Abstract
We applied a cDNA expression screening procedure with cryopreserved non-clonal CD8+ T cell populations (Lennerz et al., Proc. Natl. Acad. Sci. USA 102:16013-8, 2005) to the identification of candidate antigens for graft-versus-host disease (GvHD) and graft-versus-leukaemia (GvL) effects in allogeneic haematopoietic stem cell transplantation (allo-HSCT). In a patient–donor model system with HLA class I disparities, we identified an HLA-B*44 mismatch allele, HLA-B*4405, as the dominant target of alloreactive T cells expanded in vitro from donor peripheral blood mononuclear cells (PBMC). HLA-B*4405-reactive T cells were detectable after multiple in vitro stimulations in the patient’s post-HSCT PBMC. In a patient–donor model with full HLA compatibility, the major target antigen of donor lymphocytes stimulated in vitro with the respective patient’s pre-HSCT PBMC was restricted by HLA-A*0201 and was encoded by TRIM22-442 C, a newly detected polymorphic allele of the tripartite motif family member TRIM22 (synonym: STAF50), preferentially expressed in cells of the haematopoietic system. An arginine(R)-to-cysteine(C) exchange at position 442 generated an immunogenic T cell epitope equivalent to a minor histocompatibility antigen (mHag). TRIM22-442C-specific T cells persisted long-term in the patient’s post-HSCT PBMC. Approximately, 1.3% of Caucasians carry TRIM22.442 C in association with HLA-A*0201. In particular, the knowledge of a large and diverse panel of such mHags may be crucial for further improvement of donor selection and adoptive T cell transfer strategies. The procedure applied herein will help to accelerate and facilitate their identification.
Keywords: Alloreactivity, HLA mismatch, Minor histocompatibility antigen, Expression cloning, TRIM22, HLA-B*4405
Introduction
Allogeneic haematopoetic stem cell transplantation (allo-HSCT) can cure patients of leukaemia. The presence of T lymphocytes in the grafts proved to be essential for the beneficial effects of allo-HSCT. They are relevant mediators of graft-versus-host disease (GvHD) and graft-versus-leukaemia (GvL) effects in allo-HSCT [1, 25]. T cell depletion prevents GvHD, but increases the risk of graft rejection and leukaemia relapse. As a proof of the principle of GvL effects, Kolb et al. [13] demonstrated that the adoptive transfer of blood-derived donor lymphocytes induced complete and durable remissions in patients with relapsing leukaemia after allo-HSCT.
Although a series of relevant target antigens have been identified [9], they are not sufficient in number to explain or predict GvHD and GvL effects in a substantial proportion of patients. The knowledge of a representative spectrum of minor histocompatibility (mHags) and leukaemia-associated antigens will be required to improve donor selection and ex vivo manipulation of transplants and also open up new ways for adoptive T cell transfer.
In analogy to our work on autologous tumor models [15], we propose a cDNA expression screening procedure with cryopreserved non-clonal T cell populations for the molecular dissection of alloreactive T cell responses. We applied this procedure to the analysis of HLA class I-restricted T cell-mediated alloreactivity in two patient–donor model systems. As targets, we identified HLA allele mismatches and a new minor histocompatibility antigen encoded by a polymorphic TRIM22 allele and restricted by HLA-A*0201.
Materials and methods
Patients and donors
Patient WD was diagnosed with acute myelogenic leukaemia (AML, FAB M4) at the age of 28 years. She underwent allo-HSCT from an unrelated donor, referred to as WD donor. After 1 month, she developed a mild transitory GvHD. She is now in complete remission without GvHD for more than 6 years. According to standard serotyping, the patient and donor had identical HLA class I phenotypes (HLA-A24; -B7,44; -Cw2,7). DNA sequencing revealed a mismatch for HLA-A*24 and HLA-B*44. WD cells carried HLA-A*2402, HLA-A*2403 and HLA-B*4405, whereas WD donor cells were homozygous at the HLA-A locus for HLA-A*2402 and carried HLA-B*4403. The HLA class II phenotypes of patient WD and her donor were identical according to genotyping (not shown).
An AML (FAB M4) was diagnosed in patient VX, when he was 29 years old. Induction chemotherapy led to a partial remission. After total body irradiation, he underwent allo-HSCT from his fully HLA-matched brother (referred to as VX donor). He developed limited chronic GvHD. Cyclosporine A was given for 21 months. There was no recurrence of GvHD and the patient is in ongoing complete remission for more than 8 years.
This study was approved by the local ethics committee. Blood donors gave their written informed consent to participate.
Leukaemia cells, peripheral blood mononuclear cells and cell lines
WD leukaemia (WD-AML) cells were harvested from a therapeutic leukapheresis at diagnosis and were cryopreserved in aliquots. Peripheral blood mononuclear cells (PBMC) were separated by density centrifugation from patient WD at different time points after allo-HSCT (day 42 until day 450). PBMC were also isolated from the WD donor.
VX leukaemia cells were not available. PBMC were obtained from patient VX at complete haematological remission before he underwent allo-HSCT (VX pre-HSCT PBMC) and 5 years after allo-HSCT. PBMC were also isolated from his sibling donor (VX donor).
K562/HLA cells stably expressing HLA class I alleles after transfection were generated and maintained as described [3]. EBV-transformed B cells (EBV-B), COS-7 cells and 293T cells were cultured in RPMI 1640 supplemented with 10% FCS, 1% l-glutamine and 100 U/ml penicillin-streptomycin (Invitrogen/Gibco BRL, Breda, Netherlands).
Allogeneic mixed lymphocyte and mixed lymphocyte-leukaemia cultures, T cell cloning and peptide stimulation
Allogeneic mixed lymphocyte cultures (allo-MLCs) and lymphocyte-leukaemia cultures (allo-MLLCs) were performed on 24-well plates (Greiner, Frickenhausen, Germany; culture volume 2 ml/well). In the WD model system, donor-derived PBMC (1 × 106/well) were stimulated in allo-MLLCs with irradiated (100 Gy) WD-AML cells (1 × 106/well). The medium was AIM-V (Invitrogen/Gibco BRL) supplemented with human serum from healthy individuals (10%; referred to as AIM-V/HS). From day 3 onwards, rhIL-2 was added (250 U/ml; Proleukin; Chiron-Behring, Marburg, Germany). Responder lymphocytes were weekly restimulated. On day 28, CD8+ T cells were isolated from MLLC responders using the CD8+ T cell isolation kit (Miltenyi Biotec, Bergisch-Gladbach, Germany) and stimulated as before. On day 39, the MLLC responder population was cryopreserved in aliquots 4 days after the last stimulation. Prior to their use in assays, the aliquots were thawed and kept in AIM-V/HS with IL-2 for 2 days. This lymphocyte population was referred to as WD/1. An aliquot of WD/1 lymphocytes was cloned on day 35 by limiting dilution on 96-well plates with irradiated (100 Gy) WD-AML cells (2 × 104/well) as well as allogeneic AK-EBV-B cells and PBMC as feeders (each 2.5 × 104/well, irradiated with 100 Gy).
In the VX model system, allo-MLCs were generated by co-culturing CD8+ T cells (1 × 106/well), isolated from donor PBMC, with irradiated (35 Gy) patient VX pre-HSCT PBMC as stimulators (2 × 106/well) and irradiated (35 Gy) CD8− donor PBMC as feeders (5 × 105/well). MLC medium was AIM-V/HS supplemented with rhIL-2 (150 U/ml) and rhIL-7 (5 ng/ml; R&D, Wiesbaden-Nordenstadt, Germany) from day 3 onwards. MLC responders were weekly restimulated under the same conditions. From the second week onwards, stimulators (VX pre-HSCT PBMC) were reduced to 1 × 106/well. MLC responder cells were cryopreserved in aliquots 4 days after the final restimulation. The aliquots were thawed, restimulated once under identical conditions and applied 4–6 days later to all further testing and screening procedures. The VX-MLC population applied herein was referred to as VX/33 and was cryopreserved on day 66. VX/33 responders were cloned on round-bottom 96-well plates (Greiner; culture volume 200 μl/well) by limiting dilution on day 66, and T cell clones were propagated by weekly restimulations with irradiated patient VX pre-HSCT PBMC (1 × 104/well) and irradiated donor EBV-B lymphocytes as feeders (100 Gy; 2.5 × 104/well).
For peptide stimulation in the VX model system, sibling donor VX PBMC were loaded for 2 h in AIM-V without human serum with peptide MAVPPCCIGV (10 μg/ml). Peptide-loaded cells were irradiated (35 Gy) and applied as stimulators (2 × 106/well) of CD8+ VX post-HSCT PBMC (1 × 106/well) on 24-well plates. Irradiated CD8− PBMC (35 Gy) served as feeders (5 × 105/well). We used the same medium as for allo-MLC. Responder lymphocytes were restimulated weekly.
Cloning of HLA class I-encoding cDNAs
HLA class I-encoding cDNAs were cloned by RT-PCR into pcDNA3.1/Amp (Invitrogen/Gibco BRL) as described [6]. In particular, these were HLA-A*2402, -A*2403, -B*0702, -B*4405 and –Cw*0202 from WD leukaemia cells, HLA-B*4403 from WD donor cells, as well as HLA-A*0201, HLA-A*2402, HLA-B*1501, HLA-B*3901 and HLA-Cw*0303 from VX pre-HSCT PBMC.
Synthetic peptides
The peptides were predicted with the help of public databases (http://www.uni-tuebingen.de/uni/kxi/; http://bimas.dcrt.nih.gov/molbio/hla_bind/) and were synthesized by Dr. J.W. Drijfhout (University of Leiden, The Netherlands). The peptides were solubilized in PBS/5% DMSO and stored at −20°C. They were loaded on target cells for T cell recognition testing as described [3].
In vitro transcribed mRNA
In vitro transcription (IVT) was performed on linearized plasmids with the Ribomax large-scale RNA production system T7 (Promega, Mannheim, Germany) according to the manufacturer’s instructions. In vitro transcribed RNA was polyadenylated using the Poly (A) Polymerase (USB, Cleveland, OH, USA) according to the manufacturer’s instructions. IVT-mRNA was resuspended in water and stored at −80°C. For T cell recognition testing, IVT-mRNA was electroporated according to [28].
Construction of a cDNA library
A cDNA library was constructed from VX pre-HSCT PBMC using the pCMV-Script XR Library Construction Kit (Stratagene, Amsterdam, The Netherlands). Briefly, total RNA was isolated and poly(A+) mRNA was prepared with the Oligotex mRNA Kit (Qiagen, Hilden, Germany). mRNA was converted into cDNA using an oligo-dT primer containing an XhoI site at its 5′ end. The cDNA was ligated to EcoRI adaptors. The ligation products were then digested with XhoI and ligated into XhoI and EcoRI sites of expression vector pcDNA3.1/Amp (Invitrogen/Gibco BRL). The recombinant plasmids were electroporated into E.coli XL10-Gold Ultracompetent Cells (Stratagene, Amsterdam, The Netherlands) and the resulting library was divided into 1,920 pools of approximately 100 cDNA clones (100 × pools). Each pool was amplified and plasmid DNA was extracted with the QIAprep 96 Turbo Miniprep Kit (Qiagen).
IFN-γ ELISPOT assays
T cell recognition of peptide-loaded or transfected targets was verified with IFN-γ ELISPOT assays performed as described [3]. The assay medium was the same as that applied to lymphocyte expansion. Spot numbers were semiautomatically determined with the use of computer-assisted video image analysis (KS ELISPOT Release 4.2.0; Zeiss, Jena, Germany).
To demonstrate HLA class I-restricted T cell recognition, ELISPOT assays were performed in the presence of the following murine monoclonal antibodies: W6/32 (anti-HLA class I) [2], MA2.1 (anti–HLA-A2) [17] and B1.23.2 (anti–HLA-B/C) [14].
cDNA library screening
COS-7 cells (20,000/well) were co-transfected on MultiScreen HA plates (Millipore, Eschborn, Germany) with HLA-A*0201 cDNA in pcDNA3.1/Amp (100 ng/well) and pooled cDNA from the VX library (∼300 ng/well) using the PolyFect™ transfection reagent (0.5 μl/well; Qiagen) in 35 μl RPMI. After 20–24 h, allo-MLC responders were added to COS-7 transfectants at 5,000–10,000/well in 50 μl AIM-V/HS containing rhIL-2 (150 IU/ml) and rhIL-7 (5 ng/ml). Four days earlier, cryo-preserved MLC responder cells had been thawed and restimulated (at 1 × 106/well) in 2 ml AIM-V/HS containing irradiated VX pre-HSCT PBMC (1 × 106/well), rhIL-2 (150 IU/ml) and rhIL-7 (5 ng/ml). These T cells were incubated with COS-7 transfectants for 16–20 h at 37°C in 5% CO2. T cell recognition was verified with IFN-γ ELISPOT assays (see above).
Peptide/HLA tetramer staining
The MAVPPCCIGV/A2 tetramer (peptide MAVPPCCIGV complexed with HLA-A2.1; Proimmune Limited, Oxford, UK) was applied according to the manufacturer’s recommendations. PBMC (4 × 106) were incubated with phycoerythin-labeled tetramers for 45 min on ice in 100 μl buffer (PBS, 1% BSA, pH 7.4). Then anti-CD8 FITC (Caltag Laboratories, Burlingame, CA, USA) was added. After 15 min, the cells were washed three times and resuspended in buffer with PI (2 μg/ml). PI-negative cells were gated and 500,000 events were counted using a Coulter Epics XL flow cytometer (Beckman Coulter, Brea, CA, USA).
RT-PCR amplification of the polymorphic TRIM22 region
Total RNA was isolated from VX pre-HSCT PBMC and from donor PBMC. Reverse transcription was performed on 3 μg of total RNA using the Superscript First Strand cDNA synthesis kit (Invitrogen/Gibco BRL). A 690 bp TRIM22-cDNA fragment was amplified by PCR using the forward primer 5′-GCAGAGGCAGGATGCCAGCACGC-3′ (nucleotide positions 827–849) and the reverse primer 5′-GAGACAATGCCTGCCTCATAGTC-3′ (nucleotide positions 1494–1516) for 30 cycles (1 min at 95°C, 1 min at 65°C, and 1 min at 68°C). PCR products were sequenced to discriminate between the TRIM22-442C and -442R alleles.
Allele frequency determination with LightCycler PCR
LightCycler PCR was applied to find out, how frequently TRIM22 alleles are polymorphic at nucleotide position 1476. The forward primer 5′- CAATATGGCTACTGGGTTATAGGATT-3′ (positions 1362-1387) and the reverse primer 5′-CTGAGCGACATCCAGAGAACTTGTAGAT-3′ (positions 1548-1572) were used to amplify a 210 bp fragment of the human TRIM22 gene harbouring the polymorphic site. These amplification primers were synthesized by standard phosphoramidite chemistry (TIB MOLBIOL, Berlin, Germany). The 3′-fluorescein-labelled detection probe was a 21-mer oligonucleotide. Its sequence, 5′-CTGTGCCTCCCTGTCGTATTG-X-3′, was complementary to the antisense strand of TRIM22 with the polymorphic nucleotide located seven bases from its 3′ end. The anchor probe labelled at its 5′ end with LightCycler Red 640 and modified at its 3′ end by phosphorylation to block extension was a 31-mer binding at a distance of two bases 5′ to the detection probe (5′-LC Red640-GTTTTCCTAGACTATGAGGCAGGCATTGTCTp-3′). Both fluorophore-labelled probes were synthesized and purified by reverse-phase HPLC (TIB MOLBIOL). PCR was performed with genomic DNA isolated from PBMC of 184 individuals. PCR amplification was performed using 10 pmol of forward and reverse primers and 3 pmol of anchor and detection probe for 1 cycle as denaturation step (10 min at 95°C), 39 cycles for amplification (3″ at 95°C, 12″ at 58°C and 15″ at 72°C), 1 cycle as melting step (30″ from 48°C to 85°C at 0.1°C/s), then for 30″ at 40°C to cool the reaction.
T Cell Receptor V beta chain analysis
The T cell receptor (TCR) V beta usage of T cell clones was analysed by using the IOTest Beta Mark TCR V Repertoire kit (Beckman Coulter) or by RT-PCR with TCR V chain-specific primers [21].
Results
Allo-MLLC and allo-MLC responder populations
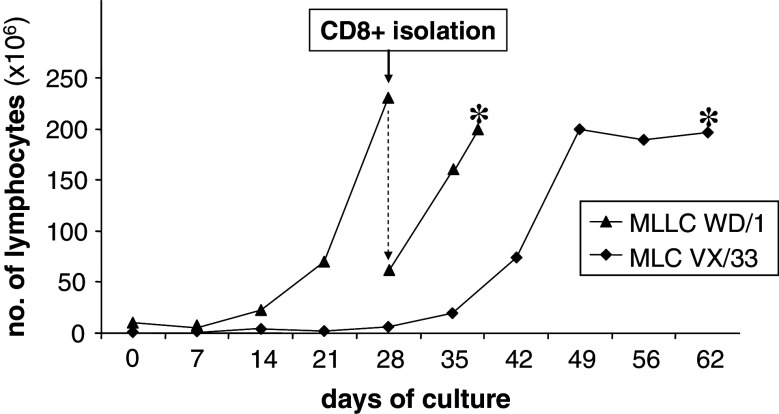
We analysed the specificity of CD8+ allo-MLC and allo-MLLC responder populations generated in allo-HSCT model systems WD and VX. According to HLA class I genotyping, patient WD and her donor differed in an HLA-B*44 and an HLA-A*24 allele mismatch, while patient VX and his sibling donor were fully HLA class I-matched. The generation of allo-MLLC WD/1 and allo-MLC VX/33 from donor-derived peripheral blood lymphocytes is detailed in Materials and methods. We regularly observed that total lymphocyte counts increased only after 2 or 3 weeks (Fig. 1). T cell expansion depended on the presence of stimulator cells (data not shown), which strongly suggested that it was antigen driven.
Fig. 1.
Expansion of allo-MLLC and allo-MLC responder populations. Allo-MLLC WD/1 and allo-MLC VX/33 were generated in model systems WD and VX from blood lymphocytes of the respective healthy donors as detailed in Materials and methods. On day 21, CD8+ responder lymphocytes were purified from allo-MLLC WD/1 responders and expanded further. * Cryopreservation
In both models, recognition of stimulator cells by alloreactive T cells was constantly detectable in IFN-γ ELISPOT assays and was almost completely blocked by an antibody against a common HLA class I determinant. The WD/1 responders were completely blocked by an antibody against a common HLA-B/C determinant, whereas VX/33 responders were blocked by an anti-HLA-A2 antibody. Patient-derived EBV-B cells were only available in the WD model and were recognized by WD/1 responders. Donor-derived EBV-B cells were not recognized in any case (not shown).
Panel testing for recognition of known targets
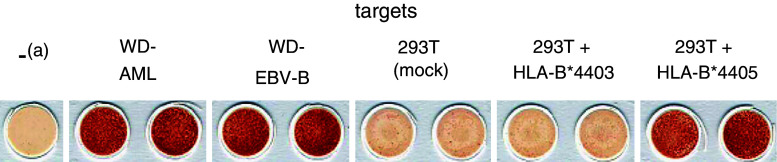
In each model system, a panel of plasmids encoding patient-derived HLA class I alleles was transiently transfected into COS-7 or 293T cells and transfectants were tested for their ability to induce IFN-γ spot formation by alloreactive T cells (not shown). In model WD, only the mismatch allele HLA-B*4405 conferred recognition by WD/1 responder lymphocytes. Figure 2 demonstrates their anti-HLA-B*4405 reactivity on mRNA transfectants.
Fig. 2.
Allo-MLLC WD/1 lymphocytes recognize the mismatch allele HLA-B*4405. The WD/1 responder population was generated from donor lymphocytes by stimulation with WD-AML cells. Shown are filters of an IFN-γ ELISPOT assay. Targets were patient-derived EBV-B cells (WD-EBV-B, 1 × 105 cells/well), stimulator leukaemia cells (WD-AML, 1 × 105 cells/well) and 293T cells (1 × 105 cells/well; mock-transfected, electroporated with IVT-mRNA encoding patient-derived HLA-B*4405, or electroporated with IVT-mRNA encoding donor-derived HLA-B*4403). Effectors (allo-MLLC WD/1) were added at 10,000 cells/well. a denotes lymphocytes only
In model VX, VX/33 responder cells derived from the fully HLA-matched sibling donor did not recognize any of the respective HLA transfectants (not shown). This indicated that the alloreactive T cells were peptide specific. We excluded targeting of the mHag HA-1 [5] (not shown). Considering the patient’s HLA class I phenotype, further known candidate mHags would have been HA-2 and HA-8 [9]. Due to the low number of candidates, we refrained from mHag typing and further antigen panel testing and directly prepared for a cDNA library screening.
cDNA library screening and expression cloning (model VX)
A cDNA library was constructed from VX pre-HSCT PBMC. We screened 1.920 100 × pools of this library with allo-MLC VX/33 (d66 + 4) responder T cells. Due to the preferential restriction of VX/33 (see above), cDNA pools were co-transfected with HLA-A*0201 into COS-7 cells. The transfectants were tested for their ability to induce IFN-γ spot formation. Only 100 × pool #609 was clearly positive and was stepwise cloned. We identified the spot-inducing cDNA clone #609.4.33. Its recognition depended on the co-transfection of HLA-A*0201 (not shown).
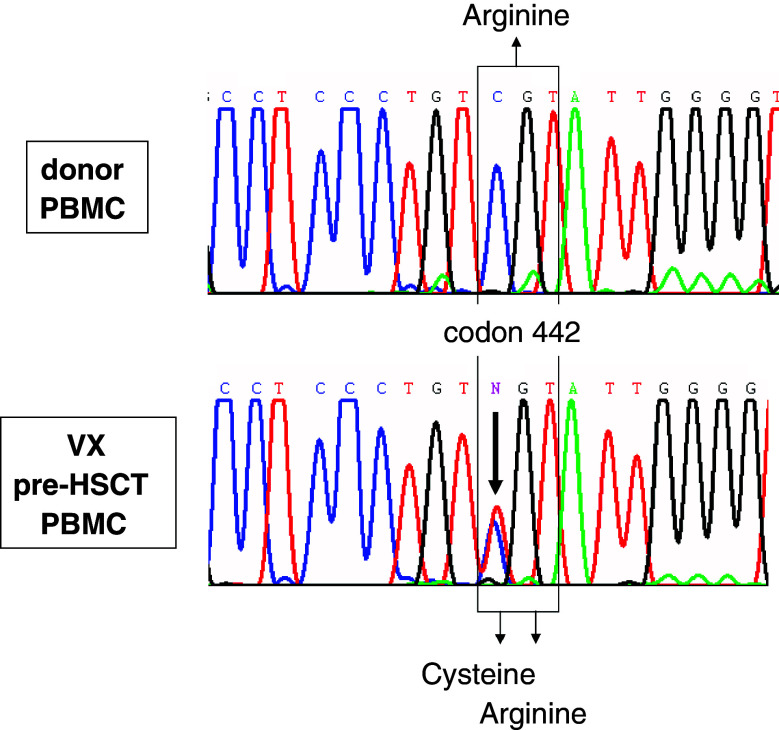
The insert of #609.4.33 (sequence available under EMBL/GenBank accession number AM040752) was 2,064 bp long and was found to contain a 5′-deleted fragment (deletion size: 826 bp) of the “tripartite binding motif 22” or TRIM22 cDNA (synonym: STAF50) [27]. Alignment with the published TRIM22 cDNA sequence (accession numbers BC035582 and NM_006074) revealed, that the #609.4.33 insert carried a cytidine-to-thymidine (C→T) exchange at the complete ORF’s nucleotide position 1324 causing an arginine(R)-to-cysteine(C) exchange at amino acid position 442. Considering that the cDNA library had been constructed from normal VX pre-HSCT haematopoietic cells, we had found a natural polymorphism of TRIM22. Indeed, pre-HSCT lymphocytes of patient VX expressed both TRIM22.442 R and TRIM22.442 C, while the donor’s lymphocytes expressed only the TRIM22.442 R allele (Fig. 3).
Fig. 3.
Expression of TRIM22.442 C and TRIM22.442 R alleles in patient VX and his sibling donor’s PBMC. Total RNA was extracted from VX pre-HSCT PBMC and from donor PBMC. RT-PCR amplification of the polymorphic TRIM22 region was performed as detailed in Materials and methods and PCR products were directly sequenced. Sequences around codon 442 are shown
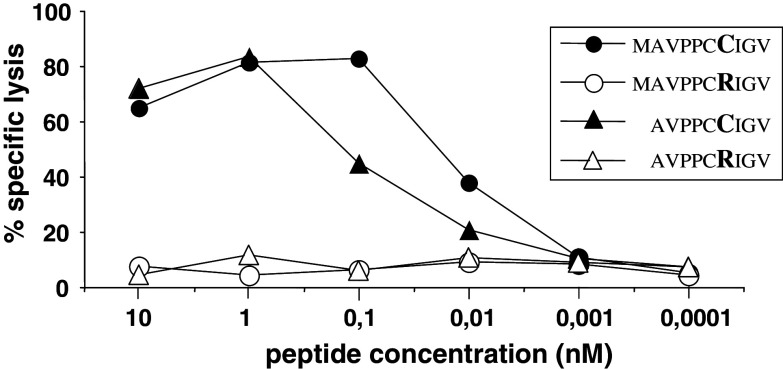
Only transfection of TRIM22.442 C cDNA, but not of TRIM22.442 R cDNA induced T cell recognition (not shown). This suggested that the polymorphic residue 442 generated an immunogenic epitope. HLA-A2.1-binding nonamer peptides (FMAVPPCRI, FMAVPPCCI, AVPPCRIGV, AVPPCCIGV) and decamer peptides (MAVPPCRIGV, MAVPPCCIGV, TLFMAVPPCR, TLFMAVPPCC) around polymorphic residue 442 were predicted with the help of public algorithms, synthesized and tested for recognition by MLC VX/33 (d66 + 4) responder T cells. The decamer peptide 436-445 (MAVPPCCIGV) and the nonamer peptide 437-445 (AVPPCCIGV) were recognized, while the corresponding TRIM22.442R peptides (MAVPPCRIGV and AVPPCRIGV) did not sensitize targets. The decamer 442C peptide was recognized at the lowest concentration. It induced half-maximal recognition at 0.01 nM concentrations in lysis assays (Fig. 4).
Fig. 4.
Recognition of TRIM22 peptides by VX/33 responder lymphocytes. Data of a 4 h 51Cr release assay are shown. Targets were 51Cr-labeled K562/A*0201 cells (2 × 103/well) pulsed with graded doses of TRIM22 peptides (sequences and concentrations as indicated, polymorphic 442 residues are indicated in bold characters). Effectors were VX/33 (d66 + 4) responder lymphocytes added at an effector-to-target cell ratio of 10:1
A LightCycler PCR was developed that allowed to discriminate between both TRIM22 alleles. With this test we detected the TRIM22.442C allele in 5 out of 184 individual blood samples (not shown).
Clonal analysis of allo-MLLC and allo-MLC responders
Clonal T cells were derived by limiting dilution from WD/1 and from VX/33 responders as detailed in Materials and methods. We analysed 26 clones from WD/1 responders. A total of 23 clones showed reactivity towards leukaemia and patient-derived EBV-B cells as well as COS-7, 293T or K562 cells transfected with HLA-B*4405. Two clones recognized leukaemia cells, but not the patient’s EBV-B cells and not HLA-B*4405-transfected cells. One T cell clone recognized targets transfected with HLA-A*2403. This indicated that anti-HLA-B*4405 reactivity dominated among WD/1 responders, whereas T cells against the other mismatch were rare and therefore not detectable in the panel test. TCR Vβ analysis revealed that the anti-HLA-B*4405 T cell response was oligoclonal and involved at least three distinct receptors (not shown).
Eight clones were derived from VX/33 responders that recognized the patient’s pre-HSCT PBMC. Seven of them were specific for TRIM22.442C as demonstrated with transfectants and peptide-loaded target cells (not shown). This meant that TRIM22.442C-specific T cells predominated among the allo-MLC responder cells in the VX model.
Frequency analyses in peripheral blood
In the WD model, blood samples were collected from the patient at different time points from day 42 to day 450 after allo-HSCT. In ex vivo IFN-γ ELISPOT analyses, reactivity against HLA-B*4405 transfectants could not be detected in any of these samples, whereas reactivity against WD-EBV-B cells derived from the patient’s pre-HSCT PBMC was continuously present from day 63 onwards. Also in the peripheral blood of the donor, HLA-B*4405-reactive T cells were not detectable ex vivo (not shown). This indicated that the frequencies of anti-HLA-B*4405 T cells in the patient’s post-day 42 blood and in the donor’s blood were below 1 in 5,000 CD8+ T cells (<0.02%), which we regard as the detection limit for IFN-γ ELISPOT assays. PBMC collected from the patient’s blood on day 83 after allo-HSCT were then stimulated with EBV-B cells derived from the patient’s blood before allo-HSCT. After 11 stimulations, T cells against the HLA-B*4405 mismatch allele were detectable among responder lymphocytes (not shown).
In the VX model, PBMC were separated from the peripheral blood of the patient 5 years after allo-HSCT (VX post-HSCT PBMC). According to chimerism analysis, these PBMC were of donor origin. They were directly stained with MAVPPCCIGV/A2 tetramers. As shown in Fig. 5, tetramer-positive T cells did not form a clearly distinct subpopulation and were detectable at a frequency level of 0.1% within ex vivo CD8+ T cells. These PBMC were then weekly stimulated with irradiated VX pre-HSCT PBMC (MLC 42) or with donor PBMC loaded with peptide MAVPPCCIGV (pIVS). As seen after double fluorescence staining of responders, TRIM22.442C-specific CD8+ T cells forming distinct subpopulations were enriched in both populations. Tetramer-positive T cells were isolated from pIVS responders. They were found to recognize VX pre-HSCT PBMC and target cells loaded with the TRIM22.442C peptide both in a 51Cr release assay and in an IFN-γ ELISPOT assay (not shown). This indicated, that TRIM22.442C-specific CD8+ T cells persisted in the patient’s peripheral blood even years after allo-HSCT.
Fig. 5.
Persistence of TRIM22.442C-specific CD8+ positive T cells in the peripheral blood of patient VX after allo-HSCT. PBMC of patient VX, isolated 5 years after allo-HSCT, were weekly stimulated with irradiated patient VX-derived pre-HSCT PBMC (MLC 42) or with irradiated donor-derived PBMC loaded with TRIM22.442C peptide MAVPPCCIGV (pIVS). Results of double fluorescence staining at indicated time points with the MAVPPCCIGV/A2 tetramer and anti-CD8 antibodies are shown
Discussion
We applied an expression screening procedure to the dissection and the molecular characterization of alloreactive T cells. In two patient–donor model systems, with and without HLA class I disparities, allo-MLLC or allo-MLC responder populations were generated from PBMC of the respective healthy donors. Stimulator cells were leukaemia cells in allo-MLLC and patient-derived pre-HSCT PBMC in allo-MLC. The usage of cryopreserved responder populations avoided permanent T cell propagation and the a priori availability of T cell clones and thereby improved the feasibility of T cell-based expression screening [15]. The procedure allowed us to verify HLA class I mismatch responses and responses to HLA-presented peptides and let us identify a new mHag recognized in association with HLA-A*0201.
Mismatch HLA alleles can evoke GvHD and graft rejection, but may also induce GvL effects. The prediction of permissive and non-permissive HLA mismatches in unrelated allo-HSCT is still an unresolved issue [11, 22]. AML patient WD and her unrelated donor were mismatched for HLA-B*44 and HLA-A*24 alleles. These disparities were serologically not detectable. HLA-B*4405-reactive T cells were readily expandable in allo-MLLCs from the donor’s PBMC and dominated in independently generated allo-MLLCs, while T cells against HLA-A*2403 only constituted a minority of responder lymphocytes. At least HLA-B*4405-reactive T cells were detectable in the patient’s post-HSCT PBMC after multiple stimulations in vitro. Clinically, the HLA-B*44 and HLA-A*24 mismatches were obviously permissive in the patient’s case, because she only experienced mild GvHD. However, mismatches of HLA-B*4402 and HLA-B*4403, the two most frequent HLA-B*44 alleles, are considered to represent a significant barrier to transplantation and have been associated with transplant rejection and GvHD [7, 12]. HLA-B*4402 and HLA-B*4403 differ in a single amino acid at position 156 within the alpha 2 domain. HLA-B*4405 is closer related to HLA-B*4402, both differing only in the amino acid position 116, located in the F pocket region of the peptide-binding groove. The impact of these HLA-B*44 polymorphisms on peptide loading and binding has been extensively studied [16, 26].
Minor histocompatibility antigens described so far were predominantly identified with T lymphocyte clones isolated from patients’ peripheral blood during acute graft rejection or GvHD following HLA-matched allo-HSCT [6]. In the VX model, we sensitized PBMC derived from the patient’s fully HLA-matched sibling donor against patient-derived pre-HSCT PBMC in vitro to generate anti-mHag T cells. We reasoned that mHag-reactive T cells might be more difficult to expand and maintain in vitro from patients’ post-HSCT PBMC during the first few months after allo-HSCT, when they are heavily immunocompromised.
Expansion of MLC responders was delayed, probably due to the low frequency of mHag-reactive T cells in the donor’s blood lymphocytes (Fig. 1). The allo-response generated in the VX model system was mainly restricted by HLA-A*0201 and found to predominantly target a peptide encoded by a new polymorphic allele of the TRIM22 gene, named TRIM22.442 C. Nonamer and decamer peptides containing the 442C residue on position six or seven, respectively, sensitized targets. The T cells’ avidity for the decamer peptide (Fig. 4) was in the same range as seen for anti-viral T cells [24]. Since homologous 442R peptides carrying the same agretopes were not recognized (Fig. 4) and TRIM22.442 R cDNA did not induce T cell recognition even in the high-efficiency COS expression system (not shown), we assume that the polymorphic residue constituted a specific epitope.
The protein encoded by the TRIM22 gene, located on chromosome 11p15, is a member of the tripartite motif (TRIM) family and is also designated as stimulated transacting factor of 50 kDa (StAF50). TRIM22 mRNA is predominantly expressed in peripheral blood leukocytes and in lymphoid tissues. Its expression is upregulated by p53 and can be induced by interferons. It is believed to mediate cell proliferation arrest in p53 and interferon (IFN) signaling pathways and to contribute to the interferons’ antiviral effects [18, 19, 27]. TRIM22 was also shown to be expressed at high levels in human immature CD34+ progenitor cells and to decline during maturation in most lineages [20]. Thus, it might be speculated that an immunogenic TRIM22 mismatch can lead to preferential killing of patient-derived haematopoiesis cells including leukaemic cells and thereby prevent graft rejection and accelerate the establishment of complete chimerism. This process might be supported by proinflammatory cytokine storms regularly occurring during the early phase after allo-HSCT.
T cells against TRIM22.442C were still detectable in the patient’s peripheral blood 5 years after allo-HSCT and 4 years after the immunosuppressive drugs were definitely discontinued (Fig. 5). The patient is still free of detectable disease and shows no sign of GvHD, although TRIM22 is supposedly expressed at basal levels in non-lymphohaematopoiesis tissues [23]. Animal experiments demonstrated that T cells against such antigens could mediate anti-leukaemic effects in the absence of GvHD [8]. As an explanation, it was argued that non-haematopoiesis tissues are less sensitive to T cell recognition than haematopoiesis cells due to lower levels of MHC or antigen expression on non-haematopoiesis cells [4, 10].
TRIM22.442C is a rare allele. We estimate that approximately 1.3% of Caucasians carry it in association with HLA-A*0201. But improvement of donor selection or adoptive T cell transfer might depend on the knowledge of a large and diverse panel of such mHags. The procedure introduced herein will help to accelerate and facilitate the search for further GvHD and GvL candidate antigens.
Acknowledgments
We thank N. Kaus, M. Fatho and B. Schuch for their excellent technical assistance and M. Jülch for performing flow cytometry. This research was supported by a grant from the “Deutsche Krebshilfe” (Project 70-2428) to C.W., G.D. and T.W. C.H. was supported by grant 70-2427-HuI from the “Deutsche Krebshilfe”. The authors declare that they have no competing financial interests.
References
- 1.Apperley JF, Jones L, Hale G, Waldmann H, Hows J, Rombos Y, et al. Bone marrow transplantation for patients with chronic myeloid leukaemia: T-cell depletion with Campath-1 reduces the incidence of graft-versus-host disease but may increase the risk of leukaemic relapse. Bone Marrow Transplant. 1986;1:53–66. [PubMed] [Google Scholar]
- 2.Barnstable CJ, Bodmer WF, Brown G, Galfre G, Milstein C, Williams AF, Ziegler A. Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens: new tools for genetic analysis. Cell. 1978;14:9–20. doi: 10.1016/0092-8674(78)90296-9. [DOI] [PubMed] [Google Scholar]
- 3.Britten CM, Meyer RG, Kreer T, Drexler I, Wolfel T, Herr W. The use of HLA-A*0201-transfected K562 as standard antigen-presenting cells for CD8(+) T lymphocytes in IFN-gamma ELISPOT assays. J Immunol Methods. 2002;259:95–110. doi: 10.1016/S0022-1759(01)00499-9. [DOI] [PubMed] [Google Scholar]
- 4.Daar AS, Fuggle SV, Fabre JW, Ting A, Morris PJ. The detailed distribution of HLA-A, B, C antigens in normal human organs. Transplantation. 1984;38:287–292. doi: 10.1097/00007890-198409000-00018. [DOI] [PubMed] [Google Scholar]
- 5.den Haan JM, Meadows LM, Wang W, Pool J, Blokland E, Bishop TL, et al. The minor histocompatibility antigen HA-1: a diallelic gene with a single amino acid polymorphism. Science. 1998;279:1054–1057. doi: 10.1126/science.279.5353.1054. [DOI] [PubMed] [Google Scholar]
- 6.Ennis PD, Zemmour J, Salter RD, Parham P. Rapid cloning of HLA-A,B cDNA by using the polymerase chain reaction: frequency and nature of errors produced in amplification. Proc Natl Acad Sci USA. 1990;87:2833–2837. doi: 10.1073/pnas.87.7.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleischhauer K, Kernan NA, O’Reilly RJ, Dupont B, Yang SY. Bone marrow allograft rejection by T lymphocytes recognizing a single amino acid difference in HLA-B44. N Engl J Med. 1990;323:1818–1822. doi: 10.1056/NEJM199012273232607. [DOI] [PubMed] [Google Scholar]
- 8.Fontaine P, Roy-Proulx G, Knafo L, Baron C, Roy DC, Perreault C. Adoptive transfer of minor histocompatibility antigen-specific T lymphocytes eradicates leukemia cells without causing graft-versus-host disease. Nat Med. 2001;7:789–794. doi: 10.1038/89907. [DOI] [PubMed] [Google Scholar]
- 9.Goulmy E. Minor histocompatibility antigens: from transplantation problems to therapy of cancer. Hum Immunol. 2006;67:433–438. doi: 10.1016/j.humimm.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 10.Griem P, Wallny HJ, Falk K, Rotzschke O, Arnold B, Schonrich G, Hammerling G, Rammensee HG. Uneven tissue distribution of minor histocompatibility proteins versus peptides is caused by MHC expression. Cell. 1991;65:633–640. doi: 10.1016/0092-8674(91)90095-G. [DOI] [PubMed] [Google Scholar]
- 11.Kawase T, Morishima Y, Matsuo K, Kashiwase K, Inoko H, Saji H, et al. High-risk HLA allele mismatch combinations responsible for severe acute graft versus host disease and implication for its molecular mechanism. Blood. 2007;110(7):2235–2241. doi: 10.1182/blood-2007-02-072405. [DOI] [PubMed] [Google Scholar]
- 12.Keever CA, Leong N, Cunningham I, Copelan EA, Avalos BR, Klein J, et al. HLA-B44-directed cytotoxic T cells associated with acute graft-versus-host disease following unrelated bone marrow transplantation. Bone Marrow Transplant. 1994;14:137–145. [PubMed] [Google Scholar]
- 13.Kolb HJ, Mittermuller J, Clemm C, Holler E, Ledderose G, Brehm G, Heim M, Wilmanns W. Donor leukocyte transfusions for treatment of recurrent chronic myelogenous leukemia in marrow transplant patients. Blood. 1990;76:2462–2465. [PubMed] [Google Scholar]
- 14.Lemonnier FA, Rebai N, Le BP, Malissen B, Caillol DH, Kourilsky FM. Epitopic analysis of detergent-solubilized HLA molecules by solid-phase radioimmunoassay. J Immunol Methods. 1982;54:9–22. doi: 10.1016/0022-1759(82)90108-9. [DOI] [PubMed] [Google Scholar]
- 15.Lennerz V, Fatho M, Gentilini C, Frye RA, Lifke A, Ferel D, et al. The response of autologous T cells to a human melanoma is dominated by mutated neoantigens. Proc Natl Acad Sci USA. 2005;102:16013–16018. doi: 10.1073/pnas.0500090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macdonald WA, Purcell AW, Mifsud NA, Ely LK, Williams DS, Chang L, et al. A naturally selected dimorphism within the HLA-B44 supertype alters class I structure, peptide repertoire, and T cell recognition. J Exp Med. 2003;198:679–691. doi: 10.1084/jem.20030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McMichael AJ, Parham P, Rust N, Brodsky FM. A monoclonal antibody that recognizes an antigenic determinant shared by HLA-A2 and B17. Hum Immunol. 1980;1:121–129. doi: 10.1016/0198-8859(80)90099-3. [DOI] [PubMed] [Google Scholar]
- 18.Nisole S, Stoye JP, Saib A. TRIM family proteins: retroviral restriction and antiviral defence. Nat Rev Microbiol. 2005;3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- 19.Obad S, Brunnstrom H, Vallon-Christersson J, Borg A, Drott K, Gullberg U. Staf50 is a novel p53 target gene conferring reduced clonogenic growth of leukemic U-937 cells. Oncogene. 2004;23:4050–4059. doi: 10.1038/sj.onc.1207524. [DOI] [PubMed] [Google Scholar]
- 20.Obad S, Olofsson T, Mechti N, Gullberg U, Drott K. Expression of the IFN-inducible p53-target gene TRIM22 is down-regulated during erythroid differentiation of human bone marrow. Leuk Res. 2007;31:995–1001. doi: 10.1016/j.leukres.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 21.Peggs K, Verfuerth S, Pizzey A, Ainsworth J, Moss P, Mackinnon S. Characterization of human cytomegalovirus peptide-specific CD8(+) T-cell repertoire diversity following in vitro restimulation by antigen-pulsed dendritic cells. Blood. 2002;99:213–223. doi: 10.1182/blood.V99.1.213. [DOI] [PubMed] [Google Scholar]
- 22.Petersdorf EW, Gooley T, Malkki M, Horowitz M. Clinical significance of donor-recipient HLA matching on survival after myeloablative hematopoietic cell transplantation from unrelated donors. Tissue Antigens. 2007;69(Suppl 1):25–30. doi: 10.1111/j.1399-0039.2006.759_2.x. [DOI] [PubMed] [Google Scholar]
- 23.Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, et al. The tripartite motif family identifies cell compartments. Embo J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slifka MK, Whitton JL. Functional avidity maturation of CD8+ T cells without selection of higher affinity TCR. Nat Immunol. 2001;2:711–717. doi: 10.1038/90650. [DOI] [PubMed] [Google Scholar]
- 25.Sullivan KM, Weiden PL, Storb R, Witherspoon RP, Fefer A, Fisher L, et al. Influence of acute and chronic graft-versus-host disease on relapse and survival after bone marrow transplantation from HLA-identical siblings as treatment of acute and chronic leukemia. Blood. 1989;73:1720–1728. [PubMed] [Google Scholar]
- 26.Thammavongsa V, Raghuraman G, Filzen TM, Collins KL, Raghavan M. HLA-B44 polymorphisms at position 116 of the heavy chain influence TAP complex binding via an effect on peptide occupancy. J Immunol. 2006;177:3150–3161. doi: 10.4049/jimmunol.177.5.3150. [DOI] [PubMed] [Google Scholar]
- 27.Tissot C, Mechti N. Molecular cloning of a new interferon-induced factor that represses human immunodeficiency virus type 1 long terminal repeat expression. J Biol Chem. 1995;270:14891–14898. doi: 10.1074/jbc.270.25.14891. [DOI] [PubMed] [Google Scholar]
- 28.Van Tendeloo VF, Ponsaerts P, Lardon F, Nijs G, Lenjou M, Van Broeckhoven C, Van Bockstaele DR, Berneman ZN. Highly efficient gene delivery by mRNA electroporation in human hematopoietic cells: superiority to lipofection and passive pulsing of mRNA and to electroporation of plasmid cDNA for tumor antigen loading of dendritic cells. Blood. 2001;98:49–56. doi: 10.1182/blood.V98.1.49. [DOI] [PubMed] [Google Scholar]