Abstract
The reduction or loss of MHC-I antigen surface expression in human and murine tumor cells is partly attributable to the dysregulation of various components of the MHC-I antigen-processing machinery. Accumulating evidence suggests that autophagy, besides its vital role in maintaining the cellular homeostasis, plays an important role in MHC-II surface expression. Here, we report that autophagy is a negative regulator of MHC-I antigen expression in B16 melanoma cells; however, in the presence of IFN-γ, it is converted to a positive regulator. We show that autophagy not only participates in the degradation of MHC-I antigen but also plays a role in the generation of MHC-I-binding peptides. For these two processes, IFN-γ interferes with MHC-I antigen degradation, rather than affecting peptide generation. Using B16 melanoma mouse model, we further show that autophagy may enhance the cytolysis of CTL to melanoma cells at the early stage of melanoma, but impairs the cytolysis at the late stage. Such different consequences may be explained by the different levels of IFN-γ during tumor progression. Taken together, our findings demonstrate that autophagy is involved in the regulation of MHC-I antigen expression, through which autophagy can play different roles in tumor immunity.
Electronic supplementary material
The online version of this article (doi:10.1007/s00262-009-0752-1) contains supplementary material, which is available to authorized users.
Keywords: MHC class I, Melanoma cell, Autophagy, IFN-γ
Introduction
Efficient tumor antigen-specific CTL priming and effector function’s exertion require cell surface presentation of tumor antigenic peptides by major histocompatibility complex MHC-I molecules [1, 2]. However, the reduction or loss of MHC-I antigen surface expression in human and murine tumor cells is a common event, which profoundly impedes antitumor immunity [3, 4]. Downregulation of MHC-I antigen surface expression can be attributable to the dysregulation of various components of the MHC-I antigen-processing machinery, including: downregulation of multiple components of the multicatalytic proteasome, in particular LMP2 and LMP7 [5]; downregulation of transporter associated with antigen processing TAP1 and TAP2 [6]; and defect of tumor antigen peptide loading in the endoplasmic reticulum (ER) [7].
Autophagy, in particular macroautophagy, refers to an evolutionarily conserved process that allows the bulk degradation of long-lived proteins and organelles [8, 9]. Though autophagy has become an important area in cancer research, its role in tumor is controversial and equally cogent arguments can be made for pro- and anti-tumor effects of this important process. The tumor suppressor role of autophagy came from the evidence of the increased tumorigenesis in Beclin 1/Atg6 haploinsufficient mice [10]. The mechanisms by which a defect in autophagy could contribute to the pathogenesis of cancer might lie in (1) autophagy effectively removes aged and damaged organelles, and defects in autophagy lead to genetic instability and tumorigenesis; (2) defects in autophagy may escape autophagy-mediated cell death thus increasing cell survival under certain circumstances. On the other hand, documents also indicate the tumor promoting effect of autophagy [11]. This may be partially ascribed to that autophagy allows tumor cells to respond to hypoxia-induced nutrient deprivation by supplying amino acids, fatty acids, and nucleotides. Therefore, the promoting or suppressing role of autophagy in tumors may depend on the context [12, 13].
Besides the roles in tumorigenesis, autophagy is also involved in the antigen-presenting processes [14, 15]. For instance, cytosolic peptides can be presented on MHC class II molecules through autophagy [16–18], since the processing of MHC class II-binding peptides depends on the autophagy-involved endosomal/lysosomal system. Nevertheless, MHC-I-binding peptides are processed through the proteasomal system [19–21]. Whether autophagy involves the process of MHC-I antigen presentation remains unclear. In the present study, we ask whether autophagy is involved in the reduction or loss of MHC-I antigen in tumor cells. We provide evidence that autophagy is a negative regulator of MHC-I antigen in B16 melanoma cells, however, in the presence of IFN-γ, it is converted to a positive regulator. The underlying mechanisms probably involve the degradation of MHC-I antigen and generation of MHC-I-binding peptides by autophagy and the regulation of IFN-γ on autophagy-mediated MHC-I antigen degradation. Our data also show that autophagy may promote the cytolysis of CTL to melanoma cells at the early stage of B16 menaloma, but impairs the cytolysis at the late stage, indicating the different roles of autophagy in tumor immunity.
Materials and methods
Cell line and mouse model
Mouse melanoma B16-F1 cell line was purchased from China Center for Type Culture Collection (CCTCC, Wuhan, China). B16 cells (2 × 105) were subcutaneously injected to the left flank of C57BL/6 mice (Chinese Academy of Medical Science, Beijing, China). The tumor tissue and spleen were used for the indicated experiments.
Reagents
Rapamycin, 3-methyladenine, wortmannin, Bafilomycin A1 and chloroquine were purchased from Sigma (St. Louis, MO, USA) for the autophagic study. Their concentrations used in vitro and in vivo were described in figure legends.
Flow cytometric analysis
Peritoneal macrophages or B16 cells were stained with PE-conjugated anti-MHC-I (mouse IgG2a), FITC-conjugated anti-CD80 (Armenian Hamster IgG), and PE-conjugated anti-CD86 (rat IgG2a) antibodies (eBioscience, San Diego, CA) or corresponding isotype controls for 30 min at 4°C. After washing, the cells were used for flow cytometric analysis (BD LSR II).
ImmunoGold electron microscopy
For transmission electron microscopy, cells were fixed and permeabilized with Fix/Perm solution (BD Parmingen, San Jose, CA, USA). Then, the cells were stained with biotin-conjugated anti-MHC-I antibody (Invitrogen, Carlsbad, CA, USA) and streptavidin-gold (Sigma). Then, the cells were further fixed with 2.5% glutaraldehyde and 1% osmic acid. Ultrathin sections of embedded cells were cut with ULTRACUT (Leica, Wetzlar, Germany). The sections were examined in a FEI Tecnai G212 transmission electron microscope (Zeiss, Thornwood, NY, USA).
Construction of B16 tumor cell line expressing Beclin 1-siRNA
The Beclin 1-siRNA sequence (cagtcaagtcttacaaggg) and its control siRNA (ctggtcaagtctacaagag) were inserted into RNAi-Ready pSIREN-RetroQ expressing vector with U6 promoter (BD Biosciences, Clontech, Palo Alto, CA). The recombinant siRNA-expressing plasmids and control plasmids were transfected into B16 tumor cells using FuGENE 6 transfection reagent (Roche, Indianapolis, IN, USA) for stable expression after selection.
siRNA transient transfection
Mouse Atg5, Atg7 and Beclin 1 siRNAs and the control siRNAs were obtained from Dharmacon (Layfayette, CO, USA). All siRNAs were transfected into macrophages or B16 cells. Briefly, 200 pmoles of synthesized oligonucleotide was mixed with 100 μl of Nucleofector solution (Amaxa, Gaithersburg, MD, USA), and transfected into cells (5 × 106) by electroporation using Nucleofector instrument (Amaxa).
Conventional RT-PCR and real time RT-PCR
Total RNA was extracted from cells with TRIzol reagent (Invitrogen) or from tissues homogenized in TRIzol according to the manufacturer’s instructions. The relative quantity of mRNA was determined by RT-PCR (One-step RT-PCR kit, Qiagen, Valencia, CA, USA). The mRNA of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the internal control. The primer sequences were as follows: LMP2, sense 5′-GTACCGTGAGGACTTGTTAGC-3′, antisense 5′-AGAGTGATGGCATCTGTGG TG-3′; LMP7, sense 5′-ATCTCCGTGTCTGCAGCATC-3′, antisense 5′-TCACTGACATCGGAACTCTC-3′; TAP1, sense 5′-ACCATGGAGGAAATCACAGC-3′, antisense 5′-AAGAAGAACCGTCCGAGAAG-3′; TAP2, sense 5′-GTATGATCACCACTACCTGCAC-3′, antisense 5′-AATCACCAGCATCGTCCTGTC-3′; Tapasin, sense 5′-AGCTTGGGATGACGATGAG-3′, antisense 5′-AGGAGGAGAAAAGCAGACAG-3′; Beclin 1, sense 5′-GAGGAAGAGGCTAACTCA G-3′, antisense 5′-CAGTGACATTGAGCTGAGTG-3′; GAPDH, sense 5′-GTGGAGATTGTTGCCATCAACG-3′, antisense 5′-CAGTGGATGCAGGGATGATGTTCTG-3′.
For real time RT-PCR assays, the cDNA sequences of all detected genes were retrieved from NCBI database. The primers were designed with the Oligo Primer Analysis 4.0 software and the sequences were blasted (http://www.ncbi.nlm.nih.gov/BLAST/). The primer sequences were shown in supplementary Table 1. 100 ng of total RNA was used for reverse transcription using Superscript II RNase H reverse transcriptase (Invitrogen) in a volume of 25 μl. Then 2 μl of cDNA was amplified with SYBR Green Universal PCR Mastermix (Bio-Rad, Richmond, CA, USA) in duplicate. For sample analysis, the threshold was set based on the exponential phase of products, and CT value for samples was determined. The resulting data were analyzed with the comparative CT method for relative gene expression quantification against house keeping gene GAPDH.
In vitro proteasome inhibitor activity assays
Proteasomes of B16 cells were isolated with the Proteasome Isolation Kit (Calbiochem, San Diego, CA, USA), and incubated for 2 h at RT in 50 mM Tris-HCl and 5 mM MgCl2 in the presence and absence of proteasome inhibitors 100 mM LAC, or 100 mM MG-115. The fluorosubstrates Z-Leu-Leu-Leu-AMC, Z-Leu-Leu-Glu-AMC, Suc-Leu-Leu-Val-Tyr-AMC, (Calbiochem) were then added to give a final concentration of 100 mM along with additional inhibitor necessary to maintain original inhibitor concentration. The fluorescence was measured at different time points to calculate the proteasome activity.
Acid treatment
B16 cells were preincubated for 2 h at 37°C in the presence or absence of proteasome inhibitors Lac or GM-115 before acid treatment. Then the cells (1 × 106) were centrifuged and the resulting pellet was resuspended gently in 15 ml of 300 mM glycine (pH 2.5) with 1% (w/v) BSA and incubated for 4 min at 37°C. The suspension was neutralized by dilution with 30 ml of cell medium containing 0.5 N NaOH and 0.2 M HEPES and centrifuged. Cells were resuspended into 200 μl of cell medium (10 ng IFN-γ) and incubated for 5 h at 37°C in the presence or absence of proteasome inhibitor to allow class I re-expression.
In situ IFN-γ analysis
Tumor tissues were isolated from animals at different time point after B16 cell injection and homogenized in PBS (0.5 ml) containing 100 μM PMSF (Sigma), 1% (vol/vol) aprotinin (Sigma), 2 μg/ml leupeptin (Sigma), and 1 μg/ml pepstatin (Sigma). The lysate was clarified by centrifugation in a microcentrifuge for 5 min at maximum speed. The supernatant was appropriately diluted for IFN-γ ELISA (R&D Systems) assay. Data represent the mean of five individual animals from each group in pictogram (pg) per tumor weight (g).
Cytotoxicity assay
B16 melanoma cells (2 × 105) were inoculated to mice. 12 days later, the splenic T cells were isolated by T cell-enrichment column and further stimulated by HSP70-B16 peptide complexes in the presence of APCs for six days as described previously [22]. Stimulated splenic T cells were cocultured with B16 cells that were pre-treated or left untreated with different agents. The flow cytometry-based method [23] was used for cytotoxicity assay.
Western blot analysis
Cell lysates or tumor tissue homogenates (30 μg of total protein) and prestained molecular weight markers were separated by SDS-PAGE followed by transfer onto nitrocellulose membranes. The membranes were blocked in TBST (Tris-buffered saline with 0.5% of Triton X-100) containing 5% nonfat milk, and probed with the indicated antibodies. After incubation with the secondary antibody conjugated with horseradish peroxidase, membranes were extensively washed, and the immunoreactivity was visualized by enhanced chemiluminescence according to the manufacturer’s protocol (ECL kit, Santa Cruz, CA, USA). All antibodies were purchased from Santa Cruze Biotechnology (Santa Cruz, CA, USA).
Statistics
Results were expressed as mean value ± SD and interpreted by ANOVA-repeated measure test. Differences were considered to be statistically significant when P < 0.05.
Results
Autophagy is a negative regulator of surface MHC-I antigen expression
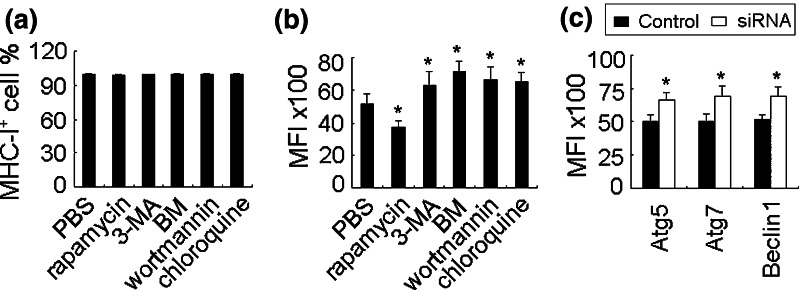
To investigate whether autophagy regulates MHC-I, we first chose macrophages for this purpose. The macrophages were isolated from peritoneal cavity of mice, which highly expressed MHC-I antigen and had autophagic activity, evaluated by flow cytometry and electron microscopy (data not shown). Then, the cells were treated in vitro with rapamycin or 3-methyladenine (3-MA) or wortmannin or bafilomycin A1 (BM) or chloroquine for 24 h. Among them, rapamycin is a specific inhibitor of mTOR and has been well established to increase the flux of autophagy [24]; 3-MA and wortmannin are inhibitors of PI-3K class III and suppress autophagosome formation [25]; bafilomycin A1 blocks the fusion of autophagosomes with lysosomes [25]; whereas chloroquine is lysosomal enzyme inhibitor that blocks the lysosomal degradation of autophagosomes [25]. The flow cytometric analysis showed that these agents did not alter the number of MHC-I antigen positive cells, but affected the mean fluorescence intensity (MFI) significantly (Fig. 1a, b). Rapamycin decreased the MFI from 5,157 to 3,748, and 3-MA, BM, wortmannin and chloroquine nevertheless increased the MFI of MHC-I antigen at different levels. In line with these data, knockdown of autophagy-related genes Atg5, Atg7 or Beclin 1 also increased MHC-I antigen expression by macrophages (Fig. 1c). Taken the functional connection between surface MHC-I antigen and costimulatory molecules, we additionally detected the molecules CD80 and CD86. We found that CD80 was not changed by autophagy, but CD86 was significantly altered (supplementary Fig. 1a, b). The inhibition of autophagy not only increased CD86 positive cell number but also increased the MFI. These data suggested that autophagy may be a negative regulator of MHC-I antigen.
Fig. 1.
Autophagy negatively regulates MHC-I antigen expression. a, b Autophagy decreases MHC-I expression in macrophage. The cultured peritoneal macrophages were treated with rapamycin (0.5 μg/ml), 3-MA (3 mM), BM (10 nM), wortmannin (100 nM), or chloroquine (25 μM) for 24 h. The MHC-I antigen positive cell proportion (a) and the mean fluorescent intensity MFI (b) were analyzed by flow cytometry. c Atg5, Atg7, and becalin 1 siRNAs were transfected into macrophages, respectively. 24 h later, the MFI of MHC-I antigen were analyzed by flow cytometry. The data in this figure are the representatives of three independent experiments. *P < 0.05, compared with control group
Autophagy synergizes with INF-γ to upregulate MHC-I antigen expression by B16 cells
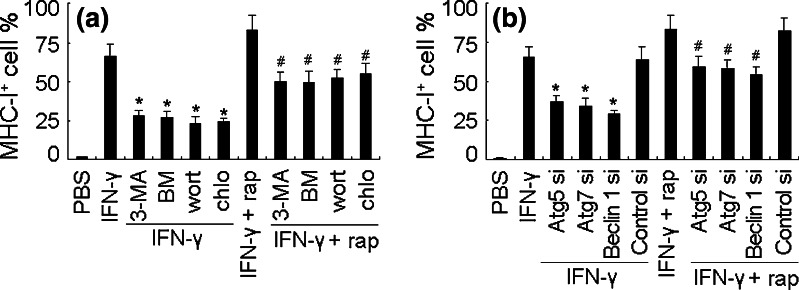
Downregulation of MHC-I antigen is a typical feature of B16 melanoma cells due to the coordinated dysregulation of the antigen-processing components [6]. Based on the above data, we here hypothesized that autophagy contributed to MHC-I antigen downregulation in B16 melanoma cells. To test this hypothesis, we took the advantage of the restoration of MHC-I antigen expression on B16 cells by IFN-γ. We treated B16 cells with rapamycin plus IFN-γ. Surprisingly, rapamycin did not show inhibitory effect, but synergized with IFN-γ to further upregulate MHC-I antigen expression (Fig. 2a). On the other hand, the suppression of autophagy by 3-MA, BM, wortmannin and chloroquine downregulated MHC-I antigen of B16 cells in the presence of IFN-γ or IFN-γ + rapamycin (Fig. 2a). The similar result was also obtained by knockdown of Atg5, Atg7 or Beclin 1 (Fig. 2b).
Fig. 2.
The synergistic effect of autophagy and INF-γ on MHC-I antigen expression. a Autophagy synergizes with INF-γ to upregulate MHC-I of B16 cells. B16 cells were treated with IFN-γ (10 ng/ml) in the presence or absence of rapamycin, 3-MA, BM, wortmannin, or chloroquine for 24 h. The MHC-I antigen positive cells were analyzed by flow cytometry. *P < 0.05, compared with IFN-γ group. # P < 0.05, compared with IFN-γ + rap group. b Atg5, Atg7, and beclin 1 siRNAs were transfected into IFN-γ-treated B16 cells, respectively. 24 h later, the MHC-I antigen positive cells were analyzed by flow cytometry. *P < 0.05, compared with IFN-γ group. # P < 0.05, compared with IFN-γ + rap group
The regulatory mechanisms of IFN-γ signaling on B16 MHC-I antigen expression via autophagic pathway
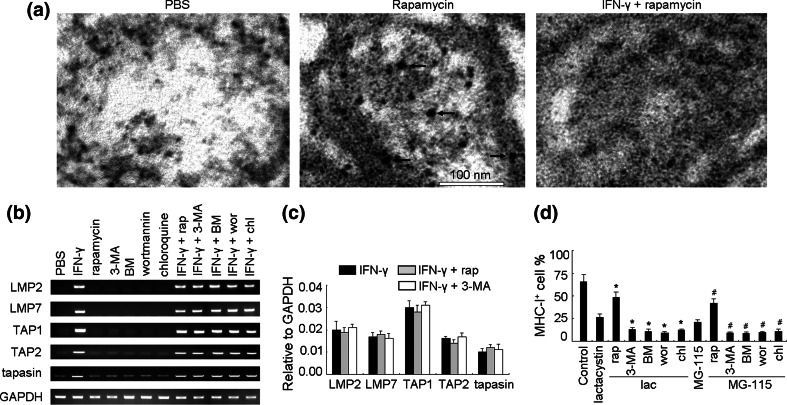
To explain the above unexpected phenomena, we performed ImmunoGold electron microscopy assay. We found that MHC-I antigen molecules were present in the autophagosomes/autophagolysosomes of B16 cells treated with rapamycin but the addition of IFN-γ inhibited such co-localization (Fig. 3a). In addition, MHC-I antigen was not observed in PBS-treated cells (Fig. 3a). This observation suggested that autophagy may be involved in the degradation of MHC-I antigen but that INF-γ can rescue the process. However, this did not explain the enhancement of IFN-γ-induced MHC-I antigen expression by autophagy activation. A possibility was that autophagy might affect antigen-processing pathways that can enhance or stabilize MHC-I antigen expression in the presence of IFN-γ. To address this possibility, we first measured the impact of autophagy on gene transcription of five proteins critical for antigen processing including the proteasome subunits LMP2 and LMP7, the peptide transporter associated with antigen processing TAP1 and TAP2, and the chaperone tapasin [26]. Our results indicated that neither autophagy inducer nor inhibitors affected the expression of any of these five genes induced by IFN-γ (Fig. 3b, c). Since activation of autophagy increases intracellular degradation, we then reasoned that autophagy might increase the generation of MHC-I-binding peptides independent of the ubiquitin-proteasome pathway. In order to address this question, we adapted a method using acid to remove surface class I peptide complexes induced by IFN-γ (supplementary Fig. 2) and then analyzed re-expression of MHC-I antigen in the presence or absence of proteasome inhibitors [19, 20]. Interestingly, although proteasome inhibitors lactacystin and MG-115 blocked 97% of cleavage activities of proteasomes (supplementary Fig. 3), substantial levels of IFN-γ-induced MHC-I antigen remained detectable on the cell surface (26 and 21%, respectively; Fig. 3d), suggesting the existence of proteasome-independent pathways for generation of MHC-I-binding peptides. Furthermore, decreased expression of MHC-I antigen by proteasome inhibitors could be partially restored in the presence of rapamycin while the autophagy inhibitors further suppressed MHC-I antigen surface expression (Fig. 3d). Taken together, these data imply that autophagy alone may degrade intracellular MHC-I antigen but that it facilitates the generation of MHC-I binding peptides in the presence of IFN-γ. As a consequence, activation of autophagy may be a necessary step for IFN-γ-induced surface expression of MHC-I antigen in B16 cells and serve as an alternative pathway for class I peptide generation.
Fig. 3.
The regulatory mechanisms of IFN-γ signaling on MHC-I expression via autophagy pathway. a ImmunoGold electron microscopy demonstrating the presence of MHC-I antigen in autophagosome of B16 cells. Bold arrows indicated gold particles. The scale bar was shown. b, c The expression of LMP2, LMP7, TAP1, TAP2, and tapasin in B16 cells under different conditions was analyzed by RT-PCR (b) and real time RT-PCR (c). d Autophagy is a source for the generation of MHC-I antigenic peptides. IFN-γ-effected B16 cells were pretreated with proteasome inhibitor Lac (100 μM) or MG-115 (50 μM) for 2 h, and treated with 300 mM glycine (pH 2.5) for 4 min to remove the surface MHC-I antigen. After washing, the cells were incubated for 5 h in the presence IFN-γ and proteasome inhibitor plus rapamycin or autophic inhibitors. The reexpression of MHC-I antigen was analyzed by flow cytometry. *P < 0.05, compared with lactacystin group. # P < 0.05, compared with GM-115 group
The influence of autophagy on CTL cytolysis of B16 cells is dependent on IFN-γ in vitro
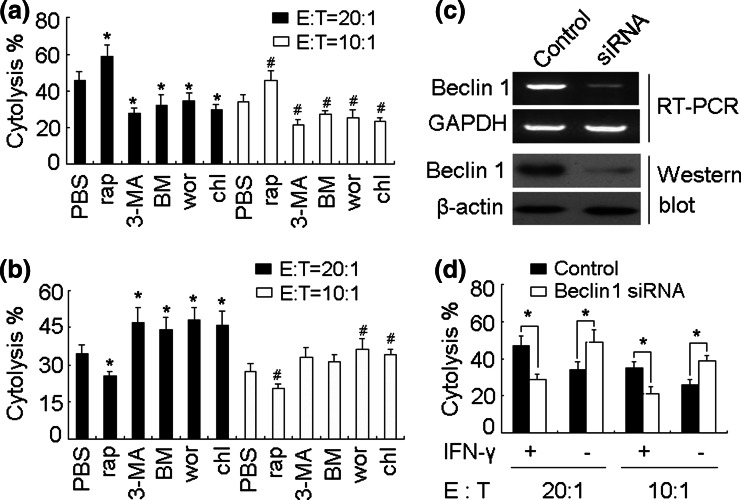
We next asked whether autophagy influenced the CTL cytolysis against B16 cells. To address this question, spleen T cells from B16 cell-inoculated C57BL/6 mice were stimulated in vitro with HSP70-B16 peptide complexes [22] in the presence of irradiated APCs for 6 days to generate B16 specific CTLs. The cytotoxicity assay was then performed. The efficient killing to IFN-γ-treated B16 cells was observed, which could be enhanced or impaired by addition of rapamycin or autophagy inhibitors (Fig. 4a). However, single rapamycin treatment decreased the cytolysis and single autophagy inhibitor treatment increased the cytolysis (Fig. 4b). Moreover, we used another approach to further confirm these data. We constructed a B16 cell line stably expressing Beclin 1 siRNA (Fig. 4c), which was used to interfere with the formation of autophagosome. This was because Beclin 1, the mammalian homolog of yeast ATG6, together with the class III phosphoinositide-3 kinase (PI3K), mainly mediates the formation of the double-membrane-bound autophagosomes containing sequestered cellular contents [25]. Consistently, under such condition, the effect of IFN-γ treatment on cytolysis was impeded (Fig. 4d). Thus, these data indicated that autophagy may positively or negatively influence the cytolysis of CTL to B16 cells dependent on the existence or absence of IFN-γ.
Fig. 4.
Autophagy influences the sensitivity of B16 cells to CTLs. a, b Autophagy affects the cytolysis to B16 cells. B16 cells (2 × 105) were inoculated to mice. 12 days later, spleen T cells were isolated and stimulated in vitro with HSP70-B16 peptide complexes in the presence of irradiated normal splenocytes as APCs for 6 days. The cells were harvested as effector cells. B16 cells were treated with rapamycin, 3-MA, BM, wortmannin, or chloroquine in the presence (a) or absence (b) of IFN-γ for 24 h, and used as target cells. The cytolysis assay was performed as described in “Materials and methods”. *P < 0.05, compared with PBS group (20:1). # P < 0.05, compared with PBS group (10:1). c siRNA silences Beclin 1 expression. After stable transfection of Beclin 1 siRNA vector into the B16 cell line, Beclin 1 mRNA and protein expression were examined by RT-PCR and Western blot. d Beclin 1 knockdown affects the cytolysis to B16 cells. The above effector cells were generated. Beclin 1 siRNA-expressing B16 cells were treated with or without IFN-γ for 24 h, and used as target cells. The cytolysis assay was performed (*P < 0.05)
Different roles of autophagy in CTL cytolysis against B16 melanoma
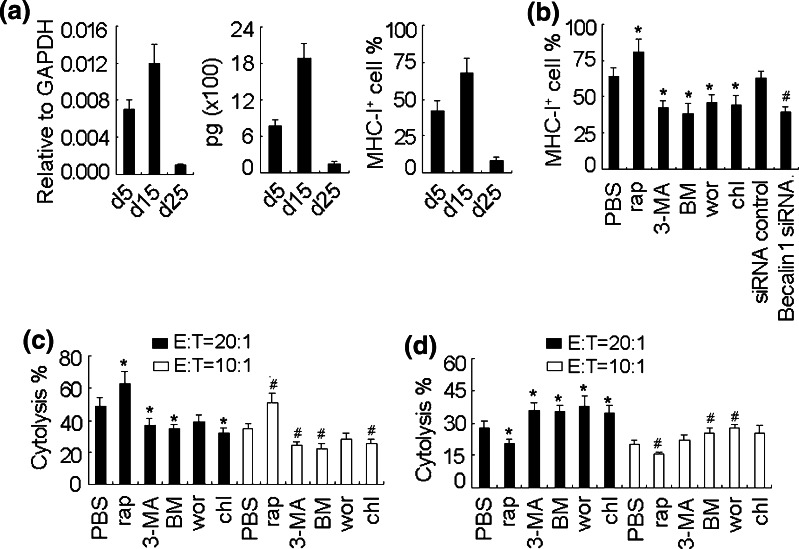
The upregulated expression of MHC-I antigen in melanoma cells should benefit the cytolysis of CTL against melanoma. We then used the B16 s.c. tumor model to study the role of autophagy in CTL cytolysis in vivo. We first measured tumor IFN-γ level, due to the synergistic effect of IFN-γ and autophagy on MHC-I antigen expression (Fig. 2). The high level of tumor IFN-γ could be detected on d5 after tumor cell inoculation and probably reached to the highest level on d15, then gradually decreased, evaluated by both real time RT-PCR and ELISA (Fig. 5a, left and middle). In parallel, the similar expression pattern of MHC-I antigen in melanoma tumor cells was observed (Fig. 5a, right). Then, we injected rapamycin to small melanoma-bearing mice (d12) in line with the high level of IFN-γ. The expression of MHC-I antigen in tumor cells was further increased 2 days after injection. In contrast, the injection of autophagy inhibitors nevertheless decreased MHC-I antigen expression (Fig. 5b). Consistently, the expression of tumor cell MHC-I antigen was decreased in beclin-1 siRNA-expressing melanoma (Fig. 5b).
Fig. 5.
Different roles of autophagy in CTL cytolysis against B16 melanoma. a The level of IFN-γ and MHC-I antigen in B16 melanoma of different time points after inoculation. B16 cells were inoculated s.c. to mice. The tumor was removed at different time points for the IFN-γ detection by real time RT-PCR (left) and ELISA (middle). The tumor cells were also isolated for MHC-I antigen analysis by flow cytometry (right). b Autophagy increases MHC-I antigen expression in tumor cells of small melanoma. B16 cells were inoculated s.c. to mice. 12 days later, mice were injected i.p. with rapamycin (100 μg/mouse), wortmannin (300 ng/mouse), chloroquine (200 μg/mouse), or BM (100 ng/mouse), or injected multiply at tumor site with 3-MA (50 μg/mouse). 24 h later, the tumors were removed and the melanoma cells were isolated for MHC-I antigen expression analysis by flow cytometry. *P < 0.05, compared with PBS group. # P < 0.05, compared with siRNA control. c, d Autophagy affects the CTL cytolysis against B16 melanoma. B16 cells (2 × 105) were inoculated to mice. 12 days later, spleen T cells were isolated and stimulated in vitro with HSP70-B16 peptide complexes in the presence of irradiated normal splenocytes as APCs for 6 days. The cells were used as effector cells. The melanoma-bearing mice of d4 or d25 after inoculation were pretreated with rapamycin, wortmannin, chloroquine, or BM, or 3-MA one day in advance. The tumor cells isolated from d14 melanoma (c) or d25 melanoma (d) were used as the target cells. The cytotoxicity assay was performed. *P < 0.05, compared with PBS group (20:1). # P < 0.05, compared with PBS group (10:1)
To clarify whether autophagy affects the CTL cytolysis against B16 melanoma, we then treated melanoma (d12) with rapamycin or autophagy inhibitors for 48 h, and performed the cytolysis assay. The spleen T cells from untreated d14 tumor-bearing mice were isolated and stimulated in vitro and used as the effector cells. The tumor cells isolated from treated melanoma were used as the target cells. The rapamycin treatment effectively strengthened the sensitivity of tumor cells to CTLs, and the treatment with autophagy inhibitors nevertheless decreased the sensitivity, leading to low cytolysis (Fig. 5c). We also tested the large melanoma. The expression of MHC-I antigen was strikingly decreased in tumor cells of large melanoma (d25) (Fig. 5a). We then isolated the spleen T cells from d25 tumor-bearing mice (untreated) and stimulated them in vitro for the use as effector cells. The impaired cytolysis to the tumor cells of large melanoma was observed, and neither rapamycin nor autophagic inhibitors had the effect on the cytolysis (supplementary Fig. 4). This might be due to the disfunctional T cells in large tumor-bearing mice. By using CTLs generated from d12 tumor-bearing mice, a decreased cytolysis was observed in the rapamycin group, but the increased cytolysis was observed in the autophagy inhibitor groups (Fig. 5d). This might be due to the downregulation of MHC-I antigen expression by autophagy in the absence of IFN-γ. Taken together, these data suggest that autophagy may enhance the CTL cytolysis to melanoma tumor cells at the early stage, but impairs the CTL cytolysis at the late stage of melanoma.
Discussion
Although autophagy has been linked to endogenous MHC class II antigen processing for presentation to CD4+ T cells [14–18], a direct role for autophagy in MHC class I antigen presentation and CD8+ T cell recognition has not been firmly established. We demonstrate in the present study that autophagy may down-regulate MHC-I antigen expression in macrophage and is involved in MHC-I antigen expression in tumor cells. Interestingly, however, in the presence of IFN-γ, autophagy increases MHC-I antigen surface expression on tumor cells leading to enhanced CTL-mediated killing. We speculate that the decrease in MHC-I antigen expression by autophagy is due to the outcome of increased intracellular degradation, however, this process may be compensated for by induction of MHC gene transcription by IFN-γ. Furthermore, as a consequence of intracellular protein degradation, more antigenic peptides may be produced through the autophagic pathway leading to enhanced MHC-I/antigen stability and expression. This latter possibility is supported by a recent study where induction of autophagy indeed enhanced the formation of autophagosomes in B16 cells that carried tumor-specific epitopes. The authors demonstrated that rapamycin-treated tumor cells or autophagosomes extracted from these cells were more immunogenic than their untreated counterparts and upon a coculture with dendritic cells; these materials could elicit strong CD8+ T cell responses, identifying autophagy as an alternative route of antigen delivery to dendritic cells for cross-presentation [27]. Our results, however, provide evidence for the first time that in the presence of IFN-γ, enhancement of autophagy can increase antigen processing and presentation by tumor cells for direct CTL recognition.
The surface expression of MHC-I antigen involves multiple steps including proteolysis by proteosome, transportation of peptides into the ER, and assembly of MHC-I-peptide complex [28]. Nevertheless, autophagy might be involved in these processes: (1) ER as a type of organelle can be degraded by autophagy; and (2) proteasome system and autophagy, the two types of machinery for protein degradation, possibly exists the crosstalk between them. Here, we indeed show that autophagy downregulates MHC-I antigen expression in macrophages, which prompts us to hypothesize that autophagy is another mechanism involving MHC-I antigen loss in B16 melanoma cells besides the downregulation of multiple components of the MHC-I antigen-processing pathway. Considering that IFN-γ is capable of correcting the loss of MHC-I antigen of B16 cells, we treated B16 cells with IFN-γ. Surprisingly, autophagy synergized with IFN-γ to induce MHC-I antigen expression, but not antagonized the effect. Such phenomena might be explained by our findings: (1) IFN-γ signaling may prevent the uptake of MHC-I antigen during autophagic process; and (2) autophagy may be a source to provide MHC-I peptides. These raise the interesting questions: (1) how IFN-γ signaling guides autophagy to selectively target molecules or organelles, such as MHC-I antigen; and (2) how autophagy captures antigen and processes them to MHC-I peptide. Addressing such questions should promote the further understanding of the biology of autophagy and the MHC-I antigen-processing machinery.
CD8+ T cell activation by MHC-I antigen-mediated pathway requires the help of costimulatory molecules, typically CD80 and CD86 [29–31]. An interesting finding in this study is that autophagy also affects the expression of costimulatory molecules besides MHC-I antigen. Autophagy seems not to affect CD80 expression, but strikingly affects CD86 expression in peritoneal macrophages. On the other hand, CD80 or CD86 was undetectable in B16 cells by flow cytometry in our study, even if we added IFN-γ or the combination of IFN-γ and rapamycin or autophagy inhibitors (data not shown). We did not elucidate the underlying mechanism here. However, the severe inhibition of CD86 expression by B16 cell-relative factors might be the reason that autophagy has no effect on CD86 expression. In our study, the deficient expression of CD80 and CD86 may impair the activation of T cells by B16 cells, but seems not to influence the effector phase. This is not the case showed in J558 plasmacytoma cells [32].
Tumorigenesis is considered to experience the immunoediting processes from immunosurveillance to equilibrium and immune escape [33–35]. Our data indicate that autophagy may be profoundly involved in these processes. At the early stage of B16 melanoma, autophagy synergizes with IFN-γ to upregulate tumor surface MHC-I antigen, thus leading to efficient CTL cytolysis. Furthermore, autophagy was recently identified as an active process for cross-presentation of tumor antigens to dendritic cells. Therefore, autophagy may augment both tumor-specific Th1 cells and CTLs for tumor immunosurveillance, and defects in autophagy could promote tumorigenesis. However, during the immune escape stage, the tumor immunosuppressive microenvironment prevents the generation of IFN-γ [36], thus leading to autophagy to exert its function of downregulating MHC-I antigen. Therefore, autophagy can further augment the tumor immunosuppression. These findings may have important clinical importance. Since the clinical cancer patients usually have advanced tumor, the use of autophagy-promoting drugs such as rapamycin might further jeopardize antitumor immunity by downregulating MHC-I antigen. However, the combination of autophagy-promoting drugs and chemo- or radiotherapy may be useful to enhance antitumor immunity. Taken together, we propose that autophagy plays different roles in tumor immunity dependent on tumor microenvironment.
Electronic Supplementary Material
Acknowledgments
This work was supported by the Program for New Century Excellent Talents in University (NCET-08-0219), National Natural Science Foundation of China (30871020).
References
- 1.Wang RF, Rosenberg SA. Human tumor antigens for cancer vaccine development. Immunol Rev. 1999;170:85–100. doi: 10.1111/j.1600-065X.1999.tb01331.x. [DOI] [PubMed] [Google Scholar]
- 2.Ochsenbein AF, Sierro S, Odermatt B, Pericin M, Karrer U, Hermans J, Hemmi S, Hengartner H, Zinkernagel RM. Roles of tumour localization, second signals and cross priming in cytotoxic T-cell induction. Nature. 2001;411:1058–1064. doi: 10.1038/35082583. [DOI] [PubMed] [Google Scholar]
- 3.McDougall CJ, Ngoi SS, Goldman IS, Godwin T, Felix J, DeCosse JJ, Rigas B. Reduced expression of HLA class I and II antigens in colon cancer. Cancer Res. 1990;50:8023–8027. [PubMed] [Google Scholar]
- 4.Hicklin DJ, Marincola FM, Ferrone S. HLA class I antigen downregulation in human cancers: T cell immunotherapy revives an old story. Mol Med Today. 1999;5:178–186. doi: 10.1016/S1357-4310(99)01451-3. [DOI] [PubMed] [Google Scholar]
- 5.Dovhey SE, Ghosh NS, Wright KL. Loss of interferon-gamma inducibility of TAP1 and LMP2 in a renal cell carcinoma cell line. Cancer Res. 2000;60:5789–5796. [PubMed] [Google Scholar]
- 6.Seliger B, Wollscheid U, Momburg F, Blankenstein T, Huber C. Characterization of the major histocompatibility complex class I deficiencies in B16 melanoma cells. Cancer Res. 2001;61:1095–1099. [PubMed] [Google Scholar]
- 7.Facoetti A, Nano R, Zelini P, Morbini P, Benericetti E, Ceroni M, Campoli M, Ferrone S. Human leukocyte antigen and antigen processing machinery component defects in astrocytic tumors. Clin Cancer Res. 2005;11:8304–8311. doi: 10.1158/1078-0432.CCR-04-2588. [DOI] [PubMed] [Google Scholar]
- 8.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kundu M, Thompson CB. Autophagy: basic principles and relevance to disease. Annu Rev Pathol. 2008;3:427–455. doi: 10.1146/annurev.pathmechdis.2.010506.091842. [DOI] [PubMed] [Google Scholar]
- 10.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato K, Tsuchihara K, Fujii S, Sugiyama M, Goya T, Atomi Y, Ueno T, Ochiai A, Esumi H. Autophagy is activated in colorectal cancer cells and contributes to the tolerance to nutrient deprivation. Cancer Res. 2007;67:9677–9684. doi: 10.1158/0008-5472.CAN-07-1462. [DOI] [PubMed] [Google Scholar]
- 12.Ogier-Denis E, Codogno P. Autophagy: a barrier or an adaptive response to cancer. Biochim Biophys Acta. 2003;1603:113–128. doi: 10.1016/s0304-419x(03)00004-0. [DOI] [PubMed] [Google Scholar]
- 13.Hippert MM, O’Toole PS, Thorburn A. Autophagy in cancer: good, bad, or both? Cancer Res. 2006;66:9349–9351. doi: 10.1158/0008-5472.CAN-06-1597. [DOI] [PubMed] [Google Scholar]
- 14.Menéndez-Benito V, Neefjes J. Autophagy in MHC class II presentation: sampling from within. Immunity. 2007;26:1–3. doi: 10.1016/j.immuni.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 15.Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol. 2007;7:767–777. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dengjel J, Schoor O, Fischer R, Reich M, Kraus M, Müller M, Kreymborg K, Altenberend F, Brandenburg J, Kalbacher H, Brock R, Driessen C, Rammensee HG, Stevanovic S. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc Natl Acad Sci USA. 2005;102:7922–7927. doi: 10.1073/pnas.0501190102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmid D, Münz C. Localization and MHC class II presentation of antigens targeted for macroautophagy. Methods Mol Biol. 2008;445:213–225. doi: 10.1007/978-1-59745-157-4_14. [DOI] [PubMed] [Google Scholar]
- 18.Riedel A, Nimmerjahn F, Burdach S, Behrends U, Bornkamm GW, Mautner J. Endogenous presentation of a nuclear antigen on MHC class II by autophagy in the absence of CRM1-mediated nuclear export. Eur J Immunol. 2008;38:2090–2095. doi: 10.1002/eji.200737900. [DOI] [PubMed] [Google Scholar]
- 19.Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/S0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 20.Goldberg AL, Cascio P, Saric T, Rock KL. The importance of the proteasome and subsequent proteolytic steps in the generation of antigenic peptides. Mol Immunol. 2002;39:147–164. doi: 10.1016/S0161-5890(02)00098-6. [DOI] [PubMed] [Google Scholar]
- 21.York IA, Mo AX, Lemerise K, Zeng W, Shen Y, Abraham CR, Saric T, Goldberg AL, Rock KL. The cytosolic endopeptidase, thimet oligopeptidase, destroys antigenic peptides and limits the extent of MHC class I antigen presentation. Immunity. 2003;18:429–440. doi: 10.1016/S1074-7613(03)00058-X. [DOI] [PubMed] [Google Scholar]
- 22.Xiao H, Huang B, Yuan Y, Li D, Han LF, Liu Y, Gong W, Wu FH, Zhang GM, Feng ZH. Soluble PD-1 facilitates 4–1BBL-triggered antitumor immunity against murine H22 hepatocarcinoma in vivo. Clin Cancer Res. 2007;13:1823–1830. doi: 10.1158/1078-0432.CCR-06-2154. [DOI] [PubMed] [Google Scholar]
- 23.Huang B, Zhao J, Li H, He KL, Chen Y, Chen SH, Mayer L, Unkeless JC, Xiong H. Toll-like receptors on tumor cells facilitate evasion of immune surveillance. Cancer Res. 2005;65:5009–5014. doi: 10.1158/0008-5472.CAN-05-0784. [DOI] [PubMed] [Google Scholar]
- 24.Takeuchi H, Kondo Y, Fujiwara K, Kanzawa T, Aoki H, Mills GB, Kondo S. Synergistic augmentation of rapamycin-induced autophagy in malignant glioma cells by phosphatidylinositol 3-kinase/protein kinase B inhibitors. Cancer Res. 2005;65:3336–3346. doi: 10.1158/0008-5472.CAN-04-3640. [DOI] [PubMed] [Google Scholar]
- 25.Yin XM, Ding WX, Gao W. Autophagy in the liver. Hepatology. 2008;47:1773–1785. doi: 10.1002/hep.22146. [DOI] [PubMed] [Google Scholar]
- 26.Garbi N, Tanaka S, van den Broek M, Momburg F, Hämmerling GJ. Accessory molecules in the assembly of major histocompatibility complex class I/peptide complexes: how essential are they for CD8(+) T-cell immune responses? Immunol Rev. 2005;207:77–88. doi: 10.1111/j.0105-2896.2005.00303.x. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Wang LX, Yang G, Hao F, Urba WJ, Hu HM. Efficient cross-presentation depends on autophagy in tumor cells. Cancer Res. 2008;68:6889–6895. doi: 10.1158/0008-5472.CAN-08-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Antoniou AN, Powis SJ, Elliott T. Assembly and export of MHC class I peptide ligands. Curr Opin Immunol. 2003;15:75–81. doi: 10.1016/S0952-7915(02)00010-9. [DOI] [PubMed] [Google Scholar]
- 29.Van Gool SW, Vandenberghe P, de Boer M, Ceuppens JL. CD80, CD86 and CD40 provide accessory signals in a multiple-step T-cell activation model. Immunol Rev. 1996;153:47–83. doi: 10.1111/j.1600-065X.1996.tb00920.x. [DOI] [PubMed] [Google Scholar]
- 30.Schlom J, Hodge JW. The diversity of T-cell co-stimulation in the induction of antitumor immunity. Immunol Rev. 1999;170:73–84. doi: 10.1111/j.1600-065X.1999.tb01330.x. [DOI] [PubMed] [Google Scholar]
- 31.Greenwald RJ, Freeman GJ, Sharpe AH. The B7 family revisited. Annu Rev Immunol. 2005;23:515–548. doi: 10.1146/annurev.immunol.23.021704.115611. [DOI] [PubMed] [Google Scholar]
- 32.Zheng P, Sarma S, Guo Y, Liu Y. Two mechanisms for tumor evasion of preexisting cytotoxic T-cell responses: lessons from recurrent tumors. Cancer Res. 1999;59:3461–3467. [PubMed] [Google Scholar]
- 33.Dunn GP, Old LJ, Schreiber RD. The three Es of cancer immunoediting. Annu Rev Immunol. 2004;22:329–360. doi: 10.1146/annurev.immunol.22.012703.104803. [DOI] [PubMed] [Google Scholar]
- 34.Kim R, Emi M, Tanabe K. Cancer immunoediting from immune surveillance to immune escape. Immunology. 2007;121:1–14. doi: 10.1111/j.1365-2567.2007.02587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reiman JM, Kmieciak M, Manjili MH, Knutson KL. Tumor immunoediting and immunosculpting pathways to cancer progression. Semin Cancer Biol. 2007;17:275–287. doi: 10.1016/j.semcancer.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dunn GP, Ikeda H, Bruce AT, Koebel C, Uppaluri R, Bui J, Chan R, Diamond M, White JM, Sheehan KC, Schreiber RD. Interferon-gamma and cancer immunoediting. Immunol Res. 2005;32:231–245. doi: 10.1385/IR:32:1-3:231. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.