Abstract
We characterized a new melanoma antigen derived from one of the multiple open reading frames (ORFs) of the meloe transcript. The meloe gene is overexpressed in melanomas as compared to other cancer cell lines and normal tissues. The corresponding transcript is rather unusual, in that it does not contain a long unique ORF but multiple short ORFs. We recently characterized a tumor epitope derived from a polypeptide (MELOE-1) encoded by the ORF1230–1370 and involved in relapse prevention of melanoma patients treated with autologous tumor infiltrating lymphocytes (TIL). Here we show that the ORF285–404 encodes a polypeptide called MELOE-2 that also generated a HLA-A2 epitope recognized by a melanoma-specific T cell clone derived from the same TIL population from which we derived the MELOE-1-specific T cell clone. We also showed that HLA-A2 melanoma cells were spontaneously recognized by the MELOE-2-specific T cell clone, and we detected the presence of MELOE-2 reactive T cells in another TIL population infused to a patient who remained relapse-free after TIL treatment. These results demonstrate that translation of meloe transcript in melanoma cells can produce at least two immunogenic polypeptides, MELOE-1 and MELOE-2, from two distinct ORFs that could be relevant target for melanoma immunotherapy.
Keywords: Antigen, Melanoma, Meloe, T lymphocyte
Introduction
Many tumor associated-antigens (TAA), targets of autologous CD8+ cytotoxic T lymphocytes (CTL), have been identified during the last 20 years (see [1] for a review). According to the pattern of expression in neoplastic and normal tissues, TAAs can be roughly subdivided into two main classes: proteins expressed specifically by tumor cells and proteins also expressed at a significant level by normal cells. In the first category are found cancer-germline antigens, proteins expressed by unconventional gene expression and mutated antigens. The main advantage of these proteins is their tumor specificity, but they are expressed by a limited fraction of tumors. TAA belonging to the second category are the differentiation antigens, mainly expressed in the melanocytic lineage, and the antigens overexpressed by various tumor tissues. Tumor antigen discovery has resulted in the molecular identification of several major histocompatibility complex HLA-restricted tumor peptides, which has opened the way for antigen-targeted immunotherapies. However, although the immunogenicity of a number of TAA has been documented in clinical trials, the therapeutical potential of only few of these antigens has been suggested. Indeed, the therapeutical potential of the Melan-A antigen in melanoma has been strongly suggested by the analysis of several active [2, 3] and passive [4–9] immunotherapy protocols targeting this antigen. Furthermore, we recently showed a correlation between the infusion of T cells reactive against a HLA-A2 epitope derived from the melanoma antigen MELOE-1, and relapse prevention of patients treated with autologous tumor infiltrating lymphocytes (TIL) [10]. This antigen is encoded by the meloe gene, located on chromosome 2 and overexpressed in melanoma cell lines, compared to other cancer cell lines and normal tissues. The structure of the corresponding transcript was rather unusual, in that it lacked a long open reading frame (ORF) but contained only multiple short ORFs. The ORF1230–1370 encodes the protein MELOE-1 that contains the HLA-A2 restricted epitope recognized by several melanoma TIL populations [10].
Here, we describe an additional antigen encoded by the ORF285–404 of the meloe cDNA, recognized by a CTL clone derived from the TIL population from which we previously derived the MELOE-1-specific CTL clone (M170). This protein was thus called MELOE-2, and we characterized a HLA-A2-restricted epitope derived from this polypeptide and recognized by the specific CTL clone. Using specific tetramers, we demonstrated the presence of MELOE-2 reactive T cells in an additional TIL population, also infused to a patient who is still relapse-free. These results show that at least two of the multiple ORFs of the meloe cDNA are translated in melanoma cells, and generate peptides able to induce an immune response in melanoma TIL populations.
Materials and methods
Cell lines and TIL cultures
Twenty T cell populations were expanded from cryopreserved samples of TIL (derived from tumor invaded lymph nodes) infused to melanoma patients included in a phase I/II protocol between 1994 and 1998. This clinical trial aimed at comparing the survival of stage III melanoma patients randomly treated by IL-2 alone or TIL + IL-2, in an adjuvant setting [11]. Ten additional HLA-A2 stage III melanoma patients, treated with TIL and IL-2 in a similar adjuvant setting but not included in the clinical trial, were also analyzed in the present study. All the patients were enrolled between December 2001 and June 2006. The randomized clinical trial was approved by the local ethics committee (CPP Ouest IV, Nantes, France) and the French security agency (Afssaps). Concerning the ten additional patients, only the Afssaps authorization was necessary. TIL samples were expanded according a procedure previously described [12, 13]. M170 TIL population containing tumor-specific T cells was cloned by limiting dilution [14], and tumor-specific T cell clones were amplified as previously described [12]. Melanoma cells lines and colorectal carcinoma cell line C4-A were established, respectively, in the Unit of cellular therapy and in our laboratory. Mouse fibrosarcoma WEHI 164 clone 13 and COS-7 cells were obtained from T. Boon (LICR, Brussels, Belgium). Renal carcinoma cell line A498 was a gift from C. Saï (INSERM U892, Nantes, France). Colorectal carcinoma cell lines (CaCo-2, Sw480, Sw707, LS174T, HTC116), breast carcinoma cell line 734-B, were gifts from M. Grégoire (INSERM U601, Nantes, France), S. Chouaib (INSERM U487, Villejuif, France) and D. Jäger (Klinik und Poliklinik für Onkologie, Zürich, Germany). Breast cancer cell line MCF-7 and melanoma cell line A375 were purchased from the ATCC. Normal melanocytes (98M09 and 01M08), were gifts from M. Regnier (L’Oréal Laboratory, Paris, France).
Functional analysis of T cells
Cytotoxic activity of MELOE-2-specific T cells was measured in a standard 4-h assay against 51Cr-labeled peptide-loaded T2 cells at an E:T ration of 10/1. Measurement of TNF produced by M170.51 T cell clone in response to tumor cells or transfected COS-7 cells was performed as previously described [15], using WEHI 164 clone 13 cells [16]. mAb against HLA class I (clone W6.32), HLA-B/C (clone B1.23.2), HLA-A2 (clone BB7.2) added to cultures in some experiments, were produced in our laboratory from hybridomas obtained from the ATCC for W6.32 and BB7.2 antibodies and from F. Lemonier (Pasteur Institute, France) for B1.23.2 antibody. For intracytoplasmic cytokine staining, after a 6-h stimulation period with melanoma cells at an E:T ration of 1:2, in presence of brefeldin A at 10 μg/mL (Sigma, St Louis, MO, USA), T cells were labeled with an APC-coupled anti-CD8 antibody (BD Biosciences, France), and fixed for 10 min at room temperature in PBS 4% paraformaldehyde (Sigma). Fixed lymphocytes were stained for cytokine production using anti-TNF-α, anti-IFN-γ, anti-GM-CSF and anti-IL2 specific antibodies (BD Biosciences, France), as previously described [17]. After staining, cells were resuspended in PBS and analyzed on a LSR flow cytometer using Cell Quest software. Concerning CD107a mobilization experiment, 105 MELOE-2-specific T cells were stimulated with 2 × 105 M6 or M170 melanoma cells for 4 h at 37°C in the presence of PE conjugated mAb specific for CD107a (BD Biosciences, France). The T cells were then stained with an APC conjugated anti-CD8 mAb and analyzed by flow cytometry.
ORFs constructs
The various ORFs from meloe sequence, and the partial sequences of the ORF285–404 were generated either by PCR, or direct annealing of the two forward and reverse complementary primers (see Table 1 for primers). Oligonucleotides were designed with EcoRI and XhoI sites included for subcloning in pcDNA3, and with a Kozak sequence (gccaccATG) for upper primer and a stop codon for lower primer. After subcloning in expression vector, sequencing was carried out by the DNA Sequencing Facility of the IFR 26 (Nantes, France).
Table 1.
Sequences of primers used ORFs subcloning
| Primers | 5′–3′ sequencesa |
|---|---|
| ORF subcloning | |
| (285–404) forward | aaaGAATTCgccaccATGAGTGAAAATGCAGGAGG |
| (285–404) reverse | aaaCTCGAGTCACTGGCACAGTGCAG |
| (486–689) forward | aaaGAATTCgccaccATGTCCGTAGGGAGACGC |
| (486–689) reverse | aaaCTCGAGTCACCCTAGAGCTGCCAAG |
| (1105–1194) forward | aaaGAATTCgccaccATGAAATTTGAATTAATTTGCAGAAC |
| (1105–1194) reverse | aaaCTCGAGTTAATTCCTAAGGCATTCATTC |
| (1230–1370) forward | aaaGAATTCgccaccATGAGTTGTGTAGGTTATCC |
| (1230–1370) reverse | aaaCTCGAGTCACAGGGATGCCGGCC |
| ORF285–404 partial sequence subcloning | |
| (285–341) forward | AATTCgccaccATGAGTGAAAATGCAGGAGGTGCCGTAGCGAGAACAGCGACAGCATTCTGCGCATTGTGAC |
| (285–341) reverse | TCGAGTCACAATGCGCAGAATGCTGTCGCTGTTCTCGCTACGGCACCTCCTGCATTTTCACTCATggtggcG |
| (285–371) forward | AATTCgccaccATGAGTGAAAATGCAGGAGGTGCCGTAGCGAGAACAGCGACAGCATTCTGCGCATTGGTGAGCCCGACTCCCCAGCCTCGGTGCCCATGAC |
| (285–371) reverse | TCGAGTCATGGGCACCGAGGCTGGGGAGTCGGGCTCACCAATGCGCAGAATGCTGTCGCTGTTCTCGCTACGGCACCTCCTGCATTTTCACTCATggtggcG |
| (345–404) forward | AATTCgccaccATGAGCCCGACTCCCCAGCCTCGGTGCCCACCGAAGCCCCCTCTGGCTGCACTGTGCCAGTGAC |
| (345–404) reverse | TCGAGTCACTGGCACAGTGCAGCCAGAGGGGGCTTCGGTGGGCACCGAGGCTGGGGAGTCGGGCTCATggtggcG |
a Italics enzyme restriction sites, lower case Kozak sequence
Synthetic peptides
Peptides were purchased from Eurogentec (Angers, France). Purity (>70 or >90% for tetramer production) was controlled by reversed-phase high-performance liquid chromatography. Peptides were lyophilized, dissolved in DMSO at 10 mg/mL and stored at −20°C.
MELOE-2C28L/A2-specific tetramer labeling
HLA-A*0201/MELOE-2 α3-mutated monomers were generated by the recombinant protein facility (IFR26, INSERM U892, Nantes, France), as previously described [18]. TIL populations and M170.51 T cell clone were co incubated for 1 h at 4°C in the dark with PE or APC-conjugated MELOE-2 tetramer (10 μg/mL) and CD8 mAb (5 μg/mL), and 104 events were analyzed on a FACSCalibur.
Immunomagnetic cell sorting and expansion of T cell-sorted populations
HLA-A*0201/MELOE-2 monomers (20 μg/mL) were incubated for 1 h at room temperature with 6 × 106 streptavidin-coated beads (Dynabeads M-280 streptavidin, DYNAL, Compiegne, France) and washed in PBS-0.1% BSA. 5 × 106 TIL were rotated for 4 h at 4°C with monomer-coated beads [18, 19]. After ten washes, bead coated cells were expanded using a polyclonal T cell stimulation protocol [12]. A panel of 24 anti-Vβ mAbs (Vβ1, −2, −3, −4, −5.1, −5.2, −5.3, −7, −7.2, −8, −9, −11, −12, −13.1, −13.2,−13.6, −14, −16, −17, −18, −20, −21.3, −22 and –23) was used to analyze the diversity of M278-sorted TIL population (Immunotech Beckman-Coulter, Marseille, France).
Results
T cell clone selection and characterization
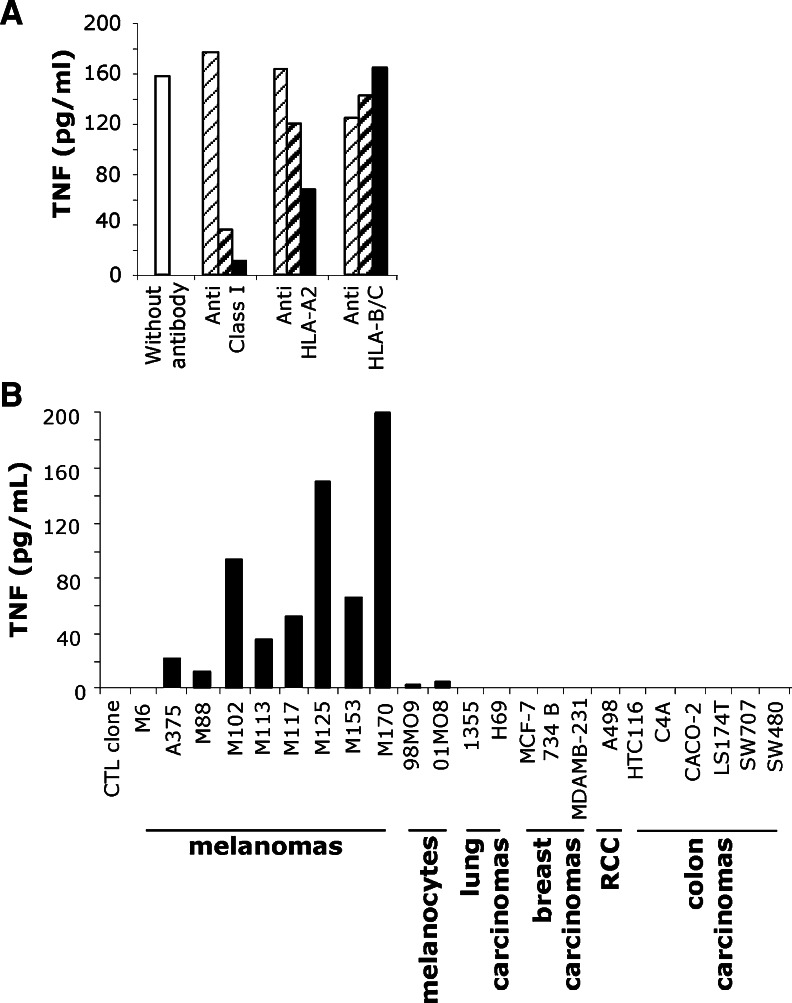
A TIL population, which had been infused in 1998 to a melanoma patient M170 who is still relapse-free, was cloned by limiting dilution. Aside from Melan-A-specific T cell clones, we derived eight CTL clones that showed a reactivity pattern consistent with the recognition of (a) new antigen(s). One of these clones, characterized in a previous study [10], was specific for a new melanoma overexpressed antigen, called MELOE-1. In the present study, we focused on another CD8+ T cell clone, hereafter referred to as M170.51. As illustrated by Fig. 1a, the recognition of the autologous melanoma cell line was restricted by the HLA-A2 molecule, since the TNF release of this T cell clone was inhibited by an anti-HLA-A2-specific antibody but not by an anti-B/C antibody. In order to determine the distribution of the target antigen, we tested M170.51 reactivity toward HLA-A2 melanocytes (98M09 and 01MO8) and various HLA-A2 tumor cell lines, including melanomas, ovarian, lung, breast, renal and colon carcinomas, using a TNF release assay. As shown in Fig. 1b, this T cell clone recognized all the HLA-A2 melanoma cell lines tested but none of the other HLA-A2 tumor cell types. In addition, M170.51 T cell clone also weakly recognized the two HLA-A2 melanocyte cell lines tested.
Fig. 1.
T cell clone selection and characterization. a TNF secretion by M170.51 T cell clone in response to the autologous melanoma cell line. 104 CTL were added to 3 × 104 M170 melanoma cells, in the presence of blocking antibodies directed against class I, A2 and B/C HLA, diluted to 1/5,000 (thin hatched bars) 1/500 (thick hatched bars) and 1/50 (black bars). b TNF response of M170.51 CTL clone to HLA-A*0201 tumor cell lines. M6 cell line, HLA-A2 negative, was used as a negative control. 09M08 and 01M08 were HLA-A*0201 melanocyte cell lines
Identification of the cDNA coding for the antigen
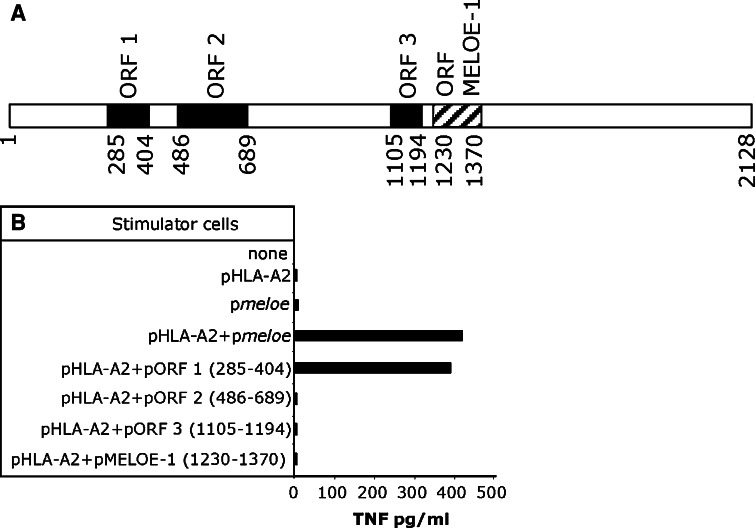
Since the tumor cell line recognition pattern was similar to that of MELOE-1-specific T cell clone previously identified [10], we tested M170.51 T cell clone for recognition of COS-7 cells co-transfected with the meloe cDNA (Fig. 2a) and the HLA-A*0201 cDNA. As shown in Fig. 2b, COS-7 cells co-transfected with those two plasmids were recognized by this CD8+ T cell clone. The meloe cDNA does not contain a long ORF, but multiple short ORFs. The ORF1230–1370 recognized by MELOE-1-specific T cell clone [10] is indicated with a hatched box on the Fig. 2a. Upon co-transfection into COS-7 cells with the HLA-A*0201 coding cDNA, we tested this ORF together with three additional ORFs (black boxes in Fig. 2a) for recognition by M170.51 CTL clone (Fig. 2b). Results showed that the ORF285–404 encoded the polypeptide, thereafter called MELOE-2, containing the epitope recognized by this T cell clone.
Fig. 2.
Characterization of the cDNA coding for the recognized antigen. a Structure of meloe cDNA. Black boxes and hatched box, illustrating the ORF coding for the MELOE-1 antigen, were tested for recognition by the CTL clone M170.51. b TNF response of MELOE-2-specific clone to COS-7 cells (E/T ratio 1/3) transfected with indicated plasmids. The specific T cell clone was added 2 days after the transfection, and its reactivity was assessed by a TNF release assay
Identification of the peptide recognized by M170.51 T cell clone
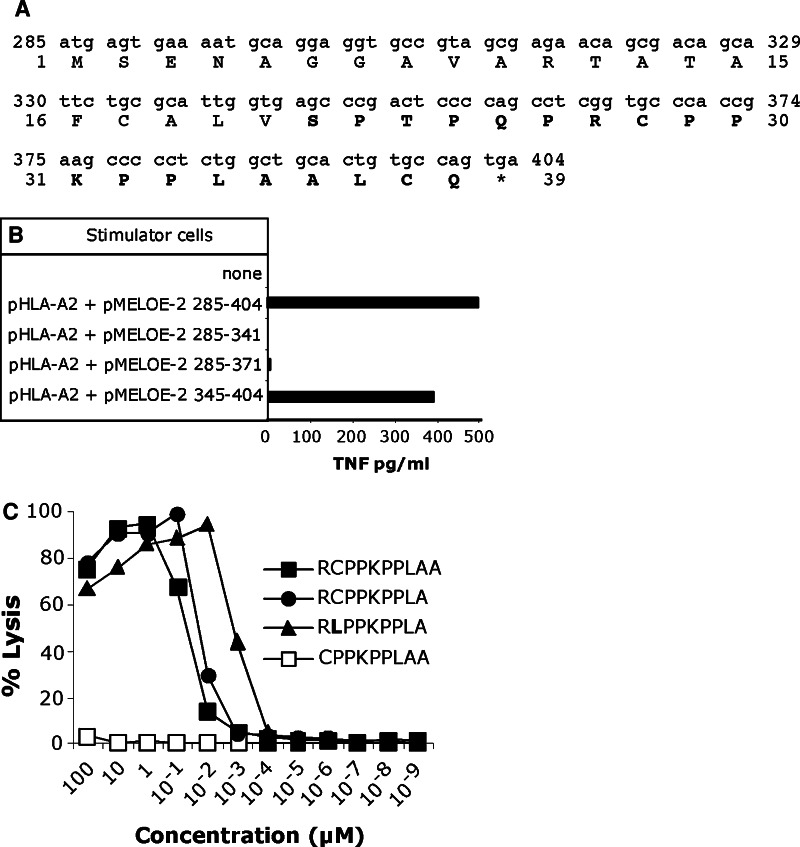
Using HLA BIMAS prediction analysis (http://www.bimas.cit.nih.gov), we could not identify peptides able to bind to the HLA-A*0201 with a high stability within the 39 amino-acid protein encoded by the ORF285–404 (Fig. 3a). In order to restrict the region that encodes the peptide recognized by M170.51 CTL clone, we tested three cDNA regions of MELOE-2 for recognition by the specific CTL clone, upon transfection into COS-7 cells (Fig. 3b). The region including 385–404 bp was recognized by M170.51 T cell clone, whereas the region 285–341 bp and the overlapping region 285–371 bp did not induce T cell clone activation. Thus, the region encoding the peptide recognized by M170.51 CTL clone was comprised between amino acids 21 and 39, indicated in bold on Fig. 3a. We tested various decapeptides located within this region and showed that MELOE-227–36 peptide “RCPPKPPLAA” was recognized by M170.51 CTL clone, with an EC50 of 7 × 10−8 M (black squares on Fig. 3c). We then tested the recognition of two additional peptides derived from this decapeptide. We observed that deletion of the arginine at the N-terminal end (position 27) completely abrogated the CTL response (open squares on Fig. 3c), whereas deletion of the alanine at the C-terminal end (position 36) slightly increased the CTL clone response (circles on Fig. 3c). In conclusion, the optimal natural nonapeptide appeared to be MELOE-227–35 (RCPPKPPLA), recognized with an EC50 of 3 × 10−8 M. However, the sequence of MELOE-227–35 peptide was not optimal for binding to the HLA-A*0201 molecule, since it lacks the dominant anchor amino acid residues (leucine or methionine) at position 2, and valine at position 9 [20, 21]. This suboptimal binding prevents us from producing a HLA-A2 tetramer folded with this natural peptide. Thus, with the aim to produce a MELOE-2-specific tetramer folded with a peptide exhibiting an improved binding to the HLA molecule, we introduced a single amino acid substitution (C28L) in the natural peptide sequence. We first checked the recognition of this modified peptide by our MELOE-2-specific CTL clone as shown in Fig. 3c, the peptide analog MELOE-2C28L was better recognized by M170.51 CTL clone, with a half maximal lysis of 10−9 M (triangles), compared to 3 × 10−8 M for the natural peptide (circles).
Fig. 3.
Characterization of meloe derived peptide recognized by M170.51 T cell clone. a Nucleotide and amino acid sequence of the ORF 285–404 of meloe isolated from M134 cDNA library. The peptidic region containing the recognized peptide is indicated in bold. b M170.51 TNF responses to COS-7 cells (E/T ratio 1/3) transfected with indicated plasmids. The specific T cell clone was added 2 days after the transfection, and its reactivity was assessed by a TNF release assay. c Cytotoxicity of M170.51 CTL clone against peptide-pulsed T2 cells. Target cells were chromium labeled for 60 min and incubated for 30 min with a range of the indicated peptides. M170.51 T cell clone was added at an E/T ratio of 10/1, and chromium release was then measured after a 4-h incubation period
Presence of MELOE-2-specific lymphocytes in another TIL population
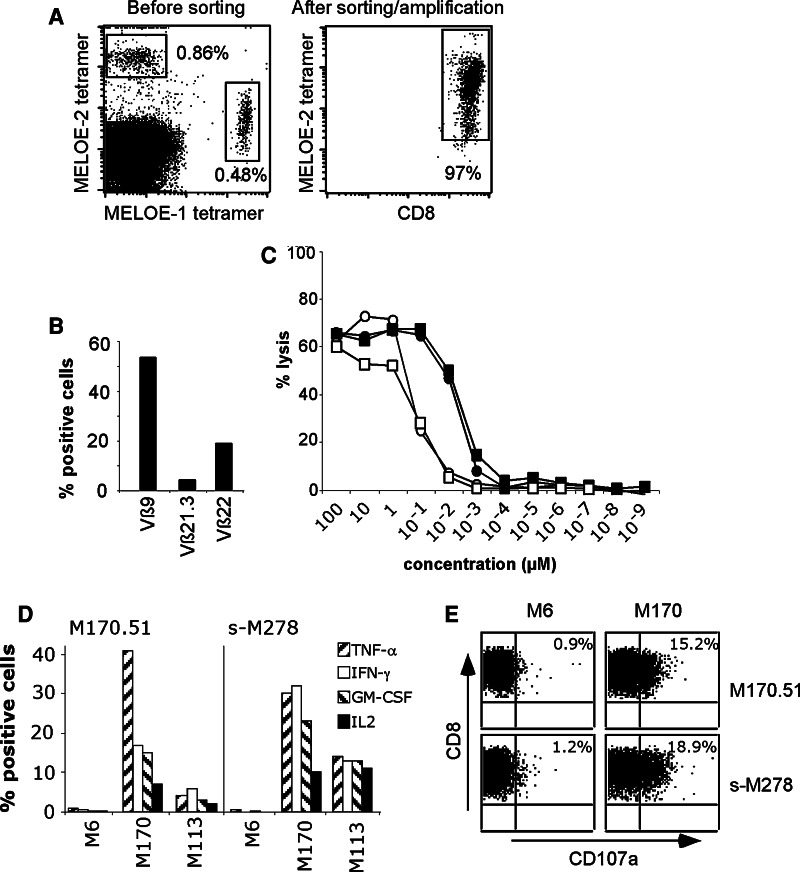
As we previously documented the existence of a correlation between the presence of MELOE-1-specific lymphocytes among TIL infused to melanoma patients and relapse prevention (P < 0.001) [10], we looked for the presence of MELOE-2-specific lymphocytes among the same 30 HLA-A2-positive TIL populations using a HLA-A2/peptide tetramer loaded with the MELOE-2C28L analog peptide. In addition to the M170 TIL population that contained less than 0.1% of MELOE-2-specific cells (data not shown), we detected the presence of MELOE-2/A2-specific T cells in another TIL population, M278, also infused to a relapse-free melanoma patient, that also contained MELOE-1-specific T cells (Fig. 4a, left panel). After multimer sorting and expansion [22], we obtained a pure MELOE-2-specific population (Fig. 4a, right panel). The diversity of TCR Vβ usage of this sorted populations was assessed with a panel of 24 anti-Vβ antibodies representing the most frequently expressed Vβ chains within a normal repertoire. In M278-sorted TIL population, three different Vβ chains were expressed above 1% (Vß9, Vß21.3 and Vß22), indicating the presence of a rather polyclonal-specific TCR repertoire (Fig. 4b). In order to assess the reactivity of this sorted population toward the natural MELOE-227–35, we measured its cytotoxicity against T2 cells loaded with the natural and modified MELOE-2 peptides. As illustrated on Fig. 4c, M278-sorted TIL population (squares) recognized the MELOE-2C28L analog (black symbols) better than the natural peptide (open symbols), with an EC50 similar to that of the M170.51 T cell clone (black and open circles). This better recognition (about tenfold) of the modified peptide is likely due to its improved affinity and/or stability for the HLA-A2 molecule. Finally, we documented the reactivity of M278-sorted TIL population on HLA-A*0201 melanoma cell lines that spontaneously express the MELOE-2 antigen, by intracellular cytokine labeling. Similar to M170.51 T cell clone, M278-sorted TIL population was able to produce TNF-α, IFN-γ, GM-CSF and IL-2 in response to HLA-A2 melanoma cell lines, as illustrated by responses toward M170 and M113 melanoma cell lines, M6 being a HLA-A2-negative melanoma cell line (Fig. 4d). As illustrated by CD107 mobilization experiment (Fig. 4e), M170.51- and M278-sorted TIL populations were also able to degranulate in response to M170 melanoma cell line, showing that MELOE-2-specific T cells could be lytic against HLA-A2 melanoma cell lines, although their lytic potential appears rather low.
Fig. 4.
Detection and analysis of MELOE-2/A2-specific CTLs in TIL infused to M278 relapse-free melanoma patient. a Labeling of A2/MELOE-227–35-specific T cells in M278 TIL population before and after multimer sorting and expansion. Left panel M278 HLA-A2 TIL population co-labeled with CD8 antibody, A2/MELOE-136–44 and A2/MELOE-227–35 tetramers. Values indicate the fraction of positive T cells among CD8-positive TIL. Right panel multimer-sorted M278 TIL population labeled with CD8 antibody and MELOE-2-specific tetramer. b Vβ chains expressed by the M278 multimer-sorted population. The expression of Vβ was evaluated by labeling the T cell population with a panel of 24 anti-Vβ mAbs. c Cytotoxicity of M170.51 CTL clone (circles) and M278-sorted TIL population (squares) against peptide-pulsed T2 cells. Target cells were chromium labeled for 60 min and incubated for 30 min with a range of the [MELOE-227–35] natural peptide (open symbols) or MELOE-2C28L analog peptide (black symbols). T cells were added at an E/T ratio of 10/1, and chromium release was then measured after a 4-h incubation period. d Production of TNF-α, IFN-γ, GM-CSF and IL-2 by the MELOE-2-specific T cell clone (M170.51) and the MELOE-2-sorted specific population (s-M278), in response to two HLA-A2-positive melanoma cell lines and a HLA-A2-negative melanoma cell line. e CD107a mobilization by M170.51 T cell clone (upper panel) and M278-sorted TIL population (lower panel), in response to a HLA-A2-negative melanoma cell line (M6) and to a HLA-A2-positive melanoma cell line (M170)
Discussion
We previously described a melanoma epitope [(MELOE-136–44)] derived from the meloe gene that is potentially involved in relapse prevention of TIL-treated melanoma patients [10]. Here we describe, in two patients, the existence of spontaneous T cell responses to an epitope derived from a polypeptide encoded by an additional ORF of the meloe cDNA. This ORF located between base pairs 285 and 404 codes for a polypeptide thereafter called MELOE-2, also recognized in the HLA-A*0201 context by a CTL clone derived from the same TIL population (M170). The MELOE-2-specific CTL clone exhibited a similar recognition profile as MELOE-1-specific CTL clone, with the recognition of all HLA-A*0201 melanoma cell lines tested, and to a lower extent of HLA-A*0201 melanocytes. Similar to MELOE-1-specific CTL clone, this T cell clone failed to recognize any of the other tumor cell lines tested (Fig. 1b), which is fully consistent with the expression profile of meloe gene [10].
The optimal natural epitope recognized by M170.51-specific CTL clone was the nonapeptide MELOE-227–35 (Fig. 3c). According to BIMAS analysis (http://www.bimas.cit.nih.gov), this peptide did not bind to the HLA-A*0201 molecule with a high stability, due to poor anchor residues. This was confirmed by our unsuccessful attempts to efficiently fold HLA-A2 complexes with this peptide. Hence, we designed a modified peptide MELOE-2C28L to include the optimal leucine residue in position 2. This peptide was more efficiently recognized by the specific CTL clone than the natural peptide (Fig. 3c). Furthermore, its enhanced binding to HLA-A2 allowed us to produce HLA-peptide tetramers and use these tetramers to look for the presence of MELOE-2-specific T cells within 30 HLA-A2 TIL populations. We thus detected a significant fraction of MELOE-2C28L/A2 tetramer-positive lymphocytes in one additional TIL population (M278) that also contained MELOE-1-specific lymphocytes (Fig. 4a, left panel). After sorting with MELOE-2C28L/A2 multimers and amplification (Fig. 4a, right panel), we provide evidence that this MELOE-2-specific population was oligoclonal as shown by diverse Vβ usage (Fig. 4b) and reactive against the MELOE-227–35 natural peptide, as shown by reactivity on T2 cells loaded with the natural peptide or on HLA-A2 melanoma cell lines (Fig. 4c–e). Although cytokine production of MELOE-2-specific T cells in response to HLA-A2 melanoma cell lines appeared significant (Fig. 4d), their lytic potential, measured by CD107a mobilization remains somewhat low, suggesting that their potential antitumor activity could rather result from an indirect activity.
These data show that the ORFs 285–404 and 1230–1370 are actually translated from the meloe mRNA and give rise to two HLA-A2 restricted epitopes recognized by melanoma-specific TILs.
Although most tumor epitopes recognized by CTLs derive from tumor associated proteins that are encoded by the main ORF of the corresponding mRNA, the use of alternative ORFs for the generation of non classically encoded tumor epitopes has previously been described. The first demonstration of alternative ORF usage was the characterization of a peptide derived from an alternative reading frame of the gp75 antigen [23]. In this case, the translation was initiated at an internal start codon and led to the production of a 24 aa polypeptide. The production of this polypeptide, recognized by melanoma TIL in the HLA-A31 context, did not occur exclusively in tumors, as lymphocytes directed against this antigen also recognized normal melanocytes. In another case reported, an epitope from the CAMEL protein was described in melanoma that originated from the translation product of an alternative ORF in the LAGE-1 gene, starting at the second ATG of the mRNA [24]. Another example of a CTL epitope derived from an alternative ORF is the NA17-A1–9 peptide produced by the translation of an intron of the N-acetylglucosaminyltransferase gene [25]. The mechanism involved in the generation of this tumor antigen was supposed to be a defect of splicing of this intronic region in tumor tissues, associated with a promoter located near the end of the relevant intron that could be activated in tumor cells. Other alternative reading frames leading to the production of tumor epitopes have been identified, such as the melanoma restricted expressed BING-4 protein encoded in a gene-rich region of the extended class II MHC [26], or frameshifts products of CDKN2A tumor suppressor gene [27]. All these non-classically generated epitopes are produced either by transcriptional mechanisms, such as the activation in transformed cells of naturally silent promoters within introns or UTRs [25, 26], or by splicing events [25] or frameshifts [23, 24, 27].
One could hypothesize that the generation of MELOE-1 and MELOE-2 polypeptides from the same messenger could result from the existence in melanomas of low levels of shorter transcripts (generated either by splicing or by the existence of a cryptic promoter) in addition to the main unspliced transcript that we previously described. To test this hypothesis, we performed in depth PCR analyses of meloe transcripts in melanoma cells, but failed to detect any other transcripts than the full length unspliced messenger (data not shown). We thus favor the hypothesis that the production of both MELOE-1 and MELOE-2 polypeptides from meloe messenger is controlled at the translational level. It could result either from re-initiation of translation, where the 40S subunit of a ribosome does not dissociate from the messenger and begins another translation at a downstream AUG codon [28–30], or to internal initiation of translation due to an IRES sequence located between MELOE-2 and MELOE-1, as previously described for several viral and cellular mRNAs [29, 31, 32] and also for a tumor antigen, MPD6 eliciting immune responses in prostate cancer patients [33].
Due to their generation at presumably low concentrations, it has been proposed that unconventionally derived tumor epitopes would be less able to induce negative selection events in the thymus [34]. Thus, unconventional epitopes may allow the expansion of a high avidity tumor-specific T cell repertoire. However, as we found CTL responses against MELOE-2-derived peptide in only two relapse-free patients, its therapeutic potential needs to be further documented.
In conclusion, our data strongly suggest that the translation of distinct ORFs of the meloe mRNA in melanoma cells, by a mechanism that remains to be formally elucidated, generates immunogenic polypeptides that could represent interesting targets for the immunotherapy of melanoma.
Acknowledgments
This work was supported by grants from the “Association pour la Recherche contre le Cancer no. 1074”, and the “INSERM”. Yann Godet was recipient of fellowship awarded by the “Association pour la Recherche contre le Cancer”. We thank the recombinant protein facility and the sequencing facility of the IFR 26 for the production of HLA/peptide complexes and sequencing experiments.
References
- 1.Vujanovic L, Butterfield LH. Melanoma cancer vaccines and anti-tumor T cell responses. J Cell Biochem. 2007;102:301–310. doi: 10.1002/jcb.21473. [DOI] [PubMed] [Google Scholar]
- 2.Jager E, Hohn H, Necker A, Forster R, Karbach J, Freitag K, Neukirch C, Castelli C, Salter RD, Knuth A, Maeurer MJ. Peptide-specific CD8+ T-cell evolution in vivo: response to peptide vaccination with Melan-A/MART-1. Int J Cancer. 2002;98:376–388. doi: 10.1002/ijc.10165. [DOI] [PubMed] [Google Scholar]
- 3.Jager E, Maeurer M, Hohn H, Karbach J, Jager D, Zidianakis Z, Bakhshandeh-Bath A, Orth J, Neukirch C, Necker A, Reichert TE, Knuth A. Clonal expansion of Melan-A-specific cytotoxic T lymphocytes in a melanoma patient responding to continued immunization with melanoma-associated peptides. Int J Cancer. 2000;86:538–547. doi: 10.1002/(SICI)1097-0215(20000515)86:4<538::AID-IJC16>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 4.Dudley ME, Wunderlich JR, Robbins PF, Yang JC, Hwu P, Schwartzentruber DJ, Topalian SL, Sherry R, Restifo NP, Hubicki AM, Robinson MR, Raffeld M, Duray P, Seipp CA, Rogers-Freezer L, Morton KE, Mavroukakis SA, White DE, Rosenberg SA. Cancer regression and autoimmunity in patients after clonal repopulation with antitumor lymphocytes. Science. 2002;298:850–854. doi: 10.1126/science.1076514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mackensen A, Meidenbauer N, Vogl S, Laumer M, Berger J, Andreesen R. Phase I study of adoptive T-cell therapy using antigen-specific CD8+ T cells for the treatment of patients with metastatic melanoma. J Clin Oncol. 2006;24:5060–5069. doi: 10.1200/JCO.2006.07.1100. [DOI] [PubMed] [Google Scholar]
- 6.Meidenbauer N, Marienhagen J, Laumer M, Vogl S, Heymann J, Andreesen R, Mackensen A. Survival and tumor localization of adoptively transferred Melan-A-specific T cells in melanoma patients. J Immunol. 2003;170:2161–2169. doi: 10.4049/jimmunol.170.4.2161. [DOI] [PubMed] [Google Scholar]
- 7.Yee C, Thompson JA, Byrd D, Riddell SR, Roche P, Celis E, Greenberg PD. Adoptive T cell therapy using antigen-specific CD8+ T cell clones for the treatment of patients with metastatic melanoma: in vivo persistence, migration, and antitumor effect of transferred T cells. Proc Natl Acad Sci USA. 2002;99:16168–16173. doi: 10.1073/pnas.242600099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benlalam H, Vignard V, Khammari A, Bonnin A, Godet Y, Pandolfino MC, Jotereau F, Dreno B, Labarriere N. Infusion of Melan-A/Mart-1 specific tumor-infiltrating lymphocytes enhanced relapse-free survival of melanoma patients. Cancer Immunol Immunother. 2007;56:515–526. doi: 10.1007/s00262-006-0204-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vignard V, Lemercier B, Lim A, Pandolfino MC, Guilloux Y, Khammari A, Rabu C, Echasserieau K, Lang F, Gougeon ML, Dreno B, Jotereau F, Labarriere N. Adoptive transfer of tumor-reactive Melan-A-specific CTL clones in melanoma patients is followed by increased frequencies of additional Melan-A-specific T cells. J Immunol. 2005;175:4797–4805. doi: 10.4049/jimmunol.175.7.4797. [DOI] [PubMed] [Google Scholar]
- 10.Godet Y, Moreau-Aubry A, Guilloux Y, Vignard V, Khammari A, Dreno B, Jotereau F, Labarriere N. MELOE-1 is a new antigen overexpressed in melanomas and involved in adoptive T cell transfer efficiency. J Exp Med. 2008;205:2673–2682. doi: 10.1084/jem.20081356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dreno B, Nguyen JM, Khammari A, Pandolfino MC, Tessier MH, Bercegeay S, Cassidanius A, Lemarre P, Billaudel S, Labarriere N, Jotereau F. Randomized trial of adoptive transfer of melanoma tumor-infiltrating lymphocytes as adjuvant therapy for stage III melanoma. Cancer Immunol Immunother. 2002;51:539–546. doi: 10.1007/s00262-002-0315-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jotereau F, Pandolfino MC, Boudart D, Diez E, Dreno B, Douillard JY, Muller JY, LeMevel B. High-fold expansion of human cytotoxic T-lymphocytes specific for autologous melanoma cells for use in immunotherapy. J Immunother. 1991;10:405–411. doi: 10.1097/00002371-199112000-00003. [DOI] [PubMed] [Google Scholar]
- 13.Pandolfino MC, Labarriere N, Tessier MH, Cassidanius A, Bercegeay S, Lemarre P, Dehaut F, Dreno B, Jotereau F. High-scale expansion of melanoma-reactive TIL by a polyclonal stimulus: predictability and relation with disease advancement. Cancer Immunol Immunother. 2001;50:134–140. doi: 10.1007/PL00006683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gervois N, Labarriere N, Le Guiner S, Pandolfino MC, Fonteneau JF, Guilloux Y, Diez E, Dreno B, Jotereau F. High avidity melanoma-reactive cytotoxic T lymphocytes are efficiently induced from peripheral blood lymphocytes on stimulation by peptide-pulsed melanoma cells. Clin Cancer Res. 2000;6:1459–1467. [PubMed] [Google Scholar]
- 15.Benlalam H, Labarriere N, Linard B, Derre L, Diez E, Pandolfino MC, Bonneville M, Jotereau F. Comprehensive analysis of the frequency of recognition of melanoma-associated antigen (MAA) by CD8 melanoma infiltrating lymphocytes (TIL): implications for immunotherapy. Eur J Immunol. 2001;31:2007–2015. doi: 10.1002/1521-4141(200107)31:7<2007::AID-IMMU2007>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 16.Espevik T, Nissen-Meyer J. A highly sensitive cell line, WEHI 164 clone 13, for measuring cytotoxic factor/tumor necrosis factor from human monocytes. J Immunol Methods. 1986;95:99–105. doi: 10.1016/0022-1759(86)90322-4. [DOI] [PubMed] [Google Scholar]
- 17.Jung T, Schauer U, Heusser C, Neumann C, Rieger C. Detection of intracellular cytokines by flow cytometry. J Immunol Methods. 1993;159:197–207. doi: 10.1016/0022-1759(93)90158-4. [DOI] [PubMed] [Google Scholar]
- 18.Bodinier M, Peyrat MA, Tournay C, Davodeau F, Romagne F, Bonneville M, Lang F. Efficient detection and immunomagnetic sorting of specific T cells using multimers of MHC class I and peptide with reduced CD8 binding. Nat Med. 2000;6:707–710. doi: 10.1038/76292. [DOI] [PubMed] [Google Scholar]
- 19.Labarriere N, Gervois N, Bonnin A, Bouquie R, Jotereau F, Lang F. PBMC are as good a source of tumor-reactive T lymphocytes as TIL after selection by Melan-A/A2 multimer immunomagnetic sorting. Cancer Immunol Immunother. 2008;57:185–195. doi: 10.1007/s00262-007-0361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falk K, Rotzschke O, Stevanovic S, Jung G, Rammensee HG. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991;351:290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- 21.Rammensee HG, Friede T, Stevanoviic S. MHC ligands and peptide motifs: first listing. Immunogenetics. 1995;41:178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- 22.Bouquie R, Bonnin A, Bernardeau K, Khammari A, Dreno B, Jotereau F, Labarriere N, Lang F. A fast and efficient HLA multimer-based sorting procedure that induces little apoptosis to isolate clinical grade human tumor specific T lymphocytes. Cancer Immunol Immunother. 2009;58:553–566. doi: 10.1007/s00262-008-0578-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang RF, Parkhurst MR, Kawakami Y, Robbins PF, Rosenberg SA. Utilization of an alternative open reading frame of a normal gene in generating a novel human cancer antigen. J Exp Med. 1996;183:1131–1140. doi: 10.1084/jem.183.3.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aarnoudse CA, van den Doel PB, Heemskerk B, Schrier PI. Interleukin-2-induced, melanoma-specific T cells recognize CAMEL, an unexpected translation product of LAGE-1. Int J Cancer. 1999;82:442–448. doi: 10.1002/(SICI)1097-0215(19990730)82:3<442::AID-IJC19>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 25.Guilloux Y, Lucas S, Brichard VG, Van Pel A, Viret C, De Plaen E, Brasseur F, Lethe B, Jotereau F, Boon T. A peptide recognized by human cytolytic T lymphocytes on HLA-A2 melanomas is encoded by an intron sequence of the N-acetylglucosaminyltransferase V gene. J Exp Med. 1996;183:1173–1183. doi: 10.1084/jem.183.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenberg SA, Tong-On P, Li Y, Riley JP, El-Gamil M, Parkhurst MR, Robbins PF. Identification of BING-4 cancer antigen translated from an alternative open reading frame of a gene in the extended MHC class II region using lymphocytes from a patient with a durable complete regression following immunotherapy. J Immunol. 2002;168:2402–2407. doi: 10.4049/jimmunol.168.5.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang J, El-Gamil M, Dudley ME, Li YE, Rosenberg SA, Robbins PF. T cells associated with tumor regression recognize frameshifted products of the CDKN2A tumor suppressor gene locus and a mutated HLA class I gene product. J Immunol. 2004;172:6057–6064. doi: 10.4049/jimmunol.172.10.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol Cell Biol. 1987;7:3438–3445. doi: 10.1128/mcb.7.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luukkonen BG, Tan W, Schwartz S. Efficiency of reinitiation of translation on human immunodeficiency virus type 1 mRNAs is determined by the length of the upstream open reading frame and by intercistronic distance. J Virol. 1995;69:4086–4094. doi: 10.1128/jvi.69.7.4086-4094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macejak DG, Sarnow P. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature. 1991;353:90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- 32.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 33.Xiong Z, Liu E, Yan Y, Silver RT, Yang F, Chen IH, Chen Y, Verstovsek S, Wang H, Prchal J, Yang XF. An unconventional antigen translated by a novel internal ribosome entry site elicits antitumor humoral immune reactions. J Immunol. 2006;177:4907–4916. doi: 10.4049/jimmunol.177.7.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayrand SM, Green WR. Non-traditionally derived CTL epitopes: exceptions that prove the rules? Immunol Today. 1998;19:551–556. doi: 10.1016/S0167-5699(98)01342-5. [DOI] [PubMed] [Google Scholar]