Abstract
Dimerization is essential for activity of human epidermal growth factor receptors (HER1/EGFR, HER2/ErbB2, HER3/ErbB3, and ErbB4) and mediates intracellular signaling events leading to cancer cell proliferation, survival, and resistance to therapy. HER2 is the preferred dimerization partner. Activation of HER signaling pathways may be blocked by inhibition of dimer formation using a monoclonal antibody (MAb) directed against the dimerization domain of HER2. The murine MAb 2C4 that specifically binds the HER2 dimerization domain was cloned as a chimeric antibody, humanized using a computer-generated model to guide framework substitutions, and variants were tested as Fabs. Pharmacokinetics and toxicology were evaluated in rodents and cynomolgus monkeys. Cloning the variable domains of MAb 2C4 into a vector containing human kappa and CH1 domains allowed construction of a mouse-human chimeric Fab. DNA sequencing of the chimeric clone permitted identification of CDR residues. The full-length IgG1 of variant F-10 was equivalent in binding to chimeric IgG1 and was designated pertuzumab (rhuMAb 2C4; Omnitarg). Pertuzumab pharmacokinetics was best described by a two-compartment model with a distribution phase of <1 day, terminal half-life of ~10 days, and volume of distribution of ~40 mL/kg that approximates serum volume. With the exception of diarrhea, pertuzumab was generally well tolerated in cynomolgus monkeys. Pertuzumab, a recombinant humanized IgG1 MAb, is the first of a new class of agents known as HER dimerization inhibitors. Inhibition of HER dimerization may be an effective anticancer strategy in tumors with either normal or elevated expression of HER2.
Keywords: Pertuzumab, 2C4, rhuMAb 2C4, HER2, HER dimerization inhibitor (HDI), Monoclonal antibody humanization
Introduction
Dimerization is essential for the activity of the transmembrane human epidermal growth factor receptors (HER1/EGFR, HER2/ErbB2, HER3/ErbB3, and ErbB4) (reviewed in Ref. [54]). Interactions between HER receptors and specific stromal ligands, the formation of different homodimers or heterodimers, and receptor cross talk, both within the HER family and with other growth factor receptors, govern the activation of specific intracellular signaling pathways, such as mitogen-activated protein kinase (MAPK) and phosphatidylinositol 3-kinase (PI3 K)/Akt [21, 39, 47]. Multiple HER ligands with different binding affinities for specific receptors have been identified, and include transforming growth factor alpha (ligand for EGFR), epidermal growth factor (EGFR), epiregulin (EGFR and HER4), beta-cellulin (EGFR), heparin-binding-EGF (EGFR and HER4), amphiregulin (EGFR), heregulin/neuregulin 1 (HER3 and HER4), and neuregulins 2,3,4 (HER4) (reviewed in Ref. [54]). Dysregulation of the complex HER signaling network is known to be associated with malignant transformation and cancer progression (reviewed in Ref. [23]). Studies have demonstrated that HER receptor dimerization mediates intracellular signaling events that lead to cancer cell proliferation, survival, and resistance to anticancer therapy in tumor cells associated with HER dysregulation [1, 2, 4, 7, 49].
The mechanisms of HER dimerization have now been elucidated through a series of elegant crystallography studies [5, 8, 17]. EGFR, HER3, and HER4 are inactive until ligand binding stabilizes the dimer-competent formation. In the inactive form, the dimerization domain (domain II) is tethered to domain IV preventing the formation of dimers with other HER receptors. Substantial domain rearrangement is triggered when ligand binds to the receptor at extracellular domains I and III. This rearrangement exposes the dimerization domain for interactions with the dimerization domain of other HER receptors that are in the active conformation. In contrast, HER2, which lacks any known ligand, is constitutively in the active conformation and structurally resembles activated ligand-bound EGFR. As a result, the dimerization domain is permanently exposed and ready to form dimers with ligand-activated HER receptors. This explains why HER2 is the preferred binding partner for other HER receptors [21]. It has also been shown that HER2-containing heterodimers are the most prevalent and the most active of all dimer combinations [21]. For example, treatment with the HER3 ligand heregulin was able to induce transformation of cells expressing HER2-HER3, but not cells that express HER3 alone [49]. Other experiments have shown that the HER2-EGFR heterodimer may be associated with greater and more prolonged proliferative signals than EGFR homodimers [21, 25, 37]. Interestingly, HER2-HER3 and HER2-EGFR heterodimers were also more effective inducers of vascular endothelial growth factor than other HER heterodimer combinations [55].
Using a stably transfected NIH 3T3 cell line expressing the HER2 gene, several IgG1κ monoclonal antibodies (MAbs) targeting HER2 were developed and shown to bind different epitopes on the extracellular domain of the receptor [14]. One of these, the MAb 2C4, was subsequently shown to bind to the putative dimerization domain (domain II) [16]. Agents specifically targeting domain II could inhibit HER dimerization, thus preventing activation of HER signaling pathways that mediate cancer cell proliferation and survival [5]. Moreover, it is logical to target the dimerization domain of HER2 because it is the most common receptor in HER dimer combinations [21]. Since the clinical utility of murine MAbs is limited by a human antimouse antibody (HAMA) response [42], 2C4 was humanized. This paper describes the humanization of 2C4 and the evaluation of pharmacokinetics and safety in preclinical models. The principal objective of this study was to create a humanized MAb that specifically binds to the extracellular dimerization domain on the HER2 receptor, therefore inhibiting HER dimerization and the resultant activation of intracellular signaling pathways associated with cancer cell proliferation and survival.
Materials and methods
Construction of chimeric and humanized 2C4 Fabs
The murine MAb 2C4 that specifically binds the extracellular domain of HER2 (ErbB2) has been described previously [14]. To construct a mouse-human chimeric Fab, total RNA was isolated from 2C4 hybridoma cells using a Stratagene RNA extraction kit and manufacturer’s protocols (Stratagene, La Jolla, CA, USA). The variable domains were amplified by reverse transcriptase polymerase chain reaction (RT-PCR), gel purified, and inserted into a pUC119-based plasmid containing a human kappa constant domain and human CH1 domain as previously described [6, 40]. Plasmids were transformed into Escherichia coli strain XL1-Blue (Stratagene, La Jolla, CA, USA) for preparation of double-stranded DNA.
For the chimera and each subsequent variant both light (L) and heavy (H) chains were completely sequenced using an automated ABI Stretch 3700 sequencer and Big Dye reagents [Applera Corp., Applied Biosystems (ABI)]. Sequences of the murine variable light (VL) and variable heavy (VH) domains (Fig. 1) [26] were used to construct a computer graphics model to determine which framework residues should be incorporated into the humanized antibody. Construction of models was performed as previously described by Carter and co-workers [6, 13].
Fig. 1.
Amino acid sequences of murine 2C4, humanized 2C4 (Fab-10) (pertuzumab), and human consensus sequences of L chain subgroup kappa I (humκI) and H chain subgroup III (hum III). CDR loops are enclosed in brackets, and differences between sequences are denoted by an asterisk (*). Residue numbering is according to Kabat et al. [26]. a VL domain. b VH domain
The vector template for humanized Fab contained a consensus human kappa subgroup I L chain, a consensus human subgroup III heavy chain (VH-CH1), an alkaline phosphatase promoter, and an f1 phage origin for production of single-stranded DNA [6, 40]. To construct the first humanized variant, the complementarity-determining region (CDR)-swap Fab-1, site-directed mutagenesis [29] was performed on a deoxyuridine-containing single-stranded DNA template prepared from E. coli strain CJ236 (BioRad, Hercules, CA, USA). Thus all six CDRs were changed to the 2C4 sequence. Plasmids for other Fab variants were constructed using Fab-1 as the template.
Chimeric 2C4 Fab and humanized Fab variants were transformed into E. coli strain 16C9, a derivative of MM294, for protein expression and cultured using AP5 modified medium for protein induction. Fabs were purified using protein G-Sepharose CL-4B (Pharmacia Biotech, Amersham Biosciences Corp., Piscataway, NJ, USA) [40], buffer exchanged into 10 mM sodium succinate, 140 mM NaCl, pH 6.0, and concentrated to a volume of 0.5 mL using a Centricon-10 (Ambicon, Millipore, Billerica, MA, USA). A 500 mL culture yielded 0.1–0.3 mg protein. Protein concentrations were determined by quantitative amino acid analysis.
Construction of chimeric and humanized 2C4 IgG1
Full-length chimeric IgG1 (chIgG1) or humanized 2C4 IgG1 (rhuMAb 2C4; pertuzumab) were generated by subcloning the murine or humanized (Fab-10, Table 1) variable domains of the L and H chains from the respective Fabs into separate pRK vectors [19]. The L and H chain plasmids were co-transfected into an adenovirus-transformed human embryonic kidney cell line 293 [20] using a high-efficiency procedure [19]. Media were changed to serum-free and harvested after 4 days. Antibodies were purified from the supernatants using protein A-Sepharose CL-4B with the buffer exchanged and concentrated, using the same buffer as for the Fab. Chinese hamster ovary (CHO) cells were transfected with di-cistronic vectors to achieve stable expression of pertuzumab (rhuMAb 2C4).
Table 1.
Binding of humanized anti-HER2 Fab variants to HER2-ECD
| Variant | Template | Changesa | Purpose | EC50 Fab-X/EC50 chimeric Fabb | ||
|---|---|---|---|---|---|---|
| Mean | SDc | n | ||||
| Chimera | Chimeric Fab | 1.0 | ||||
| Fab-1 | Human FR | Straight CDR Swap | No binding | 3 | ||
| Fab-2 | Fab-1 | Arg H71 Val | CDR-H2 conformation | 35.0 | 3.5 | 3 |
| Fab-3 | Fab-1 | Asp H73 Arg | Framework loop in VH | >10,000 | 6 | |
| Fab-4 | Fab-1 | Arg H71 Val | CDR-H2 conformation | 4.0 | 0.3 | 4 |
| Asp H73 Arg | Framework loop in VH | |||||
| Fab-5 | Fab-4 | Ala H49 Gly | CDR-H2 conformation | 1.6 | 0.6 | 3 |
| Fab-6 | Fab-4 | Phe H67 Ala | CDR-H2 conformation | 1.7 | 0.9 | 2 |
| Fab-7 | Fab-4 | Ala H49 Gly | CDR-H2 conformation | 1.4 | 0.6 | 2 |
| Phe H67 Ala | CDR-H2 conformation | |||||
| Fab-8 | Fab-4 | Leu H78 Val | CDR-H2 conformation | 2.9 | 0.6 | 2 |
| Fab-9 | Fab-4 | Asn H76 Arg | Framework loop in VH | 1.8 | 0.7 | 2 |
| Fab-10 | Fab-4 | Ile H69 Leu | CDR-H2 conformation | 1.0 | 0.3 | 4 |
H heavy chain, Arg H71 Val indicates change in the arginine at position 71 on the heavy chain to valine, Ala alanine, Arg arginine, Asp aspartic acid, Asn asparagine, Gly glycine, Ile isoleucine, Leu leucine, Phe phenylalanine, Val valine
aMurine residues are in bold type; residue numbers are according to Kabat et al. [26]
bThe murine/human chimeric Fab was used as the standard. Equivalent binding to that of the chimera is indicated by an EC 50 (50% effective concentration) Fab-X/EC50 chimeric Fab ratio of 1, while an increase in the ratio to greater than unity indicates relative binding affinity is proportionately less than the chimera. EC50 values were determined from titration curves. The EC50 for chimeric Fab was 0.13 mg/mL
cSD is standard deviation for n>2 measurements; when n=2 numbers are average difference from the mean
HER2-extracellular domain (HER2-ECD) binding assay
To measure the binding activity of Fab and immunoglobulin Gs (IgGs), NUNC maxisorp plates were coated with 1 μg/mL of HER2-ECD (Genentech Inc., South San Francisco, CA, USA) in 50 mM carbonate buffer, pH 9.6, overnight at 4°C, and then blocked with enzyme-linked immunosorbent assay (ELISA) diluent (0.5% BSA, 0.05% Polysorbate 20, PBS) at room temperature for 1 h. Serial dilutions of samples in ELISA diluent were incubated on the plates for 2 h. After washing, bound Fab or IgG molecules were detected with biotinylated murine anti-human kappa antibody (ICN 634771) followed by streptavidin-conjugated horseradish peroxidase (Sigma, St. Louis, MO, USA) using 3,3′,5,5′-tetramethyl benzidine (Kirkegaard and Perry Laboratories, Gaithersburg, MD, USA) as substrate. Absorbance was read at 450 nm. Chimera binding served as the standard. Titration curves were fit with a four-parameter nonlinear regression curve-fitting program (KaleidaGraph; Synergy Software, Reading, PA, USA). Concentrations of Fab variants corresponding to the midpoint absorbance of the titration curve of the standard were calculated and then divided by the concentration of the standard at that point.
Pharmacokinetics and toxicology in animals
All animal studies were conducted in accordance with institutional guidelines. Pertuzumab (26.7 mg/mL rhuMAb 2C4, Genentech Inc., South San Francisco, CA, USA) and vehicle (10 mM L-histidine at pH 6.0, 240 mM sucrose, 0.02% polysorbate 20) were stored at 2–8°C. All blood samples were collected without anticoagulant and allowed to clot at room temperature, and were stored frozen prior to thawing for analysis of pertuzumab serum concentrations. Mouse, rat, and cynomolgus monkey serum samples for pertuzumab concentrations were assayed using a receptor-binding ELISA. The assay uses immobilized antigen HER2-ECD (Genentech Inc., South San Francisco, CA, USA) to capture pertuzumab (rhuMAb 2C4) from serum samples [43]. Bound pertuzumab was detected with mouse anti-human Fc-horseradish peroxidase with tetramethyl benzidine used as the substrate for color development to quantify serum pertuzumab against a known standard curve. The assay has a minimum quantifiable concentration of 0.25–0.40 μg/mL of pertuzumab in mouse, rat, and cynomolgus monkey serum.
Single-dose pharmacokinetics in mice and rats
Male CD-1 mice (Charles River, Raleigh, NC, USA) weighing 0.022–0.028 kg were randomized into three groups (36 per group) that received 3, 30, or 90 mg/kg of pertuzumab by intravenous (IV) bolus. Male Sprague Dawley rats (Charles River) weighing 0.25–0.30 kg were randomized into three groups (six per group) that received 3, 30, or 90 mg/kg per animal of pertuzumab by IV bolus. For both species, blood was collected for pharmacokinetic analyses predose and at various times from 5 min to 36 days postdose.
Single-dose pharmacokinetics in monkeys
Six male and six female cynomolgus monkeys (SNBL, Everett, WA, USA), aged approximately 2–3 years and weighing 1.94–2.24 kg, were randomized into three groups (two animals per sex group) that received 15, 50, or 150 mg/kg per animal of pertuzumab by IV bolus. Blood was collected for pharmacokinetic analyses of pertuzumab at predose, and from 30 min to 28 days postdose.
Multi-dose toxicology study in monkeys
Cynomolgus monkeys were deemed the most appropriate animal model for assessing safety because of the 99% amino acid sequence homology between cynomolgus monkey and human HER2-ECD, similar binding affinity of pertuzumab to human and cynomolgus monkey HER2 (data not shown), and a lack of antibody binding to the mouse homolog (c-neu). A toxicology study was conducted in 20 males and 20 females’ cynomolgus monkeys that were healthy and free from obvious abnormalities, aged 2.5–3.0 years, and weighing 2.1–3.3 kg. Animals were randomized into four groups, vehicle or 15, 50, or 150 mg/kg per animal of pertuzumab. All doses were administered as a IV bolus, once weekly for 7 weeks. Two males and two female monkeys in the control and 150 mg/kg groups were retained after the dosing period for evaluation during a 4-week recovery period. Blood was collected for toxicokinetic analyses predose, and frequently from 45 min to 7 days following the first and seventh doses. For the second to sixth doses, blood samples were collected at peak and trough at alternating intervals. Additional blood samples were collected from the recovery animals from days 57 to 80.
Animals were monitored twice daily for general health and three times on dosing days. Detailed observations were performed once weekly and on the day of scheduled euthanasia (day 49 or 80). Body weight, temperature, blood pressure, heart rate, electrocardiogram (ECG), ophthalmic parameters, clinical pathology (including standard clinical chemistry analyses, serum creatinine kinase isoenzyme, troponin T, hematology parameters, and urinalysis), and anatomic pathology parameters (including organ weights, gross pathology, and histopathology) were assessed.
Statistical analysis of toxicology data
Toxicology data from treatment groups were compared statistically with the data from the same gender control group. With the exception of clinical pathology data, only data collected on or after the first day of treatment were analyzed statistically, using one-way analysis of variance (ANOVA) or covariance (ANCOVA) [51, 52]. Dunnett’s t-test [11] was used for control versus treated group comparisons if ANOVA or ANCOVA were significant (p≤0.05). Group comparisons were evaluated at the 5% two-tailed probability level.
Results
Humanization of 2C4 monoclonal antibody
A mouse-human chimeric Fab was constructed by cloning the variable domains of 2C4 murine MAb into a vector that contains human kappa and CH1 domains and permits expression of Fab fragments in E. coli. DNA sequencing of the chimeric clone allowed identification of the CDR residues (Fig. 1). For the purpose of this work, definitions of the CDRs based on sequence hypervariability as described by Kabat et al. [26] were used, except in the case of CDR-H1. For this CDR, the residues in the structure-based definition [9] were combined with the residues identified by sequence variability [26] and include residues H26–H35.
The consensus sequence for the human L chain subgroup kappa I and human H chain subgroup III comprised the framework for humanization (Fig. 1) [26]. This framework has been successfully used in humanization of other murine antibodies [6, 12, 40, 41, 50]. Since Fabs can be conveniently expressed and purified from E. coli, all humanized variants were initially constructed and screened for binding as Fabs. The initial variant, Fab-1, was a straight CDR swap in which the six murine CDRs were grafted onto the human framework. This Fab showed no detectable binding to HER2-ECD (Table 1).
As shown in Table 1, in Fab-2, H71 was changed from the human to the murine amino acid (Arg H71 Val) since this residue had previously been shown to affect the conformations of CDR-H1 and CDR-H2 [46, 53]. This change increased the binding from undetectable to 35-fold less than observed with the chimera. Residue H73 occurs in a framework loop adjacent to CDR-H1 and CDR-H2 and was previously shown to affect binding of other antibodies [40, 50]. In Fab-3, the residue at H73 was changed from the human to the murine amino acid (Asp H73 Arg). A slight increase in binding of Fab-3 was seen compared with Fab-1. However, when both the H71 and H73 changes were combined in Fab-4, binding was increased to within a fourfold difference from the chimera.
Several buried residues were evaluated based on their importance in previous humanizations. H49, shown to play a role in CDR-H2 conformation [40], was changed from the human to the murine amino acid (Ala H49 Gly). Phe H67, a large residue that might influence packing and conformation of CDR-H2 [50], was changed from the human to the murine amino acid (Phe H67 Ala). Each of these changes resulted in a very slight increase in binding compared with the template, Fab-4. The combination of H49 and H67 changes (Fab-7) was not improved over either mutation alone. Based on the model used, H78, which is buried and plays a role in supporting the conformation of CDR-H1 [40], was also investigated. However, binding decreased when Leu found in the human sequence at this position was changed to the murine amino acid (Leu H78 Val). None of these three changes were kept in the final humanized Fab.
A second residue in the Framework-3 loop, Asn H76, was previously observed to influence binding [12] and was therefore changed to the murine amino acid at this position (Asn H76 Arg) resulting in variant Fab-9. This change slightly increased binding affinity, but as the residue may be solvent-exposed and could contribute to immunogenicity, the human amino acid at this position (Asn) was retained. Further mutations were made to explore the importance of the L chain and other buried residues of the H chain. None of these changes improved binding (data not shown). Finally, the buried amino acid residue at position H69 in the human sequence was replaced by the murine amino acid at this position (Ile H69 Leu). This resulted in variant Fab-10 with binding equal to that of the chimeric Fab. Binding kinetics of Fab-10 and the chimeric Fab were compared using surface plasmon resonance. The studies confirmed that Fab-10 has binding equal to that of the starting chimeric antibody (Table 2) [18].
Table 2.
Binding of 2C4 Fabs to HER2-ECD using surface plasmon resonancea
| Variant | Fab bound, Rmax (RU) | k off (10−4 s−1) | k on (104 M−1 s−1) | K d (nM) |
|---|---|---|---|---|
| Chimera | 30 | 8.7±1.5 | 9.5±0.2 | 9.2±1.6 |
| Fab-10 | 25 | 9.5±0.5 | 11.2±0.1 | 8.5±0.4 |
aBinding of Fab fragments to immobilized HER2-ECD was measured with a CM5-bionsensor chip on a BIAcore-2000 system essentially as described [18]. Twofold serial dilutions of Fab from 125 nM to 1000 nM were injected at 25°C at a flow rate of 0.02 mL/min, and the net binding (in response units, RU) of Fab to a flowcell with HER2-ECD was measured versus a control flowcell with an unrelated IgG immobilized. Association (k on) and dissociation (k off) rate constants were calculated using global fitting with the BIA evaluation software, and the equilibrium dissociation constant, K d was calculated as k off/k on
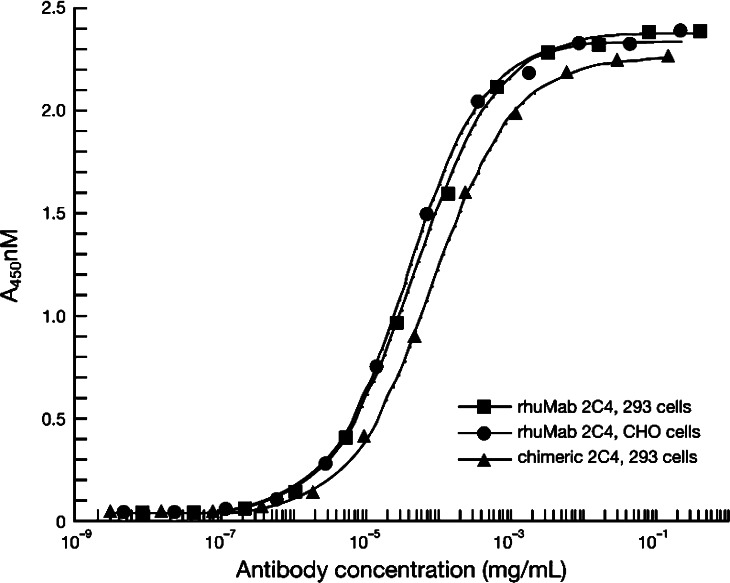
Full-length MAbs of variant Fab-10 and the chimera were constructed by fusing the respective VL and VH domains to the constant domains of human kappa L chain and human IgG1 H chain. The full-length IgG1 of variant F-10 was equivalent in binding to the chimeric IgG1 (Fig. 2) and was designated pertuzumab (rhuMAb 2C4).
Fig. 2.
Humanized 2C4 IgG1 (pertuzumab) expressed from either 293 cells (filled square) or CHO cells (filled circle) binds HER2-ECD equally well compared with the 2C4 chimeric full-length IgG1 antibody (filled triangle)
Pharmacokinetics
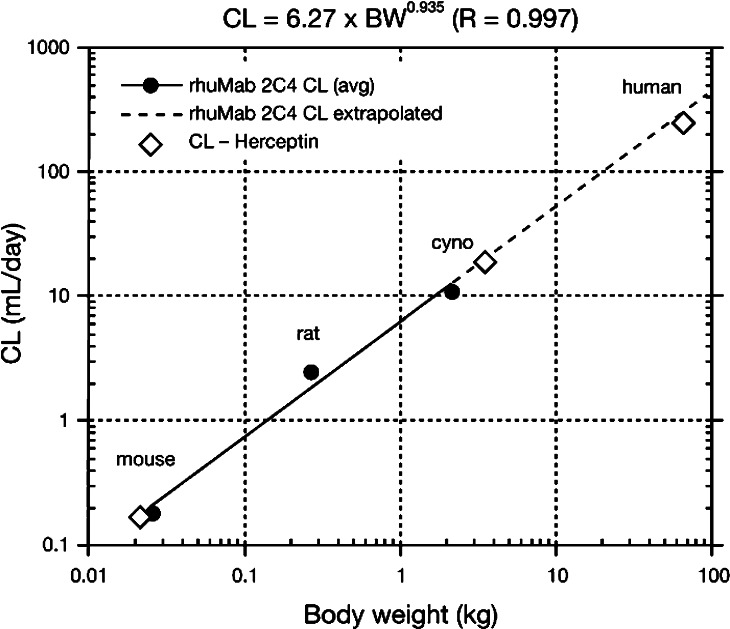
Table 3 summarizes pharmacokinetic parameters for pertuzumab. Pertuzumab serum concentration-time data from single-dose studies in mice and rats, and single-dose and multiple-dose studies in cynomolgus monkeys exhibited biphasic disposition and were fit into a two-compartment pharmacokinetic model. The distribution phase of pertuzumab was <1 day, the terminal elimination half-life was approximately 10 days, and the volume of distribution was 27–58 mL/kg. The volume of distribution of the central compartment approximated the serum volume in all models and did not change with increasing dose in multiple-dose studies in cynomolgus monkeys. However, clearance increased by approximately 30% when the dose was increased from 50 mg/kg to 150 mg/kg in these multiple-dose studies. Figure 3 shows predicted clearance in humans by allometric interspecies scaling [24].
Table 3.
Pharmacokinetic parameters for pertuzumaba
| Study type | Species/strain | No./sex | Route | Doses (mg/kg) | CL (mL/day/kg) | V c (mL/kg) | V ss (mL/kg) | αHL (day) | βHL (day) |
|---|---|---|---|---|---|---|---|---|---|
| SD PK | Mouse/CD-1 | 144/M | IV | 3, 30, 90 | 5.6–9.2 | 45–58 | 102–148 | 0.1–0.3 | 11.4–15.7 |
| SD PK | Rat/SD | 18/M | IV | 3, 30, 90 | 7.2–10.1 | 27–44 | 91–121 | 0.2–0.4 | 8.9–9.2 |
| SD PK | Monkey/Cyno | 8/M, 8/F | IV | 15, 50, 150 | 5.0–5.2 | 31–37 | 68–73 | 0.3–0.9 | 9.9–10.4 |
| MD Tox | Monkey/Cyno | 14/M, 14/F | IV | 15, 50, 150 | 5.1–7.4 | 36–41 | 64–80 | 0.5–0.6 | 8.1–10.6 |
CL clearance, V c volume of distribution of the central compartment, V ss volume of distribution at steady state, αHL initial half-life, βHL terminal half-life, SD Single-dose, MD Multi-dose, PK Pharmacokinetic, Tox Toxicology, IV Intravenous
aPharmacokinetics were characterized using a two-compartment model for the serum concentration-time data of individual animals (WinNonlin Professional Version 3.1, Pharsight Corp., Mountain View, CA, USA) using the Gauss–Newton algorithm and a reiterative weighting scheme (1/Ŷ). Serum pertuzumab concentrations found in the less-than-reportable range of the assay (LTR; <0.25 μg/mL) were not used for pharmacokinetic analysis, or in the group summary calculations. Data are ranges of group means
Fig. 3.
Interspecies scaling used to predict human clearance. The predicted CL of pertuzumab in humans by allometric scaling is approximately 330 mL/day, which is equivalent to 4.76 mL/day/kg for a 70 kg individual
Safety evaluation of pertuzumab in cynomolgus monkeys
Pertuzumab (15, 50, and 150 mg/kg IV weekly for 7 weeks) was generally well tolerated and there were no treatment-related effects on physical examination, blood pressure, heart rate, ECG, cardiac markers (creatinine kinase isoenzymes and troponin T), ophthalmic parameters, clinical pathology tests, and anatomic pathology parameters. Treatment with pertuzumab was associated with an increased incidence and occurrence of liquid and non-formed feces (Table 4). Although diarrhea was observed in all groups, including control, it was most notable in animals given 150 mg pertuzumab per kg. Diarrhea did not adversely affect body weight (except in one animal), and did not result in dehydration or changes in clinical or anatomic pathology parameters. Moreover, diarrhea was reversible, although not completely normalized, and improved by day 80 of the 4-week recovery period.
Table 4.
Incidence and frequency of diarrhea in male (M) and female (F) cynomolgus monkeys treated for 7 weeks with 15, 50, and 150 mg/kg pertuzumab IV for 7 weeks
| Pertuzumab (mg/kg) | Non-formed feces | Liquid feces | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acclimation | Treatment | Recovery | Acclimation | Treatment | Recovery | |||||||
| M | F | M | F | M | F | M | F | M | F | M | F | |
| Number of animals affected/number of animals per group | ||||||||||||
| 0 | 1/5 | 3/6 | 2/6 | 2/6 | 1/2 | 0/2 | 0/5 | 0/6 | 1/6 | 0/6 | 0/2 | 0/2 |
| 15 | 0/4 | 3/4 | 3/4 | 4/4 | – | – | 0/4 | 0/4 | 1/4 | 2/4 | – | – |
| 50 | 3/4 | 2/4 | 2/4 | 3/4 | – | – | 0/4 | 0/4 | 2/4 | 2/4 | – | – |
| 150 | 4/6 | 4/6 | 6/6 | 5/6 | 2/2 | 1/2 | 1/6 | 2/6 | 3/6 | 5/6 | 1/2 | 0/2 |
| Number of days observed (±SD) | ||||||||||||
| 0 | 0.2 (0.4) | 0.8 (1.2) | 0.7 (1.2) | 0.7 (1.2) | 1.5 (2.1) | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | 0.2 (0.4) | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) |
| 15 | 0.0 (0.0) | 3.0 (2.9) | 1.5 (1.3) | 17.0 (15.5) | – | – | 0.0 (0.0) | 0.0 (0.0) | 0.3 (0.5) | 2.0 (2.8) | – | – |
| 50 | 1.0 (0.8) | 1.3 (1.5) | 12.0 (14.0) | 8.0 (12.1) | – | – | 0.0 (0.0) | 0.0 (0.0) | 5.3 (6.4) | 1.8 (2.4) | – | – |
| 150 | 2.5 (2.6) | 6.0 (6.9) | 12.2 (9.4) | 4.2 (3.5) | 6.5 (4.9) | 0.5 (0.7) | 1.7 (4.1) | 1.0 (1.5) | 6.7 (8.9) | 11.2 (17.3) | 0.5 (0.7) | 0.0 (0.0) |
SD standard deviation
Discussion
Based on the hypothesis that inhibiting HER dimerization may block intracellular signaling pathways associated with cancer cell proliferation and survival, this work was undertaken to produce a novel anticancer therapeutic by humanizing a murine MAb that targets the dimerization domain on the HER2 receptor. The murine MAb 2C4 was successfully humanized using consensus framework sequences for L and H chains, VLκI and VHIII, to produce pertuzumab (recombinant humanized MAb 2C4), which represents the first of a new class of agents designated HER dimerization inhibitors. Results from both the toxicology and pharmacokinetic studies reported in this paper supported the initiation of human clinical studies using doses of 0.5–15 mg/kg of pertuzumab administered every 3 weeks.
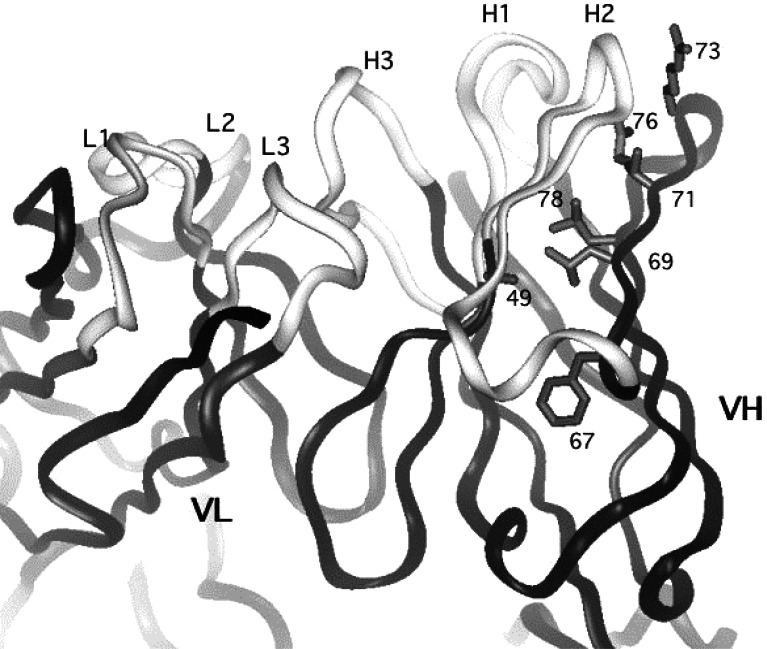
Consensus frameworks have been used successfully in several previous humanizations [6, 13, 40, 41, 50]; the current work further illustrates the utility of this approach. Transferring CDRs from the murine antibody to the human framework resulted in complete loss of binding to HER2-ECD. Remarkably, altering only three non-CDR residues in the H chain was sufficient to restore binding to that of the parent MAb. From the model, it was anticipated that two of these residues, H69 and H71, are buried and on the same side of a beta strand with their side chains directed toward CDR-H2. Thus, it was thought that they would play an important role in maintaining the conformation of this CDR. In the crystal structure of the humanized 2C4 Fab now available (Fig. 4) [48], it can be seen that this hypothesis is correct. When Leu is substituted for Ile at position H69, the packing is improved due to a better fit into a pocket between CDRH2 and framework 3. At position H71, the large human residue, Arg, would probably impinge on the Pro at position 52a on CDR H2, thereby changing the conformation of this loop. The substitution of Val, a much smaller and uncharged residue, allows CDRH2 to maintain the optimal conformation. In a sense, the pertuzumab sequence is a hybrid with respect to positions H69 and H71, since it has the closest homologue (Ile) of the favored Leu found at position H69 in subgroup III, and the favored Val from subgroup II at position 71. In the model, it appears that H73 is solvent exposed, thus it was postulated that this residue might interact directly with antigen. In the crystal structure of the pertuzumab–receptor complex [16], H73, changed to Arg in pertuzumab, can be seen to directly interact with the receptor, thus explaining the preference for Arg over Asp at this position. Position H73 is quite variable, and may play a direct role in antigen binding of other antibodies. None of the residues in the L chain were changed to derive the humanized antibody.
Fig. 4.
X-ray structure of humanized 2C4 Fab (pertuzumab) [48] (PDB accession number 1L71). The CDR loops of the light (L1-3) and heavy chains (H1-3) are indicated. Side chains of VH residues that were investigated or changed during humanization are shown (refer to Table 1 for details for specific changes and their effect on binding relative to the chimera)
Evidence from previously published in vitro and in vivo studies supports the putative mechanism of action of pertuzumab as an inhibitor of HER dimerization [2, 16]. A recent study determined the X-ray crystal structure of the soluble HER2-ECD in a complex with the antigen-binding fragment of pertuzumab [16]. The study showed that pertuzumab binds near the center of the second cysteine-rich dimerization domain (extracellular domain II), a region of the HER receptor family that is highly conserved and previously shown to be necessary for EGFR homodimerization. Experiments in which the binding site was altered by mutation indicated a steric mechanism for the ability of pertuzumab to inhibit HER2 receptor dimerization and signaling [16]. Of note, pertuzumab binds to a different extracellular domain of the HER2 receptor compared with trastuzumab, which binds at domain IV [8, 14].
In vitro studies showed that pertuzumab is far more effective than trastuzumab in disrupting the formation of HER3-heregulin-HER2 complexes in MCF-7 (low/normal HER2 expression) and SK-BR3 (high HER2 expression) breast cancer cell lines [2]. Inhibition of HER dimer formation by pertuzumab was also shown to block the heregulin-dependent phosphorylation signal of HER2 and to inhibit downstream MAPK and Akt signaling pathways in MCF-7 cells [2]. Activity was seen for intact pertuzumab and the Fab fragment [2]. In addition, pertuzumab was found to inhibit ligand-dependent receptor activation and signaling in several other cell lines, including ovarian cancer [45], androgen-independent prostate cancer [35], colon cancer [34], breast cancer [30], and lung epithelial cells [32].
In vivo experiments support the hypothesis that inhibition of HER dimerization and downstream signaling will have antiproliferative effects against a range of different tumor types. Both pertuzumab and trastuzumab inhibited breast cancer xenografts with high HER2 expression (BT474), and importantly, pertuzumab also inhibited xenografts with low levels of HER2 expression (MCF-7), achieving 59% growth inhibition compared with controls or trastuzumab (p<0.001) [2]. In the androgen-independent prostate cancer xenograft, CWRSA6, which is not responsive to trastuzumab, pertuzumab produced 82% growth inhibition compared with controls [2]. A dose-response relationship was also observed in CWRR22R androgen-independent xenografts [2]. In xenograft models of non-small cell lung cancer (NSCLC) treated with pertuzumab, tumor growth inhibition of >80% was obtained in NCI-H522 (NSCLC HER2 1+) and in Calu-3 (NSCLC HER2 3+) [33]. This degree of tumor growth inhibition was seen at trough serum concentrations of 5–25 μg/mL of pertuzumab [33].
Pertuzumab entered clinical development based on evidence of biological activity in xenograft tumor models, using a dose regimen that was supported by the preclinical and toxicology models reported in this paper. The disposition of pertuzumab in humans was expected to be similar to that of other humanized MAbs that share the same IgG1 framework, trastuzumab and bevacizumab [15, 31], because preclinical pharmacokinetics were comparable between these molecules and allometric interspecies scaling predicted similar clearance.
In a Phase Ia clinical study, patients with incurable, locally advanced, recurrent or metastatic solid tumors that had progressed during or after standard therapy were administered pertuzumab intravenously every 3 weeks at doses escalating from 0.5 mg/kg to 15 mg/kg [3]. Pharmacokinetic results of pertuzumab doses from 2 mg/kg to 15 mg/kg showed that the systemic clearance (CL) did not change with dose (grand mean = 3.42±1.20 mL/day/kg), the volume of distribution (Vd) approximated the serum volume (grand mean = 40.6±6.6 mL/day/kg) and the terminal elimination half-life was approximately 2–3 weeks (grand mean = 18.9±8.0 days). There was good agreement between pertuzumab pharmacokinetics observed in this study and expected from preclinical studies.
In comparison, a population pharmacokinetic model in patients with metastatic breast cancer estimated the clearance of trastuzumab to be 225 mL/day (3.24 mL/day/kg for a 69 kg patient) [22]. Moreover, dosing trastuzumab every 3 weeks has been shown to be feasible and effective [31]. As pertuzumab and trastuzumab appear to have comparable pharmacokinetic profiles, an every 3-week dosing schedule was recommended for clinical testing of pertuzumab.
Since pertuzumab was associated with minimal toxicity and a large safety factor in the 7-week toxicology study in cynomolgus monkeys, a starting dose of 0.5 mg/kg every 3 weeks was recommended for clinical testing. An upper dose of 15.0 mg/kg every 3 weeks was recommended by estimating the dose required to exceed serum trough concentrations of pertuzumab associated with efficacy in xenograft models (5–25 μg/mL) [33]. Pharmacokinetic simulations predicted that doses between 5.0 mg/kg and 15.0 mg/kg administered every 3 weeks would achieve targeted serum trough concentrations of 20 μg/mL in most subjects. The dose range 0.5–15.0 mg/kg represents a safety factor of 300-fold to tenfold com
pared with the highest dose of pertuzumab (150 mg/kg) used in the multi-dose toxicology studies in cynomolgus monkeys. In the 7-week toxicology study, diarrhea was the only significant toxicity and was observed in all dose groups treated with pertuzumab for 7 weeks. Diarrhea improved during the recovery period and in most cases was responsive to supportive care. Similarly, diarrhea was also observed in a chronic dosing study in which cynomolgus monkeys were treated with pertuzumab (15, 50, and 150 mg/kg IV) for 26 weeks (data not shown). Unlike in the 7-week study, diarrhea-related dehydration was observed in the 26-week study. Agents targeting the EGF receptor tyrosine kinase have also been associated with diarrhea [28, 38], although they have a different mechanism of action from pertuzumab. Pertuzumab does not bind to EGFR, but it does inhibit formation of EGFR/HER2 heterodimers [2]. EGFR may be a negative regulator of chloride secretion; therefore, inhibition of receptor activity could lead to increased fluid secretion into the intestinal lumen [27].
There was no evidence of cardiotoxicity in cynomolgus monkeys. In the clinical setting, decreased left ventricular function has been observed in patients treated the anti-HER2 MAb trastuzumab, particularly when used in combination with anthracycline therapy [44]. Evidence from preclinical studies indicates that the HER2-signaling pathway in cardiac myocytes may play an important role in the cardiotoxicity of anti-HER2 therapy [10, 36], therefore, monitoring of potential cardiac effects is mandatory in ongoing clinical studies of pertuzumab.
In the Phase Ia study, pertuzumab was generally well tolerated and no evidence of decreased left ventricular function was observed, except in one patient who had an intercurrent myocardial infarction [3]. Of note, the study also provided evidence of antitumor activity. Tumor regression was achieved in 3 of 20 patients evaluable for response (ovarian cancer, islet cell carcinoma of the pancreas, and prostate cancer). Two patients had confirmed partial responses. Stable disease lasting for more than 2.5 months was observed in 6 of 21 patients. Antibodies to pertuzumab were not detected.
In conclusion, the recombinant humanized MAb pertuzumab is the first HER dimerization inhibitor to enter clinical development. Inhibiting HER receptor dimerization by targeting the dimerization domain of HER2 prevents the activation of HER signaling pathways that mediate cancer cell proliferation and survival. Preclinical and preliminary clinical evidence suggests that inhibition of HER dimerization may be an effective anticancer strategy in tumors expressing HER2 either at normal or elevated levels.
Acknowledgments
The authors gratefully acknowledge the important contributions of Tim Breece and Charlie Schmelzer (for protein purification), Henry Lowman (for BIAcore assays), Mubashira A. Malik (for assistance with PK assays) and the Oligosynthesis and DNA Sequencing Groups at Genentech Inc.
References
- 1.Agus DB, Akita RW, Fox WD, Lofgren JA, Higgins B, Maiese K, Scher HI, Sliwkowski MX. A potential role for activated HER-2 in prostate cancer. Semin Oncol. 2000;27:76–83. [PubMed] [Google Scholar]
- 2.Agus DB, Akita RW, Fox WD, Lewis GD, Higgins B, Pisacane PI, Lofgren JA, Tindell C, Evans DP, Maiese K, Scher HI, Sliwkowski MX. Targeting ligand-activated ErbB2 signaling inhibits breast and tumor growth. Cancer Cell. 2002;2:127–137. doi: 10.1016/S1535-6108(02)00097-1. [DOI] [PubMed] [Google Scholar]
- 3.Agus DB, Gordon MS, Taylor C, Natale RB, Karlan B, Mendelson DS, Press MF, Allison DE, Sliwkowski MX, Lieberman G, Kelsey SM, Fyfe G. Phase I clinical study of pertuzumab, a novel HER dimerization inhibitor, in patients with advanced cancer. J Clin Oncol. 2005;23:2534–2543. doi: 10.1200/JCO.2005.03.184. [DOI] [PubMed] [Google Scholar]
- 4.Alimandi M, Romano A, Curia MC, Muraro R, Fedi P, Aaronson SA, Di Fiore PP, Kraus MH. Cooperative signaling of ErbB3 and ErbB2 in neoplastic transformation and human mammary carcinomas. Oncogene. 1995;10:1813–1821. [PubMed] [Google Scholar]
- 5.Burgess AW, Cho HS, Eigenbrot C, Ferguson KM, Garrett TP, Leahy DJ, Lemmon MA, Sliwkowski MX, Ward CW, Yokoyama S. An open-and-shut case? Recent insights into the activation of EGF/ErbB receptors. Mol Cell. 2003;12:541–552. doi: 10.1016/S1097-2765(03)00350-2. [DOI] [PubMed] [Google Scholar]
- 6.Carter P, Presta L, Gorman CM, Ridgway JB, Henner D, Wong WL, Rowland AM, Kotts C, Carver ME, Shepard HM. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc Natl Acad Sci USA. 1992;89:4285–4289. doi: 10.1073/pnas.89.10.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X, Yeung TK, Wang Z. Enhanced drug resistance in cells coexpressing ErbB2 with EGF receptor or ErbB3. Biochem Biophys Res Commun. 2000;277:757–763. doi: 10.1006/bbrc.2000.3731. [DOI] [PubMed] [Google Scholar]
- 8.Cho HS, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW, Jr, Leahy DJ. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. 2003;421:756–760. doi: 10.1038/nature01392. [DOI] [PubMed] [Google Scholar]
- 9.Chothia C, Lesk AM, Tramontano A, Levitt M, Smith-Gill SJ, Air G, Sheriff S, Padlan EA, Davies D, Tulip WR, Colman PM, Spinelli S, Alzari PM, Poljak RJ. Conformations of immunoglobulin hypervariable regions. Nature. 1989;342:877–883. doi: 10.1038/342877a0. [DOI] [PubMed] [Google Scholar]
- 10.Crone SA, Zhao YY, Fan L, Gu Y, Minamisawa S, Liu Y, Peterson KL, Chen J, Kahn R, Condorelli G, Ross J, Jr, Chien KR, Lee KF. ErbB2 is essential in the prevention of dilated cardiomyopathy. Nat Med. 2002;8:459–465. doi: 10.1038/nm0502-459. [DOI] [PubMed] [Google Scholar]
- 11.Dunnett CW. New tables for multiple comparisons with a control. Biometrics. 1964;20:482–491. doi: 10.2307/2528490. [DOI] [Google Scholar]
- 12.Eigenbrot C, Gonzalez T, Mayeda J, Carter P, Werther W, Hotaling T, Fox J, Kessler J. X-ray structures of fragments from binding and nonbinding versions of a humanized anti-CD18 antibody: structural indications of the key role of VH residues 59 to 65. Proteins. 1994;18:49–62. doi: 10.1002/prot.340180107. [DOI] [PubMed] [Google Scholar]
- 13.Eigenbrot C, Randal M, Presta L, Carter P, Kossiakoff AA. X-ray structures of the antigen-binding domains from three variants of humanized anti-p185HER2 antibody 4D5 and comparison with molecular modeling. J Mol Biol. 1993;229:969–995. doi: 10.1006/jmbi.1993.1099. [DOI] [PubMed] [Google Scholar]
- 14.Fendly BM, Winget M, Hudziak RM, Lipari MT, Napier MA, Ullrich A. Characterization of murine monoclonal antibodies reactive to either the human epidermal growth factor receptor or HER2/neu gene product. Cancer Res. 1990;50:1550–1558. [PubMed] [Google Scholar]
- 15.Fernando NH, Hurwitz HI. Inhibition of vascular endothelial growth factor in the treatment of colorectal cancer. Semin Oncol. 2003;30(Suppl 6):39–50. doi: 10.1016/S0093-7754(03)00124-6. [DOI] [PubMed] [Google Scholar]
- 16.Franklin MC, Carey KD, Vajdos FF, Leahy DJ, de Vos AM, Sliwkowski MX. Insights into ErbB signaling. Cancer Cell. 2004;5:317–328. doi: 10.1016/S1535-6108(04)00083-2. [DOI] [PubMed] [Google Scholar]
- 17.Garrett TP, McKern NM, Lou M, Elleman TC, Adams TE, Lovrecz GO, Kofler M, Jorissen RN, Nice EC, Burgess AW, Ward CW. The crystal structure of a truncated ErbB2 ectodomain reveals an active conformation, poised to interact with other ErbB receptors. Mol Cell. 2003;11:495–505. doi: 10.1016/S1097-2765(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 18.Gerstner RB, Carter P, Lowman HB. Sequence plasticity in the antigen-binding site of a therapeutic anti-HER2 antibody. J Mol Biol. 2002;321:851–862. doi: 10.1016/S0022-2836(02)00677-0. [DOI] [PubMed] [Google Scholar]
- 19.Gorman CM, Gies DR, McCray G. Transient production of proteins using an adenovirus transformed cell line. DNA Prot Eng Tech. 1990;2:3–10. [Google Scholar]
- 20.Graham FL, Smiley J, Russell WC, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 21.Graus-Porta D, Beerli RR, Daly JM, Hynes NE. ErbB-2, the preferred heterodimerization partner of all ErbB receptors, is a mediator of lateral signaling. EMBO J. 1997;16:1647–1655. doi: 10.1093/emboj/16.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris KA, Washington CB, Lieberman G, Lu JF, Mass R, Bruno R (2002) A population pharmacokinetic (PK) model for trastuzumab (Herceptin) and implications for clinical dosing. Proc Am Soc Clin Oncol 21 (abstr 488)
- 23.Holbro T, Civenni G, Hynes NE. The ErbB receptors and their role in cancer progression. Exp Cell Res. 2003;284:99–110. doi: 10.1016/S0014-4827(02)00099-X. [DOI] [PubMed] [Google Scholar]
- 24.Ings RMJ. Interspecies scaling and comparisons in drug development and toxicokinetics. Xenobiotica. 1990;20:120–131. doi: 10.3109/00498259009046839. [DOI] [PubMed] [Google Scholar]
- 25.Jones JT, Akita RW, Sliwkowski MX. Binding specificities and affinities of egf domains for ErbB receptors. FEBS Lett. 1999;447:227–231. doi: 10.1016/S0014-5793(99)00283-5. [DOI] [PubMed] [Google Scholar]
- 26.Kabat EA, Wu TT, Perry H, Gottesmann KS, Foeller C. Sequences ofroteins of immunological interest. 5. MD: Public Health Service, National Institutes of Health Bethesda; 1991. [Google Scholar]
- 27.Keely SJ, Uribe JM, Barrett KE. Carbachol stimulates transactivation of epidermal growth factor receptor and mitogen-activated protein kinase in T84 cells. Implications for carbachol-stimulated chloride secretion. J Biol Chem. 1998;273:27111–27117. doi: 10.1074/jbc.273.42.27111. [DOI] [PubMed] [Google Scholar]
- 28.Kris MG, Natale RB, Herbst RS, Lynch TJ, Jr, Prager D, Belani CP, Schiller JH, Kelly K, Spiridonidis H, Sandler A, Albain KS, Cella D, Wolf MK, Averbuch SD, Ochs JJ, Kay AC. Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non-small cell lung cancer: a randomized trial. JAMA. 2003;290:2149–2158. doi: 10.1001/jama.290.16.2149. [DOI] [PubMed] [Google Scholar]
- 29.Kunkel TA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee H, Akita RW, Sliwkowski MX, Maihle NJ. A naturally occurring secreted human ErbB3 receptor isoform inhibits heregulin-stimulated activation of ErbB2, ErbB3, and ErbB4. Cancer Res. 2001;61:4467–4473. [PubMed] [Google Scholar]
- 31.Leyland-Jones B, Gelmon K, Ayoub JP, Arnold A, Verma S, Dias R, Ghahramani P. Pharmacokinetics, safety, and efficacy of trastuzumab administered every three weeks in combination with paclitaxel. J Clin Oncol. 2003;21:3965–3971. doi: 10.1200/JCO.2003.12.109. [DOI] [PubMed] [Google Scholar]
- 32.Liu J, Kern JA. Neuregulin-1 activates the JAK-STAT pathway and regulates lung epithelial cell proliferation. Am J Resp Cell Mol Biol. 2002;27:306–313. doi: 10.1165/rcmb.4850. [DOI] [PubMed] [Google Scholar]
- 33.Malik MA, Totpal K, Balter I, Sliwkowski MX, Pelletier N, Reich M, Crocker L, Friess T, Bauer S, Fiebig HH, Allison DE. Dose-response studies of recombinant humanized monoclonal antibody 2C4 in tumor xenograft models. Proc Am Assoc Cancer Res. 2003;44:150. [Google Scholar]
- 34.Mann M, Sheng H, Shao J, Williams CS, Pisacane PI, Sliwkowski MX, DuBois RN. Targeting cyclooxygenase 2 and HER-2/neu pathways inhibits colorectal carcinoma growth. Gastroenterology. 2001;120:1713–1719. doi: 10.1053/gast.2001.24844. [DOI] [PubMed] [Google Scholar]
- 35.Mendoza N, Phillips GL, Silva J, Schwall R, Wickramasinghe D. Inhibition of ligand-mediated HER2 activation in androgen-independent prostate cancer. Cancer Res. 2002;62:5485–5488. [PubMed] [Google Scholar]
- 36.Negro A, Brar BK, Lee KF. Essential roles of Her2/erbB2 in cardiac development and function. Recent Prog Horm Res. 2004;59:1–12. doi: 10.1210/rp.59.1.1. [DOI] [PubMed] [Google Scholar]
- 37.Olayioye MA, Graus-Porta D, Beerli RR, Rohrer J, Gay B, Hynes NE. ErbB-1 and ErbB-2 acquire distinct signaling properties dependent upon their dimerization partner. Mol Cell Biol. 1998;18:5042–5051. doi: 10.1128/mcb.18.9.5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perez-Soler R, Chachoua A, Hammond LA, Rowinsky EK, Huberman M, Karp D, Rigas J, Clark GM, Santabarbara P, Bonomi P. Determinants of tumor response and survival with erlotinib in patients with non-small-cell lung cancer. J Clin Oncol. 2004;22:3238–3247. doi: 10.1200/JCO.2004.11.057. [DOI] [PubMed] [Google Scholar]
- 39.Pinkas-Kramarski R, Soussan L, Waterman H, Levkowitz G, Alroy I, Klapper L, Lavi S, Seger R, Ratzkin BJ, Sela M, Yarden Y. Diversification of Neu differentiation factor and epidermal growth factor signaling by combinatorial receptor interactions. EMBO J. 1996;15:2452–2467. [PMC free article] [PubMed] [Google Scholar]
- 40.Presta LG, Chen H, O’Connor SJ, Chisholm V, Meng YG, Krummen L, Winkler M, Ferrara N. Humanization of an anti-vascular endothelial growth factor monoclonal antibody for the therapy of solid tumors and other disorders. Cancer Res. 1997;57:4593–4599. [PubMed] [Google Scholar]
- 41.Presta LG, Lahr SJ, Shields RL, Porter JP, Gorman CM, Fendly BM, Jardieu PM. Humanization of an antibody directed against IgE. J Immunol. 1993;151:2623–2632. [PubMed] [Google Scholar]
- 42.Schroff RW, Foon KAA, Beatty SSM, Odham RK, Morgan AC., Jr Human anti-murine immunoglobulin response in patients receiving monoclonal antibody therapy. Cancer Res. 1985;45:879–885. [PubMed] [Google Scholar]
- 43.Sias PE, Kotts CE, Vetterlein D, Shepard M, Wong WL. ELISA for quantitation of the extracellular domain of p185HER2 in biological fluids. J Immunol Methods. 1990;132:73–80. doi: 10.1016/0022-1759(90)90400-P. [DOI] [PubMed] [Google Scholar]
- 44.Suter TM, Cook-Bruns N, Barton C. Cardiotoxicity associated with trastuzumab (Herceptin) therapy in the treatment of metastatic breast cancer. Breast. 2004;13:173–183. doi: 10.1016/j.breast.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 45.Totpal K, Balter I, Akita R, Bargiacchi F, Phillips GL, Sliwkowski MX. Targeting ErbB2/HER2’s role as a coreceptor with rhuMAb2C4 inhibits Erb/HER ligand-dependent signaling and proliferation of ovarian tumor cell lines. Proc Am Assoc Cancer Res. 2003;44:151. [Google Scholar]
- 46.Tramontano A, Chothia C, Lesk AM. Framework residue 71 is a major determinant of the position and conformation of the second hypervariable region in the VH domains of immunoglobulins. J Mol Biol. 1990;215:175–182. doi: 10.1016/S0022-2836(05)80102-0. [DOI] [PubMed] [Google Scholar]
- 47.Tzahar E, Waterman H, Chen X, Levkowitz G, Karunagaran D, Lavi S, Ratzkin BJ, Yarden Y. A hierarchical network of interreceptor interactions determines signal transduction by Neu differentiation factor/neuregulin and epidermal growth factor. Mol Cell Biol. 1996;16:5276–5287. doi: 10.1128/mcb.16.10.5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vajdos FF, Adams CW, Breece TN, Presta LG, de Vos AM, Sidhu SS. Comprehensive functional maps of the antigen-binding site of an anti-ErbB2 antibody obtained with shotgun scanning mutagenesis. J Mol Biol. 2002;320:415–428. doi: 10.1016/S0022-2836(02)00264-4. [DOI] [PubMed] [Google Scholar]
- 49.Wallasch C, Weiss FU, Niederfellner G, Jallal B, Issing W, Ullrich A. Heregulin-dependent regulation of HER2/neu oncogenic signaling by heterodimerization with HER3. EMBO J. 1995;14:4267–4275. doi: 10.1002/j.1460-2075.1995.tb00101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Werther WA, Gonzalez TN, O’Conner SJ, McCabe S, Chan B, Hotaling T, Champe M, Fox JA, Jardieu PM, Berman PW, Presta LG. Humanization of an anti-lymphocyte function-associated antigen (LFA)-1 monoclonal antibody and reengineering of the humanized antibody for binding to rhesus LFA-1. J Immunol. 1996;157:4986–4995. [PubMed] [Google Scholar]
- 51.Winer BJ (1971) Analysis of covariance. In: Statistical principles in experimental design, 2nd edn. McGraw-Hill, New York, pp 752–812
- 52.Winer BJ (1971) Design and analysis of single-factor experiments. In: Statistical principles in experimental design. 2nd edn. McGraw-Hill, New York, pp 149–260
- 53.Xiang J, Sha Y, Jia Z, Prasad L, Delbaere LT. Framework residues 71 and 93 of the chimeric B72.3 antibody are major determinants of the conformation of the heavy-chain hypervariable. J Mol Biol. 1995;253:385–390. doi: 10.1006/jmbi.1995.0560. [DOI] [PubMed] [Google Scholar]
- 54.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 55.Yen L, Benlimame N, Nie ZR, Xiao D, Wang T, Al Moustafa AE, Esumi H, Milanini J, Hynes NE, Pages G, Alaoui-Jamali MA. Differential regulation of tumor angiogenesis by distinct ErbB homo- and heterodimers. Mol Biol Cell. 2002;13:4029–4044. doi: 10.1091/mbc.E02-02-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]