Abstract
Recent studies have demonstrated expression of Fc receptor-like (FCRL) molecules, a newly identified family with preferential B-cell lineage expression, in some chronic B-cell leukemias with possible implication for classification and/or targeted immunotherapy. In this study, the expression pattern of FCRL1-5 genes was studied in 73 Iranian ALL patients and 35 normal subjects using semi-quantitative RT-PCR method. FCRL protein expression was also investigated by flow cytometry. Our results indicate significant down-regulation of all FCRL genes in ALL compared to normal subjects. Although, FCRL mRNA expression was almost exclusively confined to normal isolated B-cells compared to T-cells, but these genes were similarly expressed in B-ALL, T-ALL and different B-ALL immunophenotypic subtypes. Surface protein expression of FCRL1, 2, 4, and 5 molecules in 10 ALL and 5 normal samples confirmed the PCR results. Expression profile of FCRL molecules in different subtypes of ALL argues against their potential implication as suitable targets for classification and/or immunotherapy of ALL.
Keywords: FCRL, Acute lymphoblastic leukemia, RT-PCR, Flow cytometry, Immunophenotype
Introduction
Acute lymphoblastic leukemia (ALL) consists of distinct subtypes which display characteristic clinical, biologic and prognostic features [12]. Based on morphologic features (FAB criteria), CD markers expression (immunophenotyping) and also molecular and cytogenetic findings, leukemic cells of ALL can be classified in different subgroups with different clinical outcomes [11].
Gene profiling studies conducted in a variety of chronic and acute leukemias and lymphomas have helped to further categorize these malignancies and to establish new classifications. Gene profiling of malignant B-cells of chronic lymphocytic leukemia (CLL) has shown overexpression of some members of the Fc receptor-like (FCRL) family in this malignancy [1]. Subsequent molecular and immunocytochemical analyses have supported and extended these findings in B-CLL and some other chronic B-cell type leukemias [7, 8, 15–17, 23, 26, 29, 30]. Similar studies are lacking in ALL.
Fc receptor-like (FCRL) molecules are a family of Fc receptor homologue molecules comprising of eight structurally related members [4, 24]. Residing on chromosome 1q21-23, FCRL1-6 encode type I transmembrane glycoproteins consisting of three to nine Ig-like domains and a cytoplasmic tail containing immunoreceptor tyrosine-based activating (ITAM) and/or inhibitory (ITIM) motifs [5]. FCRL1-5 are all exclusively expressed in B cells, with the exception of FCRL3 which is also found on NK and T cells [6].
In a survey of potential roles for these molecules, it has been shown that FCRL1 might act as activating coreceptor on B cells [22], and because of presence of three ITIMs in its cytoplasmic region, FCRL4 can abrogate activating signals form B cell receptor potently than FcγRIIB [9]. FCRL3 has ITAM-like and ITIM motifs, so it might have both activating and inhibitory functions [7]. FCRL5 has the functional potential to serve as an inhibitory coreceptor on mature B cells in humans [13]. Genomic instability of 1q21-23 region and association with B cell lineage malignancies suggest that genes located in this region may have been involved in tumorigenesis [18, 27]. Differential expression of FCRL1-5 has recently been shown in various B cell malignancies, such as B-CLL [23, 29], hairy cell leukemia [16], mantle cell lymphoma (MCL) [17, 30], Burkitt’s lymphoma (BL) [7, 26] and multiple myeloma [15].
In the present study, expression of FCRL1-5 molecules has been investigated for the first time both at mRNA and protein levels in fresh leukemic cells from different subtypes of ALL to find out whether these molecules are suitable targets for classification and/or immunotherapy in this malignancy.
Materials and methods
Patients and controls
Heparinized peripheral blood (PB) and/or bone marrow (BM) samples were collected from 73 Iranian ALL patients (PB = 24 and BM = 49) attending the Hematology and Oncology Clinics of Vali-Asr and Ali-Asghar hospitals, affiliated to Tehran University of Medical Sciences and Iran University of Medical Sciences, respectively. A consent letter was taken from all patients or their parents and the study was approved by the Ethical Committee of Tehran University of Medical Sciences. Disease diagnosis was based on clinical, cytomorphologic and immunophenotypic features of BM leukemic cells [14]. Demographic features of ALL cases have been shown in Table 1. According to flow cytometric analyses of samples based on defined cluster of differentiation (CD) markers, they were classified into B-ALL (n = 54) and T-ALL (n = 16) groups, and the B-ALL group was further subdivided into Pro-B, Pre-B I, Pre-B II, and immature/mature subtypes [2]. One patient was diagnosed as mixed ALL and immunophenotypic data was not available for two more patients. Thirty-five normal subjects (16 children and 19 adults) were also included in this study. Burkitt’s lymphoma cell lines were used as positive controls for amplification of FCRL1, 3, and 5 (Ramos), FCRL2 (BL41) and FCRL4 (Raji). All cell lines were obtained from the National Cell Bank of Iran (NCBI, Pasteur Institute of Iran, Tehran, Iran).
Table 1.
Major laboratory findings in Iranian ALL patients
| No. | Patient code | Specimen | WBC × 106 cell/ml | Blast (%) | Immunophenotype | No. | Patient code | Specimen | WBC × 106 cell/ml | Blast (%) | Immunophenotype |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ND2 | BM | 9.9 | 40 | Pre-B I | 38 | ND60 | PB | 38.9 | 50 | Pre-B II |
| 2 | ND3 | BM | 8.4 | 60 | Pre-B I | 39 | ND61 | BM | 34.5 | 40 | Pre-B I |
| 3 | ND4 | PB | 4.5 | 70 | Immature/mature B | 40 | ND65 | BM | 4.1 | NI | Pre-B I |
| 4 | ND6 | BM | 5.4 | NI | Pre-B II | 41 | ND67 | BM | 245 | 90 | Pro-B |
| 5 | ND7 | BM | 90 | 50 | Pre-B I | 42 | ND68 | BM | 91.7 | 90 | T |
| 6 | ND8 | PB | 7.9 | 70 | Pre-B I | 43 | ND69 | BM | 23.2 | 93 | Pre-B I |
| 7 | ND9 | PB | 222 | 60 | T | 44 | ND71 | BM | 27.9 | 35 | Pre-B II |
| 8 | ND10 | BM | 14.2 | 32 | Pre-B I | 45 | ND73 | BM | 2.5 | NI | Immature/mature B |
| 9 | ND14 | BM | 53.7 | 80 | Pre-B I | 46 | ND74 | BM | 10.1 | NI | Pre-B II |
| 10 | ND15 | BM | 3.0 | NI | Pre-B I | 47 | ND75 | PB | 24.8 | 40 | Pre-B II |
| 11 | ND17 | BM | 5.3 | 90 | T | 48 | ND76 | BM | 46.1 | >95 | T |
| 12 | ND19 | PB | 4.8 | 80 | T | 49 | ND78 | PB | 7.7 | 35 | Pre-B I |
| 13 | ND21 | BM | 70.2 | 40 | Pre-B I | 50 | ND81 | PB | 19.4 | 30 | Pre-B I |
| 14 | ND22 | BM | 45.7 | 60 | Pro-B | 51 | ND84 | BM | 39.2 | 89 | Pre-B II |
| 15 | ND24 | BM | 14.8 | 98 | Pre-B I | 52 | ND85 | BM | 44.9 | 91 | T |
| 16 | ND25 | PB | 6.4 | 80 | Pre-B I | 53 | ND86 | BM | 72.4 | 69 | Pre-B II |
| 17 | ND26 | PB | 9.6 | 65 | Pre-B I | 54 | ND87 | BM | 2.8 | NI | Pre-B I |
| 18 | ND28 | BM | 8.4 | 90 | Pre-B I | 55 | ND88 | BM | NI | NI | Pre-B I |
| 19 | ND31 | PB | 6.8 | NI | Pre-B I | 56 | ND89 | BM | 9.6 | 70 | Pre-B I |
| 20 | ND32 | BM | 20.4 | 75 | Immature/mature B | 57 | ND91 | BM | 15.7 | 60 | Pre-B I |
| 21 | ND35 | PB | 3.1 | 52 | Pro-B | 58 | ND92 | BM | 7.9 | 85 | T |
| 22 | ND36 | BM | 8.6 | 65 | Pre-B I | 59 | ND93 | PB | 187.2 | 82 | T |
| 23 | ND39 | PB | 4 | NI | Pro-B | 60 | ND95 | PB | 5.4 | 50 | Pre-B II |
| 24 | ND40 | BM | 482 | 90 | Mixed lineage | 61 | ND96 | BM | NI | NI | NI |
| 25 | ND42 | BM | 63.5 | 70 | Pre-B I | 62 | ND97 | BM | 7 | NI | T |
| 26 | ND43 | BM | 23.1 | 90 | Pre-B I | 63 | ND98 | BM | 295 | 84 | T |
| 27 | ND46 | PB | 20.6 | 90 | Pro-B | 64 | ND99 | BM | 102 | 89 | Pre-B I |
| 28 | ND48 | BM | 47.6 | NI | Pro-B | 65 | ND100 | BM | 141.7 | 71 | Pre-B I |
| 29 | ND49 | BM | 11.9 | 80 | Pre-B II | 66 | ND101 | BM | 12.9 | NI | Pre-B II |
| 30 | ND50 | PB | 5.8 | 60 | Pre-B I | 67 | ND102 | PB | 109.5 | 72 | T |
| 31 | ND51 | PB | 33.2 | 95 | T | 68 | ND103 | PB | 3.4 | NI | Pre-B I |
| 32 | ND52 | BM | 31.1 | 55 | Pre-B II | 69 | ND104 | BM | 6.9 | 67 | Pre-B I |
| 33 | ND53 | PB | 4.4 | NI | NI | 70 | ND105 | BM | 19.1 | NI | Pre-B II |
| 34 | ND54 | BM | 145 | 60 | T | 71 | ND106 | PB | 41.5 | 33 | Pre-B II |
| 35 | ND55 | PB | 3.1 | >90 | Pre-B I | 72 | ND107 | BM | 125.4 | 92 | Pre-B II |
| 36 | ND56 | BM | 13.7 | 80 | T | 73 | ND108 | BM | 96 | 96 | T |
| 37 | ND57 | PB | 16.7 | 30 | T |
ND newly diagnosed ALL patients, WBC white blood cell count, BM bone marrow, PB peripheral blood, NI not identified
Isolation of B- and T-cells from PBMC by magnetic bead separation
Peripheral blood mononuclear cells (PBMC) were isolated by density gradient centrifugation using Histopaque (Sigma, St Louis, MO, USA). B- and T-cells were isolated from peripheral blood of 5 normal subjects using MACS negative selection kits (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer’s instruction. Briefly, PBMC were isolated and CD19+ and CD3+ cells were enriched by positive and negative selection methods, respectively, using MACS microbeads and midiMACS columns. The purity was >95% for B-cells and >90% for T-cells, as determined by flow cytometry.
RNA extraction and cDNA synthesis
Total RNA was extracted from PBMC and isolated B- and T-cells using RNA-Bee (TEL test Inc, USA) according to the manufacturer’s instruction. First-strand cDNA was synthesized as described [31].
Polymerase chain reaction (PCR)
Amplification of FCRL1-5 and β-actin genes was performed using specific primers [3, 7]. A total of 25 μl reaction mixture of PCR was prepared using 2.5 μl 10 × PCR buffer, 3.5 μl (for β-actin) or 1 μl (for FCRL1 and 4) or 1.5 μl (for FCRL2, 3, and 5) 25 mM MgCl2, 1.5 μl 10 mM dNTPs, 1 μl each primer (10 pmol/μl), 0.2 μl Taq-DNA polymerase (10 U/μl) (CinnaGen, Tehran, Iran) and 1 μl template cDNA. Each amplification reaction underwent 37 cycles (FCRL1, 3, 4 and 5), 38 cycles (FCRL2) and 29 cycles (β-actin) consisting of denaturation at 94°C for 30 s, annealing at 61.5°C (FCRL1, 3, 4 and 5), 60°C (FCRL2) and 55°C (β-actin) for 30 s, extension at 72°C for 1 min, and final extension at 72°C for 10 min. At these conditions and after adjustment of template cDNA dilution for each sample and analysis with LabWorks software, the results showed that PCR products of FCRL and β-actin were not in saturated phase. Amplified products were visualized in 1.5% agarose gel containing ethidium bromide and documented with gel documentation system (UVP, CA, USA). Amplicon sizes were 793, 430, 890, 686, 768 and 203 base pairs for FCRL1 to 5 and β-actin, respectively. The densities of FCRLs and β-actin PCR product bands were determined by Labworks 4.0 software (UVP, CA, USA), and the ratio of the two bands was calculated for each sample, as follows: (density of FCRL band/β-actin density band) × 100.
Indirect immunoflourescence
The staining was performed on leukemic B-cells from 10 ALL patients and PBMC of 5 normal subjects at surface level. After separation of the mononuclear cells and twice washing with washing buffer (PBS 0.15 M, 0.5% BSA, 0.1% NaN3), 106 cells were incubated with 2.5 μg/ml of biotinylated goat anti-human FCRL1, FCRL2, FCRL4 and FCRL5 antibodies (R&D Systems, Minneapolis, MN, USA) as primary antibodies for 45 min. Biotinylated unimmunized goat IgG (prepared in our laboratory) was included as negative control. After incubation, the cells were washed twice with washing buffer and then incubated with RPE-conjugated streptavidin (DAKO, Glostrup, Denmark) as detector, for 45 min. Cells were then washed twice before scanning by a flow cytometer (Partec, Nuremberg, Germany). Double staining of B cells from five normal subjects was performed using FITC-CD19 monoclonal antibody (clone HD37, Cytomation; DAKO, Glostrup, Denmark) and biotinylated anti-FCRL Abs to determine the frequency of FCRL expressing B cells. FITC-labelled IgG1 monoclonal antibody with irrelevant specificity (DAKO) and biotinylated unimmunized goat IgG served as negative controls to eliminate the background non-specific binding. Data analysis was performed using Flomax flow cytometry analysis software (Partec, Nuremberg, Germany).
Statistical analysis
Statistical analyses of the results were performed using Chi-Square, student’s t, and Mann–Whitney U tests, as appropriate. Analyses were conducted using the SPSS statistical package (SPSS Inc, Chicago, IL, USA). P values of less than 0.05 were considered significant.
Results and discussion
Expression profile of FCRL mRNA in ALL and normal subjects
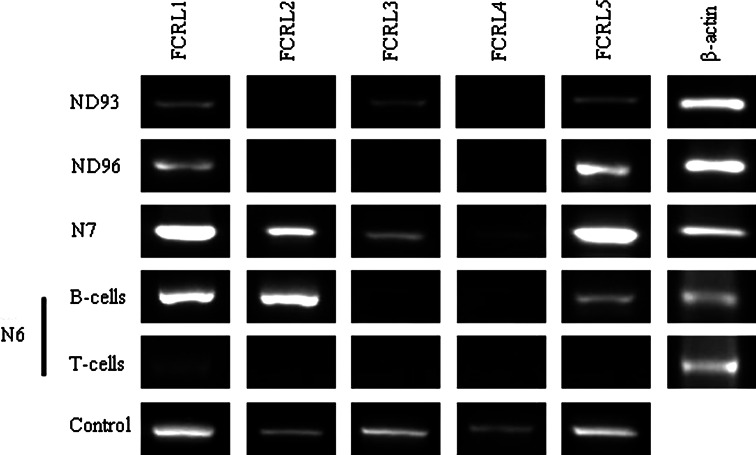
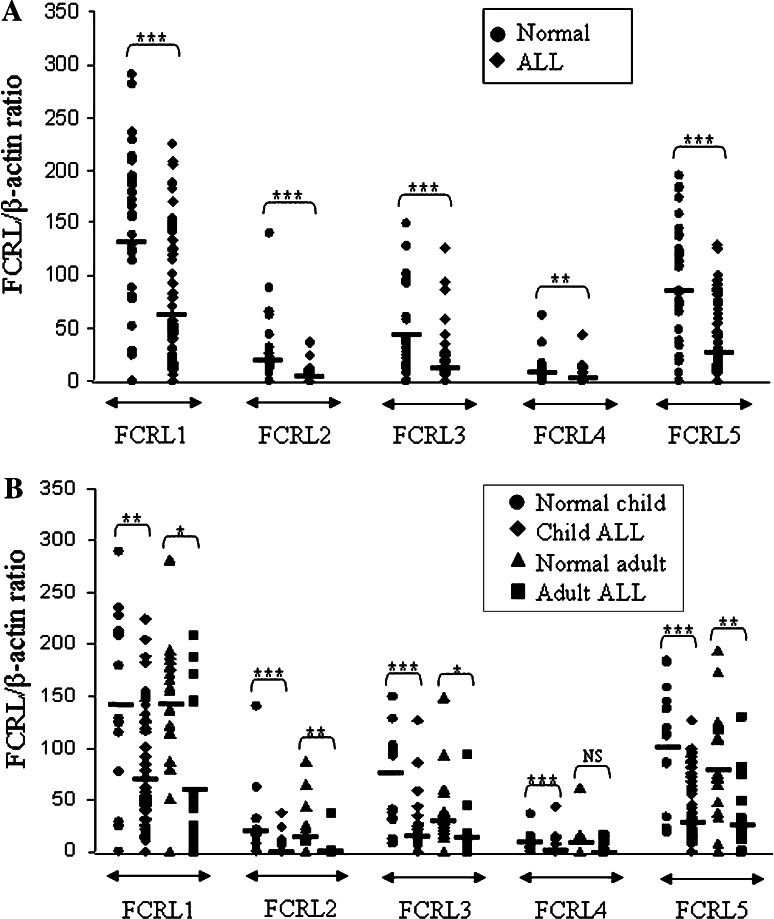
In the present study, we investigated for the first time FCRL1-5 expression profile both at mRNA and protein levels in fresh leukemic cells from patients with ALL. Using a semi-quantitative RT-PCR method relative expression of FCRL1-5 mRNA levels in BM mononuclear cells or PBMC from ALL patients (n = 73) and PBMC from normal subjects (n = 35) was determined by calculation of the ratio of FCRLs PCR amplicon band densities to that of β-actin. Representative results obtained for a number of patients are illustrated in Fig. 1. All FCRL genes were found to be significantly down-regulated in ALL patients compared to normal subjects (Fig. 2a). A similar pattern was observed both in child and adult ALL patients when compared to age-matched normal subjects, with the exception of FCRL4 expression in adult patients which was statistically not different compared to normal adult subjects (Fig. 2b). There were also no significant differences for expression of any of the FCRL1-5 genes between B-ALL (n = 54) and T-ALL (n = 16) patients. Given the exclusive constitutive expression of these molecules in B-cells, lack of significant differences between these two subtypes seems to be unexpected. FCRL1 and 5, as opposed to FCRL2, 3 and 4, were positive in most cases of either B- or T-ALL with a similar mRNA expression levels. It seems that the amplified PCR products detected for FCRL1 and 5 and to a lesser extent FCRL2, 3 and 4 in PBMC or bone marrow cells of the ALL patients, irrespective of their B- or T-lineage, are largely derived from the normal residual contaminating B-cells.
Fig. 1.
Representative RT-PCR results of FCRL1-5 genes expression in ALL patients and normal subjects. Results show FCRL1-5 expression in PBMC from 2 ALL patients (ND93 and ND 96) and one normal subject (N7) as well as purified B- and T-cells of one normal subject (N6). Control denotes FCRL positive cell lines described in “Materials and methods”. ND newly-diagnosed ALL
Fig. 2.
Distribution of FCRL1-5 mRNA levels in bone marrow mononuclear cells or PBMC from different groups of ALL patients and normal subjects. The results are expressed as the ratio of FCRL to β-actin PCR product band density in (a) all ALL patients and normal subjects and (b) different age groups of ALL patients and counterpart normal subjects. Statistically significant results are presented as: * (P values less than 0.05–0.01), ** (P values 0.01–0.001) and *** (P values less than 0.0001); NS not significant, PBMC peripheral blood mononuclear cells, horizontal bars represent mean values
There are some important methodological issues regarding our semi-quantitative RT-PCR assay. In this study representation of FCRL expression as a ratio to the housekeeping gene β-actin was intended to normalize the data and avoid variations due to differences in cDNA concentration or loading different amounts of PCR product on the electrophoresis gel. Further caution was exercised by simultaneous electrophoresis of FCRL and β-actin PCR products for each subject on the same gel. This would minimize variations due to technical shortcomings. Although real-time PCR is more demanding and preferable for quantitative measurement of FCRL mRNA expression, however, our semi-quantitative RT-PCR method has been shown to be a reliable and sensitive method for detection of a variety of tumor-associated markers [25, 28, 31]. Unfortunately, we have not been able to employ real-time PCR in this study due to some instrumental limitations.
The difference between the source of cells collected from patients and controls is also an important issue which needs clarification. The mononuclear cells isolated from BM or PB of ALL patients were predominantly immature leukemic blast B-cells (Table 1), whereas only 10–20% of the PBMC isolated from normal subjects are B-cells. Therefore, comparison of FCRL mRNA expression in these two groups of subjects may not be appropriate. Although the proper matched control for our patients’ BM samples is normal BM, ethically it has not been possible to collect BM samples, particularly from children. There are two lines of evidence which substantiate and validate the use of PB instead of BM samples for some patients in this study. First, when PB (n = 24) and BM (n = 49) ALL samples were independently and separately compared with the normal control samples, significant down-regulation of all FCRL molecules was similarly represented in both groups of patients (data not presented). The second line of evidence comes from the percent of blasts counted in all samples which shows comparable results for both groups (Table 1).
Another important issue is the age of patients and control subjects. ALL is predominantly observed in children, though adults are also affected. Despite some clinical and molecular differences reported in these two groups of patients [19, 20], detailed gene profiling studies failed to make distinction between them [21]. Nevertheless, we employed PBMC samples from both adult and child normal subjects to determine FCRL mRNA expression and compared the results with those of child and adult patients. We also compared expression levels of FCRL1-5 between the two groups of patients to find out any age-related differences. No significant differences were observed for any of the comparisons, indicating lack of association of FCRL expression with age between patients and healthy control subjects.
Expression profile of FCRL mRNA in different subtypes of ALL and isolated normal B- and T-cells
Investigation of FCRL expression in normal B-lymphocytes has shown that they are differentially expressed at various stages of differentiation. Thus, while FCRL1 is highly expressed in early and naive B-cells, FCRL2 and 3 are predominantly expressed in naive and post-germinal center cells, FCRL4 is restricted to memory B-cells and FCRL5 is broadly expressed in naive and memory B-cells as well as plasma cells [4, 6, 10]. This pattern of expression may suggest differential expression of these molecules in different subtypes of ALL.
We compared the relative expression levels of FCRL1-5 genes in different immunophenotypic subtypes of our B-ALL patients (Pro-B, Pre-B I, Pre-B II and immature/mature-B). None of the FCRL genes were found to be preferentially expressed in any of the B-ALL subtypes (Table 2).
Table 2.
Relative expression of FCRL1-5 mRNA levels in immunophenotypic subtypes of ALL and normal isolated B- and T-cells
| FCRL1 | FCRL2 | FCRL3 | FCRL4 | FCRL5 | |
|---|---|---|---|---|---|
| Pro-B (n = 6) | 45.1 (44.3) | 1.5 (3.7) | 5.9 (14.4) | 0.0 (0.0) | 32.8 (33.9) |
| Pre-B I (n = 31) | 70.8 (66.2) | 2.4 (7.9) | 15.3 (24.8) | 3.1 (8.6) | 39.5 (35) |
| Pre-B II (n = 14) | 95.3 (78.2) | 3.8 (10.1) | 18.7 (33.7) | 2.1 (5.1) | 34.1 (34.1) |
| Immature/mature-B (n = 3) | 31.6 (18.2) | 0.0 (0.0) | 2.8 (4.9) | 0.0 (0.0) | 40.6 (32.6) |
| T-ALL (n = 16) | 44.8 (38.9) | 1 (2.8) | 6.3 (10.5) | 1 (3.9) | 31 (34.7) |
| Normal B-cells (n = 6) | 170.5 (46)** | 91.2 (68)* | 11.7 (10)* | 0 | 41.5 (36)* |
| Normal T-cells (n = 6) | 7.6 (8) | 1.9 (4) | 0 | 0 | 0 |
The results represent the mean (standard deviation) ratio of FCRL PCR product band density to that of β-actin multiplied by 100. Comparison between immunophenotypic subtypes of B-ALL revealed no significant differences for any of the FCRL molecules. Significant differences between normal B- and T-cells are expressed as: * P < 0.05, ** P < 0.0001. B- and T-cells were isolated form PBMC of six normal subjects by MACS magnetic beads separation
In a recent report FCRL1 and 5, but not FCRL2, 3 or 4, were detected in Pro-B and Pre-B cells of normal bone marrow samples at protein level [29], indicating the lack of association of FCRL protein expression in ALL to the differentiation stage of the leukemic cells.
Relative mRNA expression levels of all FCRL genes were determined in isolated B-cell and T-cells enriched by magnetic beads from PBMC of 6 normal subjects. Representative results are shown in Fig. 1. Expression of FCRL molecules was mainly detectable in B-cells, with no or negligible expression in isolated T-cells (Table 2). Since constitutive expression of FCRL1-5 molecules is restricted to B-cells and other blood cells are totally negative [7, 10, 15, 22, 29, 32], with the exception of FCRL3 which is also expressed in NK and T-cells [22, 29, 32], it appears that the significant down-regulation of FCRL1, 2, 3, 4 and 5 observed in our ALL patients would have been more magnified if pure B-cells could have been isolated from the patients and normal subjects.
Expression profile of FCRL protein in leukemic cells and normal B-cells
Flow cytometric analysis of surface expression of FCRL1, FCRL2, FCRL4, and FCRL5 molecules was performed in leukemic cells from ten ALL patients (Table 3). FCRL3 specific antibody was not available for this study. While FCRL2, 4 and 5 were totally negative, only one out of 10 samples stained positive for FCRL1. Expression of FCRL protein was also studied in B-cells of five normal subjects by double staining of PBMC using FITC-CD19 MAb and biotinylated FCRL antibodies detected by RPE-streptavidin. FCRL1, 2, 4 and 5 molecules were detected at surface of B-cells from all subjects with the exception of one sample being negative for FCRL4 (Table 3).
Table 3.
Membrane expression of FCRL1, 2, 4, and 5 molecules in some ALL patients and normal subjects
| Subjects | Immunophenotype | FCRL1 | FCRL2 | FCRL4 | FCRL5 | ||||
|---|---|---|---|---|---|---|---|---|---|
| P | mRNA | P | mRNA | P | mRNA | P | mRNA | ||
| ND65-BM | Pre-B I | − | 20.8 | NI | 0 | − | 9.5 | − | 30.5 |
| ND67-BM | Pro-B | − | 57.6 | − | 0 | − | 0 | − | 37.8 |
| ND74-BM | Pre-B II | +++ | 186.6 | − | 0 | − | 0 | − | 0 |
| ND76-BM | T | − | 40.2 | − | 0 | − | 0 | − | 17.8 |
| ND86-BM | Pre-B II | − | 21.5 | − | 0 | − | 0 | − | 45.5 |
| ND93-PB | T | − | 21.7 | − | 5.3 | − | 0 | − | 20.3 |
| ND98-BM | T | − | 116.1 | − | 0 | − | 0 | − | 72.4 |
| ND99-BM | Pre-B I | − | 71.3 | − | 0 | − | 0 | − | 12.5 |
| ND100-BM | Pre-B II | − | 141.1 | − | 0 | − | 0 | − | 8.9 |
| ND107-BM | Pre-B II | − | 205.8 | − | 0 | − | 0 | − | 29.6 |
| N1 | − | ++++ | 220.2 | ++++ | 165.8 | + | NI | +++ | 29.8 |
| N2 | − | ++++ | 122.9 | ++++ | 19.1 | + | NI | ++ | 14.5 |
| N3 | − | ++++ | 157.5 | +++ | 161.4 | − | NI | + | 59.6 |
| N4 | − | ++++ | NI | ++++ | NI | ++ | NI | ++ | NI |
| N5 | − | ++++ | 230.8 | ++++ | 129.9 | ++ | NI | ++ | 107.5 |
Double staining was performed on normal PBMC using FITC-CD19 MAb and biotinylated RPE-labelled anti-FCRL antibodies. The flow cytometry results are presented as: −, 0–10%; +, 10–20%; ++, 20–40%; +++, 40–60%; ++++, >60%
ND newly diagnosed, BM bone marrow, PB peripheral blood, N normal, NI not identified, P protein
The anti-FCRL antibodies employed in our study are all polyclonal antibodies with potential cross-reactivity to other members of the FCRL family. However, as indicated by the supplier (R&D Co.), apart from the anti-FCRL1 Ab which shows minor (25%) cross-reactivity with recombinant human FCRL2 molecule, none of the other Abs cross-reacts with human FCRL proteins. The cross-reactivity of the other Abs has been stated to be less than 2–5% which is essentially negligible.
To our best of knowledge our study is the first to report FCRL1-5 expression in fresh leukemic cells of ALL at mRNA and protein levels. Expression of FCRL molecules has also been studied in a limited number of Pro- and Pre-B ALL cell lines, but all were found to be negative at mRNA and/or protein levels [7, 10, 22, 32]. There is only one published article regarding FCRL (FCRL5) expression in fresh leukemic cells from a small number (n = 8) of ALL patients [17]. In this article, expression of FCRL5 was studied at only protein level by flow cytometry, but all samples were negative. Therefore, more studies need to be performed in ALL to compare and extend our findings.
In summary, we observed down-regulation of FCRL1-5 molecules in leukemic cells from Iranian ALL patients at both mRNA and protein levels. No differences were found between any of the immunophenotypic subtypes of our B-ALL patients for any of the FCRL molecules suggesting lack of association between FCRL expression and the differentiation stage of B-ALL. Our data may also suggest unsuitability of these markers for consideration as targets for immunotherapy of ALL, as opposed to CLL and some other chronic B-cell leukemias [8, 17, 29].
Acknowledgments
This study was supported in part by grants from Tehran University of Medical Sciences and the Food and Drug Administration of the Ministry of Health and Medical Education of Iran.
Footnotes
T. Kazemi, H. Asgarian-Omran and A. Memarian have contributed equally to this study.
References
- 1.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J, Jr, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Wilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 2.Asgarian Omran H, Shabani M, Shahrestani T, Sarafnejad A, Khoshnoodi J, Vossough P, Faranoush M, Sharifian RA, Jeddi-Tehrani M, Rabbani H, Shokri F. Immunophenotypic subtyping of leukemic cells from Iranian patients with acute lymphoblastic leukaemia: association to disease outcome. Iran J Immunol. 2007;4(1):15–25. [PubMed] [Google Scholar]
- 3.Busson P, McCoy R, Sadler R, Gilligan K, Tursz T, Raab-Traub N. Consistent transcription of the Epstein-Barr virus LMP2 gene in nasopharyngeal carcinoma. J Virol. 1992;66(5):3257–3262. doi: 10.1128/jvi.66.5.3257-3262.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis RS. Fc receptor-like molecules. Annu Rev Immunol. 2007;25:525–560. doi: 10.1146/annurev.immunol.25.022106.141541. [DOI] [PubMed] [Google Scholar]
- 5.Davis RS, Dennis G, Jr, Odom MR, Gibson AW, Kimberly RP, Burrows PD, Cooper MD. Fc receptor homologs: newest members of a remarkably diverse Fc receptor gene family. Immunol Rev. 2002;190:123–136. doi: 10.1034/j.1600-065X.2002.19009.x. [DOI] [PubMed] [Google Scholar]
- 6.Davis RS, Ehrhardt GR, Leu CM, Hirano M, Cooper MD. An extended family of Fc receptor relatives. Eur J Immunol. 2005;35(3):674–680. doi: 10.1002/eji.200425886. [DOI] [PubMed] [Google Scholar]
- 7.Davis RS, Wang YH, Kubagawa H, Cooper MD. Identification of a family of Fc receptor homologs with preferential B cell expression. Proc Natl Acad Sci USA. 2001;98(17):9772–9777. doi: 10.1073/pnas.171308498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du X, Nagata S, Ise T, Stetler-Stevenson M, Pastan I. FCRL1 on chronic lymphocytic leukemia, hairy cell leukemia, and B-cell non-Hodgkin lymphoma as a target of immunotoxins. Blood. 2008;111(1):338–343. doi: 10.1182/blood-2007-07-102350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehrhardt GR, Davis RS, Hsu JT, Leu CM, Ehrhardt A, Cooper MD. The inhibitory potential of Fc receptor homolog 4 on memory B cells. Proc Natl Acad Sci USA. 2003;100(23):13489–13494. doi: 10.1073/pnas.1935944100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ehrhardt GR, Hsu JT, Gartland L, Leu CM, Zhang S, Davis RS, Cooper MD. Expression of the immunoregulatory molecule FcRH4 defines a distinctive tissue-based population of memory B cells. J Exp Med. 2005;202(6):783–791. doi: 10.1084/jem.20050879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foa R, Vitale A. Towards an integrated classification of adult acute lymphoblastic leukemia. Rev Clin Exp Hematol. 2002;6(2):181–99. doi: 10.1046/j.1468-0734.2002.00070.x. [DOI] [PubMed] [Google Scholar]
- 12.Gokbuget N, Hoelzer D. Recent approaches in acute lymphoblastic leukemia in adults. Rev Clin Exp Hematol. 2002;6(2):114–141. doi: 10.1046/j.1468-0734.2002.00068.x. [DOI] [PubMed] [Google Scholar]
- 13.Haga CL, Ehrhardt GR, Boohaker RJ, Davis RS, Cooper MD. Fc receptor-like 5 inhibits B cell activation via SHP-1 tyrosine phosphatase recruitment. Proc Natl Acad Sci USA. 2007;104(23):9770–9775. doi: 10.1073/pnas.0703354104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han X, Bueso-Ramos CE. Advances in the pathological diagnosis and biology of acute lymphoblastic leukemia. Ann Diagn Pathol. 2005;9(4):239–257. doi: 10.1016/j.anndiagpath.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 15.Hatzivassiliou G, Miller I, Takizawa J, Palanisamy N, Rao PH, Iida S, Tagawa S, Taniwaki M, Russo J, Neri A, Cattoretti G, Clynes R, Mendelsohn C, Chaganti RS, Dalla-Favera R. IRTA1 and IRTA2, novel immunoglobulin superfamily receptors expressed in B cells and involved in chromosome 1q21 abnormalities in B cell malignancy. Immunity. 2001;14(3):277–289. doi: 10.1016/S1074-7613(01)00109-1. [DOI] [PubMed] [Google Scholar]
- 16.Ise T, Maeda H, Santora K, Xiang L, Kreitman RJ, Pastan I, Nagata S. Immunoglobulin superfamily receptor translocation associated 2 protein on lymphoma cell lines and hairy cell leukemia cells detected by novel monoclonal antibodies. Clin Cancer Res. 2005;11(1):87–96. [PubMed] [Google Scholar]
- 17.Ise T, Nagata S, Kreitman RJ, Wilson WH, Wayne AS, Stetler-Stevenson M, Bishop MR, Scheinberg DA, Rassenti L, Kipps TJ, Kyle RA, Jelinek DF, Pastan I. Elevation of soluble CD307 (IRTA2/FcRH5) protein in the blood and expression on malignant cells of patients with multiple myeloma, chronic lymphocytic leukemia, and mantle cell lymphoma. Leukemia. 2007;21(1):169–174. doi: 10.1038/sj.leu.2404445. [DOI] [PubMed] [Google Scholar]
- 18.Itoyama T, Nanjungud G, Chen W, Dyomin VG, Teruya-Feldstein J, Jhanwar SC, Zelenetz AD, Chaganti RS. Molecular cytogenetic analysis of genomic instability at the 1q12–22 chromosomal site in B-cell non-Hodgkin lymphoma. Genes Chromosomes Cancer. 2002;35(4):318–328. doi: 10.1002/gcc.10120. [DOI] [PubMed] [Google Scholar]
- 19.Jabbour EJ, Faderl S, Kantarjian HM. Adult acute lymphoblastic leukemia. Mayo Clin Proc. 2005;80(11):1517–1527. doi: 10.4065/80.11.1517. [DOI] [PubMed] [Google Scholar]
- 20.Kohlmann A, Schoch C, Schnittger S, Dugas M, Hiddemann W, Kern W, Haferlach T. Pediatric acute lymphoblastic leukemia (ALL) gene expression signatures classify an independent cohort of adult ALL patients. Leukemia. 2004;18(1):63–71. doi: 10.1038/sj.leu.2403167. [DOI] [PubMed] [Google Scholar]
- 21.Kuchinskaya E, Heyman M, Grander D, Linderholm M, Soderhall S, Zaritskey A, Nordgren A, Porwit-Macdonald A, Zueva E, Pawitan Y, Corcoran M, Nordenskjold M, Blennow E. Children and adults with acute lymphoblastic leukaemia have similar gene expression profiles. Eur J Haematol. 2005;74(6):466–480. doi: 10.1111/j.1600-0609.2005.00433.x. [DOI] [PubMed] [Google Scholar]
- 22.Leu CM, Davis RS, Gartland LA, Fine WD, Cooper MD. FcRH1: an activation coreceptor on human B cells. Blood. 2005;105(3):1121–1126. doi: 10.1182/blood-2004-06-2344. [DOI] [PubMed] [Google Scholar]
- 23.Li FJ, Ding S, Pan J, Shakhmatov MA, Kashentseva E, Wu J, Li Y, Soong SJ, Chiorazzi N, Davis RS. FCRL2 expression predicts IGHV mutation status and clinical progression in chronic lymphocytic leukemia. Blood. 2008;112(1):179–187. doi: 10.1182/blood-2008-01-131359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maltais LJ, Lovering RC, Taranin AV, Colonna M, Ravetch JV, Dalla-Favera R, Burrows PD, Cooper MD, Davis RS. New nomenclature for Fc receptor-like molecules. Nat Immunol. 2006;7(5):431–432. doi: 10.1038/ni0506-431. [DOI] [PubMed] [Google Scholar]
- 25.Mikaelsson E, Danesh-Manesh AH, Luppert A, Jeddi-Tehrani M, Rezvany MR, Sharifian RA, Safaie R, Roohi A, Osterborg A, Shokri F, Mellstedt H, Rabbani H. Fibromodulin, an extracellular matrix protein: characterization of its unique gene and protein expression in B-cell chronic lymphocytic leukemia and mantle cell lymphoma. Blood. 2005;105(12):4828–4835. doi: 10.1182/blood-2004-10-3941. [DOI] [PubMed] [Google Scholar]
- 26.Nakayama Y, Weissman SM, Bothwell AL. BXMAS1 identifies a cluster of homologous genes differentially expressed in B cells. Biochem Biophys Res Commun. 2001;285(3):830–837. doi: 10.1006/bbrc.2001.5231. [DOI] [PubMed] [Google Scholar]
- 27.Offit K, Wong G, Filippa DA, Tao Y, Chaganti RS. Cytogenetic analysis of 434 consecutively ascertained specimens of non-Hodgkin’s lymphoma: clinical correlations. Blood. 1991;77(7):1508–1515. [PubMed] [Google Scholar]
- 28.Omran HA, Shabani M, Vossough P, Sharifian R, Tabrizi M, Khoshnoodi J, Jeddi-Tehrani M, Rabbani H, Shokri F. Cross-sectional monitoring of Wilms’ tumor gene 1 (WT1) expression in Iranian patients with acute lymphoblastic leukemia at diagnosis, relapse and remission. Leuk Lymphoma. 2008;49(2):281–290. doi: 10.1080/10428190701784706. [DOI] [PubMed] [Google Scholar]
- 29.Polson AG, Zheng B, Elkins K, Chang W, Du C, Dowd P, Yen L, Tan C, Hongo JA, Koeppen H, Ebens A. Expression pattern of the human FcRH/IRTA receptors in normal tissue and in B-chronic lymphocytic leukemia. Int Immunol. 2006;18(9):1363–1373. doi: 10.1093/intimm/dxl069. [DOI] [PubMed] [Google Scholar]
- 30.Rosenwald A, Wright G, Wiestner A, Chan WC, Connors JM, Campo E, Gascoyne RD, Grogan TM, Muller-Hermelink HK, Smeland EB, Chiorazzi M, Giltnane JM, Hurt EM, Zhao H, Averett L, Henrickson S, Yang L, Powell J, Wilson WH, Jaffe ES, Simon R, Klausner RD, Montserrat E, Bosch F, Greiner TC, Weisenburger DD, Sanger WG, Dave BJ, Lynch JC, Vose J, Armitage JO, Fisher RI, Miller TP, LeBlanc M, Ott G, Kvaloy S, Holte H, Delabie J, Staudt LM. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell. 2003;3(2):185–197. doi: 10.1016/S1535-6108(03)00028-X. [DOI] [PubMed] [Google Scholar]
- 31.Shabani M, Asgarian Omran H, Jeddi-Tehrani M, Vossough P, Faranoush M, Sharifian RA, Toughe GR, Kordmahin M, Khoshnoodi J, Roohi A, Tavoosi N, Mellstedt H, Rabbani H, Shokri F. Overexpression of Orphan Receptor Tyrosine Kinase Ror1 as a putative tumor-associated antigen in Iranian patients with acute lymphoblastic leukemia. Tumour Biol. 2008;28(6):318–326. doi: 10.1159/000121405. [DOI] [PubMed] [Google Scholar]
- 32.Won WJ, Foote JB, Odom MR, Pan J, Kearney JF, Davis RS. Fc receptor homolog 3 is a novel immunoregulatory marker of marginal zone and B1 B cells. J Immunol. 2006;177(10):6815–6823. doi: 10.4049/jimmunol.177.10.6815. [DOI] [PubMed] [Google Scholar]