Abstract
Ran is considered to be a promising target for tumor-specific immunotherapy because its protein is exclusively expressed in tumor tissues, though its mRNA can be expressed in most normal tissues. In our study, we obtained four candidate wild-type epitopes designated Ran1, Ran2, Ran3, and Ran4, derived from the Ran antigen with the highest predicted affinity with MHC-I, indicated by affinity prediction plots and molecular dynamics simulation. However, in vitro affinity assays of these epitopes showed only a moderate affinity with MHC-I. Thus, we designed altered peptide ligands (APLs) derived from Ran wild-type epitopes with preferred primary and auxiliary HLA-A*0201 molecule anchor residue replacement. Of the eight tested peptides, the 1Y analog had the strongest binding-affinity and lowest-dissociation rate to HLA-A*0201. Additionally, we investigated the CTLs activities induced by Ran wild-type peptides and the APLs in human PBMCs and in HLA-A*0201/Kb transgenic mice. Ran1 1Y was superior to other APLs and wild-type peptides in eliciting epitope-specific CTL immune responses both in vitro and in vivo. In summary, a wild-type epitope of the tumor-specific antigen Ran, expressed broadly in many tumors, was identified and designated Ran1. An APL of Ran1, Ran1 1Y, was further designed and verified in vitro and in vivo and found to elicit a stronger Ran-specific CTL response, indicating a potential anti-tumor application in the future.
Keywords: Epitope, Cytotoxic T lymphocyte, Altered peptide ligands, Ran, Cancer immunotherapy
Introduction
Anti-tumor vaccination is based on the existence of antigens, selectively or preferentially expressed by tumors, called tumor-associated antigens (TAAs) [1]. Cancer eradication using TAA vaccination has been demonstrated in numerous animal models [2]. Vaccination with peptides derived from TAAs designed to stimulate specific T-cells is being a practicable approach evaluated in clinical trials [3–5]. More than 170 antigenic peptides derived from 60 TAAs, many of which are widely expressed TAAs, have been expressed in the context of MHC molecules and recognized by cells in the available T-cell repertoire [6].
Ran (ras-related nuclear protein) is a small GTP-binding protein belonging to the RAS superfamily, essential for the translocation of RNA and proteins through the nuclear pore complex. Ran, mainly located in cell nucleus [7, 8] controls the cell cycle through the regulation of nucleo-cytoplasmic transport [9], mitotic spindle organization [10], and nuclear envelope formation [11–13]. Because of its increased expression in cancer cells and involvement in malignant transformation and/or the enhanced proliferation of cancer cells [14, 15], Ran was identified by Azuma et al. as a gene that codes for widely expressed TAA. Ran antigen was shown to be expressed in the majority of cancer cells at the protein level and in most normal tissues at the mRNA level. Two Ran-derived peptides were shown to be capable of inducing HLA-A33-restricted CTL activity against tumor cells in epithelial cancer patients [15]. On the other hand, HLA-A*0201 is the most common HLA-A allele in Asian populations, especially in the Chinese, with an estimated frequency of 50% [16]. Therefore, Ran represents a useful antigen target for specific immunotherapy against tumors in the clinic.
Several widely expressed TAAs or differentiation TAAs, described to date, including carcinoembryonic antigen (CEA), Survivin, Melan-A/MART-1, and gp100, represent self-proteins and as a result are poorly immunogenic due to immune tolerance. This most likely explains the failure of clinical trials in which self-proteins were used as immunogens. One potential strategy to counter this problem is altered peptide ligands (APL). Examples of these APLs are the Melan-A/MART-1 decamer 26-35 2L [17] and the gp100 209/2 M [18]. In 1993, Ruppert et al. [19] determined that a ‘canonical’ A2.1 motif could be defined as either L or M at position 2 (P2) and L, V, or I at position 9 (P9). In 2000, Tourdot et al. [20] demonstrated that at position 1 (P1), residue Y can enhance the MHC affinity for a low-affinity epitope. These results suggest that APL might be used to exploit a potential capacity of the T-cell repertoire to respond more effectively to the naïve epitope.
In the present study, candidate HLA-A*0201-binding peptides were predicted for Ran by using computer-based prediction algorithms. Four peptides with the highest predicted score were selected and synthesized. A peptide–MHC affinity assay was performed to determine their affinities to the HLA-A*0201 molecule. Subsequently, we modified these wild-type peptides to enhance their immunogenicity. Finally, the immunogenicity of each peptide was investigated in vitro (with PBMCs from healthy donors) and in vivo (with HLA-A*0201/Kb transgenic mice).
Materials and methods
Cell lines and animals
The Human TAP-deficient T2 cell line and BB7.2 cell line producing mAb against HLA-A*0201 were purchased from the American Type Culture Collection (ATCC, USA). The LB373-MEL cell line was a generous gift from Professor Francis Brasseur (Ludwig Institute for Cancer Research, Brussels Branch, Brussels, Belgium), the melanoma cell line M14 was provided by Dr. Zhongmin Zou (Institute of Combined Injury, Third Military Medical University, Chongqing, China), and the lung adenocarcinoma cell line A549 and breast carcinoma cell line MCF-7 were both provided by Dr. Guijun Huang (Institute of Respiratory Diseases, Third Military Medical University, Chongqing, China). The T2, A549, M14, and MCF-7 cell lines were all maintained in RPMI 1640 containing 10% FCS, and 100 μg/ml penicillin/streptomycin. The BB7.2 cell line was maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% FCS, 4 μg/l glucose and 100 μg/ml penicillin/streptomycin. All cell lines were kept at 37°C in a humidified atmosphere of 5% CO2 in air. Anti-HLA-A2 antibody was derived from BB7.2 (100 μl hybridoma culture supernatant/106 T2 cells) [21]. Anti-CD8 antibody was purchased from BD Biosciences Pharmingen (USA).
HLA-A*0201/Kb transgenic (Tg) mice, 8–12-weeks-old, were purchased from The Jackson Laboratory (USA). Mice were bred and maintained in specific pathogen-free (SPF) facilities. Animal experiments were performed in accordance with the guidelines of the Animal Care and Use Committee of Third Military Medical University.
Epitope prediction
The Ran sequence was scanned for HLA-A*0201-binding peptides using the prediction software BIMAS (Section of BioInformatics and Molecular Analysis, National Institutes of Health, Bethesda, MD, USA) [22] and SYFPEITHI (Institute for Immunology, University of Tübingen, Tübingen, Germany) [23]. The four 9-mer-peptides that showed the highest scores with both SYFPEITHI and BIMAS were selected for additional evaluation.
Molecular modeling
Models of HLA-A*0201 and binding 9-mer-peptides were established from the crystal structures of the Brookhaven Protein Data Bank: 3HLA for HLA-A*0201 and 3HSA for the nonameric peptides. The HLA-A*0201 model was simplified by using only 1 and 2 domains and 18 water molecules bound to them. Molecular mechanics and dynamics calculations were performed with the Discover 3.0 package. The force field parameters used in this study were the consistent valence force field. During the molecular dynamics and minimization, a dielectric constant of 1.0 was used and a 9.0 Å cut-off distance was applied to calculate the nonbinding interaction. The peptide ligand was first relaxed by 500 steps of conjugate gradient energy minimization, while maintaining in a fixed position. It was then submitted to a 100-ps molecular dynamics calculation at 300 K. During these 100 ps, no protein atom was allowed to move. The last conformation was then solvated in a 10 Å-thick TIP3P water shell. Energy minimization of the peptide ligand of the HLA-A*0201/ligand complex, followed by 200-ps molecular dynamics simulation of the full solvated HLA-A*0201/ligand pair was performed at 300 K.
Peptide synthesis
The candidate epitope peptides validated by epitope prediction and molecules modeling methods were then synthesized by Fmoc chemistry (Sangon, China), and purified by HPLC to a purity of >95%. Lyophilized peptides were dissolved in DMSO at a concentration of 20 mg/ml and stored at−70°C. The control peptide HBcAg(18–27) (FLPSDFFPSV) was synthesized and purified using the same methodology.
Peptide-binding assay
To determine whether the candidate epitopes can bind to HLA-A*0201 molecules, up-regulation of peptide-induced HLA-A*0201 molecules on T2 cells was examined according to a protocol described previously [21]. Briefly, 1 × 106 T2 cells were incubated with 50 μM of the synthesized peptides and 3 μg/ml of human β2-microglobulin (Serotec, UK) in serum-free RPMI 1640 medium for 16 h at 37°C/5% CO2. Expression of HLA-A*0201 on T2 cells was then determined with the FACS Calibur flow cytometer (Becton Dickinson, USA), by staining with primary anti-HLA-A2 Ab derived from BB7.2 and FITC-labeled goat-antimouse IgG (BD Biosciences Pharmingen, USA) secondary antibody. The data were analyzed using CellQuest software (Becton Dickinson, USA). The Fluorescence index (FI) was calculated as follows: FI = (mean FITC fluorescence with the given peptide − mean FITC fluorescence without peptide)/(mean FITC fluorescence without peptide). Samples were measured in triplicate and then mean FI was calculated.
Measurement of the peptide/HLA-A*0201 complex stability
As previously described [20], T2 cells (106/ml) were incubated overnight with 100 μM of each peptide in serum-free RPMI 1640 medium supplemented with 100 ng/ml β2-microglobin at 37°C. Thereafter, they were washed four times to remove free peptides, incubated with Brefeldin A (10 μg/ml) for 1 h to block HLA-A*0201 molecules newly expressed on the cell surface, and then washed and incubated at 37°C for 0, 2, 4, 6, or 8 h. Subsequently, the cells were stained with the BB7.2 antibody to evaluate the HLA-A*0201 molecule expression. For each time point, peptide-induced HLA-A*0201 expression was evaluated by the formula: mean fluorescence of peptide preincubated T2 cells − mean fluorescence of T2 cells treated in similar conditions in the absence of peptides. The dissociation complex50 (DC50) was defined as the time required for the loss of 50% of the stabilized HLA-A*0201/peptide complexes at time 0 h. The relative percentage of peptide complexes remaining at one time point was evaluated by the formula: mean fluorescence of peptide-induced HLA-A*0201 expression at one time point/mean fluorescence of peptide-induced HLA-A*0201 expression at time 0 h of the same peptide.
Analysis of Ran expression with RT-PCR and immunohistochemistry
RT-PCR was used to analyze the expression of Ran mRNA in all tumor cell lines, including LB373-MEL, A549, Jurkat, and MCF-7. Total RNA was isolated from tumor cell lines using Tripure Isolation Regent kit (Progema). Synthesis of cDNA was performed with 2 μg of total RNA with the aid of a reverse transcriptase kit (Progema) and oligo (dT) 18 primers. A 2 μl aliquot of RT product was amplified with PCR by using TaqDNA polymerase (Sangon, Shanghai) using standard procedures. The following oligonucleotides were used as primers: sense 5′-TATTGGTTGGTGATGGTGGT-3′; and antisense 5′-GTCGTGCTCATACTGTGCTG-3′. Both primers were synthesized commercially by Takara Company (Qingdao, China). Thirty amplification cycles were run: 1 min at 94°C; 1 min at 60°C; and 1 min at 72°C. Cycling was ceased with a final extension of 10 min at 72°C. RT-PCR products were then run on a gel and visualized with ethidium bromide. Moreover, the expressions of Ran protein in tumor and normal tissues were determined with immunohistochemistry. The primary antibody to the Ran antigen was purchased from BD and is a mouse monoclonal antibody against the human Ran protein.
Generation of CTLs in PBMCs from healthy donors
Dendritic cells (DCs) can effectively prime an antigen-specific T-cell response from autologous peripheral blood mononuclear cells (PBMCs) [24, 25]. In this study we used DCs to generate epitope-specific CTLs in PBMC from healthy donors (HLA-A*0201). The effector lymphocytes and DCs were prepared according to our published method [26].
ELISPOT assay
ELISPOT assays were performed using a commercially available kit (DIACLONE, France). T2 cells, pulsed with the indicated concentration of synthetic peptides, were used as stimulator cells. Effector cells (1 × 105) and peptide-prepulsed T2 cells (1 × 104) or the indicated amount of effector cells and tumor cells (1 × 104) were seeded into 96-well polyvinylidene Xuoride (PVDF)-backed microplates coated with monoclonal antibody specific for mouse interferon (mIFN-γ) or human IFN-γ. After incubation at 37°C for 16 h, cells were removed and plates were processed according to the manufacturer’s instructions. IFN-γ secreting T-cells were counted using the automated imaged analysis system ELISPOT reader.
Cytotoxicity assay
One week after the final restimulation, the cytolytic activity of the effector cells was determined by a standard 51Cr release assay in a V-bottom 96-well plate. MCF-7, A549, M14, and T2 cells, pulsed with each peptide, were labeled with Na51CrO4 for 1.5 h at 37°C according to our published methods [27, 28] and taken as the target cells. In order to assess autologous cytotoxicity, PHA-blast cells were prepared according to a protocol described previously [29].
Analysis of in vivo immunogenicity
HLA-A*0201/Kb mice were immunized with 100 μg of various peptides prepared in incomplete Freund’s adjuvant (IFA). As a control, mice were injected with an IFA emulsion without peptide. Eleven days after immunization, splenocytes from injected animals were cultured with 10 μg/ml of the immunizing peptide loaded with autologous splenocytes (as APCs) to expand the CTLs in vitro with mIL-2. After 5 days, the stimulated cells were used as effector cells and added to the targets.
Statistical analysis
Analysis of variance (ANOVA) and Student’s t-test were performed to determine the effects of the treatments. A difference was considered significant at P < 0.05.
Results
Identification of Ran-derived peptides binding to HLA-A*0201 molecules
The HLA-A*0201-restricted CTL epitopes of Ran were predicted by both BIMAS and SYFPEITHI software independently. The top four peptides IMFDVTSRV (designated Ran1), TLGVEVHPL (Ran2), YVATLGVEV (Ran3), and VLCGNKVDI (Ran4) with the highest predictive binding sore were selected. The peptides listed in Table 1 were synthesized with a purity of over 95% (Table 1).
Table 1.
Predicted Ran peptides
| Ran peptide | Position in Ran | Sequence | BIMAS score | BIMAS ranking | SYFPEITHI score | SYFPEITHI ranking |
|---|---|---|---|---|---|---|
| Ran1 | 88–96 | IMFDVTSRV | 1295.433 | 1 | 24 | 2 |
| Ran2 | 42–50 | TLGVEVHPL | 49.134 | 4 | 25 | 1 |
| Ran3 | 39–47 | YVATLGVEV | 27.995 | 6 | 23 | 3 |
| Ran4 | 118–126 | VLCGNKVDI | 17.736 | 8 | 23 | 4 |
The table shows the four peptides with the highest probability of binding to HLA-A2 predicted by “BIMAS” and “SYFPEITHI”
Molecular modeling showed that three potential CTL epitopes from Ran satisfied the criteria of HLA-A*0201-restricted CTL epitope with the exception of Ran4. As shown in Fig. 1 and Table 2, the three peptides bound to the HLA-A*0201 model structure possess a side chain of COOH-terminal anchor residues oriented into the binding groove at different distances from 17 to 19 Å. However, Ran4 might not be a candidate HLA-A*0201-restricted CTL epitope because of its large solvent-accessible surface area (SAS), compared with other peptides, and its inappropriate distance between P2 and P9 (>19 Å). Among all of the epitopes, Ran1 had lowest nonbond energy and represents the most stable of the peptide–MHC-I complexes. The Ran1 peptide therefore showed promising potential as a superior epitope candidate than the others at the prediction stage. Figure 1e shows a computerized depiction model of Ran1 with the HLA-A*0201 molecule. P1, P2, and P9 residues were able to orient into the binding groove of MHC-I molecule, but residues P3–P6 formed a different configuration protruding out of the binding groove that might form an antigenic surface recognizable by the T-cell receptor (TCR) on the CTL.
Fig. 1.
Molecular modeling of the candidate epitope peptides. The configuration of each candidate epitope, Ran1 (a), Ran2 (b), Ran3 (c), and Ran4 (d), was determined with molecular simulation. The side view was obtained when bound to the MHC-I molecule during simulation. The amino-acid sequences of the peptides used are listed in Table 1. Atom color-coding corresponds to the following: green, carbon; red, oxygen; blue, nitrogen; and white, hydrogen. A computerized model depicting the association of Ran (88–96) (IMFDVTSRV) with the HLA-A*0201 molecule is shown (e). The MHC model was constructed using crystallographic data of α1 and α2 heavy domains only (orange). The peptides are represented as a backbone structure (green)
Table 2.
Characteristic parameters of Ran wild-type peptides bound to HLA-A*0201 molecules
| Ran peptide | Sequence | Distance/Åa | SAS of anchor residue/Å2 b | |
|---|---|---|---|---|
| P2 | P9 | |||
| Ran1 | IMFDVTSRV | 17.63 | 3.6 (1.7%) | 4.9 (2.2%) |
| Ran2 | TLGVEVHPL | 18.20 | 3.7 (1.8%) | 4.4 (1.8%) |
| Ran3 | YVATLGVEV | 17.82 | 0.8 (8.5%) | 7.3 (3.3%) |
| Ran4 | VLCGNKVDI | 19.58 | 20.3 (9.5%) | 22.0 (8.6%) |
The adistance between P2 and P9 and the bSolvent-accessible surface area (SAS) of the anchor residue P2 and P9 were determined by the simulation method
Binding affinity of candidate epitope peptides with HLA-A*0201
A T2 cell–peptide binding test was used to evaluate the binding affinity of these peptides in vitro. As shown in Table 2, all of the peptides synthesized bound to HLA-A*0201 molecules but with different affinities. Of the four Ran peptides selected, Ran1, Ran2, and Ran3 showed moderate affinity to HLA-A*0201 (FI = 1.17, 1.04, and 1.09, respectively), whereas Ran4 had the lowest affinity (FI = 0.42).
Modification of candidate epitope peptides
Due to the intermediate affinity (FI < 1.5) of the candidate epitopes to the HLA-A*0201 molecule, previous strategies were applied to modify these candidate epitope peptides to enhance their immunogenicities [19, 20]. As residue I at P1 of Ran1, T at P1 of Ran1, and V at P2 of Ran3 were not the predominant anchor residues, we altered the naïve epitope Ran1 and Ran2 at P1 (Y) and the naïve epitope Ran3 at P2 (L, M). The T2 cell–peptide binding test was performed to evaluate the binding affinity of these peptides in vitro. Of the four candidate peptides, Ran1 1Y and Ran2 1Y were high-affinity epitopes (FI = 2.24 and 2.43, respectively), while Ran3 2L and Ran3 2 M have relatively lower-affinity to HLA-A*0201 (FI = 1.90 and 1.85, respectively). All APLs have stronger affinity than their wild-type candidate epitope peptides (Table 3).
Table 3.
Characteristics of APLs derived from Ran wild-type peptides bound to HLA-A*0201 molecules
| Wild-type peptides | FIb | DC50c | Modified peptides | Sequence | FIb | DC50c |
|---|---|---|---|---|---|---|
| Ran1 | 1.17 | 6−8 | Ran1 1Y | YMFDVTSRV | 2.24 | >8 |
| Ran2 | 1.04 | 2 | Ran2 1Y | YLGVEVHPL | 2.43 | 6–8 |
| Ran3 | 1.09 | 2 | Ran3 2L | YLATLGVEV | 1.90 | 6–8 |
| Ran3 | 1.09 | 2 | Ran3 2M | YMATLGVEV | 1.85 | 6–8 |
| Ran4 | 0.42 | 0 | – | – | – | – |
| HBcAga | 1.89 | >8 | – | FLPSDFFPSV | 1.89 | >8 |
aHBcAg(18–27) served as positive control
bFI was calculated with the following formula: FI = MFI sample − MFI background/MFI background. Background means no peptide stimulation. FI was determined as high (FI > 1.5), intermediate (1.0 < FI < 1.5), or weak (FI < 0.5)
cDC50 was defined as the time required for the loss of 50% of the HLA-A*0201/peptide complexes stabilized at time 0 was calculated
Measurement of the peptide/HLA-A*0201 complex stability
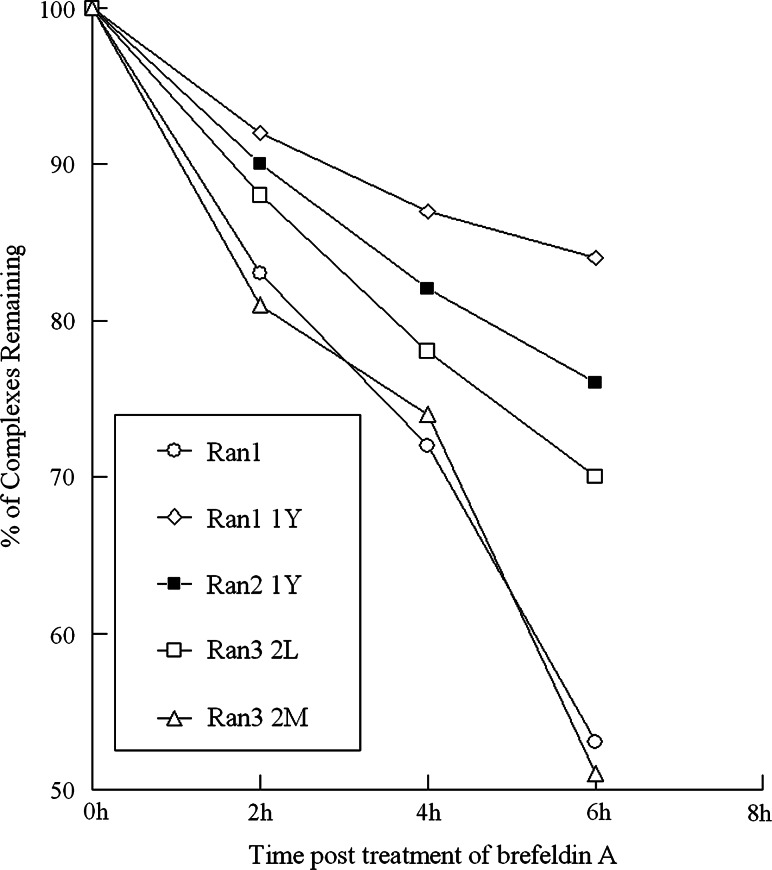
Previous studies have shown that a stable peptide–MHC complex could facilitate the formation of synapses between T-cells and APCs and the stability of peptide–MHC complex was the key factor for CTL activation [30] especially in self/tumor antigens [31]. Thus, in this study, the stability of the peptide–MHC complex for the candidate epitopes was investigated using T2-based analysis. The results showed Ran1 1Y had the longest DC50 (Table 3), whereas the DC50 of three wild-type peptides Ran2, Ran3, and Ran4 was <2 h (Table 2). Moreover, the Ran1 1Y/HLA-A*0201 complex was most stable among all of the peptide/HLA-A*0201 complexes over the 8-h observation period. As shown in Fig. 2, the remaining percentage of peptide–MHC complex was 84, 76, 70, 53, and 51% for Ran1 1Y, Ran2 1Y, Ran3 2L, Ran 3 2 M, and Ran1, respectively. Thus, among all epitope peptides, Ran1 1Y showed the highest affinity and stability with HLA-A*0201 molecules.
Fig. 2.
Comparison of the stability of the peptides complex with the HLA-A*0201 molecule (DC50 > 6 h). T2 cells were incubated overnight with peptide at a concentration of 100 μg/ml and then washed free of unbound peptide and incubated with Brefeldin A to block delivery of new class I molecules to the cell surface. At the indicated times, cells were stained for the presence of surface peptide–HLA-A*0201 complexes. Results are presented as the relative percentage of binding compared with 100% at time 0
In vitro induction of epitope-specific CTLs from PBMC
We then investigated the capacity for priming the CTL response in vitro with four wild-type candidate epitope peptides in three normal HLA-A*0201 individuals using the IFN-γ ELISPOT analysis. The data showed that only Ran1 could prime a significant amount of epitope-specific CTLs, compared with an irrelevant peptide control (Fig. 3a). Next we tested the immunogenicity of four APLs. When compared with Ran1 and other APLs-induced CTLs, Ran1 1Y-induced CTLs were shown to possess a significantly increased capacity to secrete IFN-γ by stimulation with 100, 10, or 1 μg of Ran1 loaded to T2 cells (Fig. 3b). Importantly, when T2 cells loaded with 1 μg of Ran1 peptide, Ran1 1Y-induced CTLs, they retained the greatest capability of IFN-γ secretion, equivalent of coculturing with T2 loaded with 100 or 10 μg of Ran1 peptide. However, the capacity for IFN-γ production of other three APLs established CTLs declined sharply when stimulated with T2 cell pulsed with 1 μg of the corresponding wild-type counterparts. After blocking the HLA-A*0201 or CD8 molecules with anti-HLA-A2 Ab and anti-CD8 Ab respectively, each peptide-loaded T2 cells could hardly induce any specific T-cells response and resulted in little IFN-γ secretion. Similar results were obtained by stimulation with the negative control (HBcAg18–27). These data indicated that the release of IFN-γ by these CTLs, upon recognition of T2 cells pulsed with various epitope peptides, was HLA-A2-restricted and CD8-dependent.
Fig. 3.
IFN-γ-producing CTLs primed by peptides. a The four candidate epitope peptides were used to stimulate PBMC from healthy HLA-A*0201-positive donors for 3 weeks at weekly intervals. The four kinds of primed CTLs were tested against T2 cells pulsed with Ran peptide or irrelevant peptide HBcAg18–27. b Ran1 1Y-CTL, Ran2 1Y-CTL, Ran3 2L-CTL, and Ran3 2M-CTL were tested against T2 cells prepulsed with their wild-type counterparts (100, 10, or 1 μg). Anti-HLA-A2 and anti-CD8 antibodies were, respectively, added into the culture wells containing T2 cells and peptide-induced CTLs. IFN-γ production by peptide-induced CTLs was measured using the ELISPOT assay. Experiments were repeated three times. Data represented mean ± S.D. * P < 0.05, compared with control group
Cytolytic activity of epitope-specific CTLs
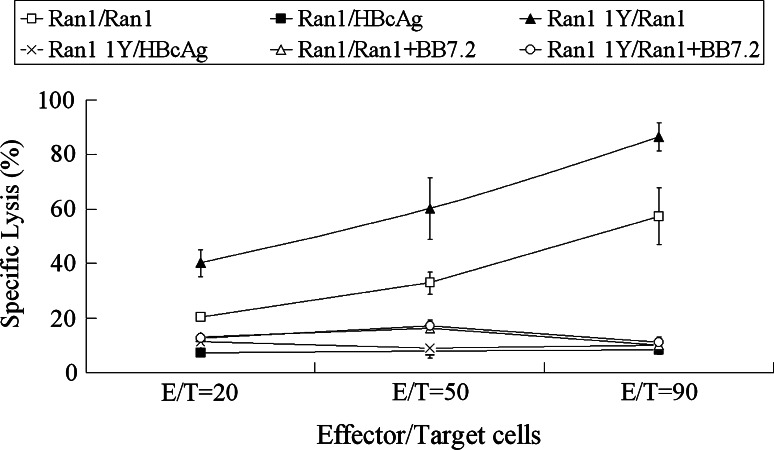
To address the question of whether IFN-γ-producing CTL lines could lyse target cells, a 51Cr release assay was performed. The expression of Ran was detected in MCF-7 [32], A549 (HLA-A*0201+), and M14 cells (HLA-A*0201−) at the mRNA and protein level by RT-PCR and immunohistochemistry, respectively (data not shown). MCF-7 cells (HLA-A*0201+, Ran+) and peptide-prepulsed T2 cells were used as target cells. Among the seven peptides tested, only peptide Ran1 and Ran1Y were able to elicit Ran-specific CTLs, which could lyse MCF-7 cells (Fig. 4). The results showed that 62.3 and 84.4% of MCF-7 cells were lysed by Ran1 and Ran1 1Y-induced CTLs at E/T ratio 90:1, respectively (Fig. 5), whereas no detectable cytolytic activity was seen in the case of the other wild-type peptides and APLs (Fig. 4). After 21 days of stimulation with Ran and Ran 1 1Y at the concentration of 10 μg/ml, the induced effectors could lyse MCF-7 and A549 cells (HLA-A*0201+, Ran+) but not M14 cells (HLA-A*0201−, Ran+; Fig. 5). To determine possible cross-reactivity of peptide-induced CTLs to normal cells, cytotoxicity was examined on autologous PHA-blast cells. As shown in Fig. 5, Ran1 and Ran1 1Y-induced CTLs exhibited no substantial cytotoxicity to these autologous cells. When anti-HLA-A2 Ab derived from BB7.2 was added to the culture wells during the cytolytic assay, anti-HLA-A2 Ab could significantly eliminate the cytotoxicity of the induced CTLs in vitro (Fig. 5). These results indicated that these effector cells recognized target cell in a HLA-A*0201-restricted manner.
Fig. 4.
Specific lysis of various cell lines by cells induced by wild-type peptides and APLs. Effectors were induced from the PBMCs of four HLA-A*0201 healthy donors as described in “Materials and methods”. 51Cr-release assays were performed to test for their cytotoxic activity against MCF-7 cells (HLA-A*0201+, Ran+), T2 cell prepulsed with wild-type peptides (HLA-A*0201+, Ran+), and T2 cells (HLA-A*0201+, Ran−) at 90:1 E/T ratio. Experiments were repeated three times. Data are represented as mean ± S.D. * P < 0.05, compared with control group
Fig. 5.
Specific lysis of various cell lines by cells induced by Ran1 and Ran1 1Y. MCF-7 (HLA-A*0201+, Ran+), A549 (HLA-A*0201+, Ran+), M14 (HLA-A*0201-, Ran+), and PHA-blast cells (HLA-A*0201+, Ran−) were incubated with or without Anti-HLA-A2 antibody from BB7.2 cells. The cytotoxic activity of the Ran1 (a) and Ran1 1Y (b) induced cells was determined against these cells at various E/T ratios by 51Cr-release assay. Experiments were repeated three times. Data are represented as mean ± S.D
In vivo induction of epitope-specific CTLs in HLA-A2.1/Kb transgenic mice
We investigated, whether Ran1 1Y would be more efficient than Ran1 in inducing immunity in vivo. T2 cells pulsed with the Ran1 peptide or irrelevant peptide HBcAg18–27 was used as target cells. The cytolytic assay showed that the CTLs primed from Ran1 or Ran1 1Y-inoculated mice could lyse T2-cells pulsed with the Ran1 peptides but not T2-cells loaded with the irrelevant peptide. According to the results of the in vitro cytolytic assay, Ran1 1Y primed CTL have a higher lytic ability than Ran1 primed CTL (86.4 vs. 57.4% at E/T ratio 90:1; Fig. 6). Moreover, anti-HLA-A2 Ab could inhibit the peptides induced splenocytes from killing the targets (Fig. 6). These results suggest that the higher immunogenicity in vivo of Ran1 1Y was achieved in an antigen-specific and HLA-A*0201-restricted fashion.
Fig. 6.
Ran1 1Y can induce stronger specific CTLs in vivo in HLA-A2.1/Kb transgenic mice than its wild-type counterpart, Ran1. The effector cells and the target cells were prepared from the splenocytes from HLA-A*0201/Kb mice and T2 cells (HLA-A*0201) pulsed with Ran1 peptide, respectively as described in “Materials and methods”. Anti-HLA-A2 antibody from BB7.2 cells were added after co-culture of the effectors and the target cells at the indicated E/T ratios, the 51Cr-release rates of the target cells were examined. T2 cells pulsed with HBcAg18–27 epitope peptide were used as a negative control. Experiments were repeated three times. Data are represented as means ± S.D
Discussion
Molecular identification and characterization of novel TAAs has rapidly evolved over the past few years. Many T-cell epitopes have been identified from TAAs, such as MAGE-1 [33], MAGE-3 [34], and TRP-2 [35]. Ran is highly expressed at both the mRNA and protein levels in most cancer cell lines or cancer tissues tested, but present at low levels in most normal tissues [15, 36]. Significantly higher expression levels of Ran proteins were demonstrated in prostate cancer tissues than in the normal tissues [37]. What’s more, Ran binding proteins, such as RanBP7 and Ran-BPM have been reported to be preferentially expressed in cancer tissues and to be associated with the increased proliferation of cancer cells [38, 39]. Furthermore, a recent study has shown that Ran was associated with the higher grade, local invasion, and metastasis of renal cancer [40]. In view of the finding that Ran and its associated proteins are strongly and selectively expressed in various cancer tissues, Ran may be an ideal target molecule for the treatment of patients with cancer.
In this study, we first predicted candidate epitopes from the Ran antigen with a combination of two different professional software programs. We obtained four candidate epitopes based on highest immunogenicity scores: Ran1, Ran2, Ran3, and Ran4. Second, we validated them using a molecular dynamics simulation method and found that one epitope Ran4 was not compatible with the HLA-A*0201-restricted CTL epitope because of the largest solvent-accessible surface area (SAS). The SAS of each residue from the epitope peptide can directly reflect the status of the packing interaction between peptide and MHC-I molecule. The larger the SAS is, the weaker the combination would be. Epitope peptides combine with MHC-I molecule mainly through weak interactions, such as hydrophobic interaction, hydrogen bonds, and salt bonds. The stronger the noncovalent bonds, the more stable the peptide–MHC-I complex. As shown in Table 3, Ran1 had the lowest nonbond energy which represents the strongest of the noncovalent bonds. Therefore, the Ran1 peptide was predicted most promising, as an epitope candidate. Third, a peptide-binding assay was used to determine the affinity of every epitope with HLA-A*0201 and showed that Ran1, Ran2, and Ran3 had moderate affinity, whereas Ran4 had the lowest affinity. This is in agreement with the results of our molecular dynamics simulation.
Our results indicated that none of the candidate epitopes have strong affinity to the HLA-A*0201 molecule. A similar phenomenon has also been observed in several other TAAs, such as Melan-A/ MART-1, gp100, and CEA. Like most other TAAs, Ran is a self-antigen and tolerance toward Ran-derived peptides with high affinity for the MHC molecules is therefore to be expected. However, Ran-derived peptides with intermediate binding affinities for MHC-I molecules might be insufficiently presented to trigger naïve CTLs, but presented at levels capable of stimulating already activated CTLs. Fortunately, a breaking of tolerance and enhancement of affinity could be achieved by immunizing with antigenic determinants provided in an optimized form defined as APL. The use of APL in vaccine therapy has now been reported in several studies i.e., gp100, and Melan-A/ MART-1. These APLs display higher affinity for binding to HLA class I molecules, thus creating a complex that can interact more efficiently with the cognate TCR [17, 18]. Furthermore, clinical trials indicate these APLs to appear to be more potent in activating naïve T-cells than their wild-type peptides, produce more clinical responses in patients, and show more regression of metastases [41, 42]. Thus, in the present study, three moderate affinity Ran epitopes were modified at the P1 and P2 positions, respectively, to achieve stronger HLA-A*0201 binding.
We further examined the affinity of APLs to HLA-A*0201 by in vitro affinity assay, which showed that the APLs had much stronger affinities to the MHC-I molecule compared to their wild-type candidate peptides. However, former studies indicated that the stability of peptide–MHC binding may facilitate the formation of the immune synapses between T-cells and APCs and warrant full T-cell activation through sustained signaling [43], and therefore result in increased peptide immunogenicity. Data herein indicates that the Ran1 1Y peptide had the lowest-dissociation rate, and formed a more stable peptide–MHC complex that persisted at cell surface for an interval sufficient to allow the induction of a strong CTL response. An in vitro CTL induction assay confirmed that the Ran1 1Y peptide could prime the more potent epitope-specific CTLs with significantly enhanced IFN-γ-producing T-cells and cytolytic activity. However, several studies have shown that some of the in vitro identified CTL epitopes had little immunogenicity in vivo and could not prime specific CTLs, partly because these peptides could not be effectively processed and presented by APCs [44, 45]. In order to prove peptide Ran1 and Ran1 1Y to be effective CTL epitopes in vivo, HLA-A*0201 transgenic mice were immunized with these two epitopes. The results showed that either Ran1 1Y or Ran1 primed CTLs in HLA-A*0201 transgenic mice could kill T2 cells pulsed with the Ran1 peptide. Moreover, Ran1 1Y primed CTLs have a relatively higher lysing ability. These results indicate that Ran1 1Y had a superior capacity to elicit immunity in vitro and in vivo, compared with its wild-type counterpart Ran1.
Despite the promising results achieved with preclinical studies of APL, this approach has not yet demonstrated a superior therapeutic potential as compared with other vaccination approaches [46]. Recent studies reported that T-cells primed by APLs with higher affinity binding to MHC might be unable to effectively cross-recognize tumor cells [47, 48]. The introduction of amino-acid substitutions in a peptide sequence may affect TCR binding to the peptide–MHC complex. This could influence the TCR repertoire of APL-induced T-cells and thus explain the lack or weak tumor cytotoxic activity observed with APLs [46]. Hence, structural modifications of peptides should be selected without altering TCR binding moieties to ensure that vaccination-primed T-cells are specific for natural antigen and efficiently recognize tumor cells [46]. In addition, novel computer-based algorithms could be used for the streamlined design of APL with retained activity on anti-tumor immunity [49]. In this study, our results confirmed that the Ran1 1Y-induced T-cells could efficiently recognize the endogenously processed epitope expressed by tumor cells in vitro. Similar results were obtained in Melan-A/MART-1, whose specific T-cells significantly cross-react with tumor in most cases analyzed [17, 50]. Nevertheless, in a recent study by Speiser et al., wild-type epitope induced T-cells showed superior tumor reactivity compared with Melan-A/MART-1 induced T-cells when CpG was used as an adjuvant [51]. This study demonstrated that wild-type epitope, if combined with potent new generation adjuvant, could still represent a valid immunization tool for future immunotherapy [51]. Thus, the actual immunogenicity and clinical efficacy of Ran1 1Y and wild-type epitope Ran1 should both be investigated in further studies.
In conclusion, our results suggest that the Ran1(88–96) IMFDVTSRV peptide derived from Ran might be capable of inducing HLA-A*0201-restricted CD8+ CTL, which would be lethal for tumor cells expressing Ran and HLA-A*0201. Furthermore, 1Y analog of Ran1, which is formed by the introduction of preferred amino-acid residues into the Ran1 wild peptide results in an increased affinity for the HLA-A*0201 allele and could induce more efficient tumor cell lysis compared with wild-type peptide. Currently, we are planning to perform preclinical trials of these two Ran epitope vaccines in a humanized animal model, the Trimera mouse, to reveal if Ran1 and its 1Y analog are capable of potential use in cancer-specific immunotherapy in patients.
Acknowledgments
This work was supported by the Major State Basic Research Development Program of China (973 Program) (No. 2007CB512401 and 2007CB512805), the National High Technology Research and Development Program of China (863 Program) (No. 2007AA021603 and 2006AA02A207), the Major Program of National Natural Science Foundation of China (No. 30490241), the General program of National Natural Science Foundation of China (No. 30771950, 30400437, 30872352, and 30571835) and CSTC (2008BB5033).
Footnotes
Fan Li, Di Yang, Yiqin Wang, and Baohua Liu contributed equally to this work.
Contributor Information
Bing Ni, Phone: +86-23-68772348, FAX: +86-23-68772348, Email: nibingxi@yahoo.com.
Yuzhang Wu, Phone: +86-23-68752680, FAX: +86-23-68752789, Email: wuyuzhang@yahoo.com.
References
- 1.Rosenberg SA. A new era for cancer immunotherapy based on the genes that encode cancer antigens. Immunity. 1999;10:281–287. doi: 10.1016/S1074-7613(00)80028-X. [DOI] [PubMed] [Google Scholar]
- 2.Rosenberg SA. Progress in human tumour immunology and immunotherapy. Nature. 2001;411:380–384. doi: 10.1038/35077246. [DOI] [PubMed] [Google Scholar]
- 3.Ribas A, Butterfield LH, Glaspy JA, Economou JS. Current developments in cancer vaccines and cellular immunotherapy. J Clin Oncol. 2003;21:2415–2432. doi: 10.1200/JCO.2003.06.041. [DOI] [PubMed] [Google Scholar]
- 4.Finn OJ. Cancer vaccines: between the idea and the reality. Nat Rev Immunol. 2003;3:630–641. doi: 10.1038/nri1150. [DOI] [PubMed] [Google Scholar]
- 5.Mine T, Gouhara R, Hida N, Imai N, Azuma K, Rikimaru T, Katagiri K, Nishikori M, Sukehiro A, Nakagawa M, Yamada A, Aizawa H, Shirouzu K, Itoh K, Yamana H. Immunological evaluation of CTL precursor-oriented vaccines for advanced lung cancer patients. Cancer Sci. 2003;94:548–556. doi: 10.1111/j.1349-7006.2003.tb01481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Novellino L, Castelli C, Parmiani G. A listing of human tumor antigens recognized by T cells: March 2004 update. Cancer Immunol Immunother. 2005;54:187–207. doi: 10.1007/s00262-004-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. doi: 10.1016/S0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- 8.Kunzler M, Hurt E. Targeting of Ran: variation on a common theme? J Cell Sci. 2001;114:3233–3241. doi: 10.1242/jcs.114.18.3233. [DOI] [PubMed] [Google Scholar]
- 9.Kierszenbaum AL, Gil M, Rivkin E, Tres LL. Ran, a GTP-binding protein involved in nucleocytoplasmic transport and microtubule nucleation, relocates from the manchette to the centrosome region during rat spermiogenesis. Mol Reprod Dev. 2002;63:131–140. doi: 10.1002/mrd.10164. [DOI] [PubMed] [Google Scholar]
- 10.Li HY, Zheng Y. Phosphorylation of RCC1 in mitosis is essential for producing a high RanGTP concentration on chromosomes and for spindle assembly in mammalian cells. Genes Dev. 2004;18:512–527. doi: 10.1101/gad.1177304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang C, Clarke PR. Chromatin-independent nuclear envelope assembly induced by Ran GTPase in Xenopus egg extracts. Science. 2000;288:1429–1432. doi: 10.1126/science.288.5470.1429. [DOI] [PubMed] [Google Scholar]
- 12.Hetzer M, Bilbao-Cortes D, Walther TC, Gruss OJ, Mattaj IW. GTP hydrolysis by Ran is required for nuclear envelope assembly. Mol Cell. 2000;5:1013–1024. doi: 10.1016/S1097-2765(00)80266-X. [DOI] [PubMed] [Google Scholar]
- 13.Vasu SK, Forbes DJ. Nuclear pores and nuclear assembly. Curr Opin Cell Biol. 2001;13:363–375. doi: 10.1016/S0955-0674(00)00221-0. [DOI] [PubMed] [Google Scholar]
- 14.Kornbluth S, Dasso M, Newport J. Evidence for a dual role for TC4 protein in regulating nuclear structure and cell cycle progression. J Cell Biol. 1994;125:705–719. doi: 10.1083/jcb.125.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Azuma K, Sasada T, Takedatsu H, Shomura H, Koga M, Maeda Y, Yao A, Hirai T, Takabayashi A, Shichijo S, Itoh K. Ran, a small GTPase gene, encodes cytotoxic T lymphocyte (CTL) epitopes capable of inducing HLA-A33-restricted and tumor-reactive CTLs in cancer patients. Clin Cancer Res. 2004;10:6695–6702. doi: 10.1158/1078-0432.CCR-04-0818. [DOI] [PubMed] [Google Scholar]
- 16.Shieh DC, Lin DT, Yang BS, Kuan HL, Kao KJ. High frequency of HLA-A*0207 subtype in Chinese population. Transfusion. 1996;36:818–821. doi: 10.1046/j.1537-2995.1996.36996420761.x. [DOI] [PubMed] [Google Scholar]
- 17.Valmori D, Fonteneau JF, Lizana CM, Gervois N, Lienard D, Rimoldi D, Jongeneel V, Jotereau F, Cerottini JC, Romero P. Enhanced generation of specific tumor-reactive CTL in vitro by selected Melan-A/MART-1 immunodominant peptide analogues. J Immunol. 1998;160:1750–1758. [PubMed] [Google Scholar]
- 18.Parkhurst MR, Salgaller ML, Southwood S, Robbins PF, Sette A, Rosenberg SA, Kawakami Y. Improved induction of melanoma-reactive CTL with peptides from the melanoma antigen gp100 modified at HLA-A*0201-binding residues. J Immunol. 1996;157:2539–2548. [PubMed] [Google Scholar]
- 19.Ruppert J, Sidney J, Celis E, Kubo RT, Grey HM, Sette A. Prominent role of secondary anchor residues in peptide binding to HLA-A2.1 molecules. Cell. 1993;74:929–937. doi: 10.1016/0092-8674(93)90472-3. [DOI] [PubMed] [Google Scholar]
- 20.Tourdot S, Scardino A, Saloustrou E, Gross DA, Pascolo S, Cordopatis P, Lemonnier FA, Kosmatopoulos K. A general strategy to enhance immunogenicity of low-affinity HLA-A2.1-associated peptides: implication in the identification of cryptic tumor epitopes. Eur J Immunol. 2000;30:3411–3421. doi: 10.1002/1521-4141(2000012)30:12<3411::AID-IMMU3411>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 21.Nijman HW, Houbiers JG, Vierboom MP, van der Burg SH, Drijfhout JW, D’Amaro J, Kenemans P, Melief CJ, Kast WM. Identification of peptide sequences that potentially trigger HLA-A2.1-restricted cytotoxic T lymphocytes. Eur J Immunol. 1993;23:1215–1219. doi: 10.1002/eji.1830230603. [DOI] [PubMed] [Google Scholar]
- 22.Parker KC, Bednarek MA, Coligan JE. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J Immunol. 1994;152:163–175. [PubMed] [Google Scholar]
- 23.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 24.Yang S, Linette GP, Longerich S, Haluska FG. Antimelanoma activity of CTL generated from peripheral blood mononuclear cells after stimulation with autologous dendritic cells pulsed with melanoma gp100 peptide G209-2M is correlated to TCR avidity. J Immunol. 2002;169:531–539. doi: 10.4049/jimmunol.169.1.531. [DOI] [PubMed] [Google Scholar]
- 25.Mellman I, Steinman RM. Dendritic cells: specialized and regulated antigen processing machines. Cell. 2001;106:255–258. doi: 10.1016/S0092-8674(01)00449-4. [DOI] [PubMed] [Google Scholar]
- 26.Tang Y, Lin Z, Ni B, Wei J, Han J, Wang H, Wu Y. An altered peptide ligand for naive cytotoxic T lymphocyte epitope of TRP-2(180–188) enhanced immunogenicity. Cancer Immunol Immunother. 2007;56:319–329. doi: 10.1007/s00262-006-0195-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu B, Chen Z, Cheng X, Lin Z, Guo J, Jia Z, Zou L, Wang Z, Hu Y, Wang D, Wu Y. Identification of HLA-A*0201-restricted cytotoxic T lymphocyte epitope from TRAG-3 antigen. Clin Cancer Res. 2003;9:1850–1857. [PubMed] [Google Scholar]
- 28.Ni B, Gao W, Zhu B, Lin Z, Jia Z, Zhou W, Zhao J, Wang L, Wu Y. Induction of specific human primary immune responses to a Semliki Forest virus-based tumor vaccine in a Trimera mouse model. Cancer Immunol Immunother. 2005;54:489–498. doi: 10.1007/s00262-004-0591-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ida K, Kawaguchi S, Sato Y, Tsukahara T, Nabeta Y, Sahara H, Ikeda H, Torigoe T, Ichimiya S, Kamiguchi K, Wada T, Nagoya S, Hiraga H, Kawai A, Ishii T, Araki N, Myoui A, Matsumoto S, Ozaki T, Yoshikawa H, Yamashita T, Sato N. Crisscross CTL induction by SYT-SSX junction peptide and its HLA-A*2402 anchor substitute. J Immunol. 2004;173:1436–1443. doi: 10.4049/jimmunol.173.2.1436. [DOI] [PubMed] [Google Scholar]
- 30.Slansky JE, Rattis FM, Boyd LF, Fahmy T, Jaffee EM, Schneck JP, Margulies DH, Pardoll DM. Enhanced antigen-specific antitumor immunity with altered peptide ligands that stabilize the MHC-peptide-TCR complex. Immunity. 2000;13:529–538. doi: 10.1016/S1074-7613(00)00052-2. [DOI] [PubMed] [Google Scholar]
- 31.Yu Z, Theoret MR, Touloukian CE, Surman DR, Garman SC, Feigenbaum L, Baxter TK, Baker BM, Restifo NP. Poor immunogenicity of a self/tumor antigen derives from peptide–MHC-I instability and is independent of tolerance. J Clin Invest. 2004;114:551–559. doi: 10.1172/JCI21695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han JF, Zhao TT, Liu HL, Lin ZH, Wang HM, Ruan ZH, Zou LY, Wu YZ. Identification of a new HLA-A*0201-restricted cytotoxic T lymphocyte epitope from CML28. Cancer Immunol Immunother. 2006;55:1575–1583. doi: 10.1007/s00262-006-0152-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Celis E, Fikes J, Wentworth P, Sidney J, Southwood S, Maewal A, Del Guercio MF, Sette A, Livingston B. Identification of potential CTL epitopes of tumor-associated antigen MAGE-1 for five common HLA-A alleles. Mol Immunol. 1994;31:1423–1430. doi: 10.1016/0161-5890(94)90158-9. [DOI] [PubMed] [Google Scholar]
- 34.van der Bruggen P, Bastin J, Gajewski T, Coulie PG, Boel P, De Smet C, Traversari C, Townsend A, Boon T. A peptide encoded by human gene MAGE-3 and presented by HLA-A2 induces cytolytic T lymphocytes that recognize tumor cells expressing MAGE-3. Eur J Immunol. 1994;24:3038–3043. doi: 10.1002/eji.1830241218. [DOI] [PubMed] [Google Scholar]
- 35.Parkhurst MR, Fitzgerald EB, Southwood S, Sette A, Rosenberg SA, Kawakami Y. Identification of a shared HLA-A*0201-restricted T-cell epitope from the melanoma antigen tyrosinase-related protein 2 (TRP2) Cancer Res. 1998;58:4895–4901. [PubMed] [Google Scholar]
- 36.Xia F, Lee CW, Altieri DC. Tumor cell dependence on Ran-GTP-directed mitosis. Cancer Res. 2008;68:1826–1833. doi: 10.1158/0008-5472.CAN-07-5279. [DOI] [PubMed] [Google Scholar]
- 37.Li P, Yu X, Ge K, Melamed J, Roeder RG, Wang Z. Heterogeneous expression and functions of androgen receptor co-factors in primary prostate cancer. Am J Pathol. 2002;161:1467–1474. doi: 10.1016/S0002-9440(10)64422-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li SR, Gyselman VG, Dorudi S, Bustin SA. Elevated levels of RanBP7 mRNA in colorectal carcinoma are associated with increased proliferation and are similar to the transcription pattern of the proto-oncogene c-myc. Biochem Biophys Res Commun. 2000;271:537–543. doi: 10.1006/bbrc.2000.2666. [DOI] [PubMed] [Google Scholar]
- 39.Emberley ED, Gietz RD, Campbell JD, HayGlass KT, Murphy LC, Watson PH. RanBPM interacts with psoriasin in vitro and their expression correlates with specific clinical features in vivo in breast cancer. BMC Cancer. 2002;2:28. doi: 10.1186/1471-2407-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abe H, Kamai T, Shirataki H, Oyama T, Arai K, Yoshida K. High expression of Ran GTPase is associated with local invasion and metastasis of human clear cell renal cell carcinoma. Int J Cancer. 2008;122:2391–2397. doi: 10.1002/ijc.23400. [DOI] [PubMed] [Google Scholar]
- 41.Loftus DJ, Squarcina P, Nielsen MB, Geisler C, Castelli C, Odum N, Appella E, Parmiani G, Rivoltini L. Peptides derived from self-proteins as partial agonists and antagonists of human CD8+ T-cell clones reactive to melanoma/melanocyte epitope MART1(27–35) Cancer Res. 1998;58:2433–2439. [PubMed] [Google Scholar]
- 42.Rosenberg SA, Yang JC, Schwartzentruber DJ, Hwu P, Marincola FM, Topalian SL, Restifo NP, Dudley ME, Schwarz SL, Spiess PJ, Wunderlich JR, Parkhurst MR, Kawakami Y, Seipp CA, Einhorn JH, White DE. Immunologic and therapeutic evaluation of a synthetic peptide vaccine for the treatment of patients with metastatic melanoma. Nat Med. 1998;4:321–327. doi: 10.1038/nm0398-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huppa JB, Gleimer M, Sumen C, Davis MM. Continuous T cell receptor signaling required for synapse maintenance and full effector potential. Nat Immunol. 2003;4:749–755. doi: 10.1038/ni951. [DOI] [PubMed] [Google Scholar]
- 44.Shen L, Rock KL. Cellular protein is the source of cross-priming antigen in vivo. Proc Natl Acad Sci U S A. 2004;101:3035–3040. doi: 10.1073/pnas.0308345101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolkers MC, Brouwenstijn N, Bakker AH, Toebes M, Schumacher TN. Antigen bias in T cell cross-priming. Science. 2004;304:1314–1317. doi: 10.1126/science.1096268. [DOI] [PubMed] [Google Scholar]
- 46.Iero M, Filipazzi P, Castelli C, Belli F, Valdagni R, Parmiani G, Patuzzo R, Santinami M, Rivoltini L. Modified peptides in anti-cancer vaccines: are we eventually improving anti-tumour immunity? Cancer Immunol Immunother. 2009;58:1159–1167. doi: 10.1007/s00262-008-0610-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rubio V, Stuge TB, Singh N, Betts MR, Weber JS, Roederer M, Lee PP. Ex vivo identification, isolation and analysis of tumor-cytolytic T cells. Nat Med. 2003;9:1377–1382. doi: 10.1038/nm942. [DOI] [PubMed] [Google Scholar]
- 48.Stuge TB, Holmes SP, Saharan S, Tuettenberg A, Roederer M, Weber JS, Lee PP. Diversity and recognition efficiency of T cell responses to cancer. PLoS Med. 2004;1:e28. doi: 10.1371/journal.pmed.0010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Houghton CS, Engelhorn ME, Liu C, Song D, Gregor P, Livingston PO, Orlandi F, Wolchok JD, McCracken J, Houghton AN, Guevara-Patino JA. Immunological validation of the EpitOptimizer program for streamlined design of heteroclitic epitopes. Vaccine. 2007;25:5330–5342. doi: 10.1016/j.vaccine.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 50.Valmori D, Gervois N, Rimoldi D, Fonteneau JF, Bonelo A, Lienard D, Rivoltini L, Jotereau F, Cerottini JC, Romero P. Diversity of the fine specificity displayed by HLA-A*0201-restricted CTL specific for the immunodominant Melan-A/MART-1 antigenic peptide. J Immunol. 1998;161:6956–6962. [PubMed] [Google Scholar]
- 51.Speiser DE, Baumgaertner P, Voelter V, Devevre E, Barbey C, Rufer N, Romero P. Unmodified self antigen triggers human CD8 T cells with stronger tumor reactivity than altered antigen. Proc Natl Acad Sci U S A. 2008;105:3849–3854. doi: 10.1073/pnas.0800080105. [DOI] [PMC free article] [PubMed] [Google Scholar]