Abstract
Tumor growth and dissemination depend partly on the reactivity of natural killer (NK) cells and T cells expressing NK-associated receptors. Their effector functions are regulated by an array of activating and inhibitory cell surface receptors with MHC class I ligand specificity, such as the killer immunoglobulin-like receptors (KIRs). Given the extensive genomic diversity of KIRs and their HLA ligands, it is reasonable to speculate that HLA, KIR gene variations and specific KIR-ligand combinations will have an impact on disease susceptibility and/or progression. Here, we discuss how KIR genotypes and KIR/HLA immunogenetic profiles may be involved in tumorigenesis, especially in malignant melanoma (MM). A hypothetical model of the impact of KIR/ligand combinations on immune responses in MM is proposed.
Keywords: KIR, HLA, Polymorphism, Malignant melanoma
Introduction
The immune surveillance concept proposed that the immune system protects the host against nascent malignancies, preventing and/or controlling tumor growth [8]. However, cancer cells succeed escape immune surveillance using several different strategies [30]. The control of tumor progression and dissemination depends to a large extent on T-cell or natural killer (NK)-cell reactivity. In the tumor–host relationship, MHC class I antigens are emerging as leading players, because of the crucial interaction of HLA molecules with T- and NK-specific receptors (NKR) present in both types of immune effector cells [1]. There are convincing evidences proving that any change in MHC pattern of malignant cells and/or in NKR expression on effector cells may modulate immune responses either enhancing or inhibiting antitumor cytolytic activity [35]. But, in what extent the diversity of immunogenetic profiles of these receptors and their ligands is related to the mechanisms of tumorigenesis is still under investigation. The present brief review will mainly focus on HLA, killer immunoglobulin-like receptor (KIR) and KIR/HLA genetic variations in malignant melanoma (MM) and the impact they may have on disease susceptibility and/or progression.
MHC genes
Genes encoding MHC molecules are highly polymorphic, and present significant variation resulting from evolutionary adaptations to an ever-changing pathogen environment. It was suggested that this extremely polymorphic region of the human genome has been generated by extensive duplications of genes with redundant functions but subtle differences in the way such a function is implemented [39]. Diversity of the genes is a result of recombinations, gene conversion and point mutations. It is likely that this evolution is pathogen-driven and polymorphic variants are subjected to natural selection. More than 2200 HLA alleles have been identified so far [http://www.anthonynolan.com/HIG/index.html]. The polymorphism is restricted not only to one locus but also to many linked loci and the patterns of haplotypes, which are held together in linkage disequilibrium. The MHC is associated with more diseases than any other region of the human genome. The highly polymorphic class I and class II loci are the major determinants of HLA-associated disease. However, data on HLA associations with solid tumors and particularly with malignant melanoma are limited (Table 1), but some studies reported the relevance of HLA class I in malignant melanoma. The presence of HLA-B40 is related to both the development and the clinical progression of melanoma [15, 31]. The distribution of HLA-A24 alleles, studied in a melanoma population for identifying individuals suitable for vaccination, revealed higher incidence of HLA-A*2402 allele in patients compared to healthy persons [4]. Homozygosity for group 2 HLA-C alleles (HLA-CLys80) seems to be associated with metastatic progression of melanoma in the Spanish population [9]. Studies on HLA class II associations and malignant melanoma were focused mainly on the DQB1 locus [22, 23]. DQB1*0301 was found to be a risk factor for development of metastatic disease in Caucasian MM patients [3, 19, 20]. In the Japanese population, such a risk factor is DQB1*0302 [16]. In addition, HLA-DRB1*1101 was predictive of melanoma recurrence and could regulate IFN-γ levels in these patients [21].
Table 1.
HLA and malignant melanoma
| HLA allele | Association | Reference |
|---|---|---|
| HLA-B40 | Predisposing/progression | [15, 31] |
| HLA-CLys80 alleles | Metastatic progression | [9] |
| DQB1*0501, *0301 | Predisposing/metastatic disease | [22] |
| DQB1*0301 | Metastatic disease | [3, 19, 20] |
| DQB1*0302 | Predisposing | [16] |
| DRB1*0802, DQA1*0101, *0401 | Protective | [16] |
| HLA-DRB1*1101 | Melanoma recurrence | [21] |
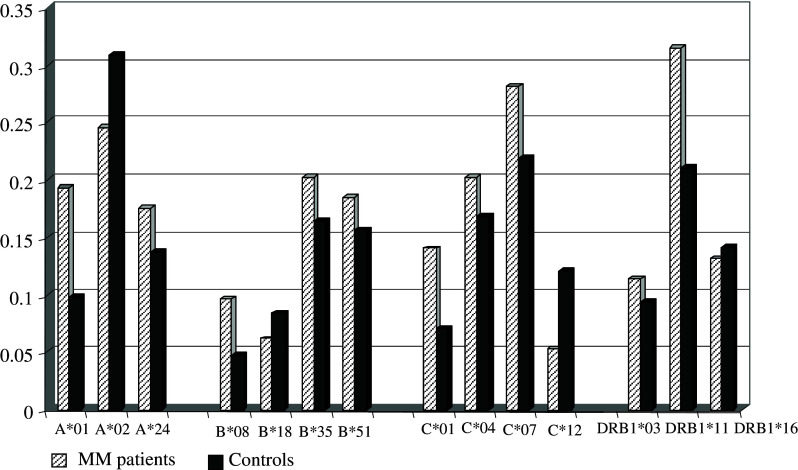
Our study in the Bulgarian population showed slightly increased frequencies of HLA-A*01, B*08, C*01 alleles in melanoma patients (Fig. 1). Similar results were observed for DRB1*11, which is in strong linkage disequilibrium with DQB1*0301 (Fig. 1). Interestingly, the DRB1*11 allele was found to be protective for different autoimmune diseases in our population [25]. Furthermore, we analyzed the relevance of extended HLA haplotypes that might be considered as more informative markers than single loci. Three haplotypes, A*01-B*35-C*04, A*01-B*08-DRB1*03, A*24-B*40-DRB1*11, conferred significant predisposition to MM in the Bulgarian population (Table 2). Therefore, we could speculate that both HLA class I and class II molecules, encoded by certain alleles could contribute to the susceptibility to develop this malignancy.
Fig. 1.
HLA allele distribution in melanoma patients and healthy subjects from the Bulgarian population. HLA-A, -B, -C, -DR allele distribution was evaluated in 50 patients with malignant melanoma and 128 healthy individuals from the Bulgarian population using PCR-SSP methods. Data presented illustrate allele frequencies showing differences between patients and controls. Slightly increased frequencies (P<0.05; Pc: NS) of HLA-A*01, B*08, C*01 and DRB1*11 alleles were observed in melanoma patients compared to healthy subjects. NS—not significant
Table 2.
HLA haplotypes associated with malignant melanoma
| Haplotype | Haplotype frequency | OR | |
|---|---|---|---|
| MM patients (n=50) | Controls (n=128) | ||
| A*01-B*35-C*04a | 0.069 | 0.000 | 19.9 |
| A*01-B*08-DRB1*03a | 0.079 | 0.019 | 4.5 |
| A*24-B*40-DRB1*11a | 0.026 | 0.000 | 7.1 |
aPc<0.05; Expectation–maximization algorithm
NK cell receptor genes
With the discovery of NK cell receptors, present on both T and NK immune effector cells and capable of recognizing MHC molecules, studies have been devoted to highlighting their relevance in tumor immunity. These receptors fall into two major structural classes, the immunoglobulin superfamily and the C-type lectin-like family, including both inhibitory and activating receptors. Genes for NK receptors in humans are clustered on two different chromosomes (reviewed in [36]). The C-type lectin genes, such as CD94 and NKG2, form natural killer complex (NKC) on chromosome 12. The immunoglobulin superfamily genes are clustered in the leukocyte receptor complex (LRC) on chromosome 19q13.4. Killer cell immunoglobulin-like receptor genes, located within LRC, are complex like the MHC, polygenic and polymorphic. Ongoing research involving KIR polymorphism is helping to develop a clear picture of KIR gene content. At the level of the human population there is evidence for balance between activating and inhibitory KIRs. The multigene KIR family, believed to be composed of 15 genes and 2 pseudogenes, is now known to exhibit allelic diversity with respect to individual genes [http://www.ebi.ac.uk/ipd/kir]. Two groups of KIR haplotypes are defined: groups A and B, which differ in number and kind of KIR genes. Four KIR genes (3DL3, 3DP1, 2DL4 and 3DL2), so-called framework genes are shared by both groups. Group A haplotypes encode inhibitory KIR2DL1, KIR2DL3 and KIR3DL1, and are devoid of stimulatory KIR genes except KIR2DS4. All other haplotypes fall into group B. Group B haplotypes generally possess more KIR genes than group A and perhaps functionally, more importantly, the gene content is biased towards activating KIRs. KIR haplotypes that are identical by gene content can be further subdivided according to allele combinations. Whereas group A haplotypes do not vary in gene content they show extensive variability on the allelic level. In contrast, group B haplotypes exhibit substantial variation in gene content but only moderate allelic polymorphism. Thus, diversity in human KIR genotype derives from three components: haplotypic gene content, allelic polymorphism, and the combination of maternal and paternal haplotypes [28]. Due to combined effects of these three components, unrelated individuals usually differ in KIR genotype, and ethnic populations have widely differing KIR genotype frequencies.
It is conceivable that KIR-gene content in the genome could contribute to the function of NK cells and T lymphocytes by modulating immune responses to malignant cells. Consequently, a genetic imbalance between activating and inhibitory KIR genes may facilitate the development of malignancies. Amheim et al. [2] showed that a genotype containing KIR2DL1, KIR2DL2, KIR2DL3, KIR2DL4, KIR3DL1, KIR3DL2, KIR3DL3 and KIR2DS4 was associated with an increased risk of cervical intraepithelial neoplasia, and the specific allele KIR2DL5*002 had an inverse association with disease in a study of Swedish women. Analysis of KIR gene distribution in melanoma patients and healthy controls in the Bulgarian population revealed that gene frequencies ranged from 32 to 100% [26]. The most frequent KIR genes in Bulgarians are: KIR2DL1, 2DL3, 3DL1 and 2DS4, followed by 2DS2 and 2DL2. The remainder (KIR2DS1, 2DS3, 3DS1) was found in less than half of the samples. Our data showed that the KIR gene frequencies observed in the control group closely resembled those found in previous studies in Caucasian populations [12, 27, 38]. Similar results for the KIR gene repertoire were obtained in our patients with MM. The phenotypic distribution of KIR genes also did not demonstrate a prevalence of a given haplotype (A or B) in the patients compared to the controls [26]. No significant differences were found for KIR gene and genotype distribution between patients with primary (stages 1 and 2) and metastatic (stages 3 and 4) melanoma. Our data indicate that the polymorphism within the KIR gene family is associated neither with the presence of MM nor with the progression of malignant disease.
KIR and HLA ligands
The extent to which KIRs act as inhibitory self receptors depends upon a person’s HLA type. KIR recognition of specific HLA class I allotypes contributes to the array of receptor–ligand interactions that determine the response of an NK cell to its target. There are only four KIRs with clearly defined specificities, all of the inhibitory type and all for HLA class I allotypes. The inhibitory KIR2DL (1, 2 and 3) recognize HLA-C allotypes. HLA-C ligand specificity is determined by the amino acid residue at position 80 in the alfa-1 helix of the HLA-C molecule, which can be either asparagine (group C1 epitope) or lysine (group C2 epitope) [5, 6, 24]. The inhibitory KIR3DL1 recognizes HLA-B Bw4 allotypes [13]. HLA-Bw4 molecules can be divided into two groups on the basis of whether isoleucine or threonine is present at position 80 (Bw480Ile and Bw480Thr, respectively). Receptor-binding and lysis-inhibition data suggest that HLA-B molecules containing Bw480Ile may be more effective ligands for KIR3DL1 than those containing Bw480Thr [11], whereas neither HLA-A Bw4 nor HLA-B Bw6 molecules bind to KIR3DL1. Other inhibitory KIRs have less well-defined HLA class I specificities, such as KIR3DL2 for certain HLA-A variants—A3, 11 [29]. Studies directly addressing whether activating KIR have the same ligands as their inhibitory homologues have identified only low-level binding of KIR2DS1 to HLA-C2 [7] and KIR2DS2 to HLA-C1 epitopes under certain circumstances [34]. A weak interaction highly restricted to HLA-Cw4 only was reported for KIR2DS4 [17, 18]. The ligand for KIR3DS1 is not known, but it shares 97% similarity in its extracellular domain to its inhibitory counterpart KIR3DL1 and might therefore recognize HLA-B Bw4 allotypes under some conditions [10, 36].
KIR/HLA immunogenotype profiles
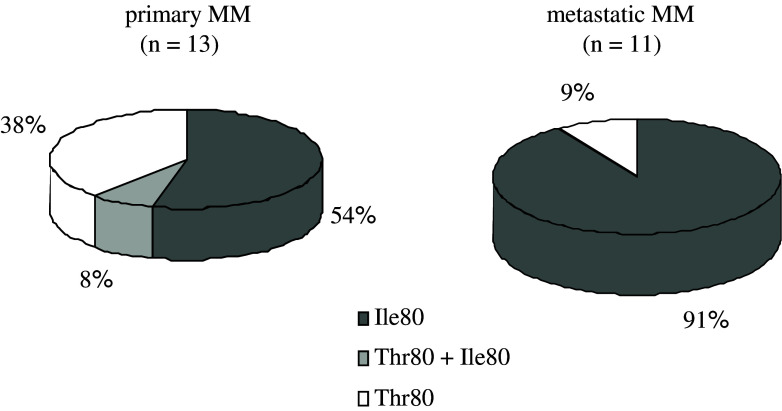
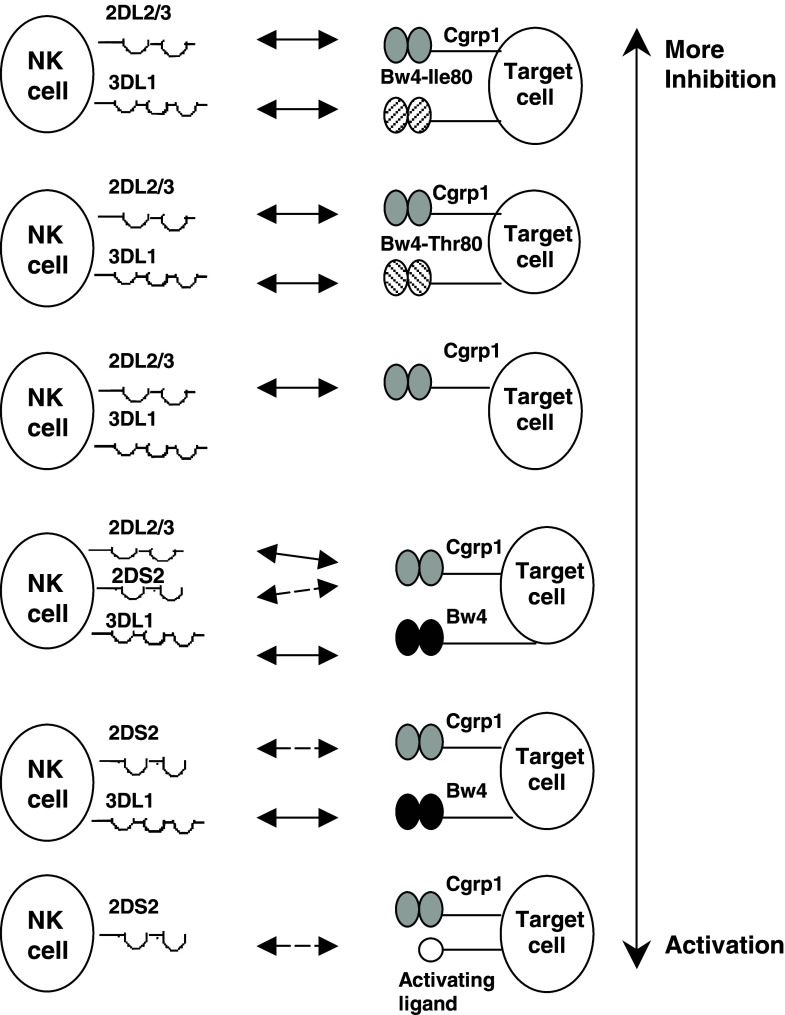
If the polymorphism of both HLA and the KIR-gene complex is considered together, then dissimilar numbers and qualities of KIR/HLA pairs appear to function in different individuals. In addition, the independent segregation of HLA and KIR genes raises the possibility that any given individual can express the receptor or the ligand only, or both receptor and ligand. Since the NK receptor/HLA ligand pairs control NK cell function, their status from “none to too many” may have a role in the immune response to different challenges [32]. Various clinical correlations, including viral infections, malignant and autoimmune diseases, point to the functional effect of the genes encoding KIR and the presence or absence of KIR/HLA ligand combinations (reviewed in [33]). Associations with different HLA/KIR combinations, involving activating or inhibitory KIRs to different extents, have been described. For example, in HPV-induced cervical cancer, specific KIR/HLA ligand pairs decrease the risk of developing neoplasia, whereas the presence of activating receptor KIR2DS1 results in increased risk of disease, particularly when the protective inhibitory combinations are missing [10]. The combined effect of HLA and KIR polymorphic genes has been also analyzed in allogeneic stem cell transplantation for treatment of hematological malignancies. The lack of HLA ligands in the recipient for donor-inhibitory KIRs can contribute to improved outcome for patients with acute myeloid leukemia (AML) [14]. It was hypothesized that alloreactive NK cells eradicate AML by killing residual leukemic cells, favor engraftment by removing host lympho-hematopoietic cells, and reduce GvHD by eliminating recipient dendritic cells [37]. Thus, KIR and HLA polymorphisms could behave as factors of susceptibility, progression or protection in malignancies. Although no differences were found for the KIR gene frequencies, our data on HLA genotyping demonstrate differences in the group 1 and group 2 HLA-C ligand distributions between melanoma patients and healthy controls [26]. HLA-C types including only HLA-CLys80 were significantly less frequent in patients than in controls. Since those individuals can use KIR2DL1 as an inhibitory receptor, a minority of melanoma patients in our study group (homozygous for HLA-CLys80) could have a limited number of functional inhibitory KIR/HLA pairs. In contrast, the majority of the patients (homozygous for HLA-CAsn80 or heterozygous for HLA-C at position 80) could use more inhibitory receptor/ligand combinations. The biological significance of KIRs in vivo depends on whether these receptors are present in individuals simultaneously with their ligands. Our data showed that the activating KIR/ligand associations were not significantly different between patients and controls [26]. In contrast, the observation of inhibitory KIR2DL2/2DL3 in association with their putative ligands was more frequent in MM patients. The analysis of the HLA-Bw4 ligands for inhibitory KIR3DL1 in the context of the presence of isoleucine or threonine at position 80 showed no difference between patients and controls [26]. However, our finding for higher incidence of more effective KIR3DL1/ligand combinations (Fig. 2) in patients with metastatic compared to those with primary malignant disease seems to support the suggestion that more effective inhibition of killer cell function via KIR3DL1/Bw4Ile80 pairs could interfere with occurrence of metastasis. Therefore, it could be speculated that the tumor dissemination and escape from immunosurveillance might be due to a prevalence of inhibitory over activating signals in melanoma patients. With this consideration in mind, a hypothetical model for the impact of KIR/HLA ligand combinations on immune responses in malignant melanoma (from inhibition to activation) is proposed (Fig. 3).
Fig. 2.
Distribution of HLA-Bw4Ile80 and Bw4Thr80 ligands in KIR3DL1/HLA-Bw4 positive patients. Although not statistically significant, our data showed higher incidence of KIR3DL1/Bw4Ile80 pairs (more effective KIR3DL1-ligands combination) in patients with metastatic compared to those with primary malignant disease
Fig. 3.
Hypothetical model for the impact of KIR/ligand combinations on immune responses in malignant melanoma. The proposed model aims to explain the effect of genetic variation at the KIR locus in combination with genes encoding their HLA ligands in malignant melanoma. The figure is based on currently available data for putative KIR ligands and on our results for KIR/ligand associations in melanoma patients, and do not illustrate all possibilities. Combinations are ordered by their ability to confer more inhibition to more activation. KIR/ligand pairs expected to result in a prevalence of inhibitory over activating signals might facilitate tumor development and dissemination
In conclusion, there may not be a direct association between KIR gene content in the genome and the presence of malignant melanoma, or melanoma progression. However, some HLA haplotypes could be regarded as predisposing to MM in the Bulgarian population. Furthermore, HLA/KIR gene combinations that seem to favor NK cell inhibition have been associated with MM and tumor dissemination. The inactivation of NK cells by self-HLA molecules might be a mechanism enabling malignant host cells to evade NK cell-mediated immunity. These data could also contribute to the immunotherapeutic strategies based on the superior antineoplastic effect against solid tumors of KIR incompatible allogeneic NK cells. Modulating the balance between activating and inhibitory signals through NK cell receptors may open a new approach to immunotherapy for cancer.
Acknowledgements
This work was supported in part by research grants from the ENACT (LSHC-CT-2004-503306) and ESTDAB (QLRT-2000-01325) EC projects. The authors thank professor Graham Pawelec for critical reading of the manuscript and valuable advices.
Footnotes
This article is a symposium paper from the conference “Progress in Vaccination against Cancer 2005 (PIVAC 5)”, held in Athens, Greece, on 20–21 September 2005.
References
- 1.Algarra I, Garcia-Lora A, Cabrera T, Ruiz-Cabello F, Garrido F. The selection of tumor variants with altered expression of classical and nonclassical MHC class I molecules: implications for tumor immune escape. Cancer Immunol Immunother. 2004;53:904–910. doi: 10.1007/s00262-004-0517-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amheim L, Dillner J, Sanjeevi CB. A population based cohort study of KIR genes and genotypes in relation to cervical intraepithelial neoplasia. Tissue Antigens. 2005;65:252–259. doi: 10.1111/j.1399-0039.2005.00359.x. [DOI] [PubMed] [Google Scholar]
- 3.Bateman AC, Turner SJ, Theaker JM, Howell WM. HLA-DQB1*0303 and *0301 alleles influence susceptibility to and prognosis in cutaneous malignant melanoma in the British Caucasian population. Tissue Antigens. 1998;52:67–73. doi: 10.1111/j.1399-0039.1998.tb03025.x. [DOI] [PubMed] [Google Scholar]
- 4.Bettinotti MP, Norris RD, Hackett JA, Thompson CO, Simonis TB, Stroncek D, Marincola FM. Frequency of human leukocyte antigen-A24 alleles in patients with melanoma determined by human leukocyte antigen-A sequence-based typing. J Immunother. 2000;23:282–287. doi: 10.1097/00002371-200003000-00013. [DOI] [PubMed] [Google Scholar]
- 5.Biassoni R, Falco M, Cambiaggi A, Costa S, Verdiani S, Pende D, Conte R, et al. Amino acid substitutions can influence the natural killer (NK)-mediated recognition of HLA-C molecules. Role of serine-77 and lysine-80 in the target cell protection from lysis mediated by “group 2” or “group 1” NK clones. J Exp Med. 1995;182:605–609. doi: 10.1084/jem.182.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biassoni R, Cantoni C, Falco M, Verdiani S, Bottino C, Vitale M, Conte R, Poggi A, Moretta A, Moretta L. The human leucocyte antigen (HLA)-C-specific ‘activatory’ or ‘inhibitory’ natural killer cell receptors display highly homologous extracellular ddomains but differ in their transmembrane and intracytoplasmic portions. J Exp Med. 1996;183:645–650. doi: 10.1084/jem.183.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biassoni R, Pessino A, Malaspina A, Cantoni C, Bottino C, Sivori S, Moretta L, Morreta A. Role of amino acid position 70 in the binding affinity of p50.1 and p58.1 receptors for HLA-Cw4 molecules. Eur J Immunol. 1997;27:3095–3099. doi: 10.1002/eji.1830271203. [DOI] [PubMed] [Google Scholar]
- 8.Burnet FM. Immunological aspects of malignant disease. Lancet. 1967;1:1171–1174. doi: 10.1016/S0140-6736(67)92837-1. [DOI] [PubMed] [Google Scholar]
- 9.Campillo JA, Martinez-Escribano JA, Muro M, Moya-Quiles R, Marin LA, Montes-Ares O, Guerra N, et al. HLA class I and class II frequencies in patients with cutaneous malignant melanoma from southeastern Spain: the role of HLA-C in disease prognosis. Immunogenetics. 2005;20:1–8. doi: 10.1007/s00251-005-0065-2. [DOI] [PubMed] [Google Scholar]
- 10.Carrington M, Wang S, Martin MP, Gao X, Schiffman M, Cheng J, Herrero R, et al. Hierarchy of resistance to cervical neoplasia mediated by combinations of killer immunoglobulin-like receptor and human leukocyte antigen loci. JEM. 2005;201:106–1075. doi: 10.1084/jem.20042158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cella M, Longo A, Ferrara GB, Strominger JL, Colonna M. NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J Exp Med. 1994;180:1235–1242. doi: 10.1084/jem.180.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crum KA, Logue SE, Curran MD, Middleton D. Development of a PCR-SSOP approach capable of defining inhibitory receptor (KIR) gene sequence repertoire. Tissue Antigens. 2000;56:313–326. doi: 10.1034/j.1399-0039.2000.560403.x. [DOI] [PubMed] [Google Scholar]
- 13.Gumperz JE, Litwin V, Philips JH, Lanier LL, Parham P. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J Exp Med. 1995;181:1133–1144. doi: 10.1084/jem.181.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu KC, Keever-Taylor CA, Wilton A, Pinto C, Heller G, Arkun K, O’Reilly RJ, et al. Improved outcome in HLA-identical sibling hematopoietic stem-cell transplantation for acute myelogenous leukemia predicted by KIR and HLA genotypes. Blood. 2005;105:4878–4884. doi: 10.1182/blood-2004-12-4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jager MJ, Volker-Dieben HJ, de Wolff-Rouendaal D, Kakebeeke-Kemme H, D’Amaro J. Possible relation between HLA and ABO type and prognosis of uveal melanoma. Doc Ophthalmol. 1992;82:43–47. doi: 10.1007/BF00156992. [DOI] [PubMed] [Google Scholar]
- 16.Kageshita T, Naruse T, Hirai S, Ono T, Horikoshi T, Nakagawa H, Tamaki K, et al. Molecular genetics analysis of HLA class II alleles in Japanese patients with melanoma. Tissue Antigens. 1997;49:466–470. doi: 10.1111/j.1399-0039.1997.tb02780.x. [DOI] [PubMed] [Google Scholar]
- 17.Katz G, Markel G, Mizrahi S, Arnon TI, Mandelboim O. Recognition of HLA-Cw4 but not HLA-Cw6 by the NK cell receptor killer cell Ig-like receptor two-domain short tail number 4. J Immunol. 2001;166:7260–7267. doi: 10.4049/jimmunol.166.12.7260. [DOI] [PubMed] [Google Scholar]
- 18.Katz G, Gazit R, Arnon TI, Gonen-Gross T, Tarcic G, Markel G, Gruda R, et al. MHC class I-independent recognition of NK-activating receptor KIR2DS4. J Immunol. 2004;173:1819–1825. doi: 10.4049/jimmunol.173.3.1819. [DOI] [PubMed] [Google Scholar]
- 19.Lee JE, Reveille JD, Ross MI, Platsoucas CD. HLA-DQB1*0301 association with increased cutaneous melanoma risk. Int J Cancer. 1994;59:510–513. doi: 10.1002/ijc.2910590413. [DOI] [PubMed] [Google Scholar]
- 20.Lee JE, Lu M, Mansfield PF, Platsoucas CD, Reveille JD, Ross MI. Malignant melanoma: relationship of HLA class II gene DQB1*0301 to decreased recurrence in AJCC on cancer stage I or II. Cancer. 1996;78:758–763. doi: 10.1002/(SICI)1097-0142(19960815)78:4<758::AID-CNCR11>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 21.Lee JE, Abdalla J, Porter GA, Bradford L, Grimm EA, Reveille JD, Mansfield PF, et al. Presence of the human leucocyte antigen class II gene DRB1*1101 predicts interferon γ levels and disease recurrence in melanoma patients. Ann Surg Oncol. 2002;9:587–593. doi: 10.1245/aso.2002.9.6.587. [DOI] [PubMed] [Google Scholar]
- 22.Lombardi ML, Mercuro O, Pirozzi G, Ionna F, Lombari V, Mozzillo N, Manzo C. Molecular analysis of HLA-DRB1 and -DQB1 polymorphism in Italian melanoma patients. J Immunotherapy. 1998;21:435–439. doi: 10.1097/00002371-199811000-00005. [DOI] [PubMed] [Google Scholar]
- 23.Lulli P, Grammatico P, Brioli G, Catricala C, Morellini M, Roccella M, Mariani B, et al. HLA-DR and -DQ alleles in Italian patients with melanoma. Tissue Antigens. 1998;51:276–280. doi: 10.1111/j.1399-0039.1998.tb03102.x. [DOI] [PubMed] [Google Scholar]
- 24.Mandelboim O, Reyburn HT, Vales-Gomez M, Pazmany L, Colonna M, Borsellino G, Strominger JL. Protection from lysis by natural killer cells of group 1 and 2 specificity is mediated by residue 80 in human histocompatibility leukocyte antigen C alleles and also occurs with empty major histocompatibility complex molecules. J Exp Med. 1996;184:913–922. doi: 10.1084/jem.184.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naumova E, Ivanova M. Putative molecular model of HLA associated autoimmune diseases in the Bulgarian population. Modern Medicine (BG) 2004;4:45–52. [Google Scholar]
- 26.Naumova E, Mihaylova A, Stoitchkov K, Ivanova M, Quin L, Toneva M. Genetic polymorphism of NK receptors and their ligands in melanoma patients: prevalence of inhibitory over activating signals. Cancer Immunol Immunother. 2005;54:172–178. doi: 10.1007/s00262-004-0575-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Norman PJ, Stephens HAF, Verity DH, Chandanayingyong D, Vaughan RW. Distribution of natural killer cell immunoglobulin-like receptor sequences in three ethnic groups. Immunogenetics. 2001;52:195–205. doi: 10.1007/s002510000281. [DOI] [PubMed] [Google Scholar]
- 28.Parham P. Immunogenetics of killer cell immunoglobulin-like receptors. Mol Immunol. 2005;42:459–462. doi: 10.1016/j.molimm.2004.07.027. [DOI] [PubMed] [Google Scholar]
- 29.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 30.Pawelec G. Tumour escape: antitumour effectors too much of a good thing? Cancer Immunol Immunother. 2004;53:262–274. doi: 10.1007/s00262-003-0469-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pellegris G, Illeni MT, Vaglini M, Rovini D, Cascinelli N, Masserini C. HLA antigens in malignant melanoma patients. Tumori. 1980;66:51–58. doi: 10.1159/000172999. [DOI] [PubMed] [Google Scholar]
- 32.Rajalingam R. Diversity of NK cell receptors and their HLA class I ligands. ASHI Quarterly. 2002;26:68–72. [Google Scholar]
- 33.Rajagopalan S, Long EO. Understanding how combinations of HLA and KIR genes influence disease. JEM. 2005;201:1025–1029. doi: 10.1084/jem.20050499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steward CA, Langier-Anfossi F, Vély F, Saulquin X, Riedmuller J, Tisserant A, Gauthier L, et al. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. PNAS. 2005;102:13224–13229. doi: 10.1073/pnas.0503594102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tarazona R, Casado JG, Soto R, DelaRosa O, Peralbo E, Rioja L, Pena J, Solana R. Expression of NK-associated receptors on cytotoxic T cells from melanoma patients: a two-edged sword? Cancer Immunol Immunother. 2004;53:911–924. doi: 10.1007/s00262-004-0507-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trowsdale J. Genetic and functional relationships between MHC and NK receptor genes. Immunity. 2001;15:363–374. doi: 10.1016/S1074-7613(01)00197-2. [DOI] [PubMed] [Google Scholar]
- 37.Velardi A, Ruggeri L, Moretta A, Moretta L. NK cells: a lesson from mismatched hematopoietic transplantation. Trends Immunol. 2002;23:438–444. doi: 10.1016/S1471-4906(02)02284-6. [DOI] [PubMed] [Google Scholar]
- 38.Witt CS, Dewing C, Sayer DC, Uhrberg M, Parham P, Christiansen FT. Population frequencies and putative haplotypes of the killer cell immunoglobulin-like receptor sequences and evidence for recombination. Transplantation. 1999;68:1784–1789. doi: 10.1097/00007890-199912150-00024. [DOI] [PubMed] [Google Scholar]
- 39.Yeager M, Hughes A. Evolution of the mammalian MHC: natural selection, recombination, and convergent evolution. Immunological Reviews. 1999;167:45–59. doi: 10.1111/j.1600-065X.1999.tb01381.x. [DOI] [PubMed] [Google Scholar]