived from the same patient. Cancer Immunol Immunother. 2001;49:621. doi: 10.1007/s002620000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shevach EM, McHugh RS, Piccirillo CA, Thornton AM. Control of T-cell activation by CD4+ CD25+ suppressor T cells. Immunol Rev. 2001;182:58. doi: 10.1034/j.1600-065X.2001.1820104.x. [DOI] [PubMed] [Google Scholar]
- 23.Smalley KS, Brafford PA, Herlyn M. Selective evolutionary pressure from the tissue microenvironment drives tumor progression. Semin Cancer Biol. 2005;15:451. doi: 10.1016/j.semcancer.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 24.Smyth MJ, Trapani JA. Lymphocyte-mediated immunosurveillance of epithelial cancers? Trends Immunol. 2001;22:409. doi: 10.1016/S1471-4906(01)01977-9. [DOI] [PubMed] [Google Scholar]
- 25.Spear BT, Kornbluth J, Strominger JL, Wilson DB. Evidence for a shared HLA-A intralocus determinant defined by monoclonal antibody 131. J Exp Med. 1985;162:1802. doi: 10.1084/jem.162.6.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun Y, Jurgovsky K, Moller P, Alijagic S, Dorbic T, Georgieva J, Wittig B, Schadendorf D. Vaccination with IL-12 gene-modified autologous melanoma cells: preclinical results and a first clinical phase I study. Gene Ther. 1998;5:481. doi: 10.1038/sj.gt.3300619. [DOI] [PubMed] [Google Scholar]
- 27.Tomlinson I, Bodmer W. Selection, the mutation rate and cancer: ensuring that the tail does not wag the dog. Nat Med. 1999;5(1):11. doi: 10.1038/4687. [DOI] [PubMed] [Google Scholar]
- 28.Thomas DA, Massague J. TGF beta directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell. 2005;8:369. doi: 10.1016/j.ccr.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 29.von Bernstorff W, Voss M, Freichel S, Schmid A, Vogel I, Jonk C, Henne-Bruns D, Kremer B, Kalthoff H. Systemic and local immunosuppression in pancreatic cancer patients. Clin Cancer Res. 2001;7:925. [PubMed] [Google Scholar]
- 30.Wang T, Niu G, Kortylewski M, Burdelya L, Shain K, Zhang S, Bhattacharya R, Gabrilovich D, Heller R, Coppola D, Dalton W, Jove R, Pardoll D, Yu H. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat Med. 2004;10:48. doi: 10.1038/nm976. [DOI] [PubMed] [Google Scholar]
- 31.Woo EY, Chu CS, Goletz TJ, Schlienger K, Yeh H, Coukos G, Rubin SC, Kaiser LR, June CH. Regulatory CD4(+)CD25(+) T cells in tumors from patients with early-stage non-small cell lung cancer and late-stage ovarian cancer. Cancer Res. 2001;61:4766. [PubMed] [Google Scholar]
- 32.Yang L, Carbone DP. Tumor-host immune interactions and dendritic cell dysfunction. Adv Cancer Res. 2004;92:13. doi: 10.1016/S0065-230X(04)92002-7. [DOI] [PubMed] [Google Scholar]
- 33.Zou W. Immunosuppressive networks in the tumour environment and their therapeutic relevance. Nat Rev Cancer. 2005;5:263. doi: 10.1038/nrc1586. [DOI] [PubMed] [Google Scholar]