Abstract
Effective T cell receptor (TCR) transfer until now required stable retroviral transduction. However, retroviral transduction poses the threat of irreversible genetic manipulation of autologous cells. We, therefore, used optimized RNA transfection for transient manipulation. The transfection efficiency, using EGFP RNA, was >90%. The electroporation of primary T cells, isolated from blood, with TCR-coding RNA resulted in functional cytotoxic T lymphocytes (CTLs) (>60% killing at an effector to target ratio of 20:1) with the same HLA-A2/gp100-specificity as the parental CTL clone. The TCR-transfected T cells specifically recognized peptide-pulsed T2 cells, or dendritic cells electroporated with gp100-coding RNA, in an IFNγ-secretion assay and retained this ability, even after cryopreservation, over 3 days. Most importantly, we show here for the first time that the electroporated T cells also displayed cytotoxicity, and specifically lysed peptide-loaded T2 cells and HLA-A2+/gp100+ melanoma cells over a period of at least 72 h. Peptide-titration studies showed that the lytic efficiency of the RNA-transfected T cells was similar to that of retrovirally transduced T cells, and approximated that of the parental CTL clone. Functional TCR transfer by RNA electroporation is now possible without the disadvantages of retroviral transduction, and forms a new strategy for the immunotherapy of cancer.
Keywords: TCR, RNA, Electroporation, Tumor-specific, IFNγ-production, Cytotoxicity
Introduction
Cytotoxic T lymphocytes (CTL) are believed to play a major role in the control of tumor growth, and are, therefore, of great importance in cellular strategies for immunotherapy of cancer [19]. Early attempts to adoptively transfer tumor-infiltrating lymphocytes (TIL) were unsatisfactory, because the transferred cells were often nonspecific and did not persist for long periods of time, most probably due to the fact that such TIL can have an anergic phenotype or are incapable of homing to tumor sites [10, 11]. Adoptive transfer of in vitro expanded autologous tumor-specific CTL has been shown to be effective in eradication of tumors in patients with metastatic melanoma [9, 13, 18, 20, 31]. Unfortunately, not all patients mount a detectable in vivo cytotoxic T cell response to their tumors. In fact, isolation and/or expansion of lytic tumor-specific T cells has only been possible in a fraction of patients, most likely due to the fact that, notably in tumor patients, the peripheral T cell repertoire is usually devoid of high-avidity tumor-specific CTL due to thymic selection [28] or other tolerance mechanisms [30]. In addition, these cells and in vitro generated tumor-specific T cells only have a limited life-span, and expansion of such T cells to therapeutic doses is often not feasible [2, 3, 30].
Alternatively, since CTL specificity is exclusively dictated by the T cell receptor (TCR), autologous T cells retrovirally transduced with a tumor-specific TCR were used for adoptive transfer. Reprogramming of T cells with a tumor specificity by retroviral transduction has already been shown in vitro for several antigens, e.g., MART-1 [7], MAGE-1 [29], MDM2 [27], gp100 [16, 24], tyrosinase [21] and NY-ESO-1 [33]. These autologous T cells were easily expanded to therapeutic doses. However, retroviral transduction poses the threat of irreversible genetic manipulation of autologous cells.
Therefore, we developed an optimized strategy in which the TCR α and β chain are transferred to T lymphocytes by electroporation of RNA, resulting in a transient expression of the TCR chains. A high transfection efficiency of EGFP RNA into CD8+ T cells was achieved upon step-wise development of the optimized protocol. CD8+ T cells transfected with RNA coding for the TCR α and β chains, originating from a gp100280–288/HLA-A2-specific CTL clone, produced IFNγ after stimulation with gp100280–288 peptide-loaded target cells, 4, 24 and 48 h after electroporation. This was in line with previously published observations [33, 34]. Moreover, these T cells produced IFNγ in response to gp100 RNA-electroporated DC.
Furthermore, these T cells efficiently lysed gp100280–288 peptide-loaded target cells, 1, 2, 3 and even 7 days after electroporation. Also cryopreserved TCR-RNA-electroporated T cells were able to produce IFNγ and showed cytotoxicity, which is essential for the generation of large batches of clinically feasible and effectively transfected T cells for cancer immunotherapy. Most importantly, the TCR-RNA-electroporated T cells also specifically lysed an HLA-A2+/gp100+, but not an HLA-A2−/gp100+ or an HLA-A2+/gp100− cell line. In addition, peptide-titration studies revealed that the lytic efficiency of TCR-RNA-transfected T cells was similar to that of T cells retrovirally transduced with the same cloned TCR, and approximated that of the parental CTL clone.
To our knowledge, this is the first paper describing the transfer of cytolytic capacity by TCR-RNA-transfection into T cells. Since this strategy overcomes disadvantages of retroviral transduction (i.e., the possible insertional mutagenesis), it provides a new and better way to generate tumor-specific cytolytic T lymphocytes for the immunotherapy of cancer.
Materials and methods
Cells and reagents
Peripheral blood mononuclear cells (PBMC) were prepared from whole blood of healthy donors (obtained following informed consent and approved by the institutional review board) by density centrifugation using Lymphoprep (Axis-Shield, Oslo, Norway). For the generation of dendritic cells (DC) and nonadherent fraction (NAF), PBMC were resuspended in autologous medium that consisted of RPMI 1640 (Cambrex, East Rutherford, NJ, USA) containing 1% heat-inactivated autologous plasma, 2 mM L-glutamine (BioWhittaker, Walkersville, MD, USA), 20 mg/l gentamicin (Sigma-Aldrich, St. Louis, MO, USA) and were transferred to tissue culture dishes (BD Falcon, Franklin Lakes, NJ, USA), at 30×106 cells/dish. Cells were incubated for 1–2 h at 37°C to allow for adherence, the NAF was removed and mature DC were generated from the adherent cells as described previously [22]. Matured DC were harvested and electroporated with gp100 RNA as described previously [22]. CD8+ T cells were isolated using anti-CD8 MACS beads (Miltenyi, Bergisch Gladbach, Germany) according to the manufacturer’s instructions. T cells were cultured in MLPC medium consisting of RPMI 1640, 10% human serum, 2 mM L-glutamine, 20 mg/l gentamicin, 10 mM HEPES, 1 mM sodium pyruvate (Sigma-Aldrich), 1% MEM nonessential amino acids (100x), supplemented with 20 U/ml IL-7. Twenty IU/ml IL-2 and 20 U/ml IL-7 were added on days 2 and 4. For the generation of phytohemagglutinin (PHA)-stimulated T-cell cultures, 2×106/ml NAF were cultured in T25 culture flasks in AIM-V medium (Invitrogen Gmbh, Karlsruhe, Germany) supplemented with 10% human serum, 1 μg/ml PHA (Sigma, St. Louis, MO, USA), 20 U/ml IL-7 and 20 IU/ml IL-2. IL-2 and IL-7 were added every 2 days.
The melanoma cell lines SK-MEL526 (HLA-A2+/gp100+), Colo 829 (HLA-A1+/A2−/gp100+) and NEMA (HLA-A2+/gp100−), and the TAP-deficient T×B cell hybrid T2-A1 (HLA-A1+/A2+) were cultured in R10 medium consisting of RPMI 1640, 2 mM L-glutamine, Penicillin–Streptomycin, 10% fetal calf serum, 2 mM HEPES and 2-βME. Gp100 expression of the melanoma cell lines was confirmed by intracellular staining with the mouse anti-gp100 mAb HMB45 (DAKO, Glostrup, Denmark) and donkey anti-mouse-PE (RDI, Concord, MA, USA). In short, cells were permeabilized with Cytofix/Cytoperm solution (BD Biosciences, Heidelberg, Germany), and stained with primary and secondary Abs according to the manufacturer’s instructions.
T cell receptor-transfected T cells were analyzed for TCR expression by flow cytometry using PE-conjugated anti-TCRVβ14 mAb (Beckman Coulter GmbH, Krefeld, Germany) (i.e., recognizing the gp100/A2-specific TCR), or PE-labeled gp100/HLA-A2 tetramer (Proimmune, Oxford, UK).
Peptides used in this study were: the HLA-A2-binding gp100209–217 analogue IMDQVPFSV, and gp100280–288 YLEPGPVTA.
Cloning of TCR genes
The cloning of the gp100-specific 296 TCR genes into the retroviral pBullet vector was described before [24]. The coding sequences of the TCR 296 α chain was re-cloned from the retroviral pBullet vector (kindly provided by Dr. R. Debets, ErasmusMC, Rotterdam) into the pGEM4Z-5′UTR-sig-MAGE-A3-DC.LAMP-3′UTR vector [4] (kindly provided by Prof. K. Thielemans, VUB, Brussels), by digesting both with NcoI and XhoI. The pGEM4Z-5′UTR-sig-MAGE-A3-DC.LAMP-3′UTR vector contains the 5′ and 3′ untranslated regions of the Xenopus laevis β-globin gene and a poly-A tail. At the 3′ end of the poly-A tail, unique NotI and SpeI sites are present to allow linearization of the plasmids before in vitro transcription. A bacteriophage T7 promotor allows the in vitro generation of mRNA. The coding sequences of the TCR β 296 chain was first amplified by PCR using the following primers:
- TCRB296BamHI
5′-CTC TGG ATC C BamHI AT GGG CCC CCA GCT CCT TGG CTA TG-3′
- HCB
5′CTC TCT CGA G XhoI GG ATC GCT AGC CTC TGG AAT CCT TTC TC-3′.
The PCR product was digested with BamHI and XhoI and cloned into the pGEM4Z-5′UTR-sig-MAGE-A3-DC.LAMP-3′UTR vector which was digested with BglII and XhoI.
In vitro transcription of TCR RNA
The pGEM4Z-gp100-64A vector was generated by cloning the gp100 gene into the pGEM4Z-64A vector [1] and deleting the SphI site (alternative starting codon) by site-directed mutagenesis. The pGEM4Z-enhanced GFP vector was kindly provided by Dr. I. Tcherepanova (Argos Therapeutics, Durham, NC, USA). For in vitro transcriptions, the pGEM4Z-enhanced GFP, pGEM4Z-TCRα296, pGEM4Z-TCRβ296 and pGEM4Z-gp100 were linearized with SpeI enzyme, purified with phenol/chloroform extraction and ethanol precipitation, and used as DNA templates [14]. The in vitro transcription was performed with T7 RNA polymerase (mMESSAGE mMACHINE T7 Ultra kit; Ambion, Austin, TX, USA) according to the manufacturer’s instructions. The transcribed RNA was recovered after DNaseI (Ambion) digestion on RNeasy columns (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s instructions. RNA quality was verified by agarose gel electrophoresis, RNA concentration was measured spectrophotometrically, and RNA was stored at −80°C in small aliquots.
RNA electroporation of T lymphocytes
CD8+ T cells were harvested from dishes and washed once with pure RPMI 1640 and once with OptiMEM without phenol red (Invitrogen GmbH) (all at room temperature). The cells were resuspended in OptiMEM at a concentration of 8×107/ml. RNA was transferred to a 4-mm cuvette (Peqlab, Erlangen, Germany) (150 μg/ml final concentration). A volume of 100–600 μl of cell suspension was added and incubated for 3 min before being pulsed in a Genepulser Xcell (Bio-Rad, Munich, Germany). Pulse conditions were square-wave pulse, 500 V, 5 ms. Immediately after electroporation, the cells were transferred to MLPC medium supplemented with the previously indicated concentrations of IL-7 and IL-2 where indicated.
Cryopreservation of T cells
Cryopreservation was performed as follows: cells were taken up in 20% HSA (Pharmacia & Upjohn, Peapack, NJ, USA) at a concentration of 20–50×106 cells/ml and stored for 10 min on ice. An equal volume of cryopreservation medium, i.e., 55% HSA (20%), 20% DMSO (Sigma-Aldrich) and 25% glucose (Glucosteril 40; Fresenius, Bad Homburg, Germany), was added to the cell suspension. Cells were then frozen at –1°C/min in a cryofreezing container (Nalgene, Rochester, NY, USA) to −80°C. Thawing was performed by holding cryotubes in a 37°C water-bath until detachment of the cells was visible. Cells were then poured into 10 ml of RPMI 1640, washed and added to a cell culture dish containing pre-warmed MLPC medium with 20 U IL-7/ml. Cells were rested for 0.5 h in a 37°C incubator before additional experiments.
Flowcytometry of TCR-transfected T lymphocytes
For surface stainings with anti-TCRVβ mAb, T cells were washed and thereafter suspended at 1×105 cells in 100 μl of cold FACS solution (Dulbecco’s PBS; BioWhittaker) containing 0.1% sodium azide (Sigma-Aldrich) and 0.2% HSA (Octapharma, Lachen, Switzerland) and incubated with mAb for 30 min. Cells were then washed and resuspended in 100 μl of cold FACS solution. Stained cells were analyzed for two-color immunofluorescence with a FACStar cell analyzer (BD Biosciences). Cell debris was eliminated from the analysis using a gate on forward and side light scatter. A minimum of 104 cells was analyzed for each sample. Results were analyzed using CellQuest software (BD Biosciences).
For tetramer staining, a total of 106 T cells were resuspended in 90 μl of RPMI 1640 supplemented with 5% pooled-serum, 10 mM HEPES, 1 mM sodium pyruvate, 1% MEM nonessential amino acids (100×), 2 mM L-glutamine and 20 mg/l gentamicin. Five hundred nanograms of tetramer was added. T cell phenotype was analyzed by flow cytometry using anti-CCR7 FITC and anti-CD45RA ECD (phycoerythrin-Texas Red). Cells were incubated for 20 min at 37°C, 5% CO2 and then cooled to 4°C. The cells were washed, and analyzed on a CYTOMICS FC500 from Beckman Coulter.
Induction and determination of IFN-γ production by TCR-transfected T lymphocytes
T cells electroporated with TCR RNA were cocultivated with irradiated (0.005 J/cm2) T2 cells which were loaded with an irrelevant peptide (gp100/A2209–217 analogue IMDQVPFSV) or the peptide recognized by the TCR (gp100/A2280–288 YLEPGPVTA)(both at 10 μM) for 1 h at 37°C. Fifteen thousand T cells were cocultivated with 15,000 T2 cells in a volume of 100 μl of RPMI 1640 (Cambrex) supplemented with 10% pooled-plasma (heat-inactivated and sterile-filtered plasma from healthy donors), 10 mM HEPES (Sigma-Aldrich), 1 mM sodium pyruvate (Sigma-Aldrich), 1% MEM nonessential amino acids 100× (Sigma-Aldrich), 2 mM L-glutamine (Cambrex), 20 mg/l gentamicin (Sigma-Aldrich) and 20 IU/ml IL-2. Supernatants were harvested after 16 h and IFN-γ production was determined using a commercially available ELISA kit according to the manufacturer’s protocol (DPC Biermann, Bad Nauheim, Germany).
Cytotoxicity assay
Cytotoxicity was tested in standard 4–6 h 51Cr-release assays. In short, T2 target cells were labeled with 100 μCi of Na512CrO4/106 cells for 1 h at 37°C/5% CO2, washed, loaded with peptides for 1 h at 37°C/5% CO2 and washed again before cocultivation with effector T cells. Peptides were loaded at a concentration of 10 μM, or as indicated. As alternative target the melanoma cell lines SK-MEL562, Colo829 and NEMA were used. Target cells were added to 96-well plates at 1,000 cells/well. Effector cells, i.e., TCR-transfected T cells, were added at an E:T ratio of 60:1, 20:1, 7:1 and 3:1. Percentage cytolysis, i.e., 51Cr release, was calculated as follows: [(measured release − background release)/(maximum release − background release)×100%].
Results
T cells are efficiently transfected with RNA
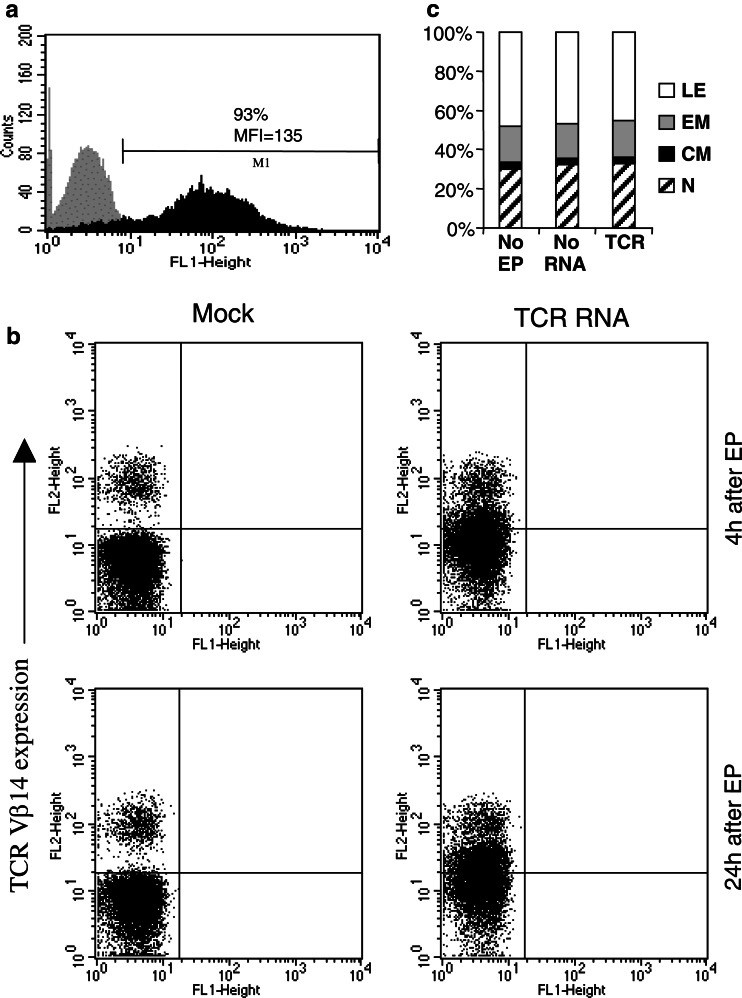
Optimized and reproducible transfection of CD8+ T cells was achieved by a step-wise development of the electroporation protocol, using EGFP RNA as a model. CD8+ T cells of several donors were transfected, according to this optimized protocol, with RNA coding for EGFP or the α and β chains of the TCR originating from CTL clone specific for HLA-A2-presented gp100 peptide (296 CTL clone). Expression levels of EGFP and TCR chains was determined by flow cytometry. Approximately 93% of the electroporated T cells expressed EGFP (Fig. 1a) (MFI=135) 4 h after electroporation, pointing to a very high transfection efficiency of T cells. A low, but significant TCR β chain expression of the gp100-specific TCR was detected with an anti-TCRVβ14 mAb on the cell membrane 4 h (P=0.0027) and 24 h (P=0.0025) after electroporation (Fig. 1b). No gp100/A2 tetramer binding was detected when TCR-transfected T cells were stained (data not shown). Mock-transfected T cells did not show any expression of EGFP and only endogenous expression of the TCR β chain (Fig. 1a, b). In addition, the T cell phenotype of nonelectroporated, mock-electroporated and TCR-RNA-electroporated T cells was determined by FACS staining for CCR7 and CD45RA. As shown in Fig. 1c, there was no influence of electroporation on the phenotype of bulk electroporated T cells compared to nonelectroporated T cells.
Fig. 1.
EGFP and TCR expression of RNA-transfected CD8+ T cells. a CD8+ T cells were electroporated with EGFP RNA and EGFP expression in these cells was determined by FACS analysis 4 h after electroporation (black histogram). CD8+ T cells electroporated without RNA served as negative control (gray histogram). b CD8+ T cells were electroporated with TCR α and β chain RNA (TCR RNA) and TCR Vβ14 surface expression on these cells was determined by FACS analysis 4 and 24 h after electroporation (EP). CD8+ T cells electroporated without RNA served as negative control (Mock). c The influence of electroporation and TCR expression on T-cell phenotype was examined by CCR7 and CD45RA staining 24 h after electroporation. Assignment of T cell phenotype was as follows: lytic effectors (LE): CD45RA+/CCR7−, effector memory (EM): CD45RA−/CCR7−, central memory (CM): CD45RA−/CCR7+, naïve (N): CD45RA+/CCR7+. All data are representative for three standardized independent experiments
Antigen-positive target cells specifically stimulate TCR-transfected T cells to produce IFNγ
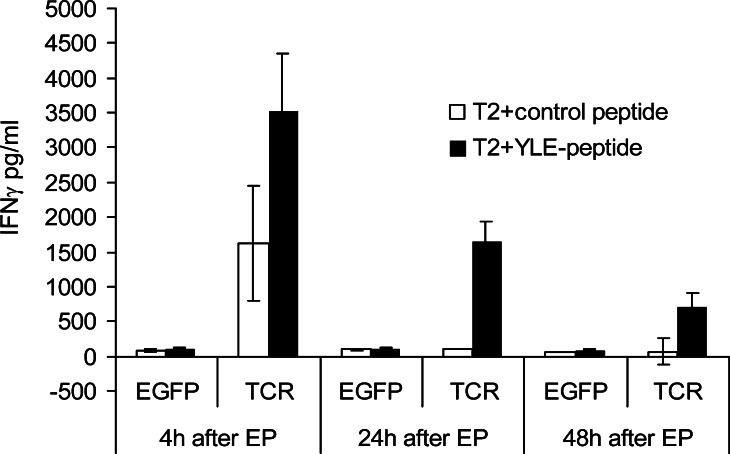
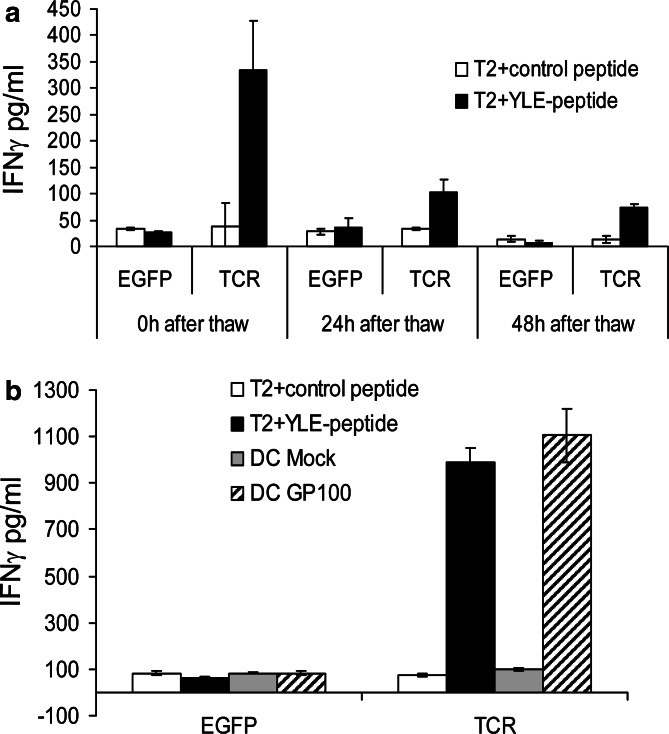
Although the measured TCR expression was low, CD8+ T cells transfected with TCR RNA were tested for their cytokine production capacity in response to target cells loaded with gp100280–288 peptide at 4, 24 and 48 h after electroporation. As shown in Fig. 2, only T cells transfected with RNA coding for the gp100/A2 specific TCR responded to T2 cells loaded with the gp100280–288 peptide with IFNγ production, while T cells electroporated with EGFP RNA were not able to produce IFNγ. Furthermore, T2 cells loaded with a control peptide (i.e., gp100209–217 analogue) were not able to induce IFNγ production by RNA-transfected T cells (Fig. 2). Even at 48 h after electroporation, there was a clear specific IFNγ production by T cells transfected with RNA coding for the gp100/A2-specific TCR (Fig. 2). Next, we tested whether TCR-transfected T cells could by cryopreserved without loss of IFNγ production capacity. Figure 3a shows that the T cells, frozen 4 h after transfection with RNA coding for the gp100/A2-specific TCR, still produced IFNγ when they were stimulated with gp100280–288-peptide-loaded T2 cells directly after thawing. Moreover, IFNγ production by TCR-transfected T cells was determined after incubation with RNA-electroporated DC. Only DC electroporated with gp100 RNA, but not mock-electroporated DC, were able to stimulate TCR-transfected T cells to produce IFNγ (Fig. 3b).
Fig. 2.
TCR-RNA-transfected T cells specifically produce IFNγ after stimulation with peptide-loaded target cells. CD8+ T cells were electroporated with EGFP RNA (EGFP) or RNA coding for the TCR α and β chain (TCR) and were used as effector cells in IFNγ-production assays, 4, 24 and 48 h after electroporation (EP). Irradiated T2 cells either loaded with a control peptide (white bars) or gp100280–288 peptide (YLE-peptide, black bars) were used as stimulator cells and IFNγ production was measured in supernatants in an ELISA and are expressed in pg/ml. Average values of triplicates ± SD are shown. The effector to stimulator cell ratio was 1:1. Data of one (out of three) representative T cell donor are shown
Fig. 3.
TCR-RNA-transfected T cells can be cryopreserved without loss of IFNγ production capacity. CD8+ T cells were electroporated with EGFP RNA (EGFP) or RNA coding for the TCR α and β chain (TCR) and were cryopreserved 4 h after electroporation. These T cells were used as effector cells in IFNγ-production assays, 0 h (a and b), 24 h (a) and 48 h (a) after thawing. Irradiated T2 cells either loaded with a control peptide (white bars) or gp100280–288 peptide (YLE-peptide, black bars) (a and b) and mock-electroporated DC (Mock, grey bars) or DC electroporated with gp100 RNA (GP100, diagonally striped bars) (b) were used as stimulator cells and IFNγ production was measured in supernatants in an ELISA and are expressed in pg/ml. Average values of triplicates ± SD are shown. The effector to stimulator cell ratio was 1:1. Data of one (out of three) representative T cell donor are shown
Peptide-loaded targets and melanoma cells are specifically lysed by TCR-transfected T cells
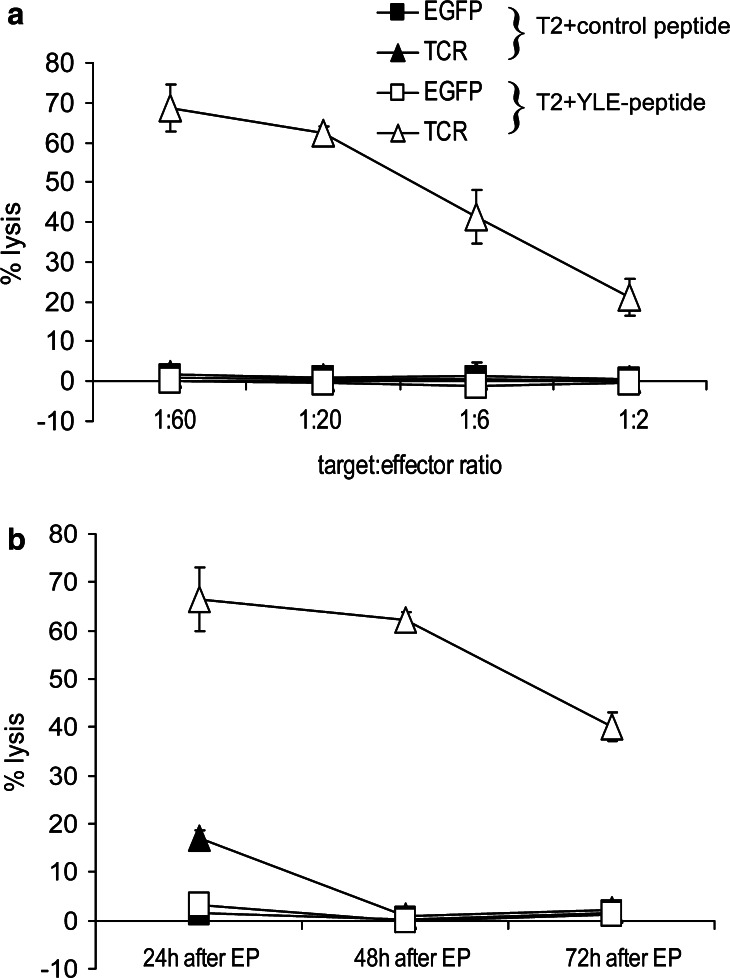
CD8+ T cells transfected with TCR RNA were tested for their cytolytic capacity on target cells loaded with gp100280–288 peptide at 24, 48 and 72 h after electroporation. As shown in Fig. 4a (representative figure at the 48 h time-point), only T2 cells loaded with the gp100280–288 peptide were lysed by TCR-RNA-transfected T cells. T2 cells loaded with a control peptide were not lysed and T cells transfected with EGFP RNA did not lyse any of the targets (Fig. 4a). A time-course of cytolysis by RNA-transfected T cells was performed (Fig. 4b). A specific lysis of T2 cells loaded with the gp100280–288 peptide at an target to effector ratio of 1:20 was observed at all time-points (Fig. 4b). At all other measured taget to effector ratios a specific lysis was seen (data not shown). Furthermore, at the highest target to effector ratio (i.e., 1:60) a specific lysis was still observed 1 week after electroporation of the T cells (data not shown), pointing to the longevity of specific lysis of TCR-RNA-electroporated T cells. Moreover, the cytolytic capacity of the TCR-RNA-electroporated T cells was preserved after cryopreservation and T cells that were PHA-stimulated and expanded before electroporation, were able to produce IFNγ in response to peptide-loaded target cells and also lysed these cells (data not shown).
Fig. 4.
TCR-RNA-transfected T cells specifically lyse peptide-loaded target cells. CD8+ T cells were electroporated with EGFP RNA (EGFP, squares) or RNA coding for the TCR α and β chain (TCR, triangles) and were used as effector cells in standard 4 h cytotoxicity assays, 24, 48 and 72 h after electroporation (EP). Irradiated T2 cells either loaded with a control peptide (closed symbols) or gp100280–288 peptide (YLE-peptide, open symbols) were used as target cells and % lysis was calculated (see Materials and methods for more details). The target to effector cell ratio was 1:60, 1:20, 1:6 and 1:2 (a, 48 h time-point is shown), or 1:20 (b, time-course is shown). Average values of triplicates ± SD are shown. Data of one (out of three) representative T cell donor are shown.
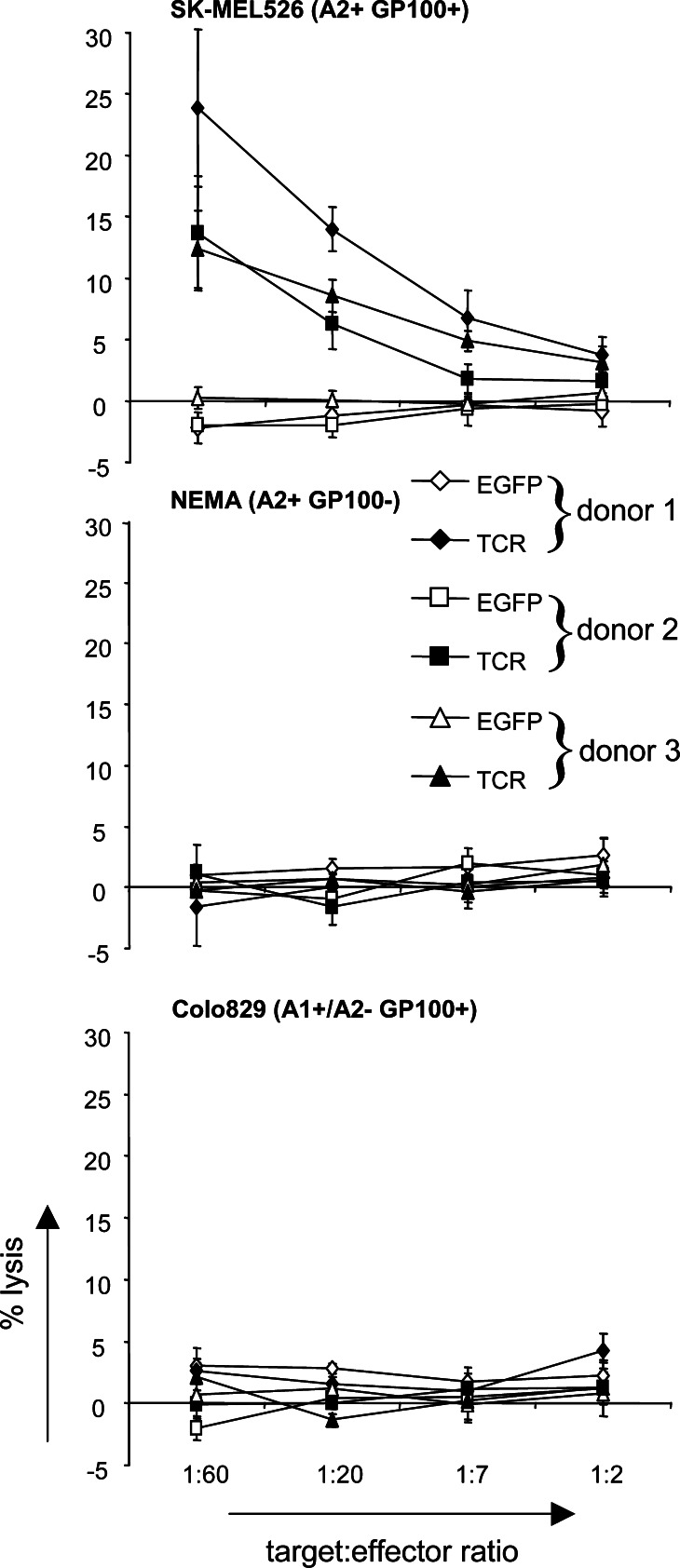
More importantly, TCR-RNA-electroporated T cells (4 h after electroporation) were also able to specifically recognize and lyse tumor cells that are gp100+ and HLA-A2+ (Fig. 5, SK-MEL526). Tumor cell lines that were gp100− but HLA-A2+ (NEMA), or gp100+ but HLA-A2− (Colo829) were not recognized by the TCR-RNA-electroporated T cells (Fig. 5). EGFP RNA-electroporated T cells did not lyse any of the target cells (Fig. 5).
Fig. 5.
TCR-RNA-transfected T cells specifically lyse a melanoma cell line. CD8+ T cells of 3 donors were electroporated with EGFP RNA (EGFP, open symbols) or RNA coding for the TCR α and β chain (TCR, closed symbols) and were used as effector cells in standard 4 h cytotoxicity assays 24 h after electroporation. The melanoma cell lines SK-MEL526 (HLA-A2+/gp100+), NEMA (HLA-A2+/gp100−) and Colo829 (HLA-A1+/A2−/gp100+) were used as target cells and percent lysis was calculated. Average values of triplicates ± SD are shown. The target to effector cell ratio was 1:60, 1:20, 1:7 and 1:2
The cytolytic efficiency of TCR-RNA-transfected T cells is similar to that of retrovirally transduced T cells and approximates that of the parental CTL clone
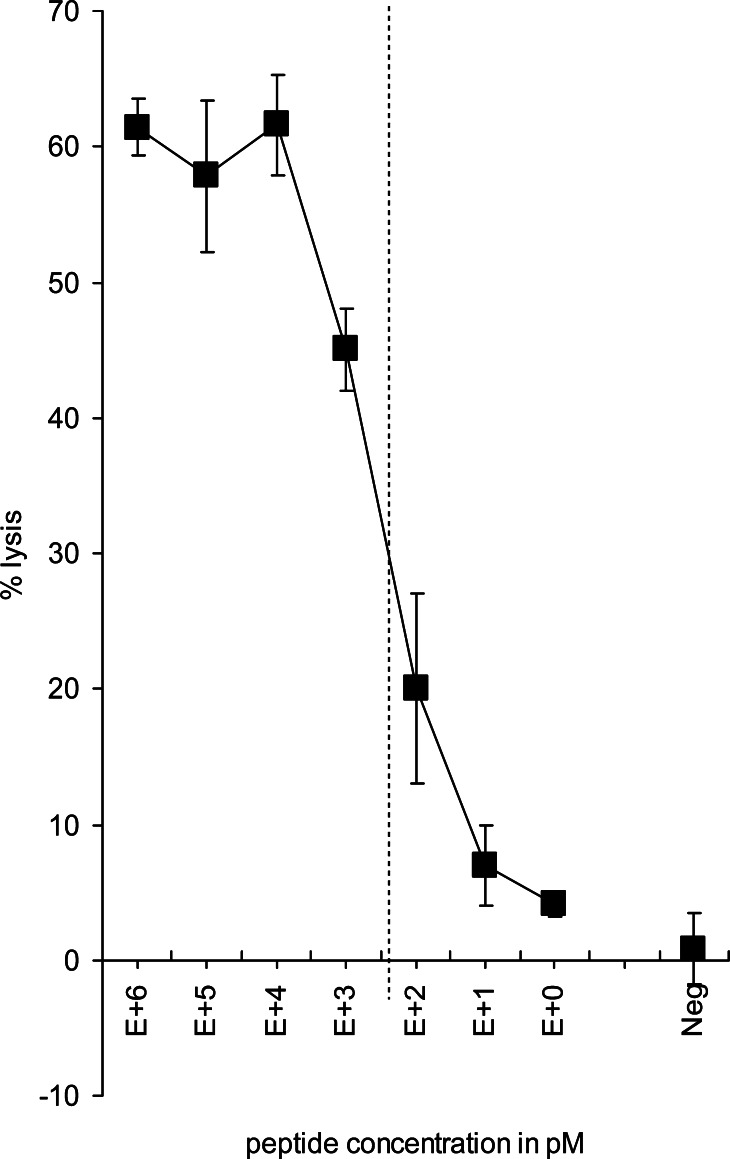
From previous published work, we know that the cytolytic efficiency of the parental 296 CTL clone was preserved following retroviral transduction of TCR genes into T cells [24]. To test the avidity of the TCR-RNA-transfected T cells, we performed a cytotoxicity assay (24 h after electroporation) in which we titrated the gp100280–288 peptide on T2 target cells (Fig. 6). The peptide concentration corresponding to 50% of the maximum lysis (i.e., ED50) of the TCR-RNA-transfected T cells ranged from 300 to 1,000 pM in three independent experiments (Fig. 6). This is in the same range as the cytolytic efficiency of the retrovirally transduced T cells (ED50=300 pM). Both approximate the cytolytic efficiency of the parental 296 CTL clone, which has an ED50 of 50 pM [24]. EGFP RNA-transfected T cells did not lyse T2 cells loaded with the gp100280–288 peptide (10 μM) (Fig. 6) and T2 cells loaded without peptide were not lysed by the TCR-RNA-transfected T cells (data not shown). Taken together, the TCR-RNA-transfected T cells lyse targets with high avidity, similar to retrovirally transduced T cells.
Fig. 6.
TCR-RNA-transfected T cells have a similar cytolytic capacity as retrovirally transduced T cells, which approximates cytolytic efficiency of the parental CTL clone. CD8+ T cells were electroporated with EGFP RNA (Neg) or RNA coding for the TCR α and β chain and were used as effector cells in cytotoxicity assays 24 h after electroporation. Irradiated T2 cells loaded with different concentrations of gp100280–288 peptide (as indicated) were used as target cells and percent lysis was calculated. The target to effector cell ratio was 1:15. The peptide concentration corresponding to 50% of the maximum lysis (ED50), used to measure the cytolytic efficiency, is indicated by the dotted line. Average values of triplicates ± SD are shown. Data of one (out of five) representative T cell donor is shown
Discussion
This paper reports on the functional characterization of CD8+ T cells that were electroporated with RNA coding for the TCR α and β chain originating from an HLA-A2/gp100280–288-specific CTL clone [24] with the aim to generate lytic effector function. Functionality of these TCR-transfected T cells was tested along several lines: (1) specific IFNγ production 4, 24 and 48 h after electroporation in response to stimulation with peptide-loaded target cells (Fig. 2), (2) specific IFNγ production after cryopreservation (i.e., 0, 24 and 48 h after thawing) in response to stimulation with peptide-loaded target cells (Fig. 3a), (3) specific IFNγ production in response to stimulation with gp100-RNA-electroporated dendritic cells (Fig. 3b), (4) specific cytolysis of peptide-loaded target cells 24, 48 and 72 h (Fig. 4) and 1 week (data not shown) after electroporation, (5) specific cytolysis of a HLA-A2+/gp100+ melanoma cell line (Fig. 5) and (6) cytolytic efficiency using peptide-loaded target cells (Fig. 6). The observed IFNγ production by TCR-RNA-transfected T cells was in line with data of Zhao et al. [33, 34]. However, to our knowledge, this is the first paper to describe transfer of cytolytic function by TCR-coding RNA transfection of T cells.
The expression level of the TCR on the T cell membrane was low, but present when determined with anti-TCRVβ mAb. No binding of HLA-A2/gp100280–288 tetramers was seen, while transfection efficiency of EGFP RNA was >90% (Fig. 1a, b). Zhao and co-workers recently published similar, high transfection efficiency with GFP RNA, but also a transfection efficiency of approximately 60% with TCR RNA determined with anti-TCRVβ mAb [33, 34]. However, this group electroporated stimulated PBL instead of purified T cells [33, 34]. Our transfection efficiency of TCR RNA did not improve when we electroporated stimulated PBL (data not shown). Nonetheless, our nonactivated TCR-RNA-electroporated CD8+ T cells were functional in both IFNγ release and cytotoxicity assays. We observed a minor shift of the peak in flow cytometry when we used anti-TCRVβ mAb (Fig. 1b). It might, therefore, be that the receptor density on the cell membrane of our TCR-RNA-electroporated T cells is low. This would also explain why we did not observe tetramer binding, since tetramers need to be bound by several TCRs at the same time to adhere firmly to the T cell membrane. If the TCR molecules are too far apart, only monovalent binding of the tetramer is possible, which results into low avidity [17]. Since a large portion of the electroporated T cells has already a lytic phenotype (Fig. 1c), probably a low number of TCRs—as well known for T cell clones—is sufficient to trigger them to lyse targets and produce IFNγ.
In the experiments described in this paper, CD8+ T cells were purified using magnetic selection, and were then electroporated directly with RNA, without prior stimulation. These cells were clearly functional, already 4 h after electroporation. This is in sharp contrast to previous publications. Smits et al. [26] showed that efficient transfection of EGFP RNA into T cells was only possible after stimulation of total PBMC with PHA for 3 days. This stimulation resulted in 50% EGFP positive CD4+ or CD8+ T cells. Stimulation of PBMC with an EBV-transformed autologous B-cell line for 3 days resulted in an transfection efficiency of approximately 5% in both CD4+ and CD8+ T cells [26]. Zhao et al. used PBL that were stimulated with OKT3 Ab and IL-2 for 3 days before electroporation [33], but very recently also unstimulated PBL [34].
The lower transfection efficiency of stimulated T cells and absence of EGFP positive T cells, when they were not stimulated, in the work of Smits and coworkers, in comparison to the data presented in this paper, might be caused by the different electroporation conditions used. Furthermore, the lower TCR-RNA-transfection efficiency presented in this paper, compared to data of Zhao and coworkers, might be due to differences in the TCR RNAs and their translation efficacy. A major difference in transfection efficiency is rather unlikely, given the identical EGFP expression of >90% in both cases. On the other hand, it might be that the magnetic selection of our CD8+ T cells, stimulated these cells (i.e., we did not work with “really” unstimulated cells), making them more accessible for RNA transfection 24 h after this selection. Furthermore, the introduction of mRNA into T cells might also activate them via TLR7/8 triggering [25].
Nonetheless, we have also tested PHA/IL-2/IL-7-stimulated T cells, which were magnetically selected for CD8 after 3 days of stimulation and then RNA electroporated, in IFNγ secretion and cytotoxicity assays. We did not observe an improvement of EGFP or TCR expression in these cells. However, these cells were as good as unstimulated TCR-transfected T cells in production of IFNγ and cytolysis in response to peptide-loaded target cells (data not shown). More importantly, by PHA/IL-2/IL-7 stimulation we could expand the number of CD8+ T cells, which might be crucial for the generation of large numbers of TCR-RNA-electroporated T cells for immunotherapy of cancer. However, retroviral transduction cannot be done with resting T cells [30]. With the method described in this paper, unstimulated T cells can also be transfected by electroporation.
Also of eminent importance to be able to use the TCR-transfected CD8+ T cells in immunotherapy of cancer, is that the specific lysis of target cells is stable over several days. As measured in cytotoxicity time-courses, the T cells were still very lytic 3 days after electroporation in all measured target to effector ratios and lytic activity could even be detected 1 week after electroporation in the highest target to effector ratio (Fig. 4 and data not shown), pointing to the longevity of specific lysis. Moreover, the cytolytic capacity of the TCR-RNA-electroporated T cells was preserved after cryopreservation, making it possible to generate several large batches of T cells for repetitive treatment at once. Furthermore, we have proved that T cells transfected with TCR RNA have a similar cytolytic efficiency as retrovirally transduced T cells and that it approximates the cytolytic efficiency of the parental CTL clone (Fig. 6 and [24]), making TCR-RNA-transfected CD8+ T cells a practicable alternative for adoptive transfer of expanded tumor-specific CTL clones.
Most importantly, TCR-RNA-transfected CD8+ T cells have several advantages over immunotherapy with retrovirally transduced T cells. The largest drawback of using retroviral transduction is that the provirus can integrate at random in the genome of the transduced cells. Thus, it can also integrate in genes involved in cell cycle control and subsequently disturb cell growth (i.e., insertional mutagenesis). Data of a genetherapeutic clinical trial in severe combined immunodeficiency (SCID), in which autologous hematopoietic stem cells were retrovirally transduced with a vector containing a gene encoding the common γ chain, which is defective in SCID patients, showed that the provirus integrated in the LMO-2 oncogene, causing leukaemia-like symptoms [6, 12, 15]. The risk of malignant transformation in mature T lymphocytes is probably substantially lower [5, 30], but cannot be excluded. The problem of possible insertional mutagenesis is completely absent when RNA is electroporated in cells and for applications where only transient expression of certain molecules is necessary, RNA electroporation is a good alternative. Since RNA is only transiently present in the transfected cell and is not integrated into the genome, this procedure cannot be scaled under gene therapy. Therefore, our TCR-RNA-transfection of T cells is much safer, considering mutagenesis, than retroviral transduction of TCR genes into T cells.
Another important aspect is that, due to the safety and simplicity of the RNA transfection (both from a technical and a regulatory point of view), a rapid screening of candidate TCRs for therapeutic usefulness is possible. To this end, we are preparing a clinical trial of intratumoral injection of TCR-transfected T cells. Although long-term expression is not achieved by transient electroporation, we are convinced that the use of TCR-RNA-transfected T cells in a clinical trial is feasible. Transient expression of the TCR may even have advantages considering the safety of adoptively transferred T cells (see below). It is in principle possible to electroporate large numbers of T cells, however, this might not be necessary when T cells are injected in the tumor. Few T cells can already cause destruction of parts of the tumor, which subsequently may cause epitope spreading by effective presentation of antigens by antigen presenting cells. One can also think of a combination-therapy, first injecting TCR-transfected T cells to induce epitope spreading and then injecting dendritic cells pre-loaded with antigen. In these cases, many T cells and/or long-term expression of the specific TCR may not be needed.
In most studies, in which TCR genes were retrovirally transduced into T cells, full-length TCR α and β chain genes were used to retarget these T cells. There is a potential hazard in using full-length TCR chain genes, because in theory, the introduced chains can pair with the endogenous TCR chains [8, 30, 32]. This alternative pairing can lead to unpredictable specificities, which may be autoreactive. This formation of self-reactive TCRs is a recognized concern for gene therapy regulatory committees. That alternative pairing takes place was shown in studies in which full-length TCR chains specific for gp100 or MDM-2 were retrovirally transferred to T cells. Only a fraction of the T cells expressing the introduced TCR β chain (i.e., 50–60% and 30–50%, respectively) were able to bind the respective MHC/peptide tetramer [24, 27].
One solution for the alternative-pairing problem is to introduce single or two chain modified TCR-based receptors, that are structurally different from full-length TCRs, resulting in exclusive pairing between the introduced TCR chains [8]. Such receptors specific for several melanoma antigens, e.g., MAGE-1 [29], gp100 [23], were already functionally introduced in T cells. However, careful comparison of full-length and modified single chain TCR specific for an OVA peptide showed that single chain TCR-transduced T cells were less efficient in response to stimulation with OVA peptide-pulsed target cells (i.e., especially when low concentrations of peptide were used) and natively expressing target cells as compared to full-length TCR-transduced T cells [32]. Therefore, to generate the most effective T cells, it would be best to stay as close as possible to nature and use full-length TCR chains for TCR transfer.
Electroporation of RNA coding for the full-length TCR α and β can also be used as alternative to overcome long-term problems with autoreactivity caused by pairing of introduced and endogenous TCR chains. Although we cannot exclude that such alternative pairing takes place in this strategy, the possibly generated autoreactive T cells will loose this autoreactivity after some time, because the introduced TCR α and β chain are only transiently expressed. When the introduced TCR α and β chain expression is diminished, only normal autologous T cells are left. This is not the case when full-length TCR chains are introduced by stable retroviral transduction, which will never loose the introduced TCR chains, causing a constantly present autoreactivity in the patient.
Apart from these advantages of TCR-RNA-transfected T cells over retrovirally transduced ones for the use in adoptive transfer, we believe that TCR-RNA-transfected T cells will also evolve in widely used “reagents” and substitutes for reportedly unstable human T cell clones, to detect and monitor specific MHC/peptide complexes on APC and target cells in vitro.
Taken together, we have shown that, by electroporation of RNA coding for full-length TCR α and β chains, into bulk CD8+ T cells, fully functional cytotoxic T cells were generated. They proved to be highly efficient and highly specific in IFNγ-release and cytotoxicity assays, over days. This new transfer method represents a safer and easier alternative to retroviral transduction and holds great promise for the adoptive immunotherapy of cancer.
Acknowledgements
The retroviral pBullet vectors containing genes coding for the TCR 296 α and β chain were kindly provided by Dr. R. Debets (ErasmusMC, Rotterdam, The Netherlands). The pGEM4Z-enhanced GFP vector was kindly provided by Dr. I. Tcherepanova (Argos Therapeutics, Durham, NC, USA). The pGEM4Z-5′UTR-sig-MAGE-A3-DC.LAMP-3′UTR vector was kindly provided by Prof. K. Thielemans (VUB, Brussels, Belgium). This study was supported by the DFG—German Research Foundation (Collaborative Research Centre SFB643, Project C1), the Interdisciplinary Clinical research Centre (IZKF project B5) of the University Hospital Erlangen and the European Union (DCVACC, contract no.: 503037).
Footnotes
Niels Schaft and Jan Dörrie contributed equally
References
- 1.Boczkowski D, Nair SK, Nam JH, Lyerly HK, Gilboa E. Induction of tumor immunity and cytotoxic T lymphocyte responses using dendritic cells transfected with messenger RNA amplified from tumor cells. Cancer Res. 2000;4:1028–1034. [PubMed] [Google Scholar]
- 2.Bolhuis RL, Gratama JW. Genetic re-targeting of T lymphocyte specificity. Gene Ther. 1998;9:1153–1155. doi: 10.1038/sj.gt.3300697. [DOI] [PubMed] [Google Scholar]
- 3.Bolhuis RL, Willemsen RA, Gratama JW. Clinical applications of redirected cytotoxicity. In: Sitkovsky MV, Henkart PA, editors. Cytotoxic cells. Philadelphia, PA: Lippincott, Williams & Wilkins; 2000. [Google Scholar]
- 4.Bonehill A, Heirman C, Tuyaerts S, Michiels A, Breckpot K, Brasseur F, Zhang Y, Van Der Bruggen P, Thielemans K. Messenger RNA-electroporated dendritic cells presenting MAGE-A3 simultaneously in HLA class I and class II molecules. J Immunol. 2004;11:6649–6657. doi: 10.4049/jimmunol.172.11.6649. [DOI] [PubMed] [Google Scholar]
- 5.Bonini C, Grez M, Traversari C, Ciceri F, Marktel S, Ferrari G, Dinauer M, Sadat M, Aiuti A, Deola S, Radrizzani M, Hagenbeek A, Apperley J, Ebeling S, Martens A, Kolb HJ, Weber M, Lotti F, Grande A, Weissinger E, Bueren JA, Lamana M, Falkenburg JH, Heemskerk MH, Austin T, Kornblau S, Marini F, Benati C, Magnani Z, Cazzaniga S, Toma S, Gallo-Stampino C, Introna M, Slavin S, Greenberg PD, Bregni M, Mavilio F, Bordignon C. Safety of retroviral gene marking with a truncated NGF receptor. Nat Med. 2003;4:367–369. doi: 10.1038/nm0403-367. [DOI] [PubMed] [Google Scholar]
- 6.Buckley RH. Gene therapy for SCID—a complication after remarkable progress. Lancet. 2002;9341:1185–1186. doi: 10.1016/S0140-6736(02)11290-6. [DOI] [PubMed] [Google Scholar]
- 7.Clay TM, Custer MC, Sachs J, Hwu P, Rosenberg SA, Nishimura MI. Efficient transfer of a tumor antigen-reactive TCR to human peripheral blood lymphocytes confers anti-tumor reactivity. J Immunol. 1999;1:507–513. [PubMed] [Google Scholar]
- 8.Debets R, Willemsen R, Bolhuis R. Adoptive transfer of T-cell immunity: gene transfer with MHC-restricted receptors. Trends Immunol. 2002;9:435–436. doi: 10.1016/S1471-4906(02)02290-1. [DOI] [PubMed] [Google Scholar]
- 9.Dudley ME, Wunderlich JR, Robbins PF, Yang JC, Hwu P, Schwartzentruber DJ, Topalian SL, Sherry R, Restifo NP, Hubicki AM, Robinson MR, Raffeld M, Duray P, Seipp CA, Rogers-Freezer L, Morton KE, Mavroukakis SA, White DE, Rosenberg SA. Cancer regression and autoimmunity in patients after clonal repopulation with antitumor lymphocytes. Science. 2002;5594:850–854. doi: 10.1126/science.1076514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Economou JS, Belldegrun AS, Glaspy J, Toloza EM, Figlin R, Hobbs J, Meldon N, Kaboo R, Tso CL, Miller A, Lau R, McBride W, Moen RC. In vivo trafficking of adoptively transferred interleukin-2 expanded tumor-infiltrating lymphocytes and peripheral blood lymphocytes. Results of a double gene marking trial. J Clin Invest. 1996;2:515–521. doi: 10.1172/JCI118443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Figlin RA, Thompson JA, Bukowski RM, Vogelzang NJ, Novick AC, Lange P, Steinberg GD, Belldegrun AS. Multicenter, randomized, phase III trial of CD8(+) tumor-infiltrating lymphocytes in combination with recombinant interleukin-2 in metastatic renal cell carcinoma. J Clin Oncol. 1999;8:2521–2529. doi: 10.1200/JCO.1999.17.8.2521. [DOI] [PubMed] [Google Scholar]
- 12.Hacein-Bey-Abina S, Le Deist F, Carlier F, Bouneaud C, Hue C, De Villartay JP, Thrasher AJ, Wulffraat N, Sorensen R, Dupuis-Girod S, Fischer A, Davies EG, Kuis W, Leiva L, Cavazzana-Calvo M. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N Engl J Med. 2002;16:1185–1193. doi: 10.1056/NEJMoa012616. [DOI] [PubMed] [Google Scholar]
- 13.Hanson HL, Donermeyer DL, Ikeda H, White JM, Shankaran V, Old LJ, Shiku H, Schreiber RD, Allen PM. Eradication of established tumors by CD8+ T cell adoptive immunotherapy. Immunity. 2000;2:265–276. doi: 10.1016/S1074-7613(00)00026-1. [DOI] [PubMed] [Google Scholar]
- 14.Heiser A, Dahm P, Yancey DR, Maurice MA, Boczkowski D, Nair SK, Gilboa E, Vieweg J. Human dendritic cells transfected with RNA encoding prostate-specific antigen stimulate prostate-specific CTL responses in vitro. J Immunol. 2000;10:5508–5514. doi: 10.4049/jimmunol.164.10.5508. [DOI] [PubMed] [Google Scholar]
- 15.Marshall E. Clinical research. Gene therapy a suspect in leukemia-like disease. Science. 2002;5591:34–35. doi: 10.1126/science.298.5591.34. [DOI] [PubMed] [Google Scholar]
- 16.Morgan RA, Dudley ME, Yu YY, Zheng Z, Robbins PF, Theoret MR, Wunderlich JR, Hughes MS, Restifo NP, Rosenberg SA. High efficiency TCR gene transfer into primary human lymphocytes affords avid recognition of melanoma tumor antigen glycoprotein 100 and does not alter the recognition of autologous melanoma antigens. J Immunol. 2003;6:3287–3295. doi: 10.4049/jimmunol.171.6.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogg GS, McMichael AJ. HLA-peptide tetrameric complexes. Curr Opin Immunol. 1998;4:393–396. doi: 10.1016/S0952-7915(98)80110-6. [DOI] [PubMed] [Google Scholar]
- 18.Parmiani G, Castelli C, Rivoltini L, Casati C, Tully GA, Novellino L, Patuzzo A, Tosi D, Anichini A, Santinami M. Immunotherapy of melanoma. Semin Cancer Biol. 2003;6:391–400. doi: 10.1016/j.semcancer.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 19.Restifo NP, Wunderlich JR. Principles of tumor immunity: biology of cellular immune responses. In: DeVita VT, Hellman S, Rosenberg SA, editors. Biologic therapy of cancer. Philadelphia, PA: Lippincott; 1996. [Google Scholar]
- 20.Rosenberg SA. A new era of cancer immunotherapy: converting theory to performance (see also p 65) CA Cancer J Clin. 1999;2:70–73. doi: 10.3322/canjclin.49.2.70. [DOI] [PubMed] [Google Scholar]
- 21.Roszkowski JJ, Lyons GE, Kast WM, Yee C, Van Besien K, Nishimura MI. Simultaneous generation of CD8+ and CD4+ melanoma-reactive T cells by retroviral-mediated transfer of a single T-cell receptor. Cancer Res. 2005;4:1570–1576. doi: 10.1158/0008-5472.CAN-04-2076. [DOI] [PubMed] [Google Scholar]
- 22.Schaft N, Dorrie J, Thumann P, Beck VE, Muller I, Schultz ES, Kampgen E, Dieckmann D, Schuler G. Generation of an optimized polyvalent monocyte-derived dendritic cell vaccine by transfecting defined RNAs after rather than before maturation. J Immunol. 2005;5:3087–3097. doi: 10.4049/jimmunol.174.5.3087. [DOI] [PubMed] [Google Scholar]
- 23.Schaft N, Lankiewicz B, Gratama JW, Bolhuis RL, Debets R. Flexible and sensitive method to functionally validate tumor-specific receptors via activation of NFAT. J Immunol Methods. 2003;1–2:13–24. doi: 10.1016/S0022-1759(03)00067-X. [DOI] [PubMed] [Google Scholar]
- 24.Schaft N, Willemsen RA, de Vries J, Lankiewicz B, Essers BW, Gratama JW, Figdor CG, Bolhuis RL, Debets R, Adema GJ. Peptide fine specificity of anti-glycoprotein 100 CTL is preserved following transfer of engineered TCRalphabeta genes into primary human T lymphocytes. J Immunol. 2003;4:2186–2194. doi: 10.4049/jimmunol.170.4.2186. [DOI] [PubMed] [Google Scholar]
- 25.Scheel B, Teufel R, Probst J, Carralot JP, Geginat J, Radsak M, Jarrossay D, Wagner H, Jung G, Rammensee HG, Hoerr I, Pascolo S. Toll-like receptor-dependent activation of several human blood cell types by protamine-condensed mRNA. Eur J Immunol. 2005;35:1557–1566. doi: 10.1002/eji.200425656. [DOI] [PubMed] [Google Scholar]
- 26.Smits E, Ponsaerts P, Lenjou M, Nijs G, Van Bockstaele DR, Berneman ZN, Van Tendeloo VF. RNA-based gene transfer for adult stem cells and T cells. Leukemia. 2004;11:1898–1902. doi: 10.1038/sj.leu.2403463. [DOI] [PubMed] [Google Scholar]
- 27.Stanislawski T, Voss RH, Lotz C, Sadovnikova E, Willemsen RA, Kuball J, Ruppert T, Bolhuis RL, Melief CJ, Huber C, Stauss HJ, Theobald M. Circumventing tolerance to a human MDM2-derived tumor antigen by TCR gene transfer. Nat Immunol. 2001;10:962–970. doi: 10.1038/ni1001-962. [DOI] [PubMed] [Google Scholar]
- 28.Theobald M, Biggs J, Hernandez J, Lustgarten J, Labadie C, Sherman LA. Tolerance to p53 by A2.1-restricted cytotoxic T lymphocytes. J Exp Med. 1997;5:833–841. doi: 10.1084/jem.185.5.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willemsen RA, Weijtens ME, Ronteltap C, Eshhar Z, Gratama JW, Chames P, Bolhuis RL. Grafting primary human T lymphocytes with cancer-specific chimeric single chain and two chain TCR. Gene Ther. 2000;16:1369–1377. doi: 10.1038/sj.gt.3301253. [DOI] [PubMed] [Google Scholar]
- 30.Xue S, Gillmore R, Downs A, Tsallios A, Holler A, Gao L, Wong V, Morris E, Stauss HJ. Exploiting T cell receptor genes for cancer immunotherapy. Clin Exp Immunol. 2005;2:167–172. doi: 10.1111/j.1365-2249.2005.02715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yee C, Thompson JA, Byrd D, Riddell SR, Roche P, Celis E, Greenberg PD. Adoptive T cell therapy using antigen-specific CD8+ T cell clones for the treatment of patients with metastatic melanoma: in vivo persistence, migration and antitumor effect of transferred T cells. Proc Natl Acad Sci USA. 2002;25:16168–16173. doi: 10.1073/pnas.242600099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang T, He X, Tsang TC, Harris DT. Transgenic TCR expression: comparison of single chain with full-length receptor constructs for T-cell function. Cancer Gene Ther. 2004;7:487–496. doi: 10.1038/sj.cgt.7700703. [DOI] [PubMed] [Google Scholar]
- 33.Zhao Y, Zheng Z, Robbins PF, Khong HT, Rosenberg SA, Morgan RA. Primary human lymphocytes transduced with NY-ESO-1 antigen-specific TCR genes recognize and kill diverse human tumor cell lines. J Immunol. 2005;7:4415–4423. doi: 10.4049/jimmunol.174.7.4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Y, Zheng Z, Cohen CJ, Gattinoni L, Palmer DC, Restifo NP, Rosenberg SA, Morgan RA (2005) High-efficiency transfection of primary human and mouse T lymphocytes using RNA electroporation. Mol Ther (Epub ahead of print) [DOI] [PMC free article] [PubMed]