Abstract
The human promonocytic cell line U937 undergoes apoptosis upon treatment with tumor necrosis factor alpha (TNF-α). This cell line has previously been shown to be very sensitive to the lytic effect of the autonomous parvovirus H-1. Parvovirus infection leads to the activation of the CPP32 ICE-like cysteine protease which cleaves the enzyme poly(ADP-ribose)polymerase and induces morphologic changes that are characteristic of apoptosis in a way that is similar to TNF-α treatment. This effect is also observed when the U937 cells are infected with a recombinant H-1 virus which expresses the nonstructural (NS) proteins but in which the capsid genes are replaced by a reporter gene, indicating that the induction of apoptosis can be assigned to the cytotoxic nonstructural proteins in this cell system. The c-Myc protein, which is overexpressed in U937 cells, is rapidly downregulated during infection, in keeping with a possible role of this product in mediating the apoptotic cell death induced by H-1 virus infection. Interestingly, four clones (designated RU) derived from the U937 cell line and selected for their resistance to H-1 virus (J. A. Lopez-Guerrero et al., Blood 89:1642–1653, 1997) failed to decrease c-Myc expression upon treatment with differentiation agents and also resisted the induction of cell death after TNF-α treatment. Our data suggest that the RU clones have developed defense strategies against apoptosis, either by their failure to downregulate c-Myc and/or by activating antiapoptotic factors.
Parvoviruses are small, single-stranded DNA viruses that infect a wide variety of animal species, including humans (80). The H-1 virus belongs to the subgroup of autonomous, replicating parvoviruses and contains a linear DNA genome of about 5 kb (15, 74). The low genetic complexity of parvoviruses renders them tightly dependent on cellular factors that are expressed as a function of cell proliferation (S phase of the cell cycle) and differentiation to achieve their lytic cycle (15). Parvoviruses are unable to force quiescent cells into the S phase. Cancer cells seem to provide parvoviruses with an environment favorable to their multiplication. Indeed, several parvoviruses exhibit a remarkable oncotropism (75). In agreement with this, it has been shown previously that many in vitro-transformed cells are sensitized to the parvovirus-induced killing compared to their untransformed counterparts, which correlates with an increased capacity of the transformants to sustain certain steps of the viral life cycle (10, 13, 77). In particular, the production and toxic activity of the nonstructural (NS) protein NS-1, which is a key effector of parvovirus replication and cytopathogenicity, can be stimulated in oncogene-transformed cells (61). This may account, at least partially, for the fact that parvoviruses can exert an oncosuppressive activity in vivo (75).
In order to investigate the mechanisms involved in parvovirus anticancer surveillance, we have recently isolated and characterized rare variants that derive from the human myeloid leukemia cell line U937 (83), are designated RU, and differ from the parental cell line in that they are resistant to H-1 virus infection (52). Of the four RU clones analyzed, three showed both a significant decrease, compared with the parental U937 cells, of the steady-state level of the c-Myc oncoprotein and a reduced tumorigenic capacity when implanted in scid mice. Moreover, all of the RU cells exhibited a constitutive production of nitric oxide (NO) and superoxide anion (O2−). Deregulated c-Myc expression in various tumor cells has been involved in their susceptibility to undergo apoptosis in response to several inducers (9), including tumor necrosis factor alpha (TNF-α) (35, 36). Furthermore, NO was reported to inhibit apoptosis in mononuclear cells (45) and B-lymphocytes (53), and superoxide anion was found to suppress Fas-mediated cell death (12). Altogether, the data prompted us to investigate whether parvovirus H-1 was able to induce apoptosis in U937 cells and, if so, whether the H-1 virus-resistant RU cells also resist known apoptotic inducers, TNF-α in particular, that efficiently lead U937 cells to apoptosis (6, 31, 64, 96, 97).
Apoptosis can be induced in response to various stimuli (92, 93), including such viruses as chicken anemia virus (38), measles virus (20), human immunodeficiency virus (58), influenza virus (85), or murine cytomegalovirus (102). Some ultrastructural features of erythroid precursors infected with parvovirus B19 suggest that this virus also triggers apoptosis in these cells (60). Other viruses have developed strategies to block the commitment of cells into the cell death program that can be viewed as host defense mechanisms against infection (5, 72), and some viral products can even have antagonistic effects on apoptosis, such as the adenovirus E1a and E1b gene products (7, 16).
Programmed cell death or apoptosis represents a complex phenomenon that is characterized morphologically by chromatin condensation, plasma membrane blebbing, and cell fragmentation into membrane-enclosed vesicles (apoptotic bodies). The underlying molecular mechanisms of apoptosis involve the expression of an increasing number of genes conserved from nematodes to insects and mammals (29, 79). Cellular genes such as p53 (71), members of the bcl-2 family (23, 73, 84, 101), or certain oncogenes modulate the commitment of cells into the apoptotic program in response to various stimuli (92, 93). Recent evidence shows the importance of the ICE/CED-3 family of cysteine proteases, now renamed caspases, in the apoptotic process (24, 55). Members of this family, for which several genes have already been identified (63), have common features and are all related to the ced-3 product of Caenorhabditis elegans (103). ICE was first described as the cysteine protease cleaving pro-interleukin-1β to generate active interleukin-1β (87). All the caspases contain the highly conserved QACRG sequence which comprises the active-site cysteine. Active cysteine proteases are produced by cleavage of a proenzyme at key aspartate residues, generating two subunits of approximately 20 and 10 kDa that can assemble into a heterotetrameric protein with two active sites (26). Several substrates for proteolytic cleavage by ICE-like proteins have been identified (for a review, see reference 55), in particular the poly(ADP-ribose) polymerase (PARP) (40, 46) that catalyzes the poly(ADP-ribosylation) of nuclear proteins when activated by DNA strand breaks (48), and, recently, Nagata and coworkers (19, 76) identified a caspase-activated DNase (CAD) that degrades DNA during apoptosis and its interacting protein inhibitor (ICAD), which is cleaved by caspase-3 and releases CAD.
Members of the TNF receptor (TNF-R) superfamily have been shown to play an important role in the activation of the apoptotic execution machinery (33, 65). TNF-α and the related FAS ligand (FASL) can trigger apoptosis in susceptible cells by activating their cell-surface receptors TNFR1 and Fas (Apo-1/CD95), respectively. The cytoplasmic regions of these receptors contain a death domain that is essential for the signal-transduction pathway. The death domain interacts with other proteins that contain this motif and associate with TNF-R (TRADD) or FAS (FADD/MORT-1) proteins (63). Recently, a new protein, MACH/FLICE/Mch5/caspase-8, was shown to interact with these receptor complexes (62) and appears to be a direct link between the signals at the membrane level and the apoptotic execution machinery (56). How caspase-8 transmits the signal to downstream targets, and especially how other caspases become activated, is not yet understood. Recent studies have pointed out the key role of mitochondria in the apoptotic execution in cell-free systems (42–44, 50, 99). Mitochondria appear to act through the release of soluble factors, in particular cytochrome c and a 50-kDa protease, whose translocation to the cytoplasm is regulated by Bcl-2 or members of this family (59).
We investigate here whether apoptosis was induced in U937 cells after parvovirus H-1 infection by measuring the cleavage of the PARP enzyme and the formation of apoptotic bodies. Results presented in this study give evidence for the participation of ICE-like enzymes, in particular caspase-3 (CPP32/apopain/Yama) as effectors of H-1 virus-induced killing. An early event during H-1 virus infection of U937 cells is the rapid decrease of their content in c-myc transcripts and proteins, a feature also observed in TNF-α-treated cells (data not shown) and interpreted in terms of a possible role of this oncoprotein in apoptosis. Furthermore, the four RU clones, selected for their resistance to parvovirus H-1 infection, were also found to resist apoptosis in response to TNF-α. Altogether, our data suggest a possible interconnection between the apoptotic pathways activated by TNF-α and parvovirus H-1.
MATERIALS AND METHODS
Cells and virus.
The human promonocytic cell line U937 (83) and four subclones designated RU (52) were cultured in RPMI 1640 medium (Gibco BRL) supplemented with 10% heat-inactivated fetal calf serum in a 5% CO2 atmosphere at 37°C.
H-1 parvovirus was propagated in NB-E cells and purified as described previously (10). H-1 virus inoculation and titration by plaque assay were performed according to published methods (10). Wild-type H-1 virus was inactivated by UV irradiation (258 nm, 170 kJ/m2). Recombinant H-1 virus expressing the green fluorescent protein (GFP) from jellyfish (104) was produced by cell cotransfection with an H-1 virus infectious plasmid in which the VP genes had been replaced by the GFP gene and with a helper plasmid expressing the capsid genes as described by Kestler et al. (40a). The recombinant virus was purified on cesium gradient as previously described (10).
Immunoblot analysis.
Cultures (106 cells) were inoculated with H-1 parvovirus at a multiplicity of infection (MOI) of 10 PFU per cell or treated with TNF-α (Sigma) (1 or 50 ng/ml) and cycloheximide (Sigma) (10 μg/ml). The rabbit polyclonal anti-TNF-α blocking antibody (IC-Chemikalien, Ismaning, Germany) was used at a concentration of 10 μg/ml of RPMI medium containing 0.1% bovine serum albumin (BSA). After being treated, cells were collected by low-speed centrifugation, suspended in 100 μl of RIPA buffer (10 mM Tris-HCl, pH 7.4; 150 mM NaCl; 1 mM EDTA; 1% Triton X-100; 0.5% sodium deoxycholate; 0.1% sodium dodecyl sulfate [SDS]) containing 3 M urea, 10 μM pepstatin, and 1 mM phenylmethylsulfonyl fluoride, and incubated for 1 h on ice. The samples were centrifuged at 18,000 × g to eliminate DNA and cellular debris. After the protein concentration was determined (Bio-Rad assay), 50 to 150 μg of the total protein was diluted in an equal volume of 100 mM Tris-HCl (pH 6.8)–5% SDS–10% 2-mercaptoethanol–20% glycerol–0.1% bromophenol blue, subjected to 8 or 0.1% SDS–15% polyacrylamide gel electrophoresis, and electrotransferred onto a nitrocellulose membrane (Amersham). Nonspecific binding sites were blocked by incubating the membrane for at least 2 h in phosphate-buffered saline (PBS) containing 5% BLOTTO powdered milk and 0.1% Tween-20 (Sigma). Blots were further incubated with the indicated antibodies and visualized with an enhanced chemiluminescence kit and horseradish peroxidase-conjugated second antibody according to the manufacturer’s recommendation (Amersham). Antibody directed against PARP (clone CII10) was a generous gift of Guy Poirier (Quebec City, Quebec, Canada). Antibody specific for CPP32 p11 (clone C-17) was purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.). Monoclonal antibodies against c-Myc (clone 9E10) and Max (C-17) were purchased from Sigma and Santa Cruz Biotechnology, respectively. The rabbit polyclonal serum SP8 directed against carboxy-terminal peptides of NS1 has been already described (21). His-tagged VP-2 protein from H-1 virus was expressed in bacteria and used to raise a rabbit polyclonal antiserum in our laboratory.
RNA extraction and Northern blotting.
Total cellular RNA was isolated at different times postinfection by the modified guanidium isothiocyanate method described by Chomczynski and Sacchi (11). Briefly, 106 infected cells were resuspended in 0.5 ml of 4 M guanidium isothiocyanate, 25 mM sodium citrate (pH 7), 0.5% sarcosyl, and 0.1 M 2-mercaptoethanol. After 0.5 ml of water-saturated phenol and 0.1 ml of chloroform were added, the sample was mixed and further incubated on ice for 20 min. The RNAs were precipitated from the aqueous phase with 1 volume of isopropanol at −20°C. After centrifugation, the RNA pellet was solubilized in formamide at 50°C for 15 min and stored at −70°C.
The RNAs were fractionated by electrophoresis on a 1% agarose-formaldehyde gel. After transfer under high salt conditions to a Hybond-N+ nylon filter (Amersham), the samples were prehybridized with salmon sperm DNA (100 μg/ml) and hybridized overnight at 42°C with a 32P-labeled randomly primed specific c-myc DNA probe (exon2, EcoRV-BglII fragment) in the presence of 50% formamide and 5% dextran sulfate. Membranes were then washed under highly stringent conditions (15 min at 42°C and 30 min at 55°C in 0.2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]–0.1% SDS) and autoradiographed.
Nucleus Hoechst staining.
At different times after infection (MOI of 5 PFU per cell) or after treatment with TNF-α (10 ng/ml), cultures (106 cells) were collected by low-speed centrifugation, suspended in 100 μl of PBS, and allowed to settle for 15 min on slides pretreated with 1 mg of poly-l-lysine (Sigma) per ml before fixation with 4% formalin. After being washed with PBS, the slides were incubated with Hoechst staining at a concentration of 75 μg/ml for 30 min at 4°C, washed twice with PBS, and examined under a fluorescent Leitz microscope at 450 nm after the samples had been excited at 330 nm.
RESULTS
Parvovirus H-1 induces apoptosis in the U937 cell line by activating members of the caspase family.
With very few exceptions, the replication of autonomous parvoviruses in cultures of permissive cell lines has been described to be accompanied by typical cytopathic changes (e.g., cell shrinking and nuclear morphologic changes) (80) that can now be considered characteristic of cells undergoing apoptosis. This prompted us to investigate the molecular mechanisms underlying H-1 virus-induced cell death in the human monocytic cell line U937. This cell line had been shown to be very sensitive to the cytopathic effect of H-1 virus, with fewer than 0.1% of cells surviving 4 days after infection at an MOI of 100 PFU/cell (52). Two criteria of apoptosis were considered in this study: morphologic changes (i.e., the appearance of apoptotic bodies) and cleavage of the enzyme PARP, a known substrate for some proteases of the caspase family that play a key role in programmed cell death (34, 89).
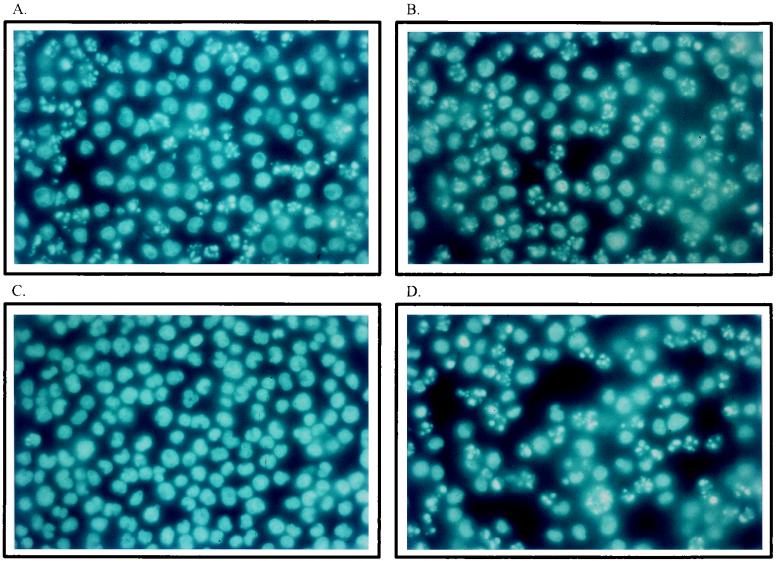
As shown in Fig. 1, typical apoptotic bodies could be observed in cultures infected with H-1 virus (5 PFU/cell), affecting 30% to more than 60% of the cells at 15 h (Fig. 1A) and 25 h (Fig. 1B) postinfection, respectively. In contrast, these morphologic alterations were hardly detectable in mock-infected cultures (Fig. 1C). U937 cells undergoing apoptosis after treatment with TNF-α (10 ng/ml), a known inducer of programmed cell death in this cell line (31, 64, 96), are shown for comparison in Fig. 1D.
FIG. 1.
Induction of morphologic changes in U937 cells infected with H-1 virus. Cultures (106 cells) were infected with H-1 virus (5 PFU/cell) (A and B), mock infected (C), or treated with TNF-α (10 ng/ml) (D) and further incubated for 15 h (A) and 25 h (B, C, and D). Cells were collected by low-speed centrifugation and suspended in PBS before being fixed in 4% formalin on poly-l-lysine-coated slides, stained with Hoechst solution, and examined by fluorescence microscopy.
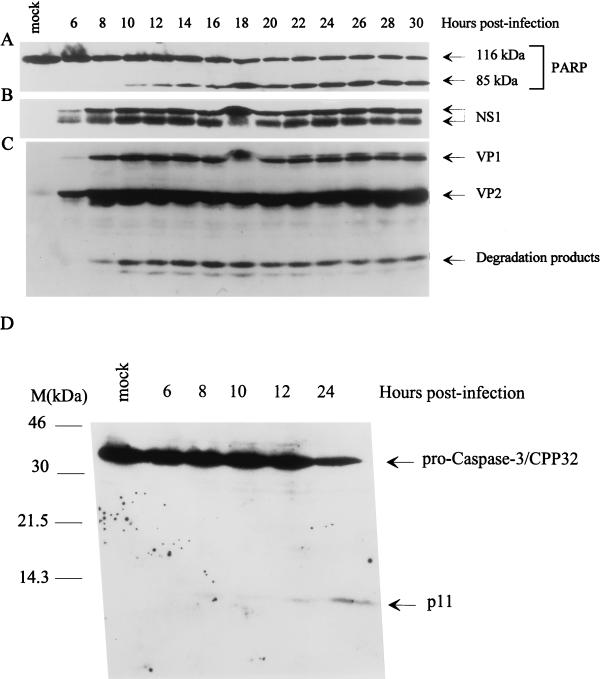
PARP, a 116-kDa enzyme implicated in DNA single-strand break repair, has been shown to be cleaved into two specific fragments (85 and 23 kDa) during the onset of apoptosis (40, 46). This cleavage is induced by members of the ICE-like cysteine protease family, in particular the CPP32/YAMA/apopain/caspase-3 (86). Since U937 cells express high levels of PARP, time course experiments were carried out to investigate whether infection with parvovirus H-1 led to the cleavage of this enzyme. At different times postinfection (MOI of 10 PFU/cell), proteins were extracted and analyzed by Western blotting with the appropriate antibodies. As shown in Fig. 2A, the cleavage of PARP could be observed at 10 h postinfection and reached its maximum after 18 h. No cleavage of PARP was detected in mock-treated cells. The production of the viral nonstructural (NS; 86 kDa) protein NS-1 in phosphorylated and unphosphorylated forms and of the capsid proteins VP-1 (88 kDa) and VP-2/3 (68 and 65 kDa, respectively) was determined at the same time intervals. As seen in Fig. 2B and C, respectively, the NS and VP proteins could be detected by Western blotting as early as 6 h postinfection, thus preceding the cleavage of PARP.
FIG. 2.
Cleavage of PARP and activation of CPP32/caspase-3 in parvovirus H-1-infected cells. Total protein extracts were prepared at different times after U937 cell infection with H-1 virus (10 PFU/cell). Aliquots (70 or 150 μg) of proteins were analyzed by Western blotting with various antibodies. (A) Monoclonal PARP antibody. The bands corresponding to full-size PARP (116 kDa) or its cleavage product (85 kDa) are indicated. (B) Polyclonal anti-NS-1 serum. The phosphorylated (upper band) and nonphosphorylated (lower band) forms of NS-1 are marked. (C) Polyclonal antiserum directed against VP-1 and VP-2 capsid proteins. (D) CPP32/caspase-3 antibody. The molecular sizes are as indicated.
We further determined whether the cleavage of PARP correlated with the activation of CPP32/caspase-3, a cysteine protease known to use PARP as a substrate in vitro (for a review, see reference 94). CPP32 is produced as a proenzyme that has an apparent molecular size of 32 kDa and is activated as a result of its cleavage into two subunits, p11 and p20, of 11 and 20 kDa, respectively. These subunits are associated in a heterotetrameric structure (22, 86, 87). As shown in Fig. 2D, the level of the proenzyme caspase-3 decreased about 8 h after infection, while the p11 fragment became visible as a weak band on the Western blot due to the relatively poor affinity of the antibody for the cleaved molecule. The p20 subunit is not recognized by the commercial antibody and could therefore not be detected in these experiments. Altogether, these results strongly suggest that H-1 virus-induced apoptosis in the U937 cell line involves the activation of at least some members of the caspase family. Apoptosis induced by H-1 infection does not seem to be restricted to the monocytic cell line U937. Indeed, although PARP cleavage could not be detected in two reference fibroblast lines, we were able to reproducibly observe other signs of apoptosis, such as a DNA ladder in the mouse A9 cells and the EJ-ras oncogene-transformed Fisher rat cells (FR-EJ4), after infection with the minute virus of mice (MVMp) and the H-1 virus, respectively (data not shown).
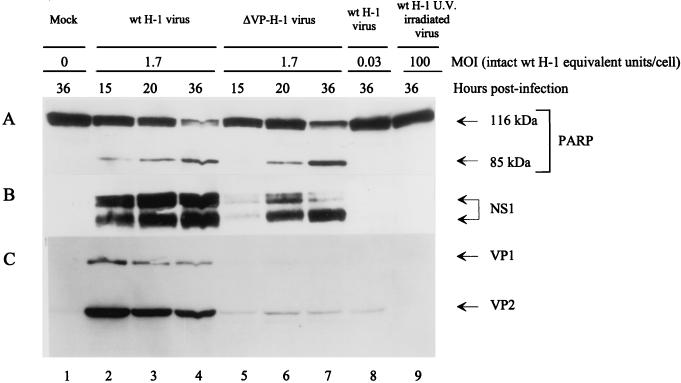
In order to determine whether H-1 virus adsorption and/or uptake is sufficient to trigger apoptosis or whether de novo synthesis of viral proteins is required, U937 cultures (106 cells) were inoculated with UV-inactivated H-1 virus (100 PFU/cell) (Fig. 3, lane 9). For comparison, parallel cultures were infected with nonirradiated H-1 virus (MOI of 1.7 PFU/cell; Fig. 3, lanes 2 to 4) or with nonirradiated recombinant H-1 virus in which the genes encoding the capsids were replaced by the GFP gene from jellyfish (104) (MOI of 1.7 PFU/cell; Fig. 3, lanes 5 to 7). A batch of cells was also infected with intact virus at a low MOI (0.03 PFU/cell; Fig. 3, lane 8), corresponding to the contamination of the recombinant virus stock with wild-type particles. After protein extraction at different times posttreatment, the cleavage of PARP and the production of viral NS-1 and VP-1/VP-2 proteins were analyzed by Western blotting. As shown in Fig. 3A, the recombinant H-1/GFP virus induced PARP cleavage (lanes 5 to 7) as efficiently as did the wild-type virus (lanes 2 to 4), whereas the UV-irradiated H-1 virus failed to do so even at the very high dose of particles equivalent to the 100 PFU/cell that was used (lane 9). The result is in keeping with the known cytotoxic activity of the parvoviral NS proteins, in particular NS-1 (8), suggesting that de novo synthesis and accumulation of NS-1 proteins, for which the wild-type and recombinant viruses are both competent, were necessary to commit cells into the apoptotic pathway, as indicated by the PARP cleavage. As illustrated in Fig. 3B, the accumulation of NS-1 proteins after infection with either wild-type H-1 virus (lanes 2 to 4) or recombinant H-1/GFP virus (lanes 5 to 7) was comparable, while the accumulation of VP polypeptides, as seen with the wild-type virus, was hardly detectable with the recombinant virus (Fig. 3C, lanes 5 to 7). This finding is in agreement with the GFP substitution for VP in the recombinant virus, since the residual level of VP production can be attributed to the emergence of a small proportion of wild-type viruses during the production of recombinant H-1/GFP virus stocks (40a). It should be stated that this wild-type contamination of the recombinant stock did not account for the ability of the latter to trigger apoptosis, since an equivalent inoculum of pure wild-type H-1 virus (MOI of 0.03 PFU/cell), which yielded the same production of VP proteins as had the recombinant stock but without any detectable NS-1 accumulation (Fig. 3B and C, lane 8), failed to induce PARP cleavage as measured by Western blotting (Fig. 3A, lane 8). Altogether, these data point to NS proteins as the major effector of the induction of PARP cleavage (Fig. 3A) and the formation of apoptotic bodies (data not shown), although an additional contribution of the capsid proteins cannot be ruled out.
FIG. 3.
Induction of the cleavage of PARP in the presence of parvoviral NS proteins. Western blot analyses were carried out with protein extracts prepared from U937 cells at different times after infection, with intact (lanes 2 to 4, lane 8) or UV-irradiated (lane 9) wild-type H-1 virus or with a recombinant H-1 virus in which the VP genes were replaced by the jellyfish GFP (lanes 5 to 7). MOIs are given as intact wild-type virus PFU equivalents per cell. (A) PARP cleavage. (B and C) Accumulation of the H-1 virus proteins NS-1 (B) and VP-1 and VP-2 (C).
It is now well established that members of the Bcl-2 protein family are key regulators of apoptosis (73). Some of these factors, such as the human Bcl-2 protein or Bcl-xl, are suppressors of cell death, whereas other members of the family, e.g., Bax and Bak, promote programmed cell death. Bcl-2 and related proteins can form channels in vitro and may participate in the regulation of mitochondrial permeability transition and in the release of apoptogenic protease activators, in particular cytochrome c and apoptosis-inducing factor from the mitochondria (44). This prompted us to investigate whether H-1 virus-induced apoptosis involved modifications in the accumulation of three members of this family: Bcl-2, Bax, and Bcl-xl. To this end, Western blotting analyses were performed with protein extracts from infected U937 cells with specific antibodies. No significant change in the steady-state levels of these proteins could be observed up to 28 h postinfection (MOI of 10 PFU/cell), suggesting that H-1 virus-induced apoptosis is not mediated by a gross alteration of their intracellular accumulation (data not shown). However, it cannot be ruled out that the parvovirus affects the posttranslational modification of these proteins or the expression of other members of the Bcl-2 family.
c-Myc is rapidly downregulated in parvovirus-induced apoptosis.
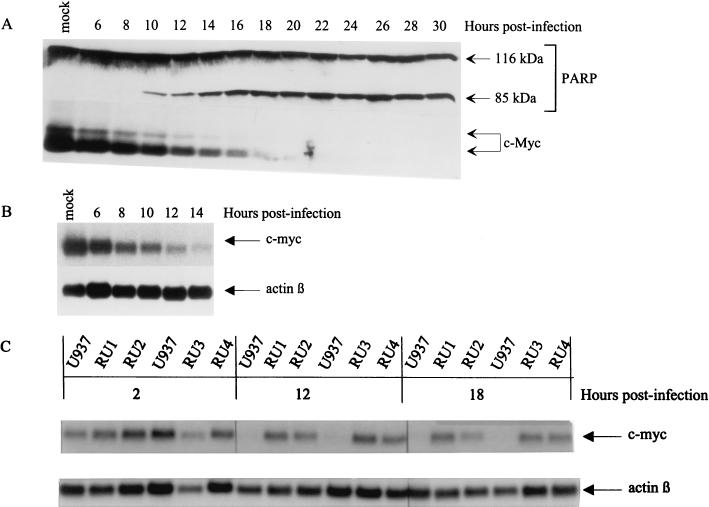
c-Myc, which is known to be involved in the regulation of cell proliferation and differentiation, was consistently found to sensitize cells to the induction of apoptosis by several treatments (3, 35). Knowing that c-Myc is overexpressed in U937 cells (18), these data prompted us to investigate the possible role of this oncoprotein in H-1 virus-induced apoptosis. It should be stated that, although the deregulation of c-Myc in the induction of apoptosis is well documented for some cell lines, there is no evidence for the requirement of the downregulation of overexpressed c-Myc during the execution of apoptosis. The rapid decrease of c-Myc in U937 cells committed to differentiation led us hypothesize that this might be also the case in U937 cells committed to apoptosis. The accumulation of c-Myc in protein extracts prepared from H-1 virus-infected U937 cells (MOI of 10 PFU/cell) at various times postinfection was analyzed by Western blotting with a specific monoclonal antibody. As shown in Fig. 4A, a significant decrease of the level of the 65-kDa and the N-terminally extended 68-kDa forms of c-Myc (81) was apparent as early as 6 h postinfection and became more pronounced with time, leading to the disappearance of a detectable c-Myc protein signal by around 18 h postinfection, when PARP cleavage was maximal. Similarly, the level of c-myc transcripts decreased with time and became undetectable by 14 h postinfection, as determined by Northern blotting analysis (Fig. 4B). This effect was specific since β-actin mRNA remained stable during the same time interval, making it unlikely that the downregulation of c-myc mRNAs reflected an overall inhibition of transcription or an enhancement of mRNA degradation in H-1 virus-infected cells. The downregulation of c-myc expression occurred concomitantly with the production of viral proteins (Fig. 2B and C) and preceded the cleavage of PARP (Fig. 2A), thereby representing an early event in parvovirus-induced apoptosis.
FIG. 4.
Downregulation of c-Myc during H-1 virus-induced apoptosis. (A) Western blot analysis of total proteins (70 μg/lane) extracted from H-1 virus-infected U937 cells (10 PFU/cell) at different times postinfection with anti-PARP (upper panel) or anti-c-Myc (lower panel) antibodies. (B) Northern blot analysis of total RNAs extracted from H-1 virus-infected U937 cells with c-myc (upper part)- or β-actin (lower part)-specific DNA probes. (C) Northern blot analysis of total RNAs extracted from H-1 virus-infected RU and U937 cells at the indicated times postinfection with c-myc (upper part)- or β-actin (lower part)-specific DNA probes.
We recently selected from the U937 line rare cell variants that resisted H-1 virus infection, of which four (RU1, RU2, RU3, and RU4) were further characterized (52). The RU derivatives were tested to determine the correlation between c-myc downregulation and H-1 virus-induced apoptosis in U937 cells. Indeed, all H-1 virus-resistant RU clones were distinguishable from the sensitive parental U937 cells because, in the former group, the level of c-myc transcripts remained unaffected up to 18 h after H-1 virus infection, as determined by Northern blotting analysis (Fig. 4C).
Resistance to the killing effect of H-1 virus cosegregates with resistance to TNF-α-induced apoptosis in U937 cell derivatives.
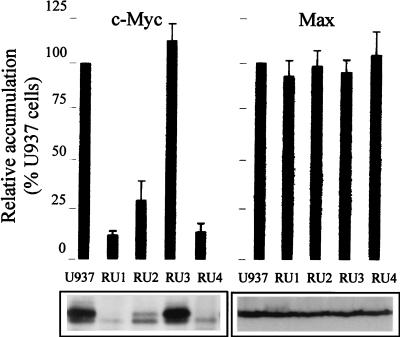
Although uninfected RU cells expressed levels of c-myc mRNAs comparable to the parental U937 cells, three of the clones (RU1, RU2, and RU4) showed a significant decrease, compared to the parental U937 cells, in the accumulation of c-Myc oncoprotein and also in their capacity to form tumors in immunodeficient mice. The transforming and pro-apoptotic properties of c-Myc are believed to require its association with the partner protein Max (2, 30, 49, 70). In contrast to c-Myc, Max expression was not significantly altered in RU cells compared with the parental U937 line (Fig. 5, lower panel). The levels of both proteins in the RU cells are given as percentages of the U937 values shown in the upper panel of Fig. 5. While being close to 100% for Max, the relative level of c-Myc was strongly reduced in the RU1 (14%), RU2 (31%), and RU4 (16%) clones. Thus, these three clones showed an imbalance between Myc and Max favoring the association of Max with itself or with other partners known to inhibit c-Myc activity (49, 57). It should also be stated that all RU clones resisted the suppressing effect of the differentiating agent TPA (12-O-tetradecanoylphorbol-13-acetate) on c-myc oncogene expression and cell proliferation (52). In addition, a subclone of U937 cells that resisted TPA-induced differentiation or programmed cell death was previously described by Hass et al. (28), in which c-myc was continuously expressed in the presence of this agent.
FIG. 5.
Steady-state levels of c-Myc and Max proteins in parental U937 cells and H-1 virus-resistant (RU) derivatives. (Upper panels) Densitometric quantification of c-Myc and Max accumulation, expressed as percentages of the U937 level. Average values and standard-deviation bars from three experiments are shown. (Lower panels) Western blot analysis of c-Myc and Max levels in uninfected U937 and RU cells, with 50 μg of total protein extracts.
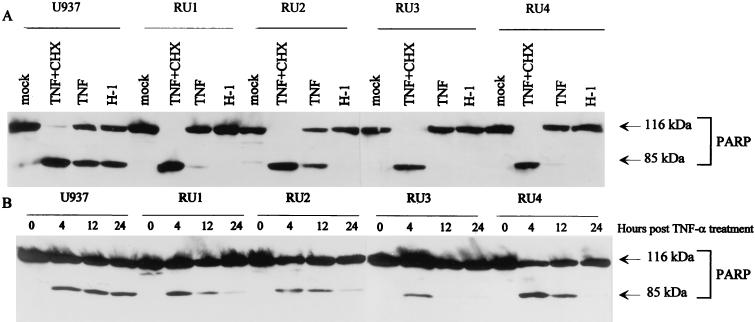
TNF-α is known to induce differentiation (78) or programmed cell death (31, 64, 96) in parental U937 cells. c-Myc has been found to sensitize some human tumor cell lines to TNF-α-induced apoptosis (35), and TNF-α-resistant fibroblasts could even be made TNF-α sensitive by the enforced expression of c-Myc (41). Therefore, the altered regulation of c-Myc expression in RU cells, in particular its resistance to downmodulating treatments, prompted us to investigate whether H-1 virus-resistant RU clones also became refractory to TNF-α-induced apoptosis. To this end, RU clones were incubated in the presence of TNF-α (50 ng/ml) either alone or with cycloheximide (10 μg/ml). Protein extracts were prepared, and the presence of cleaved PARP was measured by Western blotting as an indicator of apoptotic cell death. H-1 virus-infected cells (MOI of 10) were analyzed in parallel for comparison. As illustrated in Fig. 6A (left panel), the cells proved to be sensitive to both TNF-α (with or without cycloheximide) and H-1 virus regarding the induction of PARP cleavage, as expected from the literature (96, 97) and the above-mentioned data, respectively. In contrast, all four RU clones resisted not only H-1 virus but also TNF-α alone (or cycloheximide alone [data not shown]), though to various extents (Fig. 6A). Interestingly, TNF-α was still able to trigger the cleavage of PARP in RU cells in the presence of cycloheximide, suggesting that the death machinery is fully functional in all RU clones but that it is suppressed by a protein(s) whose synthesis is cycloheximide sensitive. It should also be stated that at early times (4 h) after treatment with TNF-α alone, a small proportion of PARP was cleaved in all RU clones (Fig. 6B), but this progressively disappeared at later times. This transience may be assigned to the instability of PARP fragments produced in low amounts in treated RU cells. Alternatively, a small fraction of the TNF-α-treated RU cell population may die through apoptosis and be eliminated over time, resulting in the overgrowth of the major resistant cells.
FIG. 6.
Comparison of the sensitivity of U937 and RU cells to TNF-α- or H-1 virus-induced apoptosis. PARP cleavage was revealed by Western blot analysis of the total protein extracts (50 μg). (Upper panel) U937 and RU cells were treated or mock treated for 24 h with TNF-α (50 ng/ml) alone, with TNF-α (50 ng/ml) plus cycloheximide (CHX) (10 μg/ml), or with H-1 virus (10 PFU/cell) as indicated at the top of each lane. (Lower panel) Cells were treated with TNF-α (1 ng/ml) for the indicated times.
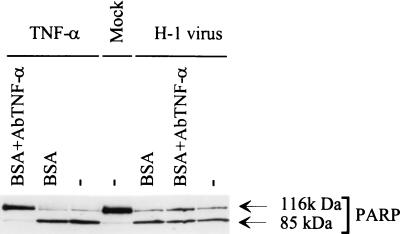
A striking parallelism was observed between H-1 virus and TNF-α regarding their abilities to induce apoptosis in the U937 cells and their failure to do so in the RU variants. This prompted us to test whether parvovirus H-1 may induce the production of TNF-α in infected cultures, leading to the death of TNF-sensitive U937 cells but sparing their TNF-resistant derivatives. Indeed, infection with several other viruses has been reported to induce the production of TNF-α (82). We were, however, unable to detect TNF-α by enzyme-linked immunosorbent assay in the medium of U937 and RU cultures at different times postinfection (data not shown). Additional experiments were carried out in which U937 cells were infected with H-1 virus (MOI of 5 PFU/cell) and further incubated in the presence of polyclonal blocking antibodies directed against TNF-α and preventing the binding of this factor to its receptor. These cultures were then tested for the cleavage of PARP by Western blotting at 24 h postinfection. As shown in Fig. 7, the anti-TNF-α serum fully inhibited the TNF-α-induced cleavage of PARP (lanes 1 and 3) but did not prevent H-1 virus from triggering this cleavage (lanes 6 and 7). Incubation with BSA did not affect the cleavage of PARP induced by either TNF-α or H-1 virus (lanes 2 and 5). These observations argue against the possibility of H-1 virus inducing apoptosis indirectly through the stimulation of TNF-α release in the medium.
FIG. 7.
Resistance of H-1 virus-induced apoptosis to blocking anti-TNF-α antibodies. PARP cleavage was analyzed by Western blotting with total protein extracts (50 μg) of U937 cells that were infected with H-1 virus (5 PFU/cell), mock treated, or treated with TNF-α (10 ng/ml) and further incubated for 24 h in the presence or absence of blocking anti-TNF-α serum. BSA (0.1%) was used as a negative control.
DISCUSSION
Many viruses have been reported to induce programmed cell death or apoptosis in infected cells (20, 38, 58, 85, 102). The autonomous parvoviruses were assumed to induce cell death by lysis of infected permissive cell lines. One study showed ultrastructural evidence of apoptosis in cultures of hematopoietic precursors infected with parvovirus B19 (60). In the present study we report sequential molecular events associated with morphologic changes characteristic of apoptosis upon parvovirus H-1 infection of the monocytic cell line U937. Hoechst staining of H-1 virus-infected U937 cells indeed reveals the typical apoptotic bodies that also appear in response to TNF-α (Fig. 1). The enzyme PARP has been demonstrated to be cleaved rapidly and specifically during apoptosis (46) by ICE-related proteases, in particular caspase-3/CPP32, which is activated in cells after treatment with various apoptotic agents. Although PARP cleavage per se does not seem to be essential for the apoptotic process (91), this event is now generally considered to be a consequence of cells committing apoptosis. We show here that H-1 virus infection triggers the cleavage of PARP and the activation of caspase-3/CPP32, which is known to use PARP as a substrate and to be involved in apoptosis.
Our data strongly suggest that the accumulation of the viral replicative NS proteins is required to trigger the cleavage of PARP and the formation of apoptotic bodies. Indeed, UV-inactivated H-1 virus is unable to induce these apoptotic markers, although it is taken up by target cells. Furthermore, recombinant H-1 virus expressing the NS proteins but lacking the capsid genes is fully competent to induce the cleavage of PARP and cell death (Fig. 3). Although NS proteins could be cytotoxic in an indirect way, through their functions in viral DNA replication and transcription (14), a direct effect is supported by recent studies with transformed cell clones which have integrated the MVMp NS protein-encoding transcription unit under the control of an inducible promoter (8, 61). While NS protein accumulation appears to be necessary for H-1-induced apoptosis, a possible synergistic effect of the capsids cannot be totally ruled out, since the recombinant H-1 virus stocks are contaminated with a low fraction of wild-type virus generated during the production. However, these contaminating wild-type viruses are present in amounts that are too small to cause a detectable cleavage of PARP in the absence of the NS-expressing recombinants (Fig. 3, lane 8). The major cytotoxic nonstructural product of parvoviruses is the protein NS-1 (8), but the mechanism of NS-1 cytotoxic activity is still elusive. In this respect, one possible target of NS-1 could be the c-myc gene. Indeed, c-Myc expression is inhibited at early times after H-1 virus infection of U937 cells (see Fig. 4), and the disappearance of both the oncoprotein and its mRNA is concomitant with the accumulation of the viral NS proteins (Fig. 4 and 1, respectively). It should be stated, however, that c-myc expression is known to be controlled at both transcriptional (100) and posttranscriptional (95) levels. Therefore, it cannot be ruled out that the observed reduction of c-myc mRNA steady-state levels in H-1 virus-infected U937 cells (Fig. 4B) results from alteration in RNA processing besides transcription initiation. The cytotoxic function of NS-1 was previously found to cosegregate with its capacity for promoter transregulation (47), leading to the suggestion that NS-1 may act at least in part by disturbing the expression of essential cellular genes.
Altogether, our observations raise the questions as to whether c-Myc is involved in H-1 virus-induced apoptosis. Although present data do not provide definite clues regarding this question, this possibility should be considered. Indeed, the c-Myc content of U937 cells decreased very rapidly after H-1 virus infection and before the onset of PARP cleavage. Furthermore, the regulation of c-Myc expression proved to be altered in four RU clones that were derived from the U937 cell line through selection for H-1 virus resistance and also failed to undergo PARP cleavage and apoptosis after treatment with TNF-α (Fig. 6A). These clones were previously shown to be impaired with regard to the downregulation of c-Myc expression and the arrest of cell proliferation in response to the differentiating agent TPA (52). Although the four RU clones displayed various steady-state levels of c-Myc oncoproteins, their respective c-Myc levels all remained unchanged after H-1 virus infection (Fig. 4C). These data support a role for c-Myc in H-1 virus-induced apoptosis in U937 cells, although it cannot be excluded that the downregulation of c-Myc is a consequence of apoptosis in this system. c-Myc, in association with its partner protein Max, is a transcription factor that is essential for the cell cycle entry and progression and is oncogenic when overexpressed (105). It has been shown that the mitotic cycle gets blocked in cells expressing NS-1, pointing to a possible link between NS toxicity and cell cycle perturbations (68, 69). It was also reported that in cells infected with MVMp or Aleutian disease virus of mink, a massive accumulation of NS-1 takes place at the G1-S transition, leading to cell cycle disturbances (66). Op De Beeck and Caillet-Fauquet (69) proposed that NS-1 might exert a direct cytostatic action by inhibiting host cell DNA replication, possibly because of NS-1-induced nicks in the chromatin. Another, nonexclusive possibility would be that upstream regulators of the cell cycle, including relevant transcription factors, are targets for NS-1. Thus, the downregulation of c-Myc upon H-1 virus infection of U937 cells might lead to their arrest in the cell G0-G1 phase, as is the case when these cells are treated with agents that induce differentiation (18). The suppression of c-Myc expression in H-1 virus-infected cells might be a signal of apoptosis resulting from a conflict between growth-arresting (c-Myc disappearance) and growth-promoting (serum) factors. Conversely, c-Myc overexpression has been consistently found to promote apoptosis by growth factor deprivation or genotoxic agents (105). Interestingly, c-Myc-associated apoptotic events share with H-1 virus-induced apoptosis the activation of a caspase-3-like protease (39) and recruit along a similar signaling pathway (32).
An alternative, but nonexclusive reason for the high sensitivity of U937 cells to the induction of apoptosis in response to H-1 virus or TNF-α may lie in their relative failure to express antiapoptotic genes upon treatment with these agents. It is noteworthy in this respect that the RU cell variants which were selected for their resistance to H-1 virus infection but proved also to survive treatment with TNF-α become sensitive to TNF-α in the presence of cycloheximide. This makes it unlikely that the failure of RU cells to respond to TNF-α-induced apoptosis is due to a deficiency in the apoptotic machinery, and it raises the possibility that the RU clones have developed an antiapoptotic defense system which requires de novo protein synthesis. Members of the Rel/NF-kB family of proteins are candidate mediators of this defense. Indeed, these transcription factors are known to counteract the apoptotic pathway triggered by TNF-α (4, 51, 88, 90). In addition, Klefstrom et al. (41) showed that c-Myc increases the cytotoxic effects of TNF-α in fibroblasts by impairing cell survival signaling via NF-κB. It is therefore conceivable that the resistance of RU cells to TNF-α is mediated, at least in part, by NF-κB or related factors. It should be stated that these transcription factors control the expression of specific genes in macrophages, including the inducible NO synthase (iNOS) gene (25, 98). We have previously demonstrated a constitutive production of NO and oxygen species in the RU clones (52), in keeping with the probable activation of NF-κB in these cells. It may be speculated that NO metabolites inhibit apoptosis in RU cells, which was proposed to occur in other systems (54) and may result from S-nitrosylation of cysteine proteases (17). Another protein candidate for the modulation of H-1 virus or TNF-α-induced apoptosis in the U937 cell system is the retinoblastoma gene product (pRB), an important regulator of the cell cycle that is also known to protect cells from apoptosis (1, 27). pRB was recently shown to be postranslationally modified by a transglutaminase-driven reaction in U937 cells undergoing apoptosis (67). This resulting pRB polymerization is accompanied by the release and subsequent degradation of the transcription factor E2F-1. It has also been demonstrated that pRB is a substrate for caspases in the context of apoptosis induced by TNF-α (37). These studies point to the importance of pRB inactivation during apoptosis. It remains to be determined whether this pRB processing occurs in H-1 virus-infected U937 cells and whether it is impaired in the H-1 virus and TNF-α-resistant RU derivatives.
In summary, the present study provides evidence for the involvement of apoptosis in the death of monocytic U937 cells infected with parvovirus H-1. The viral NS proteins are likely to be responsible for this effect. The apoptotic pathway triggered by parvovirus H-1 appears to share at least some steps with the one activated by TNF-α, as is apparent in particular from the TNF resistance of cell variants selected for their survival to virus infection. Although the precise mechanism by which H-1 virus induces apoptosis remains to be determined, it was observed that treatment with TNF-α or H-1 virus infection of U937 cells is accompanied by a rapid and drastic downregulation of c-Myc expression prior to the appearance of apoptotic signs (activation of caspase-3, PARP cleavage, and apoptotic bodies). This feature, together with the failure of TNF-α and H-1 virus-resistant cell variants to downregulate c-Myc expression, suggest that c-Myc, or factors regulated in a similar way, may play a key role in the signaling of apoptosis. In addition, cell variants resisting death inducers may emerge through the constitutive activation of antiapoptotic effector molecules.
ACKNOWLEDGMENTS
We are grateful to G. Poirier for the PARP antibodies and to A. Buerkle for his help in PARP immunoblotting. We thank J. Cornelis and J. C. Jauniaux for helpful and critical discussions, G. Balboni and A. Dege for expert technical assistance, and J. M. Vanacker for his support.
B.R. received a fellowship from the Belgian Fonds National de la Recherche Scientifique. J.-A.L.-G. was supported by a fellowship from the Commission of the European Communities (Human Capital and Mobility) and by the German Cancer Research Center.
REFERENCES
- 1.Almasan A, Yin Y, Kelly R E, Lee E Y, Bradley A, Li W, Bertino J R, Wahl G M. Deficiency of retinoblastoma protein leads to inappropriate S-phase entry, activation of E2F-responsive genes, and apoptosis. Proc Natl Acad Sci USA. 1995;92:5436–5440. doi: 10.1073/pnas.92.12.5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amati B, Land H. Myc-Max-Mad: a transcription factor network controlling cell cycle progression, differentiation and death. Curr Opin Genet Dev. 1994;4:102–108. doi: 10.1016/0959-437x(94)90098-1. [DOI] [PubMed] [Google Scholar]
- 3.Askew D S, Ashmun R A, Simmons B C, Cleveland J L. Constitutive c-myc expression in an IL-3-dependent myeloid cell line suppresses cell cycle arrest and accelerates apoptosis. Oncogene. 1991;6:1915–1922. [PubMed] [Google Scholar]
- 4.Beg A A, Baltimore D. An essential role for NF-κB in preventing TNF-α-induced cell death. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 5.Beidler D R, Tewari M, Friesen P D, Poirier G, Dixit V M. The baculovirus p35 protein inhibits Fas- and tumor necrosis factor-induced apoptosis. J Biol Chem. 1995;270:16526–16528. doi: 10.1074/jbc.270.28.16526. [DOI] [PubMed] [Google Scholar]
- 6.Bettaieb A, Record M, Come M G, Bras A C, Chap H, Laurent G, Jaffrezou J P. Opposite effects of tumor necrosis factor alpha on the sphingomyelin-ceramide pathway in two myeloid leukemia cell lines: role of transverse sphingomyelin distribution in the plasma membrane. Blood. 1996;88:1465–1472. [PubMed] [Google Scholar]
- 7.Boyd J M, Malstrom S, Subramanian T, Venkatesh L K, Schaeper U, Elangovan B, D’Sa-Eipper C, Chinnadurai G. Adenovirus E1B 19 kDa and Bcl-2 proteins interact with a common set of cellular proteins. Cell. 1994;79:341–351. doi: 10.1016/0092-8674(94)90202-x. [DOI] [PubMed] [Google Scholar]
- 8.Caillet-Fauquet P, Perros M, Brandenburger A, Spegelaere P, Rommelaere J. Programmed killing of human cells by means of an inducible clone of parvoviral genes encoding non-structural proteins. EMBO J. 1990;9:2989–2995. doi: 10.1002/j.1460-2075.1990.tb07491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Canman C E, Kastan M B. Induction of apoptosis by tumor suppressor genes and oncogenes. Semin Cancer Biol. 1995;6:17–25. doi: 10.1006/scbi.1995.0003. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y Q, de Foresta F, Hertoghs J, Avalosse B L, Cornelis J J, Rommelaere J. Selective killing of simian virus 40-transformed human fibroblasts by parvovirus H-1. Cancer Res. 1986;46:3574–3579. [PubMed] [Google Scholar]
- 11.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 12.Clement M V, Stamenkovic I. Superoxide anion is a natural inhibitor of FAS-mediated cell death. EMBO J. 1996;15:216–225. [PMC free article] [PubMed] [Google Scholar]
- 13.Cornelis J J, Becquart P, Duponchel N, Salome N, Avalosse B L, Namba M, Rommelaere J. Transformation of human fibroblasts by ionizing radiation, a chemical carcinogen, or simian virus 40 correlates with an increase in susceptibility to the autonomous parvoviruses H-1 virus and minute virus of mice. J Virol. 1988;62:1679–1686. doi: 10.1128/jvi.62.5.1679-1686.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cotmore S F, Christensen J, Nüesch J P F, Tattersall P. The NS1 polypeptide of the murine parvovirus minute virus of mice binds to DNA sequences containing the motif [ACCA]2–3. J Virol. 1995;69:1652–1660. doi: 10.1128/jvi.69.3.1652-1660.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cotmore S F, Tattersall P. The autonomously replicating parvoviruses of vertebrates. Adv Virus Res. 1987;33:91–174. doi: 10.1016/s0065-3527(08)60317-6. [DOI] [PubMed] [Google Scholar]
- 16.Debbas M, White E. Wild-type p53 mediates apoptosis by E1A, which is inhibited by E1B. Genes Dev. 1993;7:546–554. doi: 10.1101/gad.7.4.546. [DOI] [PubMed] [Google Scholar]
- 17.Dimmeler S, Haendeler J, Nehls M, Zeiher A M. Suppression of apoptosis by nitric oxide via inhibition of interleukin-1β-converting enzyme (ICE)-like and cysteine protease protein (CPP)-32-like proteases. J Exp Med. 1997;185:601–607. doi: 10.1084/jem.185.4.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Einat M, Resnitzky D, Kimchi A. Close link between reduction of c-myc expression by interferon and G0/G1 arrest. Nature (London) 1985;313:597–600. doi: 10.1038/313597a0. [DOI] [PubMed] [Google Scholar]
- 19.Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature (London) 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- 20.Esolen L M, Park S W, Hardwick J M, Griffin D E. Apoptosis as a cause of death in measles virus-infected cells. J Virol. 1995;69:3955–3958. doi: 10.1128/jvi.69.6.3955-3958.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faisst S, Faisst S R, Dupressoir T, Plaza S, Pujol A, Jauniaux J C, Rhode S L, Rommelaere J. Isolation of a fully infectious variant of parvovirus H-1 supplanting the standard strain in human cells. J Virol. 1995;69:4538–4543. doi: 10.1128/jvi.69.7.4538-4543.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faucheu C, Diu A, Chan A W, Blanchet A M, Miossec C, Herve F, Collard Dutilleul V, Gu Y, Aldape R A, Lippke J A, et al. A novel human protease similar to the interleukin-1 beta converting enzyme induces apoptosis in transfected cells. EMBO J. 1995;14:1914–1922. doi: 10.1002/j.1460-2075.1995.tb07183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gajewski T F, Thompson C B. Apoptosis meets signal transduction: elimination of a BAD influence. Cell. 1996;87:589–592. doi: 10.1016/s0092-8674(00)81377-x. [DOI] [PubMed] [Google Scholar]
- 24.Golstein P. Controlling cell death. Science. 1997;275:1081–1082. doi: 10.1126/science.275.5303.1081. [DOI] [PubMed] [Google Scholar]
- 25.Grigoriadis G, Zhan Y, Grumont R J, Metcalf D, Handman E, Cheers C, Gerondakis S. The Rel subunit of NF-κB-like transcription factors is a positive and negative regulator of macrophage gene expression: distinct roles for Rel in different macrophage populations. EMBO J. 1996;15:7099–7107. [PMC free article] [PubMed] [Google Scholar]
- 26.Gu Y, Wu J, Faucheu C, Lalanne J L, Diu A, Livingston D J, Su M S. Interleukin-1 beta converting enzyme requires oligomerization for activity of processed forms in vivo. EMBO J. 1995;14:1923–1931. doi: 10.1002/j.1460-2075.1995.tb07184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haas-Kogan D A, Kogan S C, Levi D, Dazin P, T’Ang A, Fung Y, Israel M A. Inhibition of apoptosis by the retinoblastoma gene product. EMBO J. 1995;14:461–472. doi: 10.1002/j.1460-2075.1995.tb07022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hass R, Meinhardt G, Hadam M, Bartels H. Characterization of human TUR leukemia cells: continued cell cycle progression in the presence of phorbol ester is associated with resistance to apoptosis. Eur J Cell Biol. 1994;65:408–416. [PubMed] [Google Scholar]
- 29.Hengartner M O. Programmed cell death in invertebrates. Curr Opin Genet Dev. 1996;6:34–38. doi: 10.1016/s0959-437x(96)90007-6. [DOI] [PubMed] [Google Scholar]
- 30.Henriksson M, Luscher B. Proteins of the Myc network: essential regulators of cell growth and differentiation. Adv Cancer Res. 1996;68:109–182. doi: 10.1016/s0065-230x(08)60353-x. [DOI] [PubMed] [Google Scholar]
- 31.Higuchi M, Aggarwal B B. Differential roles of two types of the TNF receptor in TNF-induced cytotoxicity, DNA fragmentation, and differentiation. J Immunol. 1994;152:4017–4025. [PubMed] [Google Scholar]
- 32.Hueber A-O, Zornig M, Lyon D, Suda T, Nagata S, Evan G I. Requirement for the CD95 receptor-ligand pathway in c-Myc-induced apoptosis. Science. 1997;278:1305–1309. doi: 10.1126/science.278.5341.1305. [DOI] [PubMed] [Google Scholar]
- 33.Itoh N, Yonehara S, Ishii A, Yonehara M, Mizushima S, Sameshima M, Hase A, Seto Y, Nagata S. The polypeptide encoded by the cDNA for human cell surface antigen Fas can mediate apoptosis. Cell. 1991;66:233–243. doi: 10.1016/0092-8674(91)90614-5. [DOI] [PubMed] [Google Scholar]
- 34.Jacobson M D. Programmed cell death: a missing link is found. Trends Cell Biol. 1997;7:801–809. doi: 10.1016/S0962-8924(97)01182-3. [DOI] [PubMed] [Google Scholar]
- 35.Janicke R U, Lee F H, Porter A G. Nuclear c-Myc plays an important role in the cytotoxicity of tumor necrosis factor alpha in tumor cells. Mol Cell Biol. 1994;14:5661–5670. doi: 10.1128/mcb.14.9.5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janicke R U, Lin X Y, Lee F H, Porter A G. Cyclin D3 sensitizes tumor cells to tumor necrosis factor-induced, c-Myc-dependent apoptosis. Mol Cell Biol. 1996;16:5245–5253. doi: 10.1128/mcb.16.10.5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Janicke R U, Walker P A, Lin X Y, Porter A G. Specific cleavage of the retinoblastoma protein by an ICE-like protease in apoptosis. EMBO J. 1996;15:6969–6978. [PMC free article] [PubMed] [Google Scholar]
- 38.Jeurissen S H, Wagenaar F, Pol J M, van der Eb A J, Noteborn M H. Chicken anemia virus causes apoptosis of thymocytes after in vivo infection and of cell lines after in vitro infection. J Virol. 1992;66:7383–7388. doi: 10.1128/jvi.66.12.7383-7388.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kagaya S, Kitanaka C, Noguchi K, Mochizuki T, Sugiyama A, Asai A, Yasuhara N, Eguchi Y, Tsujimoto Y, Kuchino Y. A functional role for death proteases in s-Myc- and c-Myc-mediated apoptosis. Mol Cell Biol. 1997;17:6736–6745. doi: 10.1128/mcb.17.11.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaufmann S H, Desnoyers S, Ottaviano Y, Davidson N E, Poirier G G. Specific proteolytic cleavage of poly(ADP-ribose) polymerase: an early marker of chemotherapy-induced apoptosis. Cancer Res. 1993;53:3976–3985. [PubMed] [Google Scholar]
- 40a.Kestler, J., B. Neeb, S. Struyf, J. Vandamme, A. D’Abramo, P. Tattersall, J. Rommelaere, C. Dinsart, and J. J. Cornelis. Unpublished data. [DOI] [PubMed]
- 41.Klefstrom J, Vastrik I, Saksela E, Valle J, Eilers M, Alitalo K. c-Myc induces cellular susceptibility to the cytotoxic action of TNF-alpha. EMBO J. 1994;13:5442–5450. doi: 10.1002/j.1460-2075.1994.tb06879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kluck R M, Bossy Wetzel E, Green D R, Newmeyer D D. The release of cytochrome c from mitochondria: a primary site for Bcl-2 regulation of apoptosis. Science. 1997;275:1132–1136. doi: 10.1126/science.275.5303.1132. [DOI] [PubMed] [Google Scholar]
- 43.Kluck R M, Martin S J, Hoffman B M, Zhou J S, Green D R, Newmeyer D D. Cytochrome c activation of CPP32-like proteolysis plays a critical role in a Xenopus cell-free apoptosis system. EMBO J. 1997;16:4639–4649. doi: 10.1093/emboj/16.15.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kroemer G, Zamzami N, Susin S A. Mitochondrial control of apoptosis. Immunol Today. 1997;18:44–51. doi: 10.1016/s0167-5699(97)80014-x. [DOI] [PubMed] [Google Scholar]
- 45.Lander H M, Sehajpal P, Levine D M, Novogrodsky A. Activation of human peripheral blood mononuclear cells by nitric oxide-generating compounds. J Immunol. 1993;150:1509–1516. [PubMed] [Google Scholar]
- 46.Lazebnik Y A, Kaufmann S H, Desnoyers S, Poirier G G, Earnshaw W C. Cleavage of poly(ADP-ribose) polymerase by a proteinase with properties like ICE. Nature (London) 1994;371:346–347. doi: 10.1038/371346a0. [DOI] [PubMed] [Google Scholar]
- 47.Legendre D, Rommelaere J. Terminal regions of the NS-1 protein of the parvovirus minute virus of mice are involved in cytotoxicity and promoter trans inhibition. J Virol. 1992;66:5705–5713. doi: 10.1128/jvi.66.10.5705-5713.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lindahl T, Satoh M S, Poirier G G, Klungland A. Post-translational modification of poly(ADP-ribose) polymerase induced by DNA strand breaks. Trends Biochem Sci. 1995;20:405–411. doi: 10.1016/s0968-0004(00)89089-1. [DOI] [PubMed] [Google Scholar]
- 49.Lindeman G J, Harris A W, Bath M L, Eisenman R N, Adams J M. Overexpressed max is not oncogenic and attenuates myc-induced lymphoproliferation and lymphomagenesis in transgenic mice. Oncogene. 1995;10:1013–1017. [PubMed] [Google Scholar]
- 50.Liu X, Kim C N, Yang J, Jemmerson R, Wang X. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- 51.Liu Z G, Hsu H, Goeddel D V, Karin M. Dissection of TNF receptor 1 effector functions: JNK activation is not linked to apoptosis while NF-κB activation prevents cell death. Cell. 1996;87:565–576. doi: 10.1016/s0092-8674(00)81375-6. [DOI] [PubMed] [Google Scholar]
- 52.Lopez-Guerrero J A, Rayet B, Tuynder M, Rommelaere J, Dinsart C. Constitutive activation of U937 promonocytic cell clones selected for their resistance to parvovirus H-1 infection. Blood. 1997;89:1642–1653. [PubMed] [Google Scholar]
- 53.Mannick J B, Asano K, Izumi K, Kieff E, Stamler J S. Nitric oxide produced by human B lymphocytes inhibits apoptosis and Epstein-Barr virus reactivation. Cell. 1994;79:1137–1146. doi: 10.1016/0092-8674(94)90005-1. [DOI] [PubMed] [Google Scholar]
- 54.Mannick J B, Miao X Q, Stamler J S. Nitric oxide inhibits Fas-induced apoptosis. J Biol Chem. 1997;272:24125–24128. doi: 10.1074/jbc.272.39.24125. [DOI] [PubMed] [Google Scholar]
- 55.Martins L M, Earnshaw W C. Apoptosis: alive and kicking in 1997. Trends Cell Biol. 1997;7:111–114. doi: 10.1016/S0962-8924(96)10053-2. [DOI] [PubMed] [Google Scholar]
- 56.Medema J P, Scaffidi C, Kischkel F C, Shevchenko A, Mann M, Krammer P H, Peter M E. FLICE is activated by association with the CD95 death-inducing signaling complex (DISC) EMBO J. 1997;16:2794–2804. doi: 10.1093/emboj/16.10.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meroni G, Reymond A, Alcalay M, Borsani G, Tanigami A, Tonlorenzi R, Nigro C L, Messali S, Zollo M, Ledbetter D H, Brent R, Ballabio A, Carrozzo R. Rox, a novel bHLHZip protein expressed in quiescent cells that heterodimerizes with Max, binds a non-canonical E box and acts as a transcriptional repressor. EMBO J. 1997;16:2892–2906. doi: 10.1093/emboj/16.10.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meyaard L, Otto S A, Jonker R R, Mijnster M J, Keet R P, Miedema F. Programmed death of T cells in HIV-1 infection. Science. 1992;257:217–219. doi: 10.1126/science.1352911. [DOI] [PubMed] [Google Scholar]
- 59.Minn A J, Velez P, Schendel S L, Liang H, Muchmore S W, Fesik S W, Fill M, Thompson C B. Bcl-x(L) forms an ion channel in synthetic lipid membranes. Nature (London) 1997;385:353–357. doi: 10.1038/385353a0. [DOI] [PubMed] [Google Scholar]
- 60.Morey A L, Ferguson D J, Fleming K A. Ultrastructural features of fetal erythroid precursors infected with parvovirus B19 in vitro: evidence of cell death by apoptosis. J Pathol. 1993;169:213–220. doi: 10.1002/path.1711690207. [DOI] [PubMed] [Google Scholar]
- 61.Mousset S, Ouadrhiri Y, Caillet-Fauquet P, Rommelaere J. The cytotoxicity of the autonomous parvovirus minute virus of mice nonstructural proteins in FR3T3 rat cells depends on oncogene expression. J Virol. 1994;68:6446–6453. doi: 10.1128/jvi.68.10.6446-6453.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Muzio M, Chinnaiyan A M, Kischkel F C, O’Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz J D, Zhang M, Gentz R, Mann M, Krammer P H, Peter M E, Dixit V M. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death-inducing signaling complex. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 63.Nagata S. Apoptosis by death factor. Cell. 1997;88:355–365. doi: 10.1016/s0092-8674(00)81874-7. [DOI] [PubMed] [Google Scholar]
- 64.Noguchi K, Naito M, Kataoka S, Yonehara S, Tsuruo T. A recessive mutant of the U937 cell line acquired resistance to anti-Fas and anti-p55 tumor necrosis factor receptor antibody-induced apoptosis. Cell Growth Differ. 1995;6:1271–1277. [PubMed] [Google Scholar]
- 65.Oehm A, Behrmann I, Falk W, Pawlita M, Maier G, Klas C, Li-Weber M, Richards S, Dhein J, Trauth B C, et al. Purification and molecular cloning of the APO-1 cell surface antigen, a member of the tumor necrosis factor/nerve growth factor receptor superfamily. Sequence identity with the Fas antigen. J Biol Chem. 1992;267:10709–10715. [PubMed] [Google Scholar]
- 66.Oleksiewicz M B, Alexandersen S. S-phase-dependent cell cycle disturbances caused by Aleutian mink disease parvovirus. J Virol. 1997;71:1386–1396. doi: 10.1128/jvi.71.2.1386-1396.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oliverio S, Amendola A, Di Sano F, Farrace M G, Fesus L, Nemes Z, Piredda L, Spinedi A, Piacentini M. Tissue transglutaminase-dependent posttranslational modification of the retinoblastoma gene product in promonocytic cells undergoing apoptosis. Mol Cell Biol. 1997;17:6040–6048. doi: 10.1128/mcb.17.10.6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Op De Beeck A, Anouja F, Mousset S, Rommelaere J, Caillet-Fauquet P. The nonstructural proteins of the autonomous parvovirus minute virus of mice interfere with the cell cycle, inducing accumulation in G2. Cell Growth Differ. 1995;6:781–787. [PubMed] [Google Scholar]
- 69.Op De Beeck A, Caillet-Fauquet P. The NS1 protein of the autonomous parvovirus minute virus of mice blocks cellular DNA replication: a consequence of lesions to the chromatin? J Virol. 1997;71:5323–5329. doi: 10.1128/jvi.71.7.5323-5329.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Packham G, Cleveland J L. c-Myc and apoptosis. Biochim Biophys Acta. 1995;1242:11–28. doi: 10.1016/0304-419x(94)00015-t. [DOI] [PubMed] [Google Scholar]
- 71.Polyak K, Xia Y, Zweier J L, Kinzler K W, Vogelstein B. A model for p53-induced apoptosis. Nature (London) 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 72.Ray C A, Black R A, Kronheim S R, Greenstreet T A, Sleath P R, Salvesen G, Pickup D J. Viral inhibition of inflammation: cowpox virus encodes an inhibitor of the interleukin-1 beta converting enzyme. Cell. 1992;69:597–604. doi: 10.1016/0092-8674(92)90223-y. [DOI] [PubMed] [Google Scholar]
- 73.Reed J C. Double identity for proteins of the Bcl-2 family. Nature (London) 1997;387:773–776. doi: 10.1038/42867. [DOI] [PubMed] [Google Scholar]
- 74.Rhode S L, III, Paradiso P R. Parvovirus genome: nucleotide sequence of H-1 and mapping of its genes by hybrid-arrested translation. J Virol. 1983;45:173–184. doi: 10.1128/jvi.45.1.173-184.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rommelaere J, Cornelis J J. Antineoplastic activity of parvoviruses. J Virol Methods. 1991;33:233–251. doi: 10.1016/0166-0934(91)90024-t. [DOI] [PubMed] [Google Scholar]
- 76.Sakahira H, Enari M, Nagata S. Cleavage of CAD inhibitor in CAD activation and DNA degradation during apoptosis. Nature (London) 1998;391:96–99. doi: 10.1038/34214. [DOI] [PubMed] [Google Scholar]
- 77.Salome N, van Hille B, Geuskens M, Rommelaere J. Partial reversion of conditional transformation correlates with a decrease in the sensitivity of rat cells to killing by the parvovirus minute virus of mice but not in their capacity for virus production: effect of a temperature-sensitive v-src oncogene. J Virol. 1989;63:4797–4807. doi: 10.1128/jvi.63.11.4797-4807.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schutze S, Scheurich P, Schluter C, Ucer U, Pfizenmaier K, Kronke M. Tumor necrosis factor-induced changes of gene expression in U937 cells. Differentiation-dependent plasticity of the responsive state. J Immunol. 1988;140:3000–3005. [PubMed] [Google Scholar]
- 79.Shen Y, Shenk T E. Viruses and apoptosis. Curr Opin Genet Dev. 1995;5:105–111. doi: 10.1016/s0959-437x(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 80.Siegl G. Biology and pathogenicity of autonomous parvoviruses. In: Berns K I, editor. The parvoviruses. New York, N.Y: Plenum Press, Inc.; 1983. pp. 297–362. [Google Scholar]
- 81.Spotts G D, Hann S R. Enhanced translation and increased turnover of c-Myc proteins occur during differentiation of murine erythroleukemia cells. Mol Cell Biol. 1990;10:3952–3964. doi: 10.1128/mcb.10.8.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sprenger H, Bacher M, Rischkowsky E, Bender A, Nain M, Gemsa D. Characterization of a high molecular weight tumor necrosis factor-alpha mRNA in influenza A virus-infected macrophages. J Immunol. 1994;152:280–289. [PubMed] [Google Scholar]
- 83.Sundstrom C, Nilsson K. Establishment and characterization of a human histiocytic lymphoma cell line (U-937) Int J Cancer. 1976;17:565–577. doi: 10.1002/ijc.2910170504. [DOI] [PubMed] [Google Scholar]
- 84.Takayama S, Sato T, Krajewski S, Kochel K, Irie S, Millan J A, Reed J C. Cloning and functional analysis of BAG-1: a novel Bcl-2-binding protein with anti-cell death activity. Cell. 1995;80:279–284. doi: 10.1016/0092-8674(95)90410-7. [DOI] [PubMed] [Google Scholar]
- 85.Takizawa T, Matsukawa S, Higuchi Y, Nakamura S, Nakanishi Y, Fukuda R. Induction of programmed cell death (apoptosis) by influenza virus infection in tissue culture cells. J Gen Virol. 1993;74:2347–2355. doi: 10.1099/0022-1317-74-11-2347. [DOI] [PubMed] [Google Scholar]
- 86.Tewari M, Quan L T, R. K O, Desnoyers S, Zeng Z, Beidler D R, Poirier G G, Salvesen G S, Dixit V M. Yama/CPP32 beta, a mammalian homolog of CED-3, is a CrmA-inhibitable protease that cleaves the death substrate poly(ADP-ribose) polymerase. Cell. 1995;81:801–809. doi: 10.1016/0092-8674(95)90541-3. [DOI] [PubMed] [Google Scholar]
- 87.Thornberry N A, Bull H G, Calaycay J R, Chapman K T, Howard A D, Kostura M J, Miller D K, Molineaux S M, Weidner J R, Aunins J, et al. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature (London) 1992;356:768–774. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- 88.Van Antwerp D J, Martin S J, Kafri T, Green D R, Verma I M. Suppression of TNF-α-induced apoptosis by NF-κB. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- 89.Villa P, Kaufmann S H, Earnshaw W C. Caspases and caspase inhibitors. Trends Biochem Sci. 1997;22:388–392. doi: 10.1016/s0968-0004(97)01107-9. [DOI] [PubMed] [Google Scholar]
- 90.Wang C Y, Mayo M W, Baldwin A S., Jr TNF- and cancer therapy-induced apoptosis: potentiation by inhibition of NF-κB. Science. 1996;274:784–787. doi: 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- 91.Wang Z, Stingl L, Morrison C, Jantsch M, Los M, Schulze-Osthoff K, Wagner E F. PARP is important for genomic stability but dispensable in apoptosis. Genes Dev. 1997;11:2347–2358. doi: 10.1101/gad.11.18.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wertz I E, Hanley M R. Diverse molecular provocation of programmed cell death. Trends Biochem Sci. 1996;21:359–364. [PubMed] [Google Scholar]
- 93.White E. Life, death, and the pursuit of apoptosis. Genes Dev. 1996;10:1–15. doi: 10.1101/gad.10.1.1. [DOI] [PubMed] [Google Scholar]
- 94.Whyte M. ICE/CED-3 proteases in apoptosis. Trends Cell Biol. 1996;6:245–248. doi: 10.1016/0962-8924(96)20025-x. [DOI] [PubMed] [Google Scholar]
- 95.Wisdom R, Lee W. Translation of c-myc mRNA is required for its post-transcriptional regulation during myogenesis. J Biol Chem. 1990;265:19015–19021. [PubMed] [Google Scholar]
- 96.Wright S C, Kumar P, Tam A W, Shen N, Varma M, Larrick J W. Apoptosis and DNA fragmentation precede TNF-induced cytolysis in U937 cells. J Cell Biochem. 1992;48:344–355. doi: 10.1002/jcb.240480403. [DOI] [PubMed] [Google Scholar]
- 97.Wright S C, Zheng H, Zhong J, Torti F M, Larrick J W. Role of protein phosphorylation in TNF-induced apoptosis: phosphatase inhibitors synergize with TNF to activate DNA fragmentation in normal as well as TNF-resistant U937 variants. J Cell Biochem. 1993;53:222–233. doi: 10.1002/jcb.240530307. [DOI] [PubMed] [Google Scholar]
- 98.Xie Q W, Kashiwabara Y, Nathan C. Role of transcription factor NF-κB/Rel in induction of nitric oxide synthase. J Biol Chem. 1994;269:4705–4708. [PubMed] [Google Scholar]
- 99.Yang J, Liu X, Bhalla K, Kim C N, Ibrado A M, Cai J, Peng T I, Jones D P, Wang X. Prevention of apoptosis by Bcl-2: release of cytochrome c from mitochondria blocked. Science. 1997;275:1129–1132. doi: 10.1126/science.275.5303.1129. [DOI] [PubMed] [Google Scholar]
- 100.Yeilding N M, Lee W M. Coding elements in exons 2 and 3 target c-myc mRNA downregulation during myogenic differentiation. Mol Cell Biol. 1997;17:2698–2707. doi: 10.1128/mcb.17.5.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yin C, Knudson C M, Korsmeyer S J, Van Dyke T. Bax suppresses tumorigenesis and stimulates apoptosis in vivo. Nature (London) 1997;385:637–640. doi: 10.1038/385637a0. [DOI] [PubMed] [Google Scholar]
- 102.Yoshida H, Sumichika H, Hamano S, He X, Minamishima Y, Kimura G, Nomoto K. Induction of apoptosis of T cells by infecting mice with murine cytomegalovirus. J Virol. 1995;69:4769–4775. doi: 10.1128/jvi.69.8.4769-4775.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yuan J, Shaham S, Ledoux S, Ellis H M, Horvitz H R. The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1 beta-converting enzyme. Cell. 1993;75:641–652. doi: 10.1016/0092-8674(93)90485-9. [DOI] [PubMed] [Google Scholar]
- 104.Zolotukhin S, Potter M, Hauswirth W W, Guy J, Muzyczka N. A “humanized” green fluorescent protein cDNA adapted for high-level expression in mammalian cells. J Virol. 1996;70:4646–4654. doi: 10.1128/jvi.70.7.4646-4654.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zornig M, Evan G I. Cell cycle: on target with Myc. Curr Biol. 1996;6:1553–1556. doi: 10.1016/s0960-9822(02)70769-0. [DOI] [PubMed] [Google Scholar]