Abstract
Tyrosinase-related protein-2 (TRP-2) is a non-mutated melanocyte differentiation antigen. The TRP-2-recognizing CD8+ T cells can evoke immune responses to melanoma in both humans and mice. Developing epitopes with amino acid replacements in their sequences might improve the low immunogenicity against this ‘self’ tumor antigen. We designed altered peptide ligands (APLs) of TRP-2(180–188) (SVYDFFVWL) with preferred primary and auxiliary HLA-A*0201 molecule anchor residue replacement. These APLs were screened for MHC-affinity by affinity prediction plots and molecular dynamics simulation, and analyzed in vitro for stability and binding-affinity to molecular HLA-A*0201. We also investigated the CTLs activities induced by TRP-2 wild-type epitope and the APLs both in vitro in human PBMCs and HLA-A2.1/Kb transgenic mice. The results indicate that TRP-2 2M analog simultaneously had stronger binding-affinity and a lower dissociation rate to HLA-A*0201, than wild-type peptide. In addition, the analog 2M was superior to other APLs and wild-type epitope in terms of immunological efficacy ex vivo as measured by the ELISPOT assays of IFN-γ and granzyme B. These results demonstrate that TRP-2 2M is an agonist epitope that can induce anti-tumor immunity superior to its wild-type epitope, and has potential application in peptide-mediated immunotherapy.
Keywords: Tyrosinase-related protein 2, Altered peptide ligands, Immunogenicity
Introduction
One of the most significant advances in the field of tumor immunology has been the identification and characterization of the tumor-associated antigens that are recognized by T cells [20]. More than 170 antigenic peptides derived from 60 human tumor antigens are expressed in the context of MHC molecules and are recognizable by cells in the available T cell repertoire, many of which are non-mutated ‘self’ antigens [17].
Of these antigens, several melanoma-associated Ags, described to date, including MART-1, gp100, tyrosinase-related protein-1 (TRP-1), and tyrosinase-related protein-2 (TRP-2), are shared antigens, recognized in an MHC-I-restricted fashion by lymphocytes from different patients. TRP-2 is a non-mutated melanocyte differentiation antigen (MDA) recognized by melanoma-reactive cytotoxic T lymphocytes (CTLs) in both humans and mice [1]. Human TRP-2 is composed of 519 amino acids and shares 84% identity with its mouse counterpart [31]. Melanoma-reactive CTLs in both humans and mice recognize the peptide, TRP-2(180–188) (SVYDFFVWL). The amino acid sequence of this peptide is identical between human and mice, thus facilitating the immunotherapeutic study for human melanoma [16]. Recent research [9] demonstrated that 51.2% of primary tumor cell lines derived from patients with glioblastoma multiforme (GBM) express TRP-2. In addition, it has been verified that an HLA-A2-restricted cytotoxic T cell clone recognizing the TRP-2(180–188) peptide (SVYDFFVWL) can specifically lyse the TRP-2 positive GBM cells in an HLA-A*0201 restricted manner [9]. Therefore, TRP-2 should be a useful antigen target for developing immunotherapeutic strategies for melanoma and glioma patients.
TRP-2(180–188)-reactive T cells with low avidity can be readily induced from the spleen cells of naïve mice when cultured with a high dose of the TRP-2(180–188) peptide, suggesting the presence of a considerable number of TRP-2(180–188)-recognizing T-cell precursors with low avidity in naïve mice. Nevertheless, the reason why significant amounts of T-cell precursors specific to the ‘self-antigen’ TRP-2 in naïve mice with failure in rejecting melanoma has not been fully elucidated. Interestingly, similar observations have been reported in humans [6]. Anti-MART1/Melan A CTL precursors are detectable in healthy donors, and the T-cell responses of melanoma patients to MART1/Melan A antigen have not been reported to correlate with the clinical outcomes [10]. However, an analog of MART1/Melan A27–35 (AAGIGILTV), MART1/Melan A27–35 1A→L, can generate CTLs with quantitative and qualitative enhanced recognition of melanoma cells expressing the naïve peptide [11]. In 1993, Ruppert et al. [22] determined that a ‘canonical’ A2.1 motif could be defined as L or M at position 2 (P2) and L, V, or I at position 9 (P9). In 2000, Tourdot et al. [26] demonstrated that at position 1 (P1) residue Y can enhance the MHC-affinity for a low affinity epitope. These results suggest that an altered peptide ligand (APL) might be used to exploit a latent capacity of the T cell repertoire to respond more effectively to the naïve epitope.
Therefore, in the present study, we applied this strategy to modify TRP-2(180–188) (SVYDFFVWL) to enhance its immunogenicity. As the residue L at P9 of naïve TRP-2 was the predominant anchor residue, we altered the naïve epitope only with P1 (Y) and/or P2 (L, M). The derived APLs were screened with three sets of software programs and analyzed by a molecular dynamics simulation method. We then investigated the immunological efficacy of the selected APLs in vitro and in vivo to provide concrete experimental evidence for potential clinical applications of these APLs.
Materials and methods
Epitope prediction, molecular modeling, and peptide synthesis
We altered TRP-2(180–188) naïve epitope (SVYDFFVWL) with P1 (Y) and/or P2 (L, M). Three sets of professional software programs, Polynomial Method [5], Quantitative Motif Method [15], and QSAR Method [34], were used to evaluate the MHC-affinity of APLs. Based on the results of the three prediction methods, the HLA-A*0201 allele-binding peptides with high scores were studied further with molecular modeling.
Models of HLA-A2.1 and binding 9-mer peptides were established from the crystal structures of the Brookhaven Protein Data Bank (USA): 3HLA for HLA-A2.1 and 3HSA for the nonameric peptides. Molecular mechanics and dynamics calculations were performed with the Discover 3.0 software package (GeoMax Information, USA). The parameters used in this study were the MMFF94 force field. During the molecular dynamics and minimization, a dielectric constant of 1.0 was used. A 9.0 Å cutoff distance was applied to calculate the non-binding interaction. The peptide ligand was first relaxed by 500 steps of conjugate gradient energy minimization, while the HLA-A2.1 molecule remained fixed, and then submitted to a 100-ps molecular dynamics calculation at 300 K. During these 100 ps, no protein atom was allowed to move. The last conformation was then solvated in a 10 Å-thick TIP3P water shell. Energy minimization of the peptide ligand, the HLA-A2.1/ligand complex, followed by 200-ps molecular dynamics simulation of the full solvated HLA-A2.1/ligand pair was performed at 300 K.
The APLs validated by epitope prediction and molecules modeling methods were then synthesized by Fmoc chemistry (Sangon, China) and purified by HPLC to a purity of > 95%. Lyophilized peptides were dissolved in DMSO at a concentration of 20 mg/ml and stored at −70°C. The wild-type peptide TRP-2(180–188) (TRP-2 WT, SVYDFFVWL), negative control peptide OVA(257–264) (SIINFKEL), and the positive control peptide HBV(18–27) (FLPSDFFPSV) were synthesized and purified by the same methods. A PADRE T helper (Th) epitope (AKFVAAWTLKAAA) was synthesized for the in vivo immunity assay.
Cell lines and animals
Human TAP-deficient T2 cell line and BB7.2 hybridoma producing anti-HLA-A2 Ab were purchased from American Type Culture Collection (ATCC, USA). The T2 cell line was maintained in RPMI 1640 supplemented with 10% fetal bovine serum and 100 μg/ml penicillin/streptomycin. BB7.2 cell line and murine melanoma cell line B16/F10 (H-2Kb+, TRP-2+) were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% FCS and 100 μg/ml of penicillin/streptomycin. Human melanoma cell LB373-MEL (HLA-A2+, TRP-2+) was a generous gift from Professor Francis Brasseur (Ludwig Institute for Cancer Research, Brussels Branch, Brussels, Belgium). The cell is cultured in Iscove’s Modified Dulbecco’s Medium (IMDM) containing penicillin (200 units/ml) and streptomycin (100 μg/ml), supplemented with FCS (10% v/v), l-arginine (116 g/ml), l-asparagine (36 μg/ml), l-glutamine (219 μg/ml), hydrocortisone (10 nM), insulin (5 μg/ml), transferrin (100 μg/ml), 17β-estradiol (10 nM), and sodium selenite (30 nM). Anti-HLA-A2 Ab was derived from BB7.2 (100 μl hybridoma culture supernatant/106 T2 cells) [14]. Anti-CD8 Ab was purchased from BD Biosciences Pharmingen (USA).
HLA-A2.1/Kb transgenic (Tg) mice, 8–12-weeks-old, were purchased from The Jackson Laboratory (USA). Mice were bred and maintained in specific pathogen-free (SPF) facilities. Animal experiments were performed in accordance with the guidelines of the Animal Care and Use Committee of Third Military Medical University.
Peptide binding assay
To determine if synthetic APLs could bind to HLA-A*0201 molecules, peptide-induced HLA-A*0201 up-regulation on T2 cells was examined according to a protocol described previously [14]. Briefly, T2 cells were incubated with 50 μM of the synthesized peptides and 3 μg/ml of human β2-microglobulin (Serotec, UK) in serum-free RPMI 1640 medium for 16 h at 37°C/5% CO2. Expression of HLA-A*0201 on T2 cells was then determined with FACS Calibur flow cytometer (Becton Dickinson, USA), by staining with anti–HLA-A2 Ab derived from BB7.2 and FITC-labeled goat-anti-mouse IgG (BD Biosciences Pharmingen, USA) used as the second antibody. The data were analyzed using CellQuest software (Becton Dickinson, USA). The fluorescence index (FI) was calculated as follows: FI = (mean FITC fluorescence with the given peptide−mean FITC fluorescence without peptide)/(mean FITC fluorescence without peptide). Samples were measured in three replications.
Measurement of the peptide/HLA-A*0201 complex stability
As previously described [26], T2 cells (106/ml) were incubated overnight with 100 μM of each peptide in serum-free RPMI 1640 medium supplemented with 100 ng/ml β2 m at 37°C. The cells were then washed four times to remove free peptides, incubated with Brefeldin A (Sigma-Aldrich, USA) (10 μg/ml) for 1 h to block HLA-A*0201 molecules newly expressed on cell surface, washed and incubated at 37°C for 0, 2, 4, 6, or 8 h. Subsequently, the cells were stained with the BB7.2 Ab to evaluate the HLA-A*0201 molecule expression. For each time point, peptide-induced HLA-A*0201 expression was evaluated by the formula: mean fluorescence of peptide preincubated T2 cells−mean fluorescence of T2 cells treated in similar conditions in the absence of peptides. The relative percentage of complexes remaining of each peptide at one time point was evaluated by the formula: mean fluorescence of peptide-induced HLA-A*0201 expression at one time point/mean fluorescence of peptide-induced HLA-A*0201 expression at time 0 h of the same peptide.
Generation of CTLs in PBMCs from healthy donors
Dendritic cells (DCs) have the unique capacity of activating naïve T cells and initiating primary T-cell response. Antigen-specific T-cell responses from peripheral blood mononuclear cells (PBMCs) can be elicited with antigenic peptide-pulsed autologous DCs [12, 30]. We obtained PBMCs from the buffy coat of heparinized whole blood samples of healthy donors (the Blood Bank of Chongqing, China) by density gradient centrifugation on the Histopaque 1077 (AXIS-SHIELD, Norway). These cells were resuspended in serum-free RPMI 1640 and allowed to adhere to six-well plates at a final concentration of 1 × 107 cells/3 ml/well. After 2 h of incubation at 37°C, non-adherent cells were gently removed with warm medium by gently pipetting. The non-adherent cells (effector lymphocytes) were cryopreserved in FCS supplemented by 10% DMSO (Sigma-Aldrich, USA).
The resultant adherent cells containing DCs were cultured in medium supplemented with 800 U/ml GM-CSF (R&D Systems, USA) and 1,000 U/ml IL-4 (R&D Systems) in 37°C/5% CO2. Every 2 days, one-half of the medium was replaced by fresh medium containing a double concentration of GM-CSF and IL-4 as indicated above. On day 5, 10 ng/ml of recombinant human tumor necrosis factor а (TNF-а) (Peprotech, USA) was added to the medium to induce phenotypic and functional maturation of DCs. After 48 h, DCs were pulsed with 20 μg/ml peptide in the presence of 3 μg/ml β2-microglobulin at 37°C for 3 h and irradiated at 30 Gy before use. The thawed 2 × 106 of non-adherent effector lymphocytes were cocultured with 2 × 105 peptide-pulsed irradiated autologous DCs in a 24-well plate in the presence of 10 ng/ml recombinant human interleukin-7 (IL-7) (Peprotech, USA). After 7 days, lymphocytes were restimulated with peptide-pulsed autologous PBMCs in medium containing 10 ng/ml IL-7 and 20 U/ml IL-2 (Peprotech, USA). About 20 U/ml of IL-2 was added 24 h later at regular intervals, 2 days after each restimulation. Lymphocytes were restimulated each week in the same manner. On the seventh day, after the three rounds of restimulation, cells were harvested and tested by ELISPOT assay.
ELISPOT assay
ELISPOT assays were performed using a commercially available kit (DIACLONE, France). T2 cells pulsed with the indicated concentration of TRP-2 WT, or irrelevant peptide HBV(18–27), were used as stimulator cells. Effector cells (1 × 105) and peptide-prepulsed T2 cells (1 × 104) or the indicated amount of effector cells and tumor cells (1 × 104) were seeded into 96-well polyvinylidene fluoride (PVDF)-backed microplates coated with monoclonal antibody specific for mouse interferon γ (mIFN-γ) or human IFN-γ and granzyme B (grB). After incubation at 37°C for 16 h, cells were removed and plates were processed according to the manufacturer’s instructions. IFN-γ or grB-secreting T cells were counted using the automated imaged analysis system ELISPOT reader.
Analysis of in vivo immunogenicity
HLA-A2.1/Kb mice were co-immunized with 100 μg of the TRP-2 WT or APLs and 50 μg/PADRE Th epitope prepared in IFA. As a control, mice were injected with an IFA emulsion without peptide. Eleven days after immunization, splenocytes from injected animals were cultured with 10 μg/ml of the immunizing peptide loaded with autologous splenocytes (as APCs) to expand CTLs in vitro with mIL-2. Five days later, peptide-specific mIFN-γ production by effector cells was measured using the ELISPOT assay.
Statistical analysis
Analysis of variance (ANOVA) and Student’s t test were performed to determine effects of the treatments. A difference was considered as significant at the significance level of P < 0.05.
Results
Epitope prediction and molecular modeling
The sequences of APLs of the CTL epitope TRP-2(180–188) (SVYDFFVWL) were initially screened by three sets of professional software programs, i.e., Polynomial Method, Quantitative Motif Method, and QSAR Method, respectively. Four APLs with high scores, TRP-2 2V→2L (SLYDFFVWL), TRP-2 2V→2M (SMYDFFVWL), TRP-2 1S2V→1Y2L (YLYDFFVWL), and TRP-2 1S2V→1Y2M (YMYDFFVWL) were finally selected. Molecular modeling showed that the four APLs derived from TRP-2(180–188) satisfied the criteria of HLA-A*0201-restricted CTL epitope. As shown in Fig. 1a and Table 1, these peptides bound to HLA-A*0201 model structure, possessed a side chain of COOH-terminal anchor residues oriented into the binding groove with different distances from 19 to 20 Å. Figure 1b shows a computer model depicting the TRP-2(180–188) (SVYDFFVWL) with the HLA-A*0201 molecule.
Fig. 1.
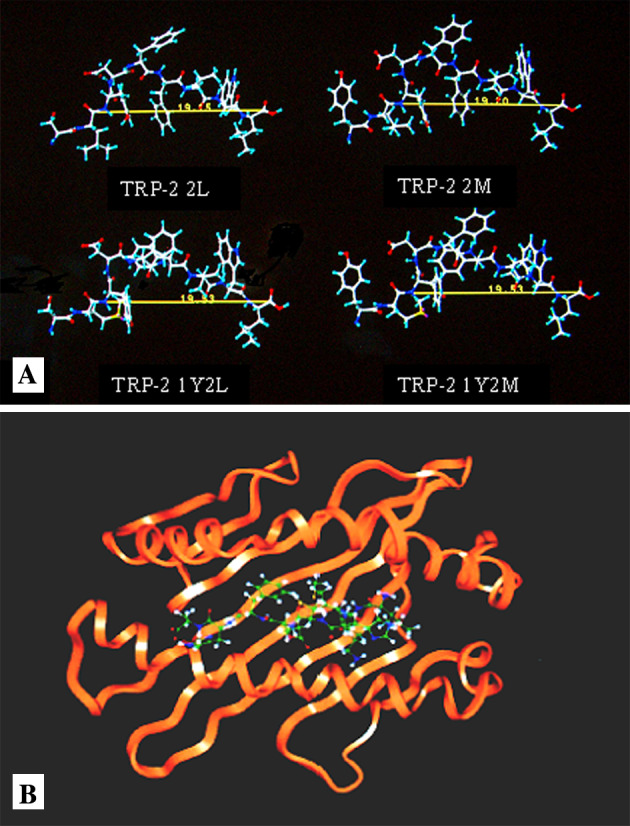
Molecular modeling of APLs of TRP-2(180–188). a Configuration of each APL determined with molecular simulation. Experimental details are described in M&M. Peptide configuration was obtained from the side view when bound to MHC molecule during the simulation. The amino acid sequence of the peptides used are listed in Table 1. Green carbon atoms, red oxygen atoms, blue nitrogen atoms, and white hydrogen atoms. b A computer model depicting association of the TRP-2(180–188) (SVYDFFVWL) with the HLA-A*0201 molecule. The MHC model was constructed by using crystallographic data of α1 and α2 heavy domains only (orange). Peptides are represented as a backbone structure (green)
Table 1.
Characteristics of TRP-2(180–188 ) APLs bound to HLA-A*0201 molecules
| Peptide | Sequence | Distance (Å) (P2–P9)a | Orientation of side chain (P9)b |
|---|---|---|---|
| TRP-2 2L | SLYDFFVWL | 19.15 | ↓ |
| TRP-2 2M | SMYDFFVWL | 19.20 | ↓ |
| TRP-2 1Y2L | YLYDFFVWL | 19.53 | ↓ |
| TRP-2 1Y2M | YMYDFFVWL | 19.53 | ↓ |
aThe distance between P2 and P9 and the orientation of the side chain at P9 was determined by the Simulation Method as described in Fig. 1
bArrows indicate the orientation of P9 residues with the respect to the floor of the binding groove of HLA-A*0201
MHC-peptide binding assay
Based on computer software predictions, the four candidate nonameric peptides with the highest estimated binding-affinity to HLA-A*0201 were selected and synthesized (Table 2). A T2 cell-peptide binding test was used to evaluate the binding affinity of these peptides in vitro. Of the four candidate peptides, TRP-2 2M and TRP-2 2L were high-affinity epitopes (FI = 2.15 and 2.49, respectively), while TRP-2 1Y2M and TRP-2 1Y2L have lower affinity to HLA-A*0201 (FI = 1.82 and 1.45, respectively). All APLs have stronger affinity than TRP-2 WT (FI = 1.32).
Table 2.
The binding affinity of TRP-2(180–188 ) APLs to HLA-A*0201 molecules
| Peptide | Sequence | Polynomial method | Quantitative motif method | QSAR method | FI |
|---|---|---|---|---|---|
| TRP-2(180–188) | SVYDFFVWL | −20.73 | 471.17 | 7.401 | 1.32 |
| TRP-2 2L | SLYDFFVWL | −19.79 | 8054.54 | 7.953 | 2.49 |
| TRP-2 2M | SMYDFFVWL | −19.48 | 4610.64 | 7.852 | 2.15 |
| TRP-2 1Y2L | YLYDFFVWL | −18.86 | 8054.54 | 8.256 | 1.45 |
| TRP-2 1Y2M | YMYDFFVWL | −18.55 | 4610.64 | 8.149 | 1.82 |
Positive control peptide HBV(18–27) and negative control peptide OVA(257–264) had FI 2.0 and 0.1, respectively
Recent studies [24, 27, 32] have suggested that a stable peptide-MHC complex could facilitate the formation of the synapses between T cells and APCs, and that the stability of peptide-MHC complex is the key factor for CTL activation. Thus, in this study, the stability of the peptide-MHC complex for wild-type peptide or APLs was investigated. The results showed that 2M/HLA-A*0201 complex was more stable than TRP-2 WT/HLA-A*0201 complex over the 8 h observation period. As shown in Fig. 2, after 6 h of treatment with brefeldin A, the remaining rate of peptide/MHC complex was 80, 71, 69, 56, and 53% for TRP-2 2M, TRP-2 1Y2M, TRP-2 1Y2L, TRP-2 WT, and TRP-2 2L, respectively. Thus, among the four APLs, TRP-2 2M peptide was the only analog that had a higher binding affinity and a higher stability to MHC molecules simultaneously than TRP-2 WT peptide.
Fig. 2.
Comparison of stability for the complex of TRP-2(180–188) wild-type peptide or APLs with HLA-A*0201 molecule. T2 cells were incubated overnight with peptide at a concentration of 100 μg/ml and then washed free of unbound peptide and incubated with brefeldin A to block delivery of new class I molecules to the cell surface. At the indicated times, cells were stained for the presence of surface peptide-HLA-A*0201 complexes. Results are presented as relative percentage of binding compared with 100% at time 0
IFN-γ production of APL-specific CTLs
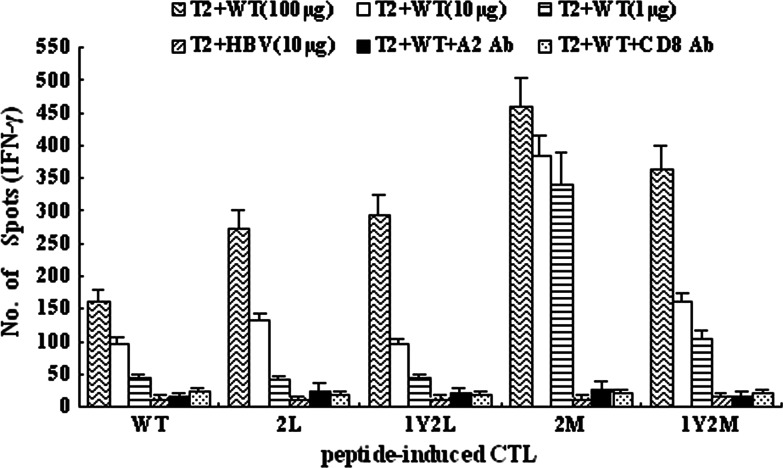
We tested the immunogenicity of the APLs in three normal HLA-A*0201 individuals to determine the capacity of prime CTL responses in vitro. The data showed that 2M analog-induced CTLs had a significantly increased capacity to secrete IFN-γ, when stimulated with 100, 10, or 1 μg of TRP-2 WT loaded to T2 cells, compared with other APLs-induced CTLs and wild-type peptide-induced CTLs (Fig. 3). Remarkably, when cocultured with 1 μg of wild-type peptide, 2M analog-induced CTLs retained almost the same level of secreting IFN-γ as cocultured with 100 or 10 μg of wild-type peptide. However, when cocultured with 1 μg of wild-type peptide, the capacity of IFN-γ production declined sharply for other APLs and wild-type peptide, compared with 100 and 10 μg of TRP-2 WT. In addition, after we used anti-HLA-A2 Ab and anti-CD8 Ab to respectively block HLA-A2 and CD8 molecules, the stimulators could hardly activate the effectors and thus these effectors produced little IFN-γ. This indicates that the release of IFN-γ by these CTLs, while recognizing T2 cells pulsed with various concentrations of WT, is in HLA-A2-restricted and CD8-dependent manners.
Fig. 3.
The TRP-2 2M analog induces more efficiently CTLs that can secrete IFN-γ when recognizing wild-type peptide. TRP-2 WT and four APLs were used to stimulate PBMC from healthy HLA-A*0201-positive donors three times at weekly intervals according to the CTL induction protocol in M&M. The five kinds of CTLs: WT-CTL, 2L-CTL, 1Y2L-CTL, 2M-CTL, and 1Y2M-CTL were tested against T2 cells prepulsed with TRP-2 WT (100, 10, or 1 μg) or HBV control peptide (10 μg), respectively. Anti-HLA-A2 and anti-CD8 mAbs were, respectively, added into the culture wells containing T2 cells, 100 μg WT peptide, and peptide-induced CTLs. Then, IFN-γ production by peptide-induced CTLs was measured using the ELISPOT assay. Results are from three independent experiments and each sample for each experiment was set with triplicate wells. Data represent means ± SD
Cytolytic activity of APL-induced CTLs
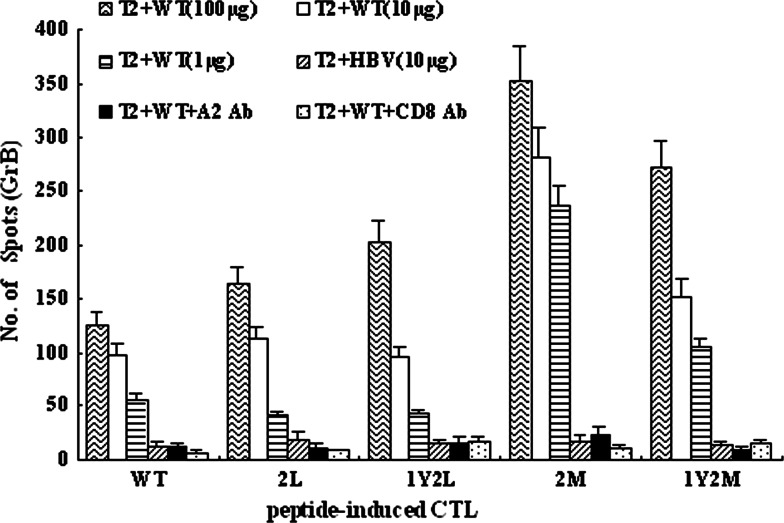
To address whether IFN-γ-producing CTL lines could lyse target cells pulsed with TRP-2 WT, granzyme B(GrB) ELISPOT assays were performed. As shown in Fig. 4, TRP-2 2M specific CTLs showed increased capacity to secrete GrB, compared with TRP-2 WT-specific CTLs and other APLs-specific CTLs. Remarkably, when cocultured with 1 μg/ml wild-type peptide, 2M-specific CTLs showed almost the same capacity of secreting GrB as cocultured with 100 and 10 μg/ml wild-type peptide. In contrast, the level of GrB secreted by CTLs induced by the analogs 2L, 1Y2L, 1Y2M, and TRP-2 WT declined dramatically when recognizing 1 μg/ml wild-type epitope loaded to T2 cells, compared with 100 or 10 μg/ml wild-type peptide (Fig. 4). This data coincided with the results of IFN-γ ELISPOT assays. In addition, after we used anti-HLA-A2 Ab and anti-CD8 Ab to block HLA-A2 and CD8 molecules, respectively, the capacity of granzyme B production declined sharply to the background level for both APLs and wild-type peptide. This indicates that the release of GrB by these CTLs, while recognizing T2 cells pulsed with WT, is in HLA-A2-restricted and CD8-dependent manners.
Fig. 4.
The TRP-2 2M analog induces more efficiently CTLs that can secrete granzyme B when recognizing wild-type peptide. TRP-2 WT and four APLs were used to stimulate PBMC from healthy HLA-A*0201-positive donors three times at weekly intervals according to the CTL induction protocol in M&M. The five kinds of CTLs: WT-CTL, 2L-CTL, 1Y2L-CTL, 2M-CTL, and 1Y2M-CTL were tested against T2 cells prepulsed with TRP-2 WT (100, 10, or 1 μg) or HBV control peptide (10 μg), respectively. Anti-HLA-A2 and anti-CD8 mAbs were, respectively, added into the culture wells containing T2 cells, 100 μg WT peptide, and peptide-induced CTLs. Then, granzyme B production by peptide-induced CTLs was measured using ELISPOT assay. Results are from three independent experiments and each sample for each experiment was set with triplicate wells. Data represent means ± SD
To simulate the in vivo effector-target cells interaction, human melanoma cell line LB373-MEL (HLA-A*0201+, TRP-2+) and murine melanoma cell line B16/F10(H-2Kb+, TRP-2+) were used further to determine if more efficient tumor lysis by APL-induced CTLs than by WT-induced CTLs could be detected.
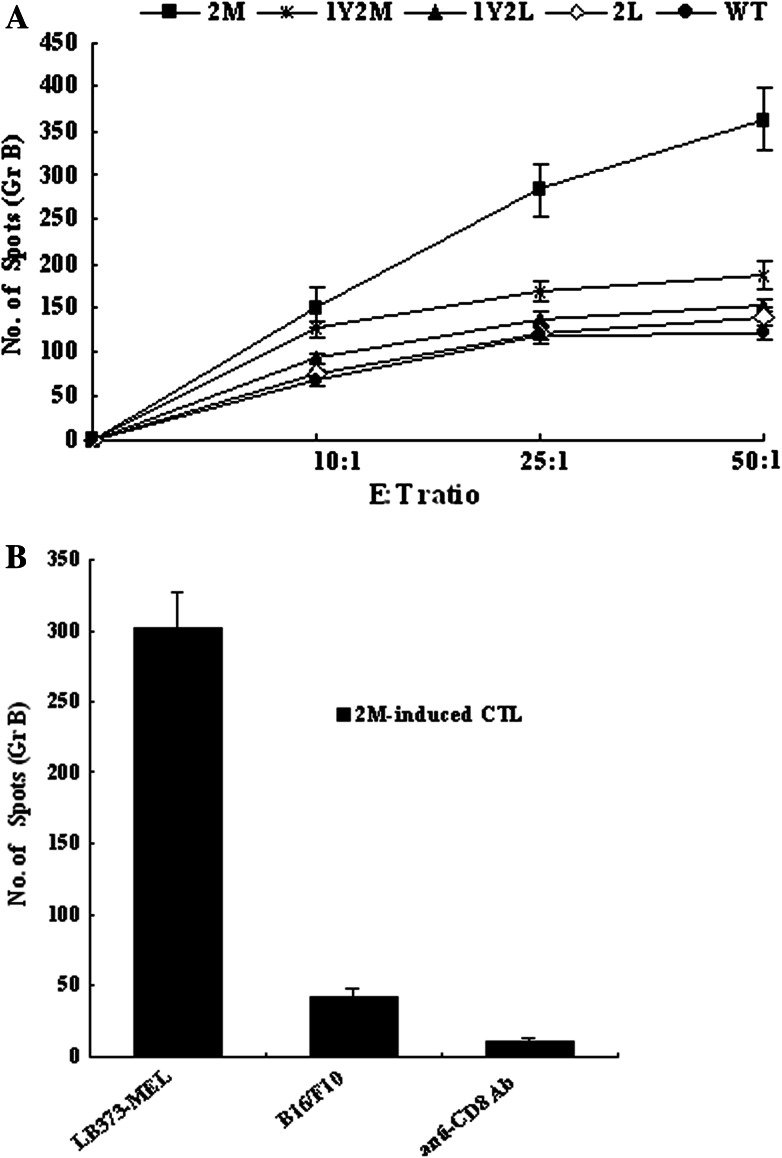
The GrB ELISPOT assay demonstrated that there was an evident increase in lysis of tumor cells for 2M induced-CTLs, compared with WT and other APLs (Fig. 5a). However, 2M-induced CTLs could not lyse the murine melanoma B16/F10 (Fig. 5b), which indicates that the 2M-induced CTLs could recognize HLA-A2-restricted epitope, and not H-2Kb-restricted epitope. In addition, the mAb against CD8 was used to block recognition of effectors and tumor cells. Our results showed that anti-CD8 mAb could significantly eliminate the cytotoxicity of the effectors against LB373-MEL melanoma cells (Fig. 5b), which implies that the peptide-induced effectors lysed the LB373-MEL cells in HLA-A2.1-restricted and CD8-dependent manners.
Fig. 5.
The TRP-2 2M-induced CTLs has more efficient tumor lysis than WT and other APLs-specific CTLs. TRP-2 WT and four APLs were used to stimulate PBMC from healthy HLA-A*0201-positive donors three times at weekly intervals according to the CTL induction protocol in M&M. a The five kinds of CTLs: WT-CTL, 2L-CTL, 1Y2L-CTL, 2M-CTL, and 1Y2M-CTL were tested against LB373-MEL at indicated E/T ratio using GrB ELISPOT assays. b The 2M-induced CTLs was tested against murine melanoma cell B16/F10 at 50:1 E/T ratio with GrB ELISPOT assay. Anti-CD8 mAb was added into the culture wells containing 2M-induced CTLs, LB373-MEL at 50:1 E/T ratio. Results from each group are from three independent experiments and each sample for each experiment was set with triplicate wells. Data represent means ± SD
Collectively, TRP-2 2M-induced CTLs could recognize wild-type peptide that loaded to T2 cells. It could release higher levels of IFN-γ and GrB and have more efficient tumor lysis than WT and other APLs, when recognizing LB373-MEL tumor cells.
In vivo induction of epitope-specific CTLs in HLA-A2.1/Kb transgenic mice
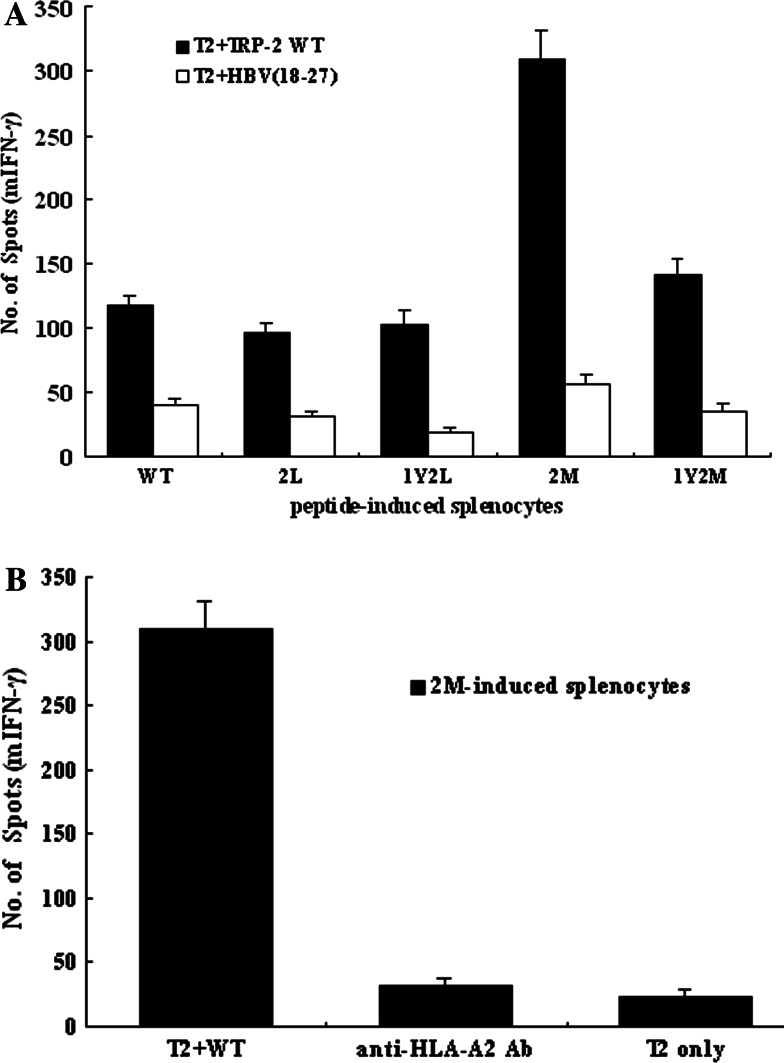
We investigated whether the TRP-2 analogs would be more efficient than the TRP-2 wild-type epitope in inducing immunity in vivo. The splenocytes from TRP-2 2M-inoculated mice show strong mIFN-γ production, while recognizing T2 cells loaded with wild-type peptide, but not T2 cells loaded with the irrelevant peptide HBV(18–27) (Fig. 6a). When anti-HLA-A2 Ab derived from BB7.2 was added to the culture wells during ELISPOT assay, anti-HLA-A2 Ab could inhibit 2M-induced splenocytes from producing mIFN-γ (Fig. 6b), which suggests that the higher immunogenicity in vivo of 2M analog was in antigen-specific and HLA-A2-restricted fashions. Therefore, the high immunogenicity of 2M peptide in vivo corresponds with a markedly increased immunogenicity of this analog in vitro.
Fig. 6.
The TRP-2 2M analog can induce stronger specific CTLs in vivo in HLA-A2.1/Kb transgenic mice than the TRP-2 wild-type epitope. HLA-A2.1/Kb mice were coimmunized with 100 μg of the TRP-2 wild-type or APLs and 50 μg of the PADRE Th epitope prepared in IFA. Mice were sacrificed 11 days after immunization, and splenocytes from the injected animals were cultured with 10 μg/ml of the immunizing peptide to expand splenocytes as described in M&M. Five days later, mIFN-γ production by peptide-induced splenocytes was measured using the ELISPOT assay. a Bulk splenocytes from TRP-2 WT or APL-immunized mice released mIFN-γ in response to T2 cells prepulsed with TRP-2 WT (10 μg) or irrelevant peptide HBV(18–27). b The inhibition effect of anti-HLA-A2 Ab on the production of mIFN-γ by 2M analog-induced splenocytes. The anti-HLA-A2 mAb was added to the culture wells containing T2 cells, WT peptide, and 2M-induced splenocytes. Then, mIFN-γ production by peptide-specific splenocytes was measured using the ELISPOT assay. Results from each group are from three independent experiments and each sample for each experiment was set with triplicate wells. Data represent means ± SD
Discussion
T lymphocytes have been shown to have no single ligand specificity, but to recognize a large array of peptides, termed altered peptide ligands(APL) [8]. These APLs can mediate a number of different outcomes in interacting with T cells, ranging from inducing selective immunological functions (partial agonist) to completely turning off their functional capacity (antagonist). In addition to suboptimal ligands, T cell receptors (TCRs) can also be triggered by agonist ligands, which are analogs that enhance T-cell stimulation by inducing immunological functions that cannot be triggered by the cognate ligand [3, 13, 19, 28]. The use of agonist peptides in vaccine therapy has now been reported in three recent clinical trials: the melanoma Ag gp100(209–217) 2M analog, CAP1-6D agonist and MART1/Melan A27–35 1L super-agonist [2]. The three agonists appear to be more potent in activating naïve T cells than its wild-type peptide, produce more clinical responses in melanoma patients, and show more regression of metastases [11, 21, 33]. This approach was applied in our study that amino acid replacements were carried out in the sequence of TRP-2(180–188) (SVYDFFVWL), mainly in MHC contact positions.
Tt is interesting to note that the analog peptides made by double substitution were not as efficient in binding to the HLA-A*0201 molecule as single substitution, i.e., analogs 2M and 2L have stronger binding ability than 1Y2M or 1Y2L. Similar phenomenon was reported that analog PSA-3 (Y154/L155) was not as efficient in binding to the HLA-A*0201 molecule as the naïve PSA-3 peptide when assayed by binding to T2-A2 cells [25].
In order to clarify why the double substitution was not as efficient in binding with the HLA-A*0201 molecule as single substitution, molecular modeling was performed to study the difference between the two binding models after single and double substitution.
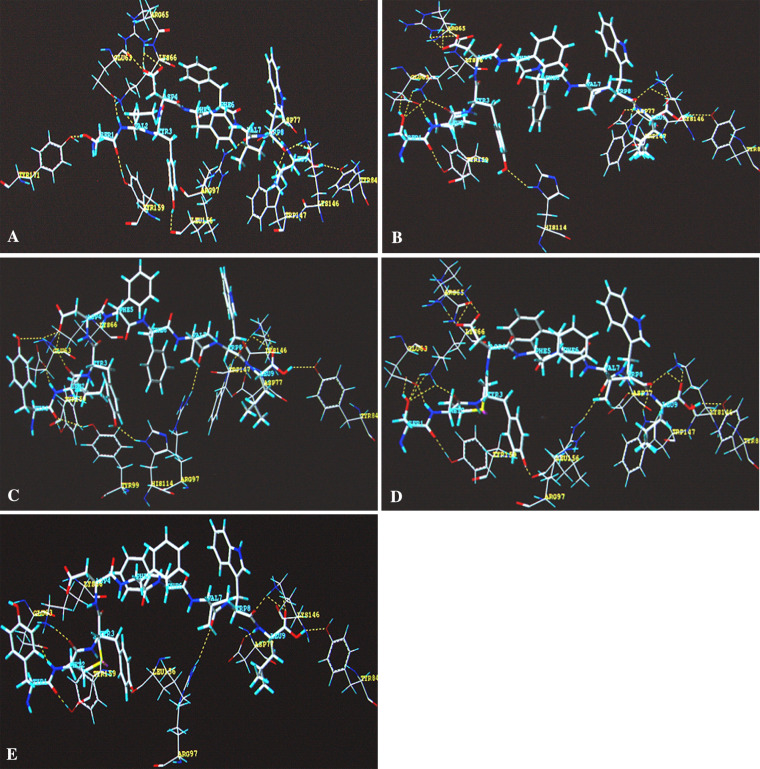
Generally, the interaction between receptor and ligand is based on the hydrophobic property, stereo property, electronic property, and hydrogen bond binding property of the ligand. The only difference between single substitution and double substitution in this study lies in the first amino acid of the peptide, which is 1S and 1Y, respectively. There is not much difference between the two amino acids in the hydrophobic property, stereo property, or electronic property. How then can we explain the significant difference in experimental binding data? After analysis of the binding models of the peptide binding and HLA-A* 0201 molecule, the results showed a great difference in hydrogen bond binding property between the single substitution peptide and the double substitution peptide (Table 3). Molecular modeling showed that single amino acid substitution at P2 enhanced the interaction between the first amino acid 1S and the HLA-A*0201 molecule. Strong hydrogen bonds were formed between 1S (TRP-2 2L and TRP-2 2M) and the residues of HLA-A*0201 (GLU63, LYS66), whereas the substituted residues at P2 (L, M) themselves did not change the hydrogen bond formation between residues at P2 and HLA-A*0201 (Table 3, Fig. 7). However, when replaced at both P1 and P2 (TRP-2 1Y2L, TRP-2 1Y2M), conformation of the Y residue at P1 could not form strong hydrogen bond interaction with the residues of the HLA-A*0201 molecule as shown in Table 3 and Fig. 7a–e.
Table 3.
H-bond formation between peptides and HLA-A*0201 molecule
| Name | Sequence | Donor and receptor of H-bond | H-bond | |
|---|---|---|---|---|
| Mimotope | HLA-A*0201 | |||
| TRP-2(180–188) | SVYDFFVWL | S1 | Tyr 171, Tyr 159 | 2 |
| V2 | Glu 63, Lys 66 | 2 | ||
| TRP-2 2L | SLYDFFVWL | S1 | Glu 63, Tyr 159 | 4 |
| L2 | Glu 63 | 2 | ||
| TRP-2 2M | SMYDFFVWL | S1 | Glu 63, Lys 66, Tyr 159 | 5 |
| M2 | Glu 63, Lys 66, | 2 | ||
| TRP-2 1Y2L | YLYDFFVWL | Y1 | Glu 63, Lys 66 | 3 |
| L2 | Glu 63, | 2 | ||
| TRP-2 1Y2M | YMYDFFVWL | Y1 | Tyr 159 | 1 |
| M2 | Glu 63 | 2 | ||
Fig. 7.
The binding models of each peptide. The hydrogen bonds (yellow broken line) are shown in the figure. The amino acid residues of the peptides are indicated in blue character and the residues of HLA-A*0201, interacting with peptide, are shown in yellow character. a–e shows the peptide TRP-2(180–188) (SVYDFFVWL), TRP-2 2L (SLYDFFVWL), TRP-2 1Y2L (YLYDFFVWL), TRP-2 2M (SMYDFFVWL), and TRP-2 1Y2M (YMYDFFVWL), respectively
Another interesting observation is that although TRP-2 2L had the stronger binding capacity than the naïve and other analogs, the 2L/HLA-A*0201 complex was not as stable as other analogs, and was even lower than the wild-type peptide. The mechanism might be “fast on, fast off” [29]. HLA affinity of peptide is a commonly used method to screen CTL epitope, however, recent research shows that immunogenicity of peptide appears to correlate more closely with the dissociation rate than with peptide binding affinity [27].
Thus, in this study, the two methods were used to discriminate the potential agonist APLs for TRP-2(180–188) (SVYDFFVWL). The 2M/HLA-A*0201 complex had retained 81% at 6 h after brefeldin A treatment. Therefore, 2M analog had both high binding affinity and stability to molecular HLA-A*0201.
Correlating the peptide binding affinity and peptide/MHC complex stability, we found that 2M analog had a significantly increased capacity to elicit IFN-γ from CTL lines compared with other APLs and wild-type peptide. Granzyme B (GrB), released from activated CTLs, is a key mediator of target cell death via the granule-mediated pathway. Therefore, the release of GrB by cytolytic lymphocytes upon effector-target interaction may be a more specific indicator of CTL cytotoxic ability than IFN-γ secretion [18, 23]. 2M-specific CTLs showed an increased capacity to lyse wild-peptide TRP-2(180–188) pulsed T2 cells and human melanoma cell lines LB373-MEL, compared with other APLs-specific CTLs and TRP-2 WT-specific CTLs. Splenocytes from 2M analog-immunized HLA-A2.1/Kb transgenic mice produced higher levels of type I cytokine (mIFN-γ) than other APLs and the wild-type peptide. These results indicated that 2M analog had a superior capacity to elicit the immunity in vitro and in vivo, compared with wild-type TRP-2(180–188) and other APLs.
Recent studies have suggested that a stable peptide-MHC complex may facilitate the formation of the immune synapses between T cells and APCs [24]. The altered peptide may thus allow full T cell activation through sustained signaling and therefore an increased peptide immunogenicity [7], while the naïve peptide forms an unstable complex that could fail to fully sustain signaling. In our study, 2M analog, that had a slower dissociation rate and formed a more stable MHC-peptide complex, persisted at the cell surface for a time sufficient to allow the induction of a strong CTL response. The in vitro and in vivo experimental results supported this hypothesis.
In conclusion, these findings indicate that TRP-2 2M analog, which is formed by introduction of preferred amino acid residues into the TRP-2(180–188) wild peptide results in an increased affinity for the HLA-A*0201 allele and could induce more efficient melanoma cell line lysis compared with the wild-type peptide. Continuing studies are investigating whether 2M analog could induce effective immune response in a preclinical model, such as B16-AAD animal model [4]. These results will reveal if 2M analog is capable of rescuing weakly responding or anergic CTLs in melanoma patients.
Acknowledgments
This study was supported by the State Key Basic Research Program of China (No.2001CB510001), the National Natural Science Foundation of China (No. 30490240, 30300315, 30471579), and the Outstanding Young Scientist Foundation of China (No.30325020).
References
- 1.Bloom MB, Perry-Lalley D, Robbins PF, Li Y, el Gamil M, Rosenberg SA, Yang JC. Identification of tyrosinase-related protein 2 as a tumor rejection antigen for the B16 melanoma. J Exp Med. 1997;185:453–459. doi: 10.1084/jem.185.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brinkman JA, Fausch SC, Weber JS, Kast WM. Peptide-based vaccines for cancer immunotherapy. Expert Opin Biol Ther. 2004;4:181–198. doi: 10.1517/14712598.4.2.181. [DOI] [PubMed] [Google Scholar]
- 3.Dressel A, Chin JL, Sette A, Gausling R, Hollsberg P, Hafler DA. Autoantigen recognition by human CD8 T cell clones: enhanced agonist response induced by altered peptide ligands. J Immunol. 1997;159:4943–4951. [PubMed] [Google Scholar]
- 4.Engelhard VH, Bullock TN, Colella TA, Sheasley SL, Mullins DW. Antigens derived from melanocyte differentiation proteins: self-tolerance, autoimmunity, and use for cancer immunotherapy. Immunol Rev. 2002;188:136–146. doi: 10.1034/j.1600-065X.2002.18812.x. [DOI] [PubMed] [Google Scholar]
- 5.Gulukota K, Sidney J, Sette A, De DeLisi C. Two complementary methods for predicting peptides binding major histocompatibility complex molecules. J Mol Biol. 1997;267:1258–1267. doi: 10.1006/jmbi.1997.0937. [DOI] [PubMed] [Google Scholar]
- 6.Harada M, Yamada H, Tatsugami K, Nomoto K. Evidence of the extrathymic development of tyrosinase-related protein-2-recognizing CD8 + T cells with low avidity. Immunology. 2001;104:67–74. doi: 10.1046/j.1365-2567.2001.01278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huppa JB, Gleimer M, Sumen C, Davis MM. Continuous T cell receptor signaling required for synapse maintenance and full effector potential. Nat Immunol. 2003;4:749–755. doi: 10.1038/ni951. [DOI] [PubMed] [Google Scholar]
- 8.Kersh GJ, Allen PM. Essential flexibility in the T-cell recognition of antigen. Nature. 1996;380:495–498. doi: 10.1038/380495a0. [DOI] [PubMed] [Google Scholar]
- 9.Liu G, Khong HT, Wheeler CJ, Yu JS, Black KL, Ying H. Molecular and functional analysis of tyrosinase-related protein (TRP)-2 as a cytotoxic T lymphocyte target in patients with malignant glioma. J Immunother. 2003;26:301–312. doi: 10.1097/00002371-200307000-00002. [DOI] [PubMed] [Google Scholar]
- 10.Loftus DJ, Castelli C, Clay TM, Squarcina P, Marincola FM, Nishimura MI, Parmiani G, Appella E, Rivoltini L. Identification of epitope mimics recognized by CTL reactive to the melanoma/melanocyte-derived peptide MART-1(27–35) J Exp Med. 1996;184:647–657. doi: 10.1084/jem.184.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loftus DJ, Squarcina P, Nielsen MB, Geisler C, Castelli C, Odum N, Appella E, Parmiani G, Rivoltini L. Peptides derived from self-proteins as partial agonists and antagonists of human CD8 + T-cell clones reactive to melanoma/melanocyte epitope MART1(27–35) Cancer Res. 1998;58:2433–2439. [PubMed] [Google Scholar]
- 12.Mellman I, Steinman RM. Dendritic cells: specialized and regulated antigen processing machines. Cell. 2001;106:255–258. doi: 10.1016/S0092-8674(01)00449-4. [DOI] [PubMed] [Google Scholar]
- 13.Nicholson LB, Waldner H, Carrizosa AM, Sette A, Collins M, Kuchroo VK. Heteroclitic proliferative responses and changes in cytokine profile induced by altered peptides: implications for autoimmunity. Proc Natl Acad Sci USA. 1998;95:264–269. doi: 10.1073/pnas.95.1.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nijman HW, Houbiers JG, Vierboom MP, van der Burg SH, Drijfhout JW, D’Amaro J, Kenemans P, Melief CJ, Kast WM. Identification of peptide sequences that potentially trigger HLA-A2.1-restricted cytotoxic T lymphocytes. Eur J Immunol. 1993;23:1215–1219. doi: 10.1002/eji.1830230603. [DOI] [PubMed] [Google Scholar]
- 15.Parker KC, Bednarek MA, Coligan JE. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J Immunol. 1994;152:163–175. [PubMed] [Google Scholar]
- 16.Parkhurst MR, Fitzgerald EB, Southwood S, Sette A, Rosenberg SA, Kawakami Y. Identification of a shared HLA-A*0201-restricted T-cell epitope from the melanoma antigen tyrosinase-related protein 2 (TRP2) Cancer Res. 1998;58:4895–4901. [PubMed] [Google Scholar]
- 17.Renkvist N, Castelli C, Robbins PF, Parmiani G. A listing of human tumor antigens recognized by T cells. Cancer Immunol Immunother. 2001;50:3–15. doi: 10.1007/s002620000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rininsland FH, Helms T, Asaad RJ, Boehm BO, Tary-Lehmann M. Granzyme B ELISPOT assay for ex vivo measurements of T cell immunity. J Immunol Methods. 2000;240:143–155. doi: 10.1016/S0022-1759(00)00191-5. [DOI] [PubMed] [Google Scholar]
- 19.Rogers PR, Grey HM, Croft M. Modulation of naive CD4 T cell activation with altered peptide ligands: the nature of the peptide and presentation in the context of costimulation are critical for a sustained response. J Immunol. 1998;160:3698–3704. [PubMed] [Google Scholar]
- 20.Rosenberg SA. Development of cancer immunotherapies based on identification of the genes encoding cancer regression antigens. J Natl Cancer Inst. 1996;88:1635–1644. doi: 10.1093/jnci/88.22.1635. [DOI] [PubMed] [Google Scholar]
- 21.Rosenberg SA, Yang JC, Schwartzentruber DJ, Hwu P, Marincola FM, Topalian SL, Restifo NP, Dudley ME, Schwarz SL, Spiess PJ, Wunderlich JR, Parkhurst MR, Kawakami Y, Seipp CA, Einhorn JH, White DE. Immunologic and therapeutic evaluation of a synthetic peptide vaccine for the treatment of patients with metastatic melanoma. Nat Med. 1998;4:321–327. doi: 10.1038/nm0398-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruppert J, Sidney J, Celis E, Kubo RT, Grey HM, Sette A. Prominent role of secondary anchor residues in peptide binding to HLA-A2.1 molecules. Cell. 1993;74:929–937. doi: 10.1016/0092-8674(93)90472-3. [DOI] [PubMed] [Google Scholar]
- 23.Shafer-Weaver K, Sayers T, Strobl S, Derby E, Ulderich T, Baseler M, Malyguine A. The Granzyme B ELISPOT assay: an alternative to the 51Cr-release assay for monitoring cell-mediated cytotoxicity. J Transl Med. 2003;1:14. doi: 10.1186/1479-5876-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slansky JE, Rattis FM, Boyd LF, Fahmy T, Jaffee EM, Schneck JP, Margulies DH, Pardoll DM. Enhanced antigen-specific antitumor immunity with altered peptide ligands that stabilize the MHC-peptide-TCR complex. Immunity. 2000;13:529–538. doi: 10.1016/S1074-7613(00)00052-2. [DOI] [PubMed] [Google Scholar]
- 25.Terasawa H, Tsang KY, Gulley J, Arlen P, Schlom J. Identification and characterization of a human agonist cytotoxic T-lymphocyte epitope of human prostate-specific antigen. Clin Cancer Res. 2002;8:41–53. [PubMed] [Google Scholar]
- 26.Tourdot S, Scardino A, Saloustrou E, Gross DA, Pascolo S, Cordopatis P, Lemonnier FA, Kosmatopoulos K. A general strategy to enhance immunogenicity of low-affinity HLA-A2. 1-associated peptides: implication in the identification of cryptic tumor epitopes. Eur J Immunol. 2000;30:3411–3421. doi: 10.1002/1521-4141(2000012)30:12<3411::AID-IMMU3411>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 27.van der Burg SH, Visseren MJ, Brandt RM, Kast WM, Melief CJ. Immunogenicity of peptides bound to MHC class I molecules depends on the MHC-peptide complex stability. J Immunol. 1996;156:3308–3314. [PubMed] [Google Scholar]
- 28.Vergelli M, Hemmer B, Kalbus M, Vogt AB, Ling N, Conlon P, Coligan JE, McFarland H, Martin R. Modifications of peptide ligands enhancing T cell responsiveness imply large numbers of stimulatory ligands for autoreactive T cells. J Immunol. 1997;158:3746–3752. [PubMed] [Google Scholar]
- 29.Vertuani S, Sette A, Sidney J, Southwood S, Fikes J, Keogh E, Lindencrona JA, Ishioka G, Levitskaya J, Kiessling R. Improved immunogenicity of an immunodominant epitope of the HER-2/neu protooncogene by alterations of MHC contact residues. J Immunol. 2004;172:3501–3508. doi: 10.4049/jimmunol.172.6.3501. [DOI] [PubMed] [Google Scholar]
- 30.Yang S, Linette GP, Longerich S, Haluska FG. Antimelanoma activity of CTL generated from peripheral blood mononuclear cells after stimulation with autologous dendritic cells pulsed with melanoma gp100 peptide G209–2M is correlated to TCR avidity. J Immunol. 2002;169:531–539. doi: 10.4049/jimmunol.169.1.531. [DOI] [PubMed] [Google Scholar]
- 31.Yokoyama K, Suzuki H, Yasumoto K, Tomita Y, Shibahara S. Molecular cloning and functional analysis of a cDNA coding for human DOPAchrome tautomerase/tyrosinase-related protein-2. Biochim Biophys Acta. 1994;1217:317–321. doi: 10.1016/0167-4781(94)90292-5. [DOI] [PubMed] [Google Scholar]
- 32.Yu Z, Theoret MR, Touloukian CE, Surman DR, Garman SC, Feigenbaum L, Baxter TK, Baker BM, Restifo NP. Poor immunogenicity of a self/tumor antigen derives from peptide-MHC-I instability and is independent of tolerance. J Clin Invest. 2004;114:551–559. doi: 10.1172/JCI200421695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zaremba S, Barzaga E, Zhu M, Soares N, Tsang KY, Schlom J. Identification of an enhancer agonist cytotoxic T lymphocyte peptide from human carcinoembryonic antigen. Cancer Res. 1997;57:4570–4577. [PubMed] [Google Scholar]
- 34.Zhihua L, Yuzhang W, Bo Z, Bing N, Li W. Toward the quantitative prediction of T-cell epitopes: QSAR studies on peptides having affinity with the class I MHC molecular HLA-A*0201. J Comput Biol. 2004;11:683–694. doi: 10.1089/cmb.2004.11.683. [DOI] [PubMed] [Google Scholar]