Abstract
Melan-A/MART1 is a melanocytic differentiation antigen recognized on melanoma tumor cells by CD8+ and CD4+ T cells. In this study, we describe a new epitope of this protein recognized in the context of HLA-Cw*0701 molecules by a CD8+ tumor infiltrating lymphocyte (TIL) clone. This CD8+ TIL clone specifically recognized and killed a fraction of melanoma cells lines expressing Melan-A/MART1 and HLA-Cw*0701. We further show that the Melan-A/MART151–61 peptide is the optimal peptide recognized by this clone. Together, these data significantly enlarge the fraction of melanoma patients susceptible to benefit from a Melan-A/MART1 vaccine approach.
Keywords: CD8+ T cells, Cancer, Melanoma, Melan-A/MART1, HLA-Cw*0701
Introduction
Characterization of tumor-associated antigens (TAA)-derived peptides efficiently presented by tumor cells and recognized by tumor reactive TIL is critical for the development of TAA-specific immunotherapies. TAA identification has been especially effective for melanoma tumors, evidencing two major classes of melanoma-associated Ags (MAA): tumor-specific proteins unexpressed in normal tissues and melanocytic differentiation Ags, like Melan-A/MART1, which are also expressed in normal melanocytes [27].
Among melanoma-associated antigens recognized by CD8+ and CD4+ T cells, Melan-A/MART-1 is one of the most consistently expressed by both a high fraction of melanoma tumor cells and melanoma tumor samples [8, 21]. Melan-A/MART1 epitopes recognized by CD8+ T cells and CD4+ T cells have been described [2, 4, 8, 13, 20, 23, 35]. Furthermore, others and we found that the Melan-A/MART126–35 peptide is the epitope most frequently recognized by CD8+ TIL from HLA-A2+ melanoma patients [1, 19, 22, 31]. Since HLA-A2 is the prevalent HLA class I allele within the Caucasian population, vaccines, and adoptive therapy trials targeting this Melan-A/MART1 epitope have been performed [3, 9, 24, 26, 32, 33]. However, despite the induction of specific T cells responses, clinical efficacy of these treatments remained disappointing. One possible explanation is that these treatments might have induced a tumor escape by selecting antigen- or HLA-loss variants [17, 34]. The simultaneous targeting of multiple epitopes presented by diverse HLA molecules and capable of stimulating CD8+ and CD4+ T cells may at least partially counteract this tumor escape mechanism. Hence, the identification of new epitopes from commonly expressed Ags restricted by various frequently expressed HLA alleles is necessary.
In this study, we identified a HLA-Cw*0701 restricted Melan-A/MART1 epitope presented by HLA-Cw*0701+ melanoma tumor cells and recognized by a CD8+ TIL. This epitope is located in the region 51–73 of the protein, already shown to contain HLA-DR4 [35] and HLA-DQ5 [23] restricted epitopes. These data widen the fraction of melanoma patients susceptible to benefit from a Melan-A/MART1 specific vaccine treatment.
Materials and methods
Culture medium
Culture medium RPMI1640 (Gibco BLR, Gaithersburg, MD, USA) was supplemented with penicillin–streptomycin (10 μg/ml) and l-Glutamine (2 mM) (Life Technologies, Cergy-Pontoise, France) and 8% pooled human serum (pHS), or 10% fetal calf serum (FCS, Eurobio, Les Ulis, France).
Cell lines
B lymphocyte cell lines (BLCLs): Crep, Hob, Dab, Do 45, Hen, Hour, M.H, and UPN221 were a kind gift from Dr H. Vié (INSERM U601, Nantes). Melanoma cell lines M6, M67, M74, M113, M125, M134, M153, M171, M199, and M200 were established in our laboratory. All these cell lines were maintained with RPMI 1640 containing 10% FCS.
Peptides
Melan-A/MART151–73 (RNGYRALMDKSLHVGTQCALTRR) was purchased from Epytop (Nimes, France). Melan-A/MART152–73, 51–62, 51–61 were purchased from Genepep (Montpellier, France). Melan-A/MART151–60, 51–59 were purchased from NeoMPS (Strasbourg, France). All peptides were at least 80% pure. Lyophilized peptides were diluted in DMSO and stored at −80°C.
Detection of Melan-A/MART151–63 and Melan-A/MART151–73 CD8+ T cell responses among TIL of melanoma patients
TIL from melanoma patients were obtained as previously described [1]. TIL were cultured with or without 10 μM peptide in presence of 10 μg/ml of brefeldin-A. After 6 h, cells were fixed and IFN-γ production by CD8+ T cells was assessed by IFN-γ intracytoplasmic and CD8 surface staining.
IFN-γ cells sorting
Specific T cells were isolated using an IFN-γ catching assay [25], according to the manufacturer’s instructions (Miltenyi Biotech, Hamburg, Germany). Briefly, 3 × 106 TIL were incubated in 200 μl of RPMI1640 8% pHS with 10 μM of Melan-A/MART151–63 peptide at 37°C, 5% CO2, for 3 h. Cells were subsequently labeled with a bispecific CD45/IFN-γ catching Ab and incubated for 45 min at 37°C. After several washes, the IFN-γ producing cells were stained with a second IFNγ-PE detection mAb and separated by anti-PE mAb conjugated with magnetic beads. The isolated cells were then amplified and cloned.
Cloning of T cells
Cells from polyclonal cultures containing specific T cells were cloned by limiting dilution as previously described [11]. Briefly, T cells were plated in U-bottom 96-well plates with irradiated feeder cells, at concentrations of 1, 0.6 or 0.3 T cells/well. Irradiated (35 grey) feeder cells consisted of 1 × 105 allogenic PBMCs and 1 × 104 BLCL cells/well. The stimulatory medium consisted of RPMI1640 8% pHS containing 150 U/ml IL-2 and 1 μg/ml Phytohemagglutinin-l (PHA-l, Sigma). After 5 days, half of the volume of medium (75 μl/well) was replaced with culture medium containing 8% pHS and 150 U/ml IL-2 without PHA-L. After 8 or 9 days, proliferating clones could be seen by microscopy and were maintained with less than 1.5 × 106 T cells/ml. After 2 weeks, aliquots of each clone (60 μl) were tested for specificity by TNF production assay. Specific clones were restimulated as above beginning with 10,000 T cell clone/wells. After 2 weeks, clones reached a resting state and were frozen for further expansion. For all experiments, clones were used at least 14 days after the start of expansion.
IFN-γ intracellular staining
T cell clones or polyclonal TIL populations (1 × 105) were cultured with or without Melan-A/MART151–63 peptide in the presence of 10 μg/ml of Brefeldin A (Sigma) for 6 h at 37°C. In some experiments mAb against HLA class I (clone W6.32), HLA-B/C (clone B1.23.2), HLA-A2 (clone BB7.2) produced in our laboratory or IgG2A control mAb (Immunotech) were added to cultures. Finally, cells were fixed for 10 min at room temperature with PBS containing 4% paraformaldehyde (Sigma). In some experiments, cells were stained with PE conjugated mAb specific for CD8 molecule (Beckman Coulter) before fixation. IFN-γ intracellular staining of fixed cells was performed as described by Jung et al. [18]. Briefly, cells were stained with 5 μg/ml of FITC conjugated mAb specific for IFN-γ (clone 4S.B3, BD Pharmingen) and then washed. Antibody dilutions and washes were made with PBS containing 0.1% of bovine serum albumin and 0.1% of saponin at room temperature. After staining, cells were resuspended in PBS and analyzed on a FACScan (Beckton & Dickinson, Franklin Lake, NJ, USA) with a gate set on T cells on the FSC-SSC dot plot.
TNF production assay
T cell clones (1 × 104) were cultured for 6 h at 37°C with 3 × 104 unpulsed or Melan-A/MART151–63 peptide pulsed B-LCL, with 3 × 104 unpulsed or Melan-A/MART151–63 peptide pulsed transfected COS-7 cells or with 3 × 104 untreated or IFN-γ treated (48 h, 500 IU/ml) melanoma cell lines unpulsed or pulsed with Melan-A/MART151–63 peptide. Then cultures supernatants were collected and TNF released by T cells was measured using a colorimetric assay based on cytotoxicity to WEHI 164 clone 13 [14]. TNF production is considered positive when more than 10 pg/ml of TNF was released.
Transfection of COS-7 cells
The COS cells were transfected with plasmids containing the appropriate HLA-B or HLA-C allele cDNA (corresponding to each patient’s alleles) and, in some conditions, cotransfected with plasmid containing the cDNA coding for Melan-A/MART1. cDNA were kindly provided by T. Boon (LICR, Brussels, Belgium) or cloned in our laboratory. COS-7 cells transfection was performed by the DEAE–dextran–chloroquine method [6, 8]. In brief, 1.5 × 104/well COS-7 cells were plated in flat-bottomed 96-wells plates 24 h before transfection. Cells were transfected with 100 ng of plasmid coding for an HLA alone or cotransfected with 100 ng of plasmid coding for Melan-A/MART1. Each transfection condition was performed in duplicate. After 24 h, transfected COS cells were unpulsed or pulsed with Melan-A/MART151–63 peptide for 1 h and then washed twice. The transfected cells were then used for experiments.
Melanoma cell lines phenotype
Untreated or IFN-γ treated (48 h, 500 IU/ml) melanoma cell lines (1 × 105) were stained with mAb specific for HLA class I (clone W6.32) or HLA B/C molecules (clone B1.23.2). For Melan-A/MART1 intracellular staining, cells were fixed with PBS containing 4% paraformaldehyde 10 min at room temperature and stained with Melan-A/MART1 specific mAb (clone A103, Dakocytomation, Denmark). Melanoma cell lines were then washed and incubated with PE conjugated goat anti-mouse mAb (Beckman Coulter). After staining, cells were resuspended in PBS and analyzed on a FACScan (Beckton & Dickinson, Franklin Lake, NJ, USA) with a gate set on melanoma cells on the FSC-SSC dot plot.
51Cr release assay
Target cells (melanoma cell lines) were pulsed for 1 h with Na2 51CrO4 (NEN life science, Paris, France) and then washed. 1 × 103 target cells were mixed with effector T cells at effector to target ratio 20/1. After 4 h of incubation at 37°C, 25 μl of supernatant was harvested, and added to 100 μl scintillation cocktail (optiphase supermix, Wallac, UK) before liquid scintillation counting. The % specific lysis was calculated as follows: (sample release − spontaneous release/maximum release − spontaneous release) × 100. The spontaneous release was calculated from targets incubated with culture medium, and the maximum release from targets lysed with RPMI containing 2.5% Triton X-100. Lysis was considered positive when more than 10% specific lysis was observed.
Results
Presence of spontaneous CD8+ T cell responses to Melan-A/MART151–63 and Melan-A/MART151–73 among the TIL of a melanoma patient
Based on the work of Storkus’ group [35] and on our previous study [23], we looked for the presence of CD4+ and CD8+ T cells specific for Melan-A/MART151–63, and Melan-A/MART151–73 peptide among tumor-infiltrating lymphocytes (TIL) populations of six patients. We found that 0.5 and 0.3% of CD8 positive TIL from one patient, M74, responded to the Melan-A/MART151–63, and to the Melan-A/MART151–73 peptide, respectively (Fig. 1a). So, we performed an IFN-γ positive cell sorting of TIL after stimulation by Melan-A/MART151–63. The sorted TIL were expanded and tested for their response to specific peptides by CD8 surface and IFN-γ intracellular staining after an “autopresentation” assay. We observed an enrichment of CD8+ specific T cells with 14% of CD8+ TIL responding to the Melan-A/MART151–63 peptide and 5% to Melan-A/MART151–73 peptide (Fig. 1b).
Fig. 1.
Enrichment of CD8+ T cells specific for Melan-A/MART151–63 and Melan-A/MART151–73 peptide from a polyclonal TIL population. a T cells were obtained from TIL of a melanoma patient. T cells were cultured with or without Melan-A/MART151–63 or Melan-A/MART151–73 peptide in presence of Brefeldin A at 37°C. After 6 h, cells were stained for surface CD8, fixed, and then stained for intracytoplasmic IFN-γ. Fluorescence was analyzed by flow cytometry with a gate set on CD8+ T cells. b CD8+ T cells specific for Melan-A/MART151–63 peptide were enriched by IFN-γ magnetic beads cell sorting and were expanded during 2 weeks. The sorted TIL were then cultured with or without Melan-A/MART151–63 and Melan-A/MART151–73 peptide in presence of Brefeldin A at 37°C during 6 h. Cells were stained for surface CD8, fixed, and then stained for intracytoplasmic IFN-γ. Fluorescence was analyzed by flow cytometry with a gate set on CD8 + T cells
HLA-Cw*0701 molecules present Melan-A/MART151–63 peptide to specific CD8+ T cells
To determine the HLA class I restriction of Melan-A/MART151–63 reactive CD8+ T cells, we cloned under limiting dilution the enriched population as previously described [11]. We obtained ten clones, which produced TNF in response to Melan-A/MART151–63 peptide. Three out of these ten clones could be expanded and cryopreserved. These three TIL clones were tested for Vβ expression and we found that 100% of cells of the three clones expressed Vβ13.1 (data not shown). The clone M74.19 was further expanded for additional experiments. We confirmed that this clone was CD4–CD8+ (data not shown), and by autopresentation followed by IFN-γ intracytoplasmic staining, that it was able to respond to Melan-A/MART151–63 peptide (Fig. 2a). Anti-HLA class I and anti-HLA-B/C, but not anti HLA-A2 and IgG2A control specific mAb, inhibited the response of clone M74.19 in a concentration-dependent manner (Fig. 2a). Therefore, this clone recognizes a Melan-A/MART1 peptide in the HLA-B (HLA-B*1402 and HLA-B*0801) or HLA-C context (HLA-Cw*0701 and HLA-Cw*0801).
Fig. 2.
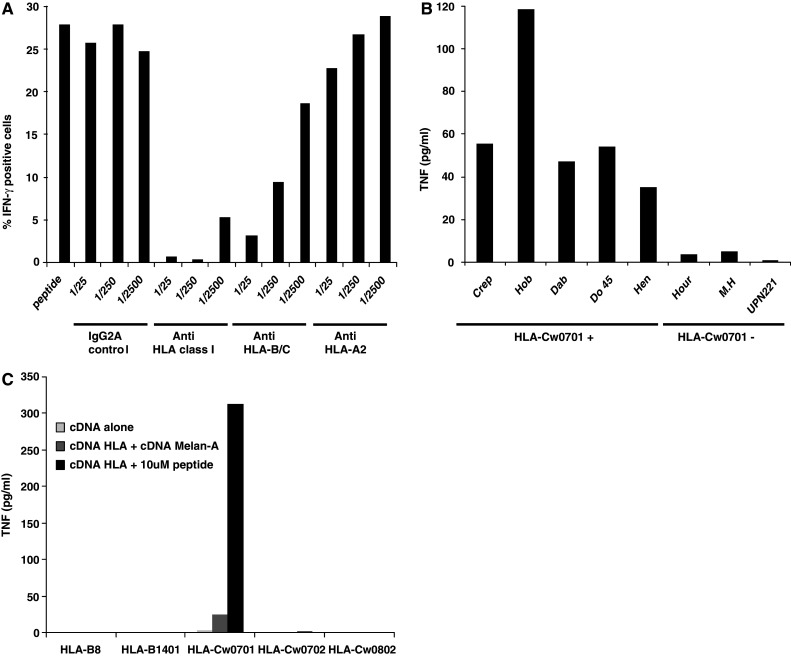
The Melan-A/MART151–63 specific CD8+ T cell clone, M74.19, is restricted by the HLA-Cw*0701 molecules. a M74.19 CD8+ T cell clone was cultured alone or with 10 μM of Melan-A/MART151–63 peptide in presence of Brefeldin A at 37°C. Inhibitory mAb against HLA class I, HLA-B/C or HLA-A2 molecules or isotype control mAb were added to cultures at different concentrations. After 6 h, cells were fixed, permeabilized and stained for intracytoplasmic IFN-γ. Fluorescence was analyzed by flow cytometry. b M74.19 T cell clone was cultured alone or with 10 μM Melan-A/MART151–63 peptide pulsed HLA-Cw*0701+ or HLA-Cw*0701− B-LCL. After 6 h, TNF was measured in co-culture supernatants. c COS-7 cells were co-transfected with plasmid encoding the HLA-B8, HLA-B*1401, HLA-Cw*0701, HLA-Cw*0702 or HLA-Cw*0802 and with or without plasmid encoding Melan-A/MART1. 24 h after transfection, COS-7 cells were either unpulsed or pulsed with 10 μM of Melan-A/MART151–63 peptide and washed. Then, they were co-cultured with M74.19 CD8+ T cell clone. After 6 h, TNF was measured in co-culture supernatants
To identify the restricting HLA allele, we first used B lymphocytes cell lines (B-LCL) expressing one or several donor HLA alleles to present the peptide to the clone M74.19: Crep, Hob, Do 45, Hen (HLA-B*0801+ and HLA-Cw*0701+), Dab (HLA-Cw7+), Hour (HLA-Cw*0702+), M.H (HLA-Cw*0801+), and UPN221 (HLA-Cw*0704+). All, and only, HLA-Cw*0701 expressing B-LCL were able to stimulate TNF production from the clone M74.19, suggesting that this allele is the restriction element (Fig. 2b). To confirm this result, we tested the capacity of COS-7 cells transfected with plasmids encoding donor HLA-B or HLA-C molecules with, or without plasmids encoding Melan-A/MART1, to stimulate the clone M74.19. Transfected cells were unpulsed or pulsed with 10 μM of Melan-A/MART151–63 peptide. Only COS-7 cells co-transfected with the HLA-Cw*0701 and Melan-A/MART1 plasmids were able to induce a response of the clone in the absence of peptide addition (Fig. 2c). That was strongly increased by peptide addition. Therefore, the Melan-A/MART1 specific TIL clone is restricted by HLA-Cw*0701 and the recognized peptide can be presented by the endogenous pathway of COS-7 cells transfected with the Melan-A/MART1 cDNA. Furthermore, clone M74.19 is unable to recognize the peptide in the HLA-Cw*0702 context, despite minor differences of sequence in the α2 and α3 region of this molecule as compared to HLA-Cw*0701 molecule.
Mapping of the minimal Melan-A/MART1 peptide presented by HLA-Cw*0701 molecules
To determine the minimal peptide sequence able to stimulate the M74.19 clone, we tested a panel of overlapping synthetic peptides located in the Melan-A/MART151–73 region for their ability to induce TNF secretion by this clone in an autopresentation assay. Removal of the N-terminal arginine 51 abrogated the clone response (Fig. 3). In contrast, removal of C-terminal residues localized between position 73 and 61, increased response of the clone. This response decreased with Melan-A/MART151–60 peptide and was suppressed by the removal of lysine 60. Therefore, Melan-A/MART151–61 peptide is the peptide optimally recognized by this clone. However, other peptides contained within the region 51–73 represent potential epitopes for this clone.
Fig. 3.
Melan-A/MART151–61 peptide is the optimal epitope recognized by M74.19 CD8+ T cell clone. M74.19 CD8+ T cell clone was cultured with different concentrations of a panel of truncating peptides located in Melan-A/MART151–73 region. After 6 h, TNF was measured in co-culture supernatants
The Melan-A/MART1 epitope recognized by M74.19 CD8+ T cell clone is presented by Melan-A/MART1+ HLA-Cw*0701+ melanoma tumor cells
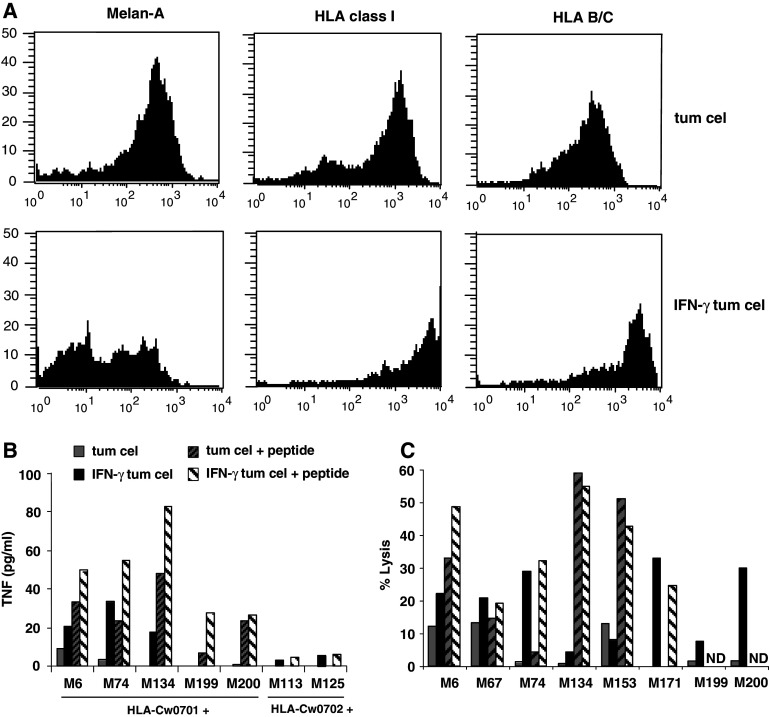
To address the presentation of the M74.19 clone epitope by melanoma tumor cells, we then tested a panel of melanoma cell lines for their ability to stimulate the clone. Because HLA-Cw expression is spontaneously low and is increased by IFN-γ [16, 36], we also tested the same cell lines pre-treated by IFN-γ for 48 h. All these cell lines were first analyzed for HLA-B/C and for Melan-A/MART1 expression. As shown for the autologous M74 cell line, as a representative result (Fig. 4a), all melanoma cell lines expressed HLA-B/C molecules and this expression was increased by IFN-γ treatment. All cell lines also expressed the Melan-A/MART1 protein, but IFN-γ decreased this expression as reported previously [16].
Fig. 4.
The Melan-A/MART1 epitope recognized by M74.19 CD8+ T cell clone is presented by Melan-A/MART1+ HLA-Cw*0701+ melanoma tumor cells. a Untreated, or IFN-γ treated melanoma tumor cell line M74 was stained with unlabeled primary mAb produced in mouse, specific for HLA class I or HLA B/C molecule or Melan-A/MART1 protein. Then melanoma tumor cell lines were stained with PE conjugated goat anti-mouse mAb. Fluorescence was analyzed by flow cytometry, b M74.19 CD8+ T cell clone was cultured with untreated, or IFN-γ treated melanoma tumor cell lines pulsed or not with Melan-A/MART151–63 peptide. After 6h, TNF was measured in co-culture supernatants. c Cytolytic activity of M74.19 CD8+ T cell clone against untreated or IFN-γ treated melanoma tumor cell lines unpulsed or pulsed with Melan-A51–61 peptide, was assessed by 4 h 51Cr release assay. ND not done
Two out of five HLA-Cw*0701+ melanoma cell lines, including the autologous cell line M74, induced TNF secretion by the clone and this stimulation was stronger upon pre-treatment by IFN-γ (Fig. 4b). Another HLA-Cw*0701+ cell line, M134, could stimulate the clone, but only after IFN-γ treatment. In contrast, two out of these five cell lines, M199 and M200, remain unable to stimulate the clone even after IFN-γ treatment. However, as expected, upon peptide loading, the five cell lines, but not HLA-Cw*0702+ cell lines, could induce TNF secretion by the clone.
Finally, we asked whether the clone was able to lyse HLA-Cw*0701+ melanoma cell lines. Three melanoma cell lines: M6, M67, and M153 out of eight were killed at a significant, but rather low level (Fig. 4c). IFN-γ pre-treatment significantly increased the lysis of two of these cell lines: M6 and M67, whereas it decreased the lysis of M153 cell line. Furthermore, IFN-γ pre-treatment induced the lysis of three cell lines, which were not killed without IFN-γ pre-treament: M74, M171, and M200. Lysis of melanoma cell lines can be further increased by the addition of Melan-A51–61 peptide. Altogether, the cytotoxic and TNF responses of the clone to melanoma cell lines were well correlated. This strongly suggests that a fraction of HLA-Cw*0701+ melanoma cell lines spontaneously present the clone epitope and that this presentation is increased by IFN-γ.
Discussion
During the past years, the majority of identified Melan-A/MART1 epitopes presented on melanoma tumor cells in association with HLA class I or class II molecules were located in the region 25–40 of the protein [2, 4, 7, 8, 30]. This study and others show that region 51–73 of Melan-A/MART1 also contains several tumor epitopes recognized by CD4+ and CD8+ T cells [23, 35]. In this study, while trying to detect CD4+ and CD8+ T cells specific for Melan-A/MART151–73 among TIL of melanoma patients, we obtained a HLA-Cw*0701 restricted specific CD8+ T cell clone from one patient. Using this clone, we demonstrated the existence of a HLA-Cw*0701 restricted Melan-A epitope in this region. Although, we did not formally identify the natural epitope presented by melanoma cells, we showed that it starts with arginine residue 51 and ends between residue 60 and 73. One could hypothesize that the response of TIL to the long Melan-A/MART151–73 peptide could be due to the processing of this peptide into smaller peptides like Melan-A/MART151–63 or Melan-A/MART151–61 (Fig. 1b). We rather think that this response resulted from the direct presentation of the long peptide, since the presence of brefeldin A in this assay prevents transport of HLA-Cw*0701/peptide complexes to the cell surface. Other unidentified epitopes may be present in this region, like the Melan-A/MART153–62 peptide recognized by T cell in an unknown HLA context after vaccination of melanoma patients with killed allogenic melanoma cells loaded dendritic cells [28]. Characterization of other epitopes from the Melan-A/MART151–73 region would be useful for development of Melan-A/MART1-based immunotherapy.
HLA-Cw*0701 allele is expressed by about 17% of the European population (http://www.ncbi.nlm.nih.gov/mhc/ihwg.cgi?ID=9&cmd=PRJOV). Therefore, epitopes from Melan-A51–73 could be used to stimulate CD8+ tumor reactive T cell responses from a significant fraction of melanoma patients. Since two other HLA-Cw7 restricted epitopes have been identified from MAGE-A2, -A3, -A6, and -A12 protein expressed by a significant fraction of melanoma tumors [5, 15, 29], multiepitopic vaccination with peptide from Melan-A/MART1 and MAGE proteins might be performed in HLA-Cw7+ melanoma patients to decrease the risk of antigen loss variant selection.
The recognition of HLA-Cw*0701+, Melan-A/MART1+ melanoma cells by the CD8+ T cell clone M74.19 was rather weak. Furthermore, we detected a CD8+ T cell response against this epitope only in 1 TIL population (M74) out of 13 HLA-Cw*0701+ melanoma patients tested (data not shown). Recognition of this epitope is largely increased by IFN-γ treatment of melanoma tumor cells, which induces an increase of HLA class I molecules expression. This result suggests that an in vivo recognition of this epitope on the surface of melanoma tumor cells may depend on the presence of IFN-γ in the tumor microenvironment that could be produced by other tumor reactive TIL. However, we also show that IFN-γ decreases Melan-/MART-1 expression by melanoma tumor cells. Other cytokines like IFN-β may be better candidates to increase tumor cell recognition by increasing not only HLA class I expression, but also melanoma antigen expression [10]. Other factors such as the weak expression on cell surface of HLA-C molecules compared to HLA-A or HLA-B expression [36], or the decrease or loss of HLA class I expression during cancer progression [12] may explain the absence of response in 12 out of 13 HLA-Cw*0701+ melanoma patients tested.
In conclusion, we reported here that Melan-A/MART1 contains a CD8+ T cell epitope within the region 51–73 efficiently presented by HLA-Cw*0701+ melanoma tumor cells. Such epitope may be helpful to design and to evaluate Melan-A/MART1 based anti-tumoral vaccination strategies.
Acknowledgments
This work was supported by grants from the «Ligue Nationale contre le Cancer» (labelisation 2003–2007), and by a grant from INCA “Thérapie adoptive cellulaire du cancer”.
Abbreviations
- B-LCL
B lymphocyte cell line
- PHS
Pooled human serum
- TIL
Tumor infiltrating lymphocytes
References
- 1.Benlalam H, Labarriere N, Linard B, Derre L, Diez E, Pandolfino MC, Bonneville M, Jotereau F. Comprehensive analysis of the frequency of recognition of melanoma-associated antigen (MAA) by CD8 melanoma infiltrating lymphocytes (TIL): implications for immunotherapy. Eur J Immunol. 2001;31:2007–2015. doi: 10.1002/1521-4141(200107)31:7<2007::AID-IMMU2007>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 2.Benlalam H, Linard B, Guilloux Y, Moreau-Aubry A, Derre L, Diez E, Dreno B, Jotereau F, Labarriere N. Identification of five new HLA-B*3501-restricted epitopes derived from common melanoma-associated antigens, spontaneously recognized by tumor-infiltrating lymphocytes. J Immunol. 2003;171:6283–6289. doi: 10.4049/jimmunol.171.11.6283. [DOI] [PubMed] [Google Scholar]
- 3.Benlalam H, Vignard V, Khammari A, Bonnin A, Godet Y, Pandolfino MC, Jotereau F, Dreno B, Labarriere N. Infusion of Melan-A/Mart-1 specific tumor-infiltrating lymphocytes enhanced relapse-free survival of melanoma patients. Cancer Immunol Immunother. 2007;56:515–526. doi: 10.1007/s00262-006-0204-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bioley G, Jandus C, Tuyaerts S, Rimoldi D, Kwok WW, Speiser DE, Tiercy JM, Thielemans K, Cerottini JC, Romero P. Melan-A/MART-1-specific CD4 T cells in melanoma patients: identification of new epitopes and ex vivo visualization of specific T cells by MHC class II tetramers. J Immunol. 2006;177:6769–6779. doi: 10.4049/jimmunol.177.10.6769. [DOI] [PubMed] [Google Scholar]
- 5.Breckpot K, Heirman C, De Greef C, van der Bruggen P, Thielemans K. Identification of new antigenic peptide presented by HLA-Cw7 and encoded by several MAGE genes using dendritic cells transduced with lentiviruses. J Immunol. 2004;172:2232–2237. doi: 10.4049/jimmunol.172.4.2232. [DOI] [PubMed] [Google Scholar]
- 6.Brichard V, Van Pel A, Wolfel T, Wolfel C, De Plaen E, Lethe B, Coulie P, Boon T. The tyrosinase gene codes for an antigen recognized by autologous cytolytic T lymphocytes on HLA-A2 melanomas. J Exp Med. 1993;178:489–495. doi: 10.1084/jem.178.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castelli C, Storkus WJ, Maeurer MJ, Martin DM, Huang EC, Pramanik BN, Nagabhushan TL, Parmiani G, Lotze MT. Mass spectrometric identification of a naturally processed melanoma peptide recognized by CD8+ cytotoxic T lymphocytes. J Exp Med. 1995;181:363–368. doi: 10.1084/jem.181.1.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coulie PG, Brichard V, Van Pel A, Wolfel T, Schneider J, Traversari C, Mattei S, De Plaen E, Lurquin C, Szikora JP, Renauld JC, Boon T. A new gene coding for a differentiation antigen recognized by autologous cytolytic T lymphocytes on HLA-A2 melanomas. J Exp Med. 1994;180:35–42. doi: 10.1084/jem.180.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dudley ME, Wunderlich JR, Robbins PF, Yang JC, Hwu P, Schwartzentruber DJ, Topalian SL, Sherry R, Restifo NP, Hubicki AM, Robinson MR, Raffeld M, Duray P, Seipp CA, Rogers-Freezer L, Morton KE, Mavroukakis SA, White DE, Rosenberg SA. Cancer regression and autoimmunity in patients after clonal repopulation with antitumor lymphocytes. Science. 2002;298:850–854. doi: 10.1126/science.1076514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunn IS, Haggerty TJ, Kono M, Durda PJ, Butera D, Macdonald DB, Benson EM, Rose LB, Kurnick JT. Enhancement of human melanoma antigen expression by IFN-beta. J Immunol. 2007;179:2134–2142. doi: 10.4049/jimmunol.179.4.2134. [DOI] [PubMed] [Google Scholar]
- 11.Fonteneau JF, Larsson M, Somersan S, Sanders C, Munz C, Kwok WW, Bhardwaj N, Jotereau F. Generation of high quantities of viral and tumor-specific human CD4+ and CD8+ T-cell clones using peptide pulsed mature dendritic cells. J Immunol Methods. 2001;258:111–126. doi: 10.1016/S0022-1759(01)00477-X. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Lora A, Algarra I, Garrido F. MHC class I antigens, immune surveillance, and tumor immune escape. J Cell Physiol. 2003;195:346–355. doi: 10.1002/jcp.10290. [DOI] [PubMed] [Google Scholar]
- 13.Godefroy E, Scotto L, Souleimanian NE, Ritter G, Old LJ, Jotereau F, Valmori D, Ayyoub M. Identification of two Melan-A CD4(+) T cell epitopes presented by frequently expressed MHC class II alleles. Clin Immunol. 2006;121(1):54–62. doi: 10.1016/j.clim.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 14.Hansen MB, Nielsen SE, Berg K. Re-examination and further development of a precise and rapid dye method for measuring cell growth/cell kill. J Immunol Methods. 1989;119:203–210. doi: 10.1016/0022-1759(89)90397-9. [DOI] [PubMed] [Google Scholar]
- 15.Heidecker L, Brasseur F, Probst-Kepper M, Gueguen M, Boon T, Van den Eynde BJ. Cytolytic T lymphocytes raised against a human bladder carcinoma recognize an antigen encoded by gene MAGE-A12. J Immunol. 2000;164:6041–6045. doi: 10.4049/jimmunol.164.11.6041. [DOI] [PubMed] [Google Scholar]
- 16.Hofbauer GF, Geertsen R, Laine E, Burg G, Dummer R. Impact of interferons on the expression of melanoma-associated antigens in melanoma short-term cell cultures. Melanoma Res. 2001;11:213–218. doi: 10.1097/00008390-200106000-00002. [DOI] [PubMed] [Google Scholar]
- 17.Ikeda H, Lethe B, Lehmann F, van Baren N, Baurain JF, de Smet C, Chambost H, Vitale M, Moretta A, Boon T, Coulie PG. Characterization of an antigen that is recognized on a melanoma showing partial HLA loss by CTL expressing an NK inhibitory receptor. Immunity. 1997;6:199–208. doi: 10.1016/S1074-7613(00)80426-4. [DOI] [PubMed] [Google Scholar]
- 18.Jung T, Schauer U, Heusser C, Neumann C, Rieger C. Detection of intracellular cytokines by flow cytometry. J Immunol Methods. 1993;159:197–207. doi: 10.1016/0022-1759(93)90158-4. [DOI] [PubMed] [Google Scholar]
- 19.Kawakami Y, Dang N, Wang X, Tupesis J, Robbins PF, Wang RF, Wunderlich JR, Yannelli JR, Rosenberg SA. Recognition of shared melanoma antigens in association with major HLA-A alleles by tumor infiltrating T lymphocytes from 123 patients with melanoma. J Immunother (1997) 2000;23:17–27. doi: 10.1097/00002371-200001000-00004. [DOI] [PubMed] [Google Scholar]
- 20.Kawakami Y, Eliyahu S, Sakaguchi K, Robbins PF, Rivoltini L, Yannelli JR, Appella E, Rosenberg SA. Identification of the immunodominant peptides of the MART-1 human melanoma antigen recognized by the majority of HLA-A2-restricted tumor infiltrating lymphocytes. J Exp Med. 1994;180:347–352. doi: 10.1084/jem.180.1.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Labarriere N, Diez E, Pandolfino MC, Viret C, Guilloux Y, Le Guiner S, Fonteneau JF, Dreno B, Jotereau F. Optimal T cell activation by melanoma cells depends on a minimal level of antigen transcription. J Immunol. 1997;158:1238–1245. [PubMed] [Google Scholar]
- 22.Labarriere N, Pandolfino MC, Raingeard D, Le Guiner S, Diez E, Le Drean E, Dreno B, Jotereau F. Frequency and relative fraction of tumor antigen-specific T cells among lymphocytes from melanoma-invaded lymph nodes. Int J Cancer. 1998;78:209–215. doi: 10.1002/(SICI)1097-0215(19981005)78:2<209::AID-IJC15>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 23.Larrieu P, Ouisse LH, Guilloux Y, Jotereau F, Fonteneau JF. A HLA-DQ5 restricted Melan-A/MART-1 epitope presented by melanoma tumor cells to CD4+ T lymphocytes. Cancer Immunol Immunother. 2007;56:1565–1575. doi: 10.1007/s00262-007-0300-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mackensen A, Herbst B, Chen JL, Kohler G, Noppen C, Herr W, Spagnoli GC, Cerundolo V, Lindemann A. Phase I study in melanoma patients of a vaccine with peptide-pulsed dendritic cells generated in vitro from CD34(+) hematopoietic progenitor cells. Int J Cancer. 2000;86:385–392. doi: 10.1002/(SICI)1097-0215(20000501)86:3<385::AID-IJC13>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 25.Manz R, Assenmacher M, Pfluger E, Miltenyi S, Radbruch A. Analysis and sorting of live cells according to secreted molecules, relocated to a cell-surface affinity matrix. Proc Natl Acad Sci USA. 1995;92:1921–1925. doi: 10.1073/pnas.92.6.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meidenbauer N, Marienhagen J, Laumer M, Vogl S, Heymann J, Andreesen R, Mackensen A. Survival and tumor localization of adoptively transferred Melan-A-specific T cells in melanoma patients. J Immunol. 2003;170:2161–2169. doi: 10.4049/jimmunol.170.4.2161. [DOI] [PubMed] [Google Scholar]
- 27.Novellino L, Castelli C, Parmiani G. A listing of human tumor antigens recognized by T cells. Cancer Immunol Immunother. 2005;54:187–207. doi: 10.1007/s00262-004-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palucka AK, Ueno H, Connolly J, Kerneis-Norvell F, Blanck JP, Johnston DA, Fay J, Banchereau J. Dendritic cells loaded with killed allogeneic melanoma cells can induce objective clinical responses and MART-1 specific CD8+ T-cell immunity. J Immunother (1997) 2006;29:545–557. doi: 10.1097/01.cji.0000211309.90621.8b. [DOI] [PubMed] [Google Scholar]
- 29.Panelli MC, Bettinotti MP, Lally K, Ohnmacht GA, Li Y, Robbins P, Riker A, Rosenberg SA, Marincola FM. A tumor-infiltrating lymphocyte from a melanoma metastasis with decreased expression of melanoma differentiation antigens recognizes MAGE-12. J Immunol. 2000;164:4382–4392. doi: 10.4049/jimmunol.164.8.4382. [DOI] [PubMed] [Google Scholar]
- 30.Schneider J, Brichard V, Boon T, Meyer zum Buschenfelde KH, Wolfel T. Overlapping peptides of melanocyte differentiation antigen Melan-A/MART-1 recognized by autologous cytolytic T lymphocytes in association with HLA-B45.1 and HLA-A2.1. Int J Cancer. 1998;75:451–458. doi: 10.1002/(SICI)1097-0215(19980130)75:3<451::AID-IJC20>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 31.Seiter S, Monsurro V, Nielsen MB, Wang E, Provenzano M, Wunderlich JR, Rosenberg SA, Marincola FM. Frequency of MART-1/MelanA and gp100/PMel17-specific T cells in tumor metastases and cultured tumor-infiltrating lymphocytes. J Immunother (1997) 2002;25:252–263. doi: 10.1097/00002371-200205000-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vignard V, Lemercier B, Lim A, Pandolfino MC, Guilloux Y, Khammari A, Rabu C, Echasserieau K, Lang F, Gougeon ML, Dreno B, Jotereau F, Labarriere N. Adoptive transfer of tumor-reactive Melan-A-specific CTL clones in melanoma patients is followed by increased frequencies of additional Melan-A-specific T cells. J Immunol. 2005;175:4797–4805. doi: 10.4049/jimmunol.175.7.4797. [DOI] [PubMed] [Google Scholar]
- 33.Yee C, Thompson JA, Byrd D, Riddell SR, Roche P, Celis E, Greenberg PD. Adoptive T cell therapy using antigen-specific CD8+ T cell clones for the treatment of patients with metastatic melanoma: in vivo persistence, migration, and antitumor effect of transferred T cells. Proc Natl Acad Sci USA. 2002;99:16168–16173. doi: 10.1073/pnas.242600099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yee C, Thompson JA, Roche P, Byrd DR, Lee PP, Piepkorn M, Kenyon K, Davis MM, Riddell SR, Greenberg PD. Melanocyte destruction after antigen-specific immunotherapy of melanoma: direct evidence of t cell-mediated vitiligo. J Exp Med. 2000;192:1637–1644. doi: 10.1084/jem.192.11.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zarour HM, Kirkwood JM, Kierstead LS, Herr W, Brusic V, Slingluff CL, Jr, Sidney J, Sette A, Storkus WJ. Melan-A/MART-1(51–73) represents an immunogenic HLA-DR4-restricted epitope recognized by melanoma-reactive CD4(+) T cells. Proc Natl Acad Sci USA. 2000;97:400–405. doi: 10.1073/pnas.97.1.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zemmour Parham J P. Distinctive polymorphism at the HLA-C locus: implications for the expression of HLA-C. J Exp Med. 1992;176:937–950. doi: 10.1084/jem.176.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]