Abstract
Tumors escape from immune surveillance by producing immunosuppressive cytokines and proapototic factors, including TGF-β and galectin-1 (Gal-1). Since immunosuppressive mechanisms might act in concert to confer tumor-immune privilege, we investigated the potential cross talk between TGF-β and Gal-1 in highly metastatic mammary adenocarcinoma (LM3) cells. While Gal-1 treatment was not capable of regulating TGF-β synthesis, a pronounced and dose-dependent increase in Gal-1 expression was observed when tumor cells were treated with TGF-β1. This effect was also observed in the murine lung adenocarcinoma LP07 and in the human breast adenocarcinoma MCF-7 cell lines. TGF-β1-mediated upregulation of Gal-1 expression was specifically mediated by TβRI and TβRII, since it was abrogated when LM3 cells were infected with retroviral vectors expressing the dominant negative forms of these receptors. In addition, gal-1 gene sequence analysis revealed the presence of three putative binding sites for Smad4 and Smad3 transcription factors, consistent with the ability of TGF-β1 to trigger a Smad-dependent signaling pathway in these cells. Thus, TGF-β1 may trigger a Smad-dependent pathway to control Gal-1 expression, suggesting that distinct mechanisms might cooperate in tilting the balance toward an immunosuppressive environment at the tumor site.
Keywords: Tumor-immune escape, Immunosuppression, Tumor progression, Galectin-1, TGF-β
Introduction
Tumor metastasis is a multistep process involving changes in cell adhesion, increased invasiveness, angiogenesis and evasion of immune responses [30]. Recent progress toward an improved understanding of the interactions between tumor cells and the host immune system has led to the realization that tumor cells have devised multiple strategies to evade immune attack [11, 12]. These strategies include the synthesis and secretion of immunosuppressive cytokines and pro-apoptotic factors, such as transforming growth factor-β (TGF-β) and galectin-1 (Gal-1), which might act in concert to counteract T-cell effector responses [13, 40, 44].
TGF-β belongs to a large superfamily of cytokines that includes the TGF-βs, activins, and bone morphogenetic proteins [25]. In mammals, there are three isoforms of TGF-β (TGF-β1, TGF-β2, TGF-β3), which are encoded by different genes but may exhibit distinct functions [25]. The TGF-βs present a conserved signaling mechanism, which starts with the binding of active TGF-β to the membrane constitutively active serine/threonine kinase TβRII, which transphosphorylates and activates the type I receptor TβRI (Alk5 in humans) [25, 43]. TβRI in turn phosphorylates the receptor-regulated Smad2 and Smad3 proteins, which form complexes with a common partner, Smad4. Activated Smad complexes then enter the nucleus, where they regulate the transcription of target genes [25, 43].
TGF-β plays a pivotal role at different steps of tumor metastasis, including cell invasion, angiogenesis, and tumor-immune escape [1, 14, 16, 45]. Recent evidence indicates that TGF-β acts on cytotoxic T lymphocytes (CTL) to specifically inhibit the expression of different cytolytic gene products; namely perforin, granzyme A, granzyme B, Fas ligand, and IFN-γ, which are collectively responsible for CTL-mediated tumor cytotoxicity [44]. Consistently, blockade of TGF-β signaling allows the generation of a potent antitumor immune response [14]. In addition, we have previously demonstrated that TGF-β signaling exhibits a tumor-promoting role in an experimental model of metastatic mammary adenocarcinoma [10].
Galectins, a family of highly conserved carbohydrate-binding proteins, are characterized by their ability to recognize N-acetyllactosamine sequences, which can be displayed on both N- and O-glycans on cell surface glycoconjugates [8]. Expression of galectin-1 (Gal-1), a prototype member of this family, has been well documented in many different tumor types including melanoma, glioblastoma, lung adenocarcinoma and mammary carcinoma [9, 19, 49]. In general, such expression correlates with the aggressiveness of these tumors and the acquisition of a metastatic phenotype [23]. There is increasing evidence that Gal-1 plays important roles in several aspects of cancer biology including neoplastic transformation [33], tumor cell adhesion [48], homotypic cell aggregation [45], invasiveness [4, 5], and inflammation [34, 38]. In addition, we have recently demonstrated that Gal-1 may also function as a soluble mediator employed by tumor cells to evade immune responses [40]. This effect has been extrapolated to human tumors as a link between tumor hypoxia and tumor-immune privilege [21].
In the present study we investigated the potential cross talk between TGF-β and Gal-1, two critical mediators of tumor progression and tumor-immune escape.
Materials and methods
Cell culture and tumor cell line
The BALB/c mammary tumor LM3 cell line was established from primary cultures of the spontaneous mammary adenocarcinoma M3, as described [47]. LM3 cells show a highly metastatic in vivo behavior when inoculated into syngeneic mice [47]. The LP07 cell line was established from primary cultures of the murine spontaneous lung carcinoma P07 as described [46]. The human mammary tumor line MCF-7 [3] was obtained from American Type Culture Collection (Rockville, Maryland). Cells were cultured at 37°C in a humidified atmosphere of 5% CO2, in minimum essential medium (MEM) (Gibco) with non-essential aminoacids, 2 mM l-glutamine and 80 μg/ml gentamicin, supplemented with 5% fetal bovine serum (FBS, Gibco). Hoechst´s staining periodically tested cells for mycoplasma contamination.
Recombinant proteins and antibodies
Recombinant Gal-1 was produced and purified as previously described [15]. Briefly, E. coli BL21 (DE3) were transformed with expression plasmids constructed using pET expression systems (Novagen; Madison, WI) and production of Gal-1 was induced by the addition of 1 mM isopropyl-β-d-thiogalactoside. Soluble fractions were obtained for subsequent purification by affinity chromatography on an asialofetuin-agarose column and stored at -20°C in 4 mM 2-ME to keep its sugar-binding activity under reducing conditions. Lipopolysaccharide content of the purified samples was tested using a Gel Clot Limulus Test (Cape Cod Inc. East Falmouth, MA). Human recombinant TGF-β1 (R&D Systems) and TGF-β3 (Calbiochem) proteins were used. The following antibodies were employed: anti-TGF-β′1 anti-TGF-β3 and anti-Smad4 were purchased from Santa Cruz Biotechnology; anti-Smad2 and anti-phospho-Smad2 were from Cell Signaling; anti-Smad4/DPC4 was from BD Biosciences; anti-β-actin and TRICT-conjugated anti-mouse were from Sigma; FITC- conjugated anti-rabbit was from Pierce, HRP-conjugated anti-rabbit and HRP-conjugated anti-mouse antibodies were from Vector. The anti-Gal-1 antibody was obtained and used essentially as described [36].
Western blot analysis
Analysis of protein expression was performed by western blot, as previously described [36]. Briefly, subconfluent monolayers of LM3 cells were washed with PBS and treated for 24 h with different concentrations (0.4, 4 and 40 ng/ml) of TGF-β1 or TGF-β3, or alternatively with different doses of recombinant Gal-1 (4 and 40 μg/ml). In the case of LP07 and MCF-7 cells subconfluent monolayers were washed with PBS and treated for 24 h with different concentrations (0.4 and 4 ng/ml) of TGF-β1. Following treatments, cells were washed and lysed in a buffer containing 50 mM Tris–HCl, 5 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 0.15 M NaCl, and protease inhibitors (Sigma). Protein content was measured using the Micro-BCA kit (Pierce). Western blot analyses for Gal-1 and TGF-β were performed essentially as described [36]. Equal amounts of protein from cell lysates (30 μg) were resolved on a 15% SDS-PAGE. After electrophoresis, proteins were electroblotted onto nitrocellulose membranes (Amersham) and incubated with an affinity-purified rabbit anti-Gal-1 antibody (1:4,000 dilution), or with rabbit anti-TGF-β1 or anti-TGF-β3 antibodies (1:200 dilution) directed against a region inside the mature TGF-β molecule. Membranes were then incubated with a HRP-labelled anti-rabbit IgG (Vector) and developed using ECL (Amersham). Films were analyzed with the Scion Image Analysis software, and the intensity of each band was recorded and expressed as arbitrary units (AU). Equal loading was checked by Ponceau S staining or by incubating the membranes with an anti-β-actin monoclonal antibody.
Subcellular localization of Gal-1
Immunofluorescence staining and confocal microscopy were used to determine subcellular localization of Gal-1. Briefly, LM3 cells were seeded on coverslips and once reached subconfluency were treated with 4 ng/ml TGF-β1 for the indicated times. Then, cells were fixed with paraformaldehyde and permeabilized with 0.05% Triton X-100 (Sigma) in PBS for 30 min. After blocking for 1 h, the coverslips were incubated with a primary anti-Gal-1 policlonal antibody (1:50), and then with a FICT-conjugated anti-rabbit (1:100) secondary antibody, for 1 h at room temperature. Photographs were taken in a confocal microscope (Eclipse E800, Nikon C1 Confocal Microscope) at a magnification of ×200 and ×600. As a control omission of the first antibody was performed to discriminate background staining.
Expression and functionality of dominant negative mutants of TGF-β receptors
LM3 cells were stably transduced with retroviral vectors expressing human wild type TβRI (p-BMN-Alk5 wt-IRES-EGFP), dominant negative forms of TβRI (pBMN-TβRI-K232R-IRES-EGFP) and TβRII (pGabe-TβRII-K277R-EGFP) generated as described [2, 27]. Retroviral infection was performed as described [2, 29]. Briefly, 105 cells were infected with different retroviruses, using 10 μg/ml Polybrene (hexadimethrine bromide, Sigma). Medium was replenished 16 h after infection, and cells were allowed to grow for additional 24–48 h before further treatments. After infection, EGFP-positive cells were selected by flow cytometry. Cells overexpressing these dominant negative receptor forms were not capable of transducing TGF-β-mediated signaling [29]. LM3 cells transduced with the empty vector (LM3/BMN) were used as controls.
Expression and activation of Smad proteins
The ability of TGF-β to induce Smad phosphorylation was studied in LM3 cells treated for 30 min with 4 ng/ml TGF-β1. Western blot analysis was performed as described above. Membranes were incubated with a rabbit anti-phospho-Smad2 (1:500) or with a mouse anti-Smad4/DPC4 (1:500). Membranes were then incubated with HRP-conjugated anti-rabbit (1:2,000) or anti-mouse (1:2,000) antibodies.
Subcellular localization of the Co-Smad, Smad4 was determined by immunofluorescence staining. Briefly, LM3 cells were seeded on coverslips and when reached subconfluency, cells were treated with 4 ng/ml TGF-β1 for 30 min. Then, cells were fixed with paraformaldehyde and permeabilized with 0.05% Triton X-100 (Sigma) in PBS for 30 min. After blocking for 1 h, the coverslips were incubated with a primary anti-Smad4 monoclonal antibody (1:100), and then with a TRICT-conjugated anti-mouse (1:200) secondary antibody, for 1 h at room temperature. Photographs were taken in a fluorescence microscope (Eclipse E400, Nikon) at a magnification of ×400. As a control omission of the first antibody was performed to discriminate background staining.
Computer-assisted sequence analysis of Smad binding sites
For prediction of putative binding sites for transcription factors of the Smad family, sequence analysis of the whole gal-1 gene (from position −3,120 to position +7,281) was performed using the MatInspector professional software (Genomatix) .
Statistical analysis
In all cases, data are representative of at least three independent experiments. Statistical analyses were performed using Student´s t test. Results were considered of biological significance when P < 0.05.
Results
In search for a potential cross talk between TGF-β1 and Gal-1, two critical mediators of tumor progression and tumor-immune escape, we investigated the reciprocal regulation of these two soluble mediators in the LM3 metastatic mammary adenocarcinoma cell line derived from a primary culture of the spontaneous mammary adenocarcinoma M3 [47].
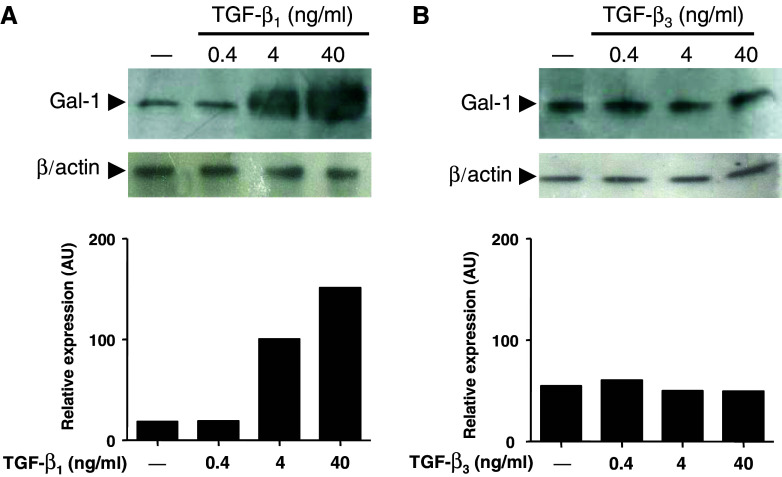
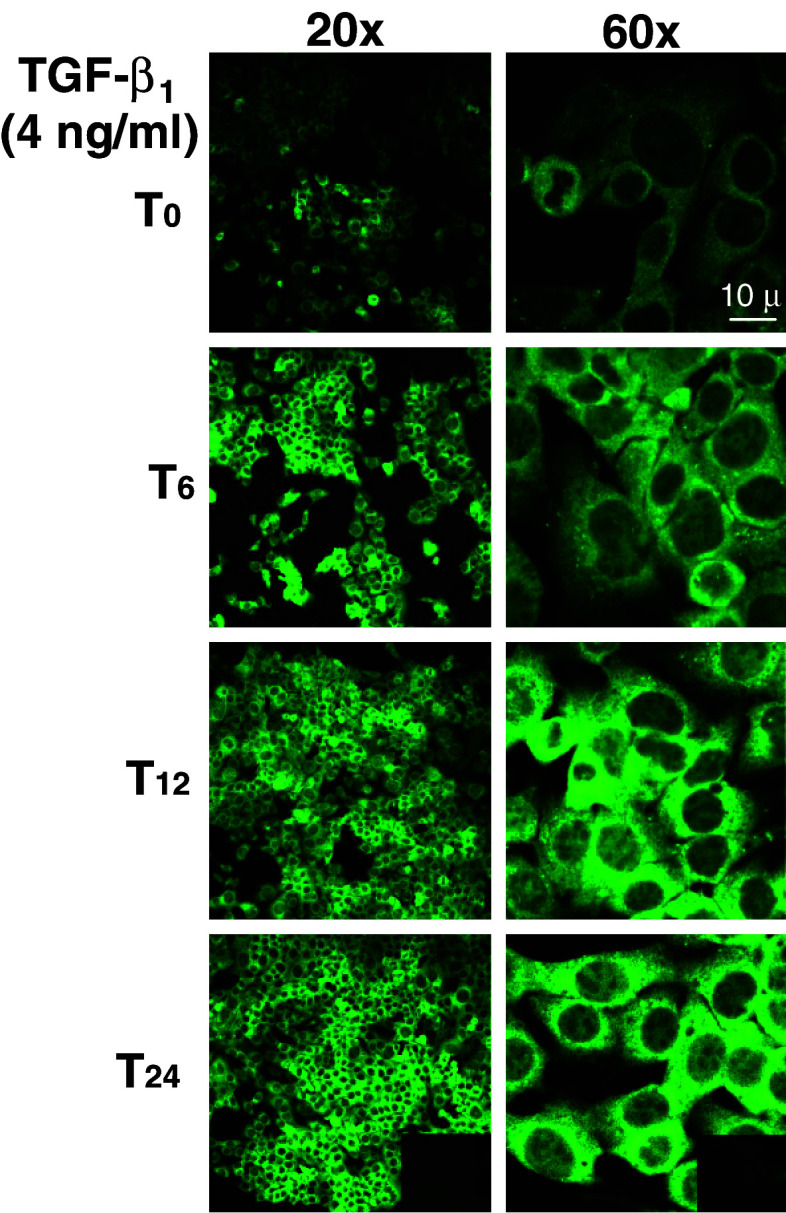
Subconfluent monolayers of LM3 cells were treated for 24 h with increasing concentrations of TGF-β1 or TGF-β3. Cells were washed, lysed and analyzed for Gal-1 expression by western blot analysis (Fig. 1). Interestingly, treatment of LM3 cells with TGF-β1 resulted in a pronounced and dose-dependent increase in Gal-1 expression level (Fig. 1a; lanes 2–4). In contrast, no changes in Gal-1 expression were observed in LM3 cells in response to TGF-β3 (Fig. 1b; lanes 2–4), suggesting that TGF-β1 specifically regulates Gal-1 expression in mammary adenocarcinoma cells. To illustrate morphologically in a time-sequence manner the subcellular distribution of Gal-1, LM3 cells were treated with TGF-β1 for 6, 12 and 24 h, fixed, stained and analyzed by immunofluorescence and confocal microscopy. As shown in Fig. 2 in the absence of TGF-β1 (T0) LM3 cells displayed a weak and diffused Gal-1 staining mostly in the cytoplasm. Treatment of LM3 cells with TGF-β1 (4 ng/ml) clearly increased Gal-1 staining in the cytoplasm in a time-dependent fashion. Interestingly, the weak Gal-1 staining detected in the nucleus was not affected by TGF-β1 treatment.
Fig. 1.
TGF-β1, but not TGF-β3, regulates Gal-1 expression in LM3 mammary adenocarcinoma cells. LM3 cells were treated for 24 h with different concentrations (0.4, 4 and 40 ng/ml) of TGF-β1 (a) or TGF-β3 (b). Following treatments, cells were lysed and analyzed for Gal-1 expression by western blot. Equal amounts of protein cell lysates (30 μg) were resolved by 15% SDS-PAGE, electroblotted onto nitrocellulose membranes and probed with the anti-Gal-1 polyclonal Ab as described in Materials and methods. Equal loading was checked by incubating the membranes with an anti-β-actin monoclonal antibody. The immunoreactive protein bands indicated by the arrow (a, b) were semiquantified by densitometry using the Scion Image software. Data are representative of three independent experiments
Fig. 2.
TGF-β1 upregulates Gal-1 expression in the cytoplasm of LM3 cells. Subcellular distribution of Gal-1 upon TGF-β1 treatment was evaluated by immunofluorescence staining and confocal microscopy. LM3 cells were cultured in coverslips and treated for 6, 12 and 24 h with 4 ng/ml TGF-β1 or left untreated (first panel in each column). Cells were then fixed, permeabilized, blocked and incubated with an anti-Gal-1 rabbit policlonal Ab and a secondary FICT-conjugated anti-rabbit polyclonal Ab. Photographs were taken in a confocal microscope (Eclipse E800, Nikon). Inset: no staining was detected in the absence of the primary Ab. Magnification: ×200 (left panels) and ×600 (right panels)
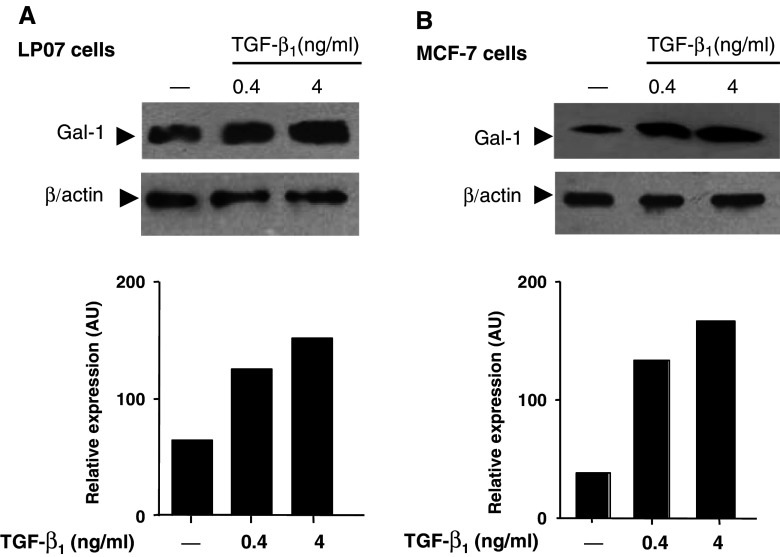
To evaluate whether TGF-β1 may regulate Gal-1 expression in another tumor cells, we selected a murine lung carcinoma cells line (LP07) and a human mammary adenocarcinoma line (MCF-7). As shown in Fig. 3a and b, treatment with TGF-β1 up-regulated Gal-1 expression in both lung LP07 and breast MCF-7 carcinoma cell lines in a dose dependent manner. These results indicate that TGF-β1-induced regulation of Gal-1 expression could be a widespread event and may not be restricted to a particular tumor cell line. In contrast, TGF-β3 had no significant effect on Gal-1 expression in both cell lines tested (data not shown), further confirming our results in LM3 cells.
Fig. 3.
TGF-β1 upregulates Gal-1 expression in LP07 and MCF-7 tumor cells. LP07 (a) and MCF-7 (b) cells were treated for 24 h with different concentrations (0.4 and 4 ng/ml) of TGF-β1. Following treatments, cells were lysed and analyzed for Gal-1 expression by western blot. Equal amounts of protein cell lysates (30 μg) were resolved by 15% SDS-PAGE, electroblotted onto nitrocelulose membranes and probed with the anti-Gal-1 polyclonal Ab as described in Materials and methods. Equal loading was checked by incubating the membranes with an anti-β-actin monoclonal antibody. The immunoreactive protein bands indicated by the arrow (a, b) were semiquantified by densitometry using the Scion Image software. Data are representative of three independent experiments
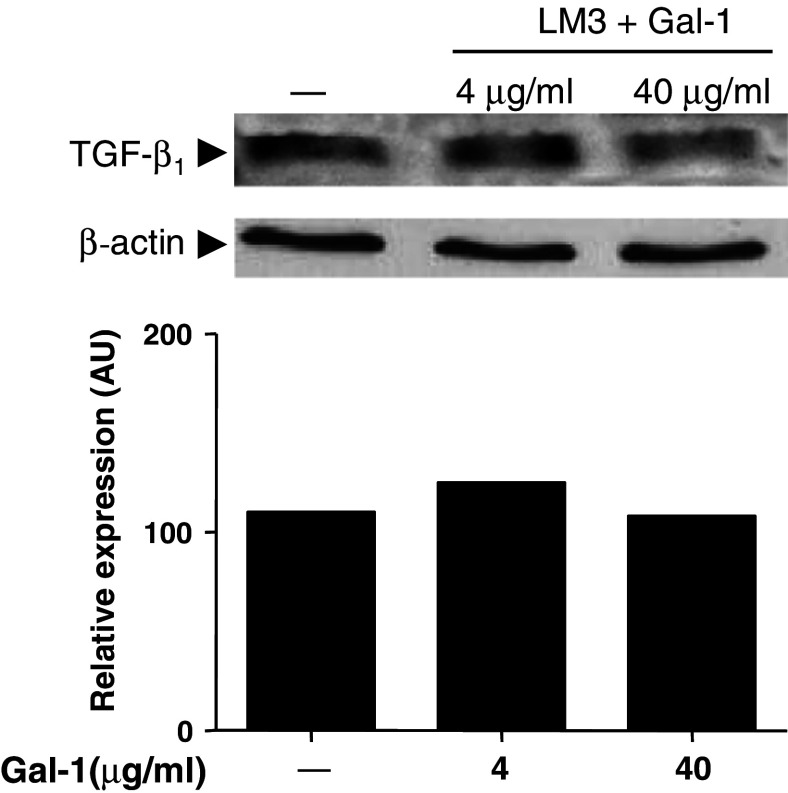
To examine the possibility of a bidirectional regulation between TGF-β1 and Gal-1, LM3 cells were treated with increasing concentrations of recombinant Gal-1. As shown in Fig. 4, Gal-1 was not able to significantly modulate TGF-β1 expression in LM3 cells. These results suggest that TGF-β1 might act as an upstream mediator in the regulation of tumor-induced immunosuppression.
Fig. 4.
Galectin-1 does not modulate TGF-β1 expression in LM3 cells. Subconfluent cells were treated for 24 h with 4 or 40 μg/ml Gal-1. Following different treatments, cells lysates were analyzed for TGF-β1 expression by western blot. Equal amounts of protein were resolved by 15% SDS-PAGE, electroblotted onto nitrocellulose membranes and probed with a specific anti-TGF-β1 antibody, as described in Materials and methods. The expression of β-actin was used as a loading control. The immunoreactive protein bands indicated by the arrow were semiquantified by densitometry using the Scion Image software. Data are representative of three different experiments
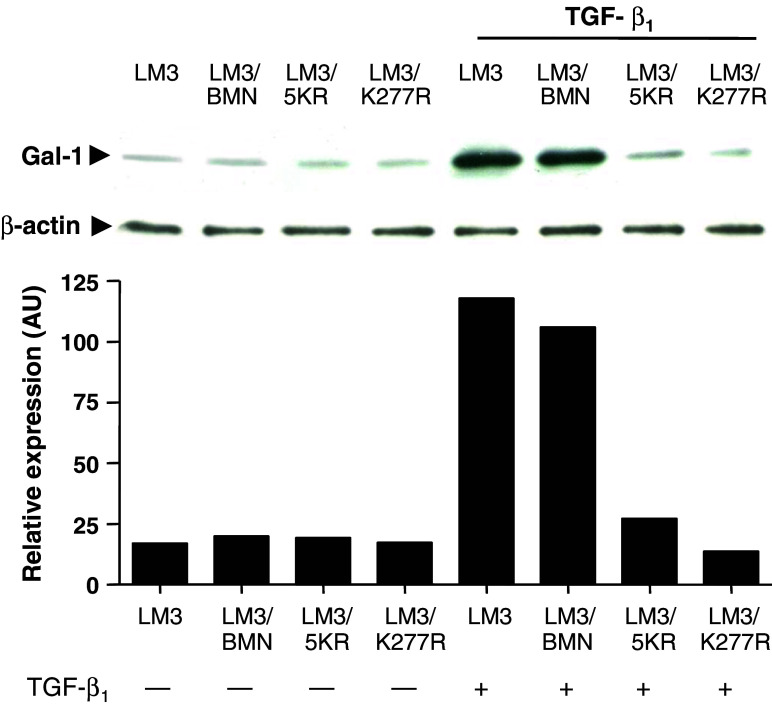
To investigate whether TGF-β1 is able to regulate Gal-1 expression through TGF-β signaling receptors, LM3 cells were stably transduced with retroviral vectors expressing the dominant negative forms (dn) of TβRI (LM3/5KR) or TβRII (LM3/ K277R) as described in Materials and methods. LM3/BMN cells, transduced with a retroviral vector expressing wt TβRI, were used as a control. These dominant negative forms, dnTβRI and dnTβRII were not able to transduce TGF-β-mediated signaling [29] and impaired the tumorigenic ability of LM3 cells (Daroqui et al., personal communication). Up-regulated expression of Gal-1 in LM3 cells clearly involved TGF-β1 binding and signaling through its specific TβRI and TβRII receptors, since this effect was completely abrogated when LM3 cells were genetically engineered to overexpress inactive forms of these kinase receptors (Fig. 5, lanes 7, 8). Moreover, no changes in Gal-1 expression were observed in response to TGF-β1 both in LM3/BMN cells and in the parental LM3 cells, expressing functional TGF-β receptors (Fig. 5, lanes 5, 6). Thus, TGF-β1 triggers a specific receptor-mediated signal to modulate Gal-1 expression in mammary adenocarcinoma cells.
Fig. 5.
TGF-β1 regulates Gal-1 expression in LM3 cells through TβRI and TβRII receptors. LM3 cells were stably transduced with vectors expressing dominant negative forms of TβRI (LM3/5KR) and TβRII (LM3/K277R) as described in Materials and methods. Parental LM3 cells as well as LM3 cells overexpressing wt Alk5 (LM3/BMN) were used as control. LM3 cells expressing either dnTβRI or dnTβRII were treated or not with 4 ng/ml TGF-β1, lysed and analyzed for Gal-1 expression by western blot, as described in Materials and methods. The immunoreactive protein bands were semiquantified by densitometry using the Scion Image software. Blots representative of three independent experiments are shown
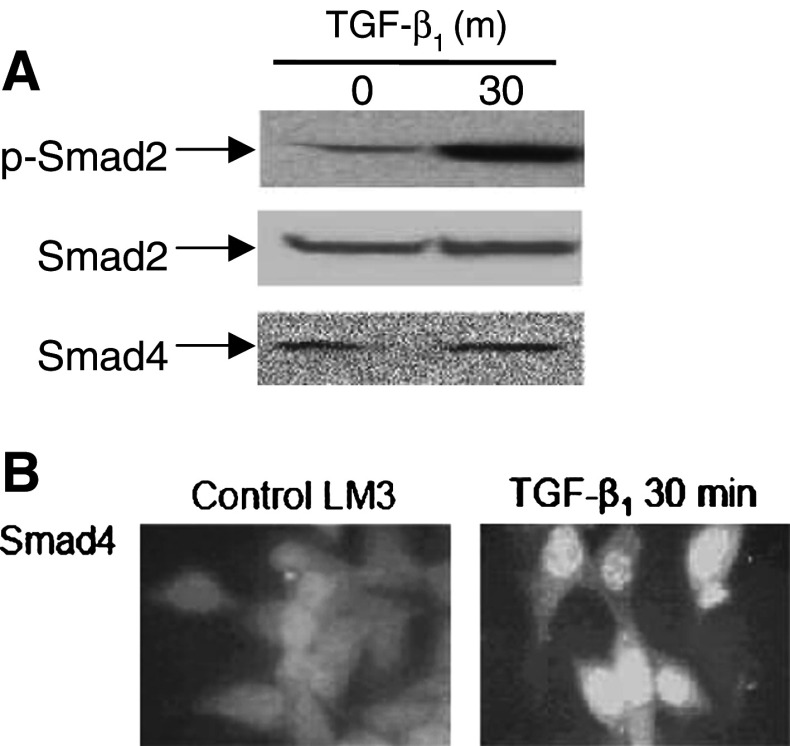
Since LM3 cells express both signaling TGF-β receptors and these receptors are functionally active in LM3 cells [10], we next analyzed whether TGF-β1 may trigger a Smad signaling pathway in these tumor cells. As shown in Fig. 6a, TGF-β1 (4 ng/ml) was able to induce Smad2 phosphorylation in LM3 cells at 30 min treatment, without affecting the total Smad2 expression level. Furthermore, TGF-β1 induced nuclear translocation of Smad4 in LM3 cells as early as 30 min following treatment (Fig. 6b).
Fig. 6.
TGF-β1 triggers a Smad-dependent signaling pathway in LM3 cells. a LM3 cells were treated for 30 min with 4 ng/ml TGF-β1, and cell lysates were subjected to western blot analysis using an anti-phospho-Smad2, an anti-Smad2 or an anti-Smad4/DPC4 antibodies, as described in Materials and methods. Data shown are representative of three independent experiments. b Subcellular localization of Smad4 was evaluated by immunofluorescence staining in LM3 cells treated for 30 min with 4 ng/ml TGF-β1. Cells cultured in coverslips were fixed, permeabilized, blocked and incubated with an anti-Smad4 monoclonal antibody and with a secondary TRICT-conjugated antibody as described above. Photographs were taken in a fluorescence microscope (Eclipse E400, Nikon). Magnification: ×400
To explore whether TGF-β1 might regulate Gal-1 expression through the Smads signaling pathway, we searched for potential regulatory binding sites for the Smad transcription factors in the gal-1 gene by computer-assisted gene sequence analysis. The entire gal-1 gene was scanned for putative binding sites for TGF-β-induced Smads using the MatInspector software. Interestingly, we identified the presence of a putative Smad4 binding site at the position –2.801 with similarity of 1.000 in the core and 0.992 in the matrix (Table 1). In addition, we found the presence of two putative Smad3 binding sites with a similarity of 1.000 in the core and 0.996 and 1.000 in the matrix in the positions −810 and +281, respectively. These findings suggest that TGF-β1 may trigger a Smad-dependent pathway to control Gal-1 expression in mammary adenocarcinoma cells.
Table 1.
Identification of putative binding sites for the Smad complex in the gal-1 gene by computer-assisted gene sequence analysis
| Transcription factor | Core similarity | Matrix similarity | Strand | Position |
|---|---|---|---|---|
| Smad4 | 1.000 | 0.992 | − | −2801 |
| Smad3 | 1.000 | 0.996 | + | −810 |
| Smad3 | 1.000 | 1.000 | + | +281 |
Discussion
In recent years, one of the most important insights into tumor immunity was provided by the identification of negative regulatory pathways (e.g., CTLA-4, PD-1/ PD-L1, Jak2/ Stat3) and immunosuppressive mediators (e.g., TGF-β, Gal-1) which might act in concert to counteract tumor effector mechanisms [11, 12]. These mechanisms are suggested to co-operate in advanced stages of cancer to limit the ability of the immune system to restrain the tumor and the effectiveness of immunotherapy strategies to successfully eradicate malignant cells [32]. It has been proposed that simultaneous blockade of different inhibitory pathways may engender successful cancer immunotherapy [31]. However, the potential cross talk between these mechanisms has not yet been examined. Our results suggest that Gal-1 is a novel transcriptional target of TGF-β signaling in LM3 mammary adenocarcinoma cells. However, further studies are required using reporter gene analysis and chromatin immunoprecipitation (ChIP) assays to directly demonstrate the transcriptional activity of TGF-β signaling in Gal-1 gene expression. Importantly, we were also able to demonstrate TGF-β1 regulation of Gal-1 expression in other tumor cell lines including the murine lung carcinoma line LP07 and the human breast adenocarcinoma cell line MCF-7 demonstrating that TGF-β1-induced Gal-1 upregulation is not specific of a particular tumor cell line. Further supporting our findings, it has been previously shown that TGF-β1 can upregulate Gal-1 mRNA expression in PAC1 rat pulmonary arterial smooth muscle cells and in human vascular smooth muscle cells [28]. Interestingly, while TGF-β1 regulates Gal-1 expression in LM3 cells, this sugar-binding protein does not influence the expression of TGF-β1, suggesting that Gal-1 expression and its associated immunosuppressive activity might be influenced by the levels of TGF-β1 in the tumor microenvironment, specially taking into account the high levels of TGF-β1 found in several tumor models [16].
TGF-β can regulate a wide variety of target genes, which play critical roles in different pathways of tumor progression including tumor suppression (e.g., BRCA1) [41], tumor cell motility and invasiveness (e.g., CUTL1) [27], cell growth and apoptosis (e.g., MAPK, TNF-α and Wnt) [17], and cancer immunoediting (e.g., perforin, granzymes, Fas ligand, and IFN-γ) [44]. In addition, recent evidence indicates that TGF-β1 can regulate gene expression through Smad-dependent or independent mechanisms, indicating that Smad4 may induce transcriptional regulation of genes involved in tumor growth and metastasis, while promoting the silencing of genes involved in tumor-suppressive functions [22]. Here we show that TGF-β1 markedly upregulates the expression of Gal-1, a critical regulator of tumor cell adhesion [45, 48], migration [4, 5], proliferation [18], and tumor-immune escape [23, 40]. Our results are also consistent with the tumor promoting activity showed by TGF-β in our experimental system [10].
Although compelling evidence indicates a critical role for Gal-1 in the modulation of tumor growth and metastasis [23], there is still scarce information about local signals (i.e., cytokines and hormonal context) and molecular pathways involved in the regulation of Gal-1 gene expression within the tumor microenvironment [7]. In this sense, the limited information regarding the regulation of Gal-1 gene expression in tumor cells is restricted to the effects of differentiating stimuli and chemotherapeutic agents including sodium butyrate [13], retinoid acid [24], thyroid stimulating hormone [6] and cyclophosphamide [37, 50].
Our results provide the first evidence that TGF-β1 signaling can control Gal-1 expression in tumor cells. It has been suggested that the combined use of different immunotherapy strategies might be beneficial for the treatment of cancer. In this regard, simultaneous blockade of the TGF-β and Gal-1 signaling pathways in vivo [20, 35, 42] might have critical implications for cancer therapy. These results are particularly interesting in terms of recent findings suggesting that multiple mechanisms of immune evasion can coexist in different tumor cell type [26, 39]. Our findings indicate that Gal-1 is another potential therapeutic target of TGF-β1, suggesting that blockade of TGF-β1 signaling might also influence protein–glycan interactions within the tumor microenvironment, with critical implications for tumor cell growth, invasiveness and tumor-immune escape.
Acknowledgments
The authors wish to thank Drs. J. Hirabayashi and K.I. Kasai for providing the plasmid pET21a/Gal1. This work was supported in part by grants from Mizutani Foundation for Glycoscience (Japan), University of Buenos Aires (UBACYT-M091), Agencia de Promoción Científica y Tecnológica (PICT 2003-05-13787) and Fundación Sales to G.A.R., and by grants from the University of Buenos Aires (UBACYT-M068) Agencia de Promoción Científica y Tecnológica (PICT 2003-05-14088) to E.B.K.J. G.A.R, L.P. and E.B.K.J. are researchers of CONICET. J.M.I., N.R., MS and M.A.T. thank CONICET and M.C.D. and P.V. thanks ANPCyT and Fundación Bunge & Born for the fellowships granted.
Footnotes
C. M. Daroqui, Juan M. Ilarregui, Natalia Rubinstein, and Mariana Salatino contributed equally to this work. E. Bal de Kier Joffé and, Gabriel A. Rabinovich jointly supervised this study and should be considered as senior authors.
References
- 1.Akhurst RJ, Derynck R. TGF-beta signaling in cancer—a double-edged sword. Trends Cell Biol. 2001;11:S44–S51. doi: 10.1016/s0962-8924(01)02130-4. [DOI] [PubMed] [Google Scholar]
- 2.Bakin AV, Rinehart C, Tomlinson AK, Arteaga CL. p38 mitogen-activated protein kinase is required for TGF β-mediated fibroblastic transdifferentiation and cell migration. J Cell Sci. 2002;115:3193–3206. doi: 10.1242/jcs.115.15.3193. [DOI] [PubMed] [Google Scholar]
- 3.Brooks SC, Locke ER, Soule HD. Estrogen receptor in a human cell line (MCF-7) from breast carcinoma. J Biol Chem. 1973;248:6251–6253. [PubMed] [Google Scholar]
- 4.Camby I, Belot N, Lefranc F, Sadeghi N, de Launoit Y, Kaltner H, Musette S, Darro F, Danguy A, Salmon I, Gabius HJ, Kiss R. Galectin-1 modulates human glioblastoma cell migration into the brain through modifications to the actin cytoskeleton and levels of expression of small GTPases. J Neuropathol Exp Neurol. 2002;61:585–596. doi: 10.1093/jnen/61.7.585. [DOI] [PubMed] [Google Scholar]
- 5.Camby I, Decaestecker C, Lefranc F, Kaltner H, Gabius HJ, Kiss R. Galectin-1 knocking down in human U87 glioblastoma cells alters their gene expression pattern. Biochem Biophys Res Commun. 2005;335:27–35. doi: 10.1016/j.bbrc.2005.07.037. [DOI] [PubMed] [Google Scholar]
- 6.Chiariotti L, Benvenuto G, Salvatore P, Veneziani BM, Villone G, Fusco A, Russo T, Bruni CB. Expression of the soluble lectin L-14 gene is induced by TSH in thyroid cells and suppressed by retinoic acid in transformed neural cells. Biochem Biophys Res Commun. 1994;199:540–546. doi: 10.1006/bbrc.1994.1262. [DOI] [PubMed] [Google Scholar]
- 7.Chiariotti L, Salvatore P, Frunzio R, Bruni CB. Galectin genes: regulation of expression. Glycoconj J. 2004;19:441–449. doi: 10.1023/B:GLYC.0000014073.23096.3a. [DOI] [PubMed] [Google Scholar]
- 8.Cooper DN. Galectinomics: finding themes in complexity. Biochim Biophys Acta. 2002;1572:209–231. doi: 10.1016/s0304-4165(02)00310-0. [DOI] [PubMed] [Google Scholar]
- 9.Danguy A, Camby I, Kiss R. Galectins and cancer. Biochim Biophys Acta. 2002;1572:285–293. doi: 10.1016/s0304-4165(02)00315-x. [DOI] [PubMed] [Google Scholar]
- 10.Daroqui MC, Puricelli LI, Urtreger AJ, Elizalde PV, Lanuza GM, de Bal Kier Joffe E. Involvement of TGF-beta(s)/T(beta)Rs system in tumor progression of murine mammary adenocarcinomas. Breast Cancer Res Treat. 2003;80:287–301. doi: 10.1023/A:1024910332621. [DOI] [PubMed] [Google Scholar]
- 11.Dunn GP, Old LJ, Schreiber RD. The immunobiology of cancer immunosurveillance and immunoediting. Immunity. 2004;21:137–148. doi: 10.1016/j.immuni.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 12.Gabrilovich D, Pisarev V. Tumor escape from immune response: mechanisms and targets of activity. Curr Drug Targets. 2003;4:525–536. doi: 10.2174/1389450033490849. [DOI] [PubMed] [Google Scholar]
- 13.Gillenwater A, Xu XC, Estrov Y, Sacks PG, Lotan D, Lotan R. Modulation of galectin-1 content in human head and neck squamous carcinoma cells by sodium butyrate. Int J Cancer. 1998;75:217–224. doi: 10.1002/(SICI)1097-0215(19980119)75:2<217::AID-IJC9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 14.Gorelik L, Flavell RA. Immune-mediated eradication of tumors through the blockade of transforming growth factor-beta signaling in T cells. Nat Med. 2001;7:1118–1122. doi: 10.1038/nm1001-1118. [DOI] [PubMed] [Google Scholar]
- 15.Hirabayashi J, Hashidate T, Arata Y, Nishi N, Nakamura T, Hirashima M, Urashima T, Oka T, Futai M, Muller WE, Yagi F, Kasai K. Oligosaccharide specificity of galectins: a search by frontal affinity chromatography. Biochim Biophys Acta. 2002;1572:232–254. doi: 10.1016/s0304-4165(02)00311-2. [DOI] [PubMed] [Google Scholar]
- 16.Kaklamani VG, Pasche B. Role of TGF-beta in cancer and the potential for therapy and prevention. Expert Rev Anticancer Ther. 2004;4:649–661. doi: 10.1586/14737140.4.4.649. [DOI] [PubMed] [Google Scholar]
- 17.Kloth JN, Fleuren GJ, Oosting J, de Menezes RX, Eilers PH, Kenter GG, Gorter A. Substantial changes in gene expression of Wnt, MAPK and TNFalpha pathways induced by TGF-beta1 in cervical cancer cell lines. Carcinogenesis. 2005;26:1493–1502. doi: 10.1093/carcin/bgi110. [DOI] [PubMed] [Google Scholar]
- 18.Kopitz J, von Reitzenstein C, Andre S, Kaltner H, Uhl J, Ehemann V, Cantz M, Gabius HJ. Negative regulation of neuroblastoma cell growth by carbohydrate-dependent surface binding of galectin-1 and functional divergence from galectin-3. J Biol Chem. 2001;276:35917–35923. doi: 10.1074/jbc.M105135200. [DOI] [PubMed] [Google Scholar]
- 19.Lahm H, Andre S, Hoeflich A, Kaltner H, Siebert HC, Sordat B, von der Lieth CW, Wolf E, Gabius HJ. Tumor galectinology: insights into the complex network of a family of endogenous lectins. Glycoconj J. 2004;20:227–238. doi: 10.1023/B:GLYC.0000025817.24297.17. [DOI] [PubMed] [Google Scholar]
- 20.Laping NJ, Grygielko E, Mathur A, Butter S, Bomberger J, Tweed C, Martin W, Fornwald J, Lehr R, Harling J, Gaster L, Callahan JF, Olson BA. Inhibition of transforming growth factor (TGF)-beta1-induced extracellular matrix with a novel inhibitor of the TGF-beta type I receptor kinase activity: SB-431542. Mol Pharmacol. 2002;62:58–64. doi: 10.1124/mol.62.1.58. [DOI] [PubMed] [Google Scholar]
- 21.Le QT, Shi G, Cao H, Nelson DW, Wang Y, Chen EY, Zhao S, Kong C, Richardson D, O’Byrne KJ, Giaccia AJ, Koong AC. Galectin-1: a link between tumor hypoxia and tumor immune privilege. J Clin Oncol. 2005;23:8932–8941. doi: 10.1200/JCO.2005.02.0206. [DOI] [PubMed] [Google Scholar]
- 22.Levy L, Hill CS. Smad4 dependency defines two classes of transforming growth factor {beta} (TGF-{beta}) target genes and distinguishes TGF-{beta}-induced epithelial-mesenchymal transition from its antiproliferative and migratory responses. Mol Cell Biol. 2005;25:8108–8125. doi: 10.1128/MCB.25.18.8108-8125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu F, Rabinovich G. Galectins as modulators of tumour progression. Nature Rev Cancer. 2005;5:29–41. doi: 10.1038/nrc1527. [DOI] [PubMed] [Google Scholar]
- 24.Lu Y, Lotan D, Lotan R. Differential regulation of constitutive and retinoic acid-induced galectin-1 gene transcription in murine embryonal carcinoma and myoblastic cells. Biochim Biophys Acta. 2000;1491:13–9. doi: 10.1016/s0167-4781(00)00055-5. [DOI] [PubMed] [Google Scholar]
- 25.Massague J. TGF-beta signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 26.Mendez R, Ruiz-Cabello F, Rodriguez T, Del Campo A, Paschen A, Schadendorf D, Garrido F (2006) Identification of different tumor escape mechanisms in several metastases from a melanoma patient undergoing immunotherapy. Cancer Immunol Immunother (Epub ahead of print) [DOI] [PMC free article] [PubMed]
- 27.Michl P, Ramjaun AR, Pardo OE, Warne PH, Wagner M, Poulsom R, D’Arrigo C, Ryder K, Menke A, Gress T, Downward J. CUTL1 is a target of TGF(beta) signaling that enhances cancer cell motility and invasiveness. Cancer Cell. 2005;7:521–532. doi: 10.1016/j.ccr.2005.05.018. [DOI] [PubMed] [Google Scholar]
- 28.Moiseeva EP, Javed Q, Spring EL, de Bono DP. Galectin-1 is involved in vascular smooth muscle cell proliferation. Cardiovasc Res. 2000;45:493–502. doi: 10.1016/S0008-6363(99)00276-X. [DOI] [PubMed] [Google Scholar]
- 29.Oft M, Heider KH, Beug H. TGFbeta signaling is necessary for carcinoma cell invasiveness and metastasis. Curr Biol. 1998;8:1243–52. doi: 10.1016/S0960-9822(07)00533-7. [DOI] [PubMed] [Google Scholar]
- 30.Pantel K, Brakenhoff RH. Dissecting the metastatic cascade. Nat Rev Cancer. 2004;4:448–456. doi: 10.1038/nrc1370. [DOI] [PubMed] [Google Scholar]
- 31.Pardoll D, Allison J. Cancer immunotherapy: breaking the barriers to harvest the crop. Nat Med. 2004;10:887–892. doi: 10.1038/nm0904-887. [DOI] [PubMed] [Google Scholar]
- 32.Pawelec G. Tumour escape from the immune response: the last hurdle for successful immunotherapy of cancer? Cancer Immunol Immunother. 1999;48:343–345. doi: 10.1007/s002620050584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paz A, Haklai R, Elad-Sfadia G, Ballan E, Kloog Y. Galectin-1 binds oncogenic H-Ras to mediate Ras membrane anchorage and cell transformation. Oncogene. 2001;20:7486–7493. doi: 10.1038/sj.onc.1204950. [DOI] [PubMed] [Google Scholar]
- 34.Rabinovich GA, Daly G, Dreja H, Tailor H, Riera CM, Hirabayashi J, Chernajovsky Y. Recombinant galectin-1 and its genetic delivery suppress collagen-induced arthritis via T cell apoptosis. J Exp Med. 1999;190:385–398. doi: 10.1084/jem.190.3.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rabinovich GA, Baum LG, Tinari N, Paganelli R, Natoli C, Liu FT, Iacobelli S. Galectins and their ligands: amplifiers, silencers or tuners of the inflammatory response? Trends Immunol. 2002;23:313–320. doi: 10.1016/S1471-4906(02)02232-9. [DOI] [PubMed] [Google Scholar]
- 36.Rabinovich GA, Rubinstein N, Matar P, Rozados V, Gervasoni S, Scharovsky GO. The antimetastatic effect of a single low dose of cyclophosphamide involves modulation of galectin-1 and Bcl-2 expression. Cancer Immunol Immunother. 2002;50:597–603. doi: 10.1007/s00262-001-0238-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rabinovich GA, Rubinstein N, Toscano MA. Role of galectins in inflammatory and immunomodulatory processes. Biochim Biophys Acta. 2002;1572:274–284. doi: 10.1016/s0304-4165(02)00314-8. [DOI] [PubMed] [Google Scholar]
- 38.Rabinovich GA, Cumashi A, Bianco GA, Ciavardelli D, Iurisci I, D’Egidio M, Piccolo E, Tinari N, Nifantiev N, Iacobelli S. Synthetic lactulose amines: novel class of anticancer agents that induce tumor-cell apoptosis and inhibit galectin-mediated homotypic cell aggregation and endothelial cell morphogenesis. Glycobiology. 2006;16:210–220. doi: 10.1093/glycob/cwj056. [DOI] [PubMed] [Google Scholar]
- 39.Real LM, Jimenez P, Kirkin A, Serrano A, Garcia A, Canton J, Zeuthen J, Garrido F, Ruiz-Cabello F. Multiple mechanisms of immune evasion can coexist in melanoma tumor cell lines derived from the same patient. Cancer Immunol Immunother. 2001;49:621–628. doi: 10.1007/s002620000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rubinstein N, Alvarez M, Zwirner NW, Toscano MA, Ilarregui JM, Bravo A, Mordoh J, Fainboim L, Podhajcer OL, Rabinovich GA. Targeted inhibition of galectin-1 gene expression in tumor cells results in heightened T cell-mediated rejection; A potential mechanism of tumor-immune privilege. Cancer Cell. 2004;5:241–251. doi: 10.1016/S1535-6108(04)00024-8. [DOI] [PubMed] [Google Scholar]
- 41.Satterwhite DJ, Matsunami N, White RL. TGF-beta1 inhibits BRCA1 expression through a pathway that requires pRb. Biochem Biophys Res Commun. 2000;276:686–692. doi: 10.1006/bbrc.2000.3510. [DOI] [PubMed] [Google Scholar]
- 42.Sorme P, Kahl-Knutsson B, Wellmar U, Magnusson BG, Leffler H, Nilsson UJ. Design and synthesis of galectin inhibitors. Methods Enzymol. 2003;363:157–169. doi: 10.1016/S0076-6879(03)01050-4. [DOI] [PubMed] [Google Scholar]
- 43.ten Dijke P, Hill CS. New insights into TGF-beta-Smad signalling. Trends Biochem Sci. 2004;29:265–273. doi: 10.1016/j.tibs.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 44.Thomas DA, Massague J. TGF-beta directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell. 2005;8:369–380. doi: 10.1016/j.ccr.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 45.Tinari N, Kuwabara I, Huflejt ME, Shen PF, Iacobelli S, Liu FT. Glycoprotein 90K/MAC-2BP interacts with galectin-1 and mediates galectin-1-induced cell aggregation. Int J Cancer. 2001;91:167–172. doi: 10.1002/1097-0215(200002)9999:9999<::AID-IJC1022>3.3.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 46.Urtreger AJ, Diament MJ, Ranuncolo SM, Del Vidal C M, Puricelli LI, Klein SM, de Bal Kier Joffe ED. New murine cell line derived from a spontaneous lung tumor induces paraneoplastic syndromes. Int J Oncol. 2001;18:639–647. doi: 10.3892/ijo.18.3.639. [DOI] [PubMed] [Google Scholar]
- 47.Urtreger A, Ladeda V, Puricelli L, Rivelli A, Vidal MC, Lustig ES, de Bal Kier Joffé E. Modulation of fibronectin expression and proteolytic activity associated with the invasive and metastatic phenotype in two murine mammary cell lines. Int J Oncol. 1997;11:489–496. doi: 10.3892/ijo.11.3.489. [DOI] [PubMed] [Google Scholar]
- 48.van den Brule F, Califice S, Castronovo V. Expression of galectins in cancer: a critical review. Glycoconj J. 2004;19:537–542. doi: 10.1023/B:GLYC.0000014083.48508.6a. [DOI] [PubMed] [Google Scholar]
- 49.van den Brule FA, Buicu C, Baldet M, Sobel ME, Cooper DN, Marschal P, Castronovo V. Galectin-1 modulates human melanoma cell adhesion to laminin. Biochem Biophys Res Commun. 1995;209:760–767. doi: 10.1006/bbrc.1995.1564. [DOI] [PubMed] [Google Scholar]
- 50.Zacarias-Fluck MF, Rico MJ, Gervasoni SI, Ilarregui JM, Toscano MA, Rabinovich GA, Scharovsky GO (2006) Low dose cyclophosphamide modulates galectin-1 expression and function in an experimental rat lymphoma model. Cancer Immunol Immunother (Epub ahead of print) [DOI] [PMC free article] [PubMed]